Abstract
The let-7 microRNA (miRNA) plays important roles in human liver development and disease such as hepatocellular carcinoma, liver fibrosis and hepatitis wherein oxidative stress accelerates the progression of these diseases. To date, the role of the let-7 miRNA family in modulation of heme oxygenase 1 (HMOX1), a key cytoprotective enzyme, remains unknown. Our aims were to determine whether let-7 miRNA directly regulates Bach1, a transcriptional repressor of the HMOX1 gene, and whether indirect up-regulation of HMOX1 by let-7 miRNA attenuates oxidant injury in human hepatocytes. The effects of let-7 miRNA on Bach1 and HMOX1 gene expression in Huh-7 and HepG2 cells were determined by real-time qRT-PCR, Western blot, and luciferase reporter assays. Dual luciferase reporter assays revealed that let-7b, let-7c, or miR-98 significantly decreased Bach1 3’-untranslated region (3’-UTR)-dependent luciferase activity but not mutant Bach1 3’-UTR-dependent luciferase activity, whereas mutant let-7 miRNA containing base complementarity with mutant Bach1 3’-UTR restored its effect on mutant reporter activity. let-7b, let-7c, or miR-98 down-regulated Bach1 protein levels by 50–70%, and subsequently up-regulated HMOX1 gene expression by 3–4 fold, compared with non-specific controls. Furthermore, Huh-7 cells transfected with let-7b, let-7c or miR-98 mimic showed increased resistance against oxidant injury induced by tert-butyl-hydroperoxide (tBuOOH), whereas the protection was abrogated by over-expression of Bach1. In conclusion, let-7 miRNA directly acts on the 3’-UTR of Bach1 and negatively regulates expression of this protein, and thereby up-regulates HMOX1 gene expression. Over-expression of the let-7 miRNA family members may represent a novel approach to protecting human hepatocytes from oxidant injury.
Keywords: let-7, HMOX1, Bach1, microRNA, oxidative stress, Huh-7 cell
1. INTRODUCTION
Heme oxygenase 1 (HMOX1) is a key cytoprotective enzyme, catalyzing heme degradation to produce ferrous iron, carbon monoxide (CO) and biliverdin, which is further reduced to bilirubin in liver, with carbon monoxide, biliverdin and bilirubin acting as antioxidants [1–5]. The induction of HMOX1 has been observed as an important event in response to oxidative stress, and the molecular mechanisms underlying regulation of HMOX1 gene expression by oxidative stress, heme and other non-heme metalloporphyrins have been extensively investigated. Among key transcription factors associated with regulating HMOX1 gene expression, we and others have demonstrated that Bach1, a member of the cap n’ collar family of zinc-containing basic leucine zipper (bZip) proteins, plays a key role in tonic repression of HMOX1 gene [6–12]. It does so by forming antagonizing heterodimers with the Maf-related oncogene family. These heterodimers bind to Maf recognition elements (MAREs) and suppress expression of genes [e.g., HMOX1 and NAD(P)H:quinone oxidoreductase (NQO1)][6, 8, 9, 13]. Down-regulation of Bach1 enhances HMOX1 gene expression, and thus has emerged as an effective approach to up-regulate HMOX1 and NQO1 gene expression in response to oxidative stress [12–14]. Our recent studies suggest that microRNAs (miRNAs), a new class of small non-coding regulatory RNAs (~22 nt), play a critical role in regulating Bach1 gene expression at a post-transcriptional level, indirectly enhancing HMOX1 gene expression in human hepatocytes [12, 14]. Bach1 contains a 3’-untranslated region (UTR) 3313 nucleotides (nt) in length which provides a basis for candidate miRNAs binding. However, to date, miRNAs that are involved in modulating Bach1 gene expression are largely unknown.
The let-7 miRNA family is one of the first two miRNAs discovered in Caenorhabdities elegans (C. elegans), and the first known human miRNA, consisting of let-7a, b, c, d, e, f, g, i and miR-98 miRNAs[15, 16]. Besides a pivotal role for the let-7 miRNA family in embryonic development and cell maturation [17–20], the let-7 miRNA family was found to play an important role in human liver development [21] and liver disease such as hepatocellular carcinoma [22, 23], hepatic fibrosis [24] and hepatitis [25], conditions known to be accelerated by oxidative stress and for which, in consequence, antioxidant therapy has been proposed as a treatment approach. Thus far, the role of let-7 miRNA in regulating Bach1 and HMOX1, and the biological function of let-7 miRNA in defending against oxidative stress in liver have not been investigated. We performed in silico analysis and discovered two highly conserved sites in the 3’-UTR of Bach1 mRNA that perfectly matched the seed region of let-7 miRNA.
In the present work, we aimed to study whether the let-7 miRNA family directly regulates Bach1, and whether indirect up-regulation of HMOX1 by let-7 miRNA attenuates oxidant injury in human hepatocytes. Our focus was on the five representative let-7 miRNA family members, let-7a, let-7b, let-7c, let-7e and miRNA-98. Our findings show that let-7 miRNA directly acts on the 3’-UTR of Bach1, translationally represses expression of this protein, and thereby up-regulates HMOX1, suggesting that over-expression of let-7 miRNA family members may represent a therapeutic approach for protection against oxidant injury induced by chronic liver diseases.
2. MATERIALS AND METHODS
2.1 miRNA target predications
Three algorithms,TargetScanHuman5.1 (http://www.targetscan.org), PicTar (http://pictar.mdcberlin.de) and miRanda (http://www.microrna.org) were used to predict putative binding sites in the Bach1 mRNA for members of the let-7 miRNA family.
2.2 Cell culture
Human hepatoma cell lines, Huh-7 (Japan Health Research Resources Bank, Osaka, Japan) and HepG2 (American Type Culture Collection, Manassas, VA) were cultured with Dulbecco’s modified Eagle medium (DMEM) supplemented with 100 units/mL penicillin, 100 µg/mL streptomycin, and 10% (v/v) fetal bovine serum (FBS) from Invitrogen (Carlsbad, CA) [26]. All cells were maintained in a humidified atmosphere of 95% room air and 5% CO2 at 37 °C.
2.3 Reporter constructs, co-transfection and luciferase reporter activity assays
pLSV40-Rluc and pLSV40-GL3/Bach1 reporter constructs were kindly provided by B. R. Cullen (Duke University, Durham, NC) [27]. pLSV40-Rluc contains a cDNA (Rluc) encoding Renilla luciferase as an internal control reporter and pLSV40-GL3/Bach1 firefly luciferase reporter construct contains the full-length 3’-UTR of Bach1 mRNA [27]. Constructs were confirmed by restriction enzyme digestion and sequencing. Mutant pLSV40-GL3/Bach1-Mut was generated by GENEWIZ, Inc. (South Plainfield, NJ).
Co-transfection of miRNA mimics and reporter constructs were performed using Lipofectamine 2000 from Invitrogen (Carlsbad, CA) according to manufacturer’s protocol. Briefly, cells were co-transfected with 0.4 µg/mL pLSV40-GL3/Bach1 or mutant pLSV40-GL3/Bach1-Mut, 0.4 µg/mL of pLSV40-Rluc, and 10–50 nM miRNA mimics. 24–48 h after transfection, cells were harvested and lysed and luciferase reporter activities were measured using Dual-Glo® Luciferase Assay System. Firefly luciferase activity was normalized to Renilla luciferase activity and total protein, determined using the bicinchoninic acid (BCA) protein assay kit. Values for cells without miRNA mimic transfection were set equal to 1.
2.4 miRNA microarray analysis
The RNA samples were Poly (A) tailed and ligated to biotinylated signal molecules using the FlashTag™ Biotin RNA labeling Kit (Genisphere, LLC, Hatfield, PA). An Enzyme Linked Oligosorbent Assay (ELOSA) QC assay was performed to verify labeling prior to array hybridization. Hybridization, washing, staining and scanning were performed using Affymetrix GeneChip® system instruments (Affymetrix, Santa Clara, CA). Affymetrix GeneChip® Operating Software (GCOS) version 1.4 was used to analyze microarray image data and to compute intensity values. Affymetrix CEL files containing raw, probe-level signal intensities were analyzed using Partek Genomics Suite (Partek, St. Louis, MO). Robust multichip averaging (RMA) was used for background correction, quantile normalization and probeset summarization with median polish [28]. Statistical difference was calculated by two-way ANOVA analysis with false discovery rate (FDR). Cluster and Treeview software was used to perform miRNA hierarchical cluster analysis and visualization [29].
2.5 Transfections with miRNA mimics and inhibitors
The miRIDIAN miRNA mimics for let-7b, let-7c, miR-98, customized mutant let-7b, and miRNA mimic negative control (MMNC), and the miRIDIAN miRNA inhibitors for let-7a, let-7c, let-7e and miRNA inhibitor negative control (MINC) were obtained from Dharmacon (Lafayette, CO). Transfection of miRNA mimics and inhibitors were performed as described previously [12]. Briefly, Huh-7 or HepG2 cells were transfected with 10–50 nM of tested miRNA mimics or inhibitors using Lipofectamine 2000 from Invitrogen (Carlsbad, CA) according to manufacturer’s protocol. After 24–48 h transfection, cells were harvested for appropriate subsequent assays.
2.6 Human Bach1 expression vector and transfection
The human Bach1 expression vector [30], constructed by subcloning the human Bach1 full length cDNA into the mammalian expression vector pCMV6-XL4 (OriGene, Rockville, MD), was obtained from OriGene. For transfection of Bach1 expression vector pCMV-Bach1, Huh-7 cells were plated in 24-well plates one day prior to transfection and transfected at 50% confluence. Cells were transfected with 0.4 µg/well of pCMV-Bach1 using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions.
2.7 Real-time quantitative RT-PCR
Total RNA from treated cells was extracted and cDNA was synthesized. Real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was performed using a CFX96™ six-channel Real-Time PCR Detection System from Bio-Rad (Hercules, CA) and iQ™ SYBR Green Supermix Real-Time PCR kit (Bio-Rad) as described previously [11, 12]. Samples without template and without reverse transcriptase were included as negative controls, which, as expected, produced negligible signals (Ct values>35). Fold-change values were calculated by comparative Ct analysis after normalizing for the invariant quantity mRNA of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in the same samples. For analysis of miRNA levels, small RNAs from treated cells were isolated using mirVANA™ Paris miRNA Isolation Kit (Ambion, Austin, TX). 15 ng of small RNAs were converted to cDNA using TaqMan MicroRNA Reverse Transcription Kit and primers. 1.33 ul of 1:10 diluted cDNAs was used for real time qPCR in a total volume of 20 ul containing 1× TaqMan Universal PCR Master Mix and specific assay for selected mature miRNAs (Applied Biosystems, Carlsbad, CA). Fold-change values were calculated by comparative Ct analysis after normalizing for the invariant quantity of RNU 48 in the same samples.
2.8 Western blots
Western blots (WB) were performed as reported previously [11, 12, 26, 31]. In brief, total proteins (30–50 µg) were separated on 4–15% gradient sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). After electrophoretic transfer onto ImmunBlot polyvinylidene fluoride (PVDF) membrane, membranes were blocked in PBS containing 5% nonfat dry milk and 0.1% Tween-20, and then incubated overnight with primary antibody at 4°C. The membranes were then incubated for 1 hour with secondary antibodies (dilution 1:10,000). A Kodak 1DV3.6 computer-based imaging system (Rochester, NY) was used to measure the relative density of each specific band and perform densitometric analysis of specific band intensities obtained after WB.
2.9 Cell viability assay
Effects of let-7b, let-7c and miR-98 on tert-butyl hydroperoxide (tBuOOH) induced cytotoxicity was measured using CellTiter-Glo® Reagent by determining the number of viable cells based on quantitation of the ATP present, which signaled the presence of metabolically active cells. Huh-7 cells were transfected for 48 h with 50 nM of let-7b or mimic negative control by Lipofectamine 2000. Cells were then incubated with various concentrations of tBuOOH for 4 or 6 h. CellTiter-Glo® Reagent was added for CellTiter-Glo luminescent cell viability assay. The luminescence was read on a Synergy HT microplate reader from Bio-Tek (Winooski, VT) with integration time set for 0.25 to 1 second. Decreases in luminescence were taken as an index of cellular cytotoxicity.
2.10 Cellular reactive oxygen species (ROS) production assay
Huh-7 cells were transfected with let-7b mimic or mimic negative control by using Lipofectamine 2000, and then treated with tBuOOH to induce levels of intracellular ROS. ROS levels were measured using DCF assay as described previously [31]. Intracellular ROS levels were measured as an increase in fluorescence of the oxidized product of 2’,7’-dichlorofluorescein diacetate (DCF-DA) on a Synergy HT Multi-Detection Microplate Reader (BioTek, Winooski, VT) at the excitation and emission wavelengths of 488 nm and 525 nm, respectively.
2.11 Statistical analysis
Initial analysis showed that results were normally distributed. Therefore, parametric statistical procedures were used. The Student’s t-test or ANOVA was used (as appropriate) to analyze the differences between samples. Values of P <0.05 were considered statistically significant. Experiments were repeated at least three times with similar results. All experiments included at least triplicate samples for each treatment group. Statistical analyses were performed with JMP 4.0.4 software from SAS Institute (Cary, NC).
3. RESULTS
3.1 In silico analysis of putative binding sites for let-7 miRNA in the 3’-UTR of Bach1 mRNA
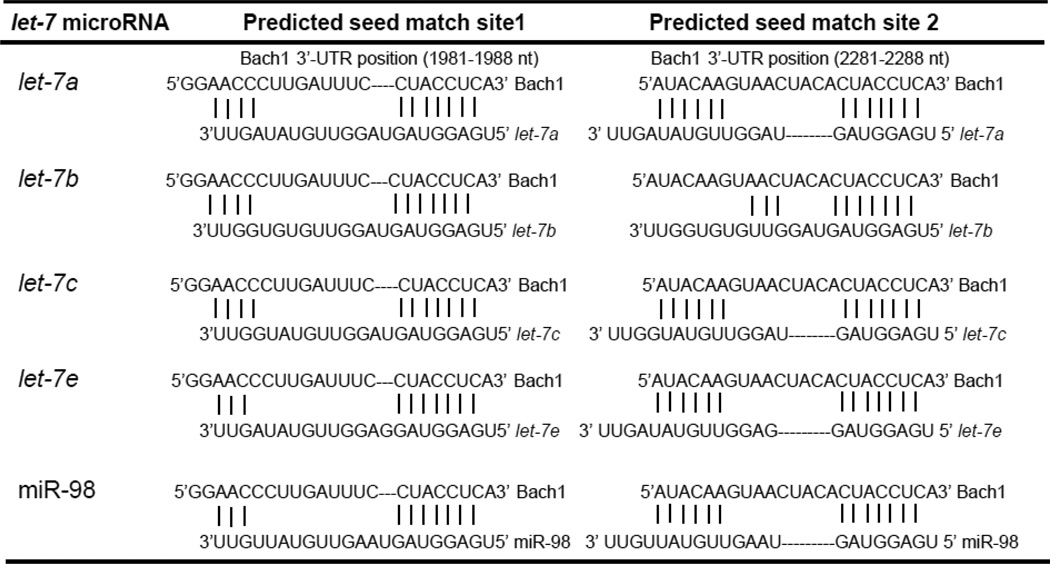
We employed three algorithms, TargetScan5.1, PicTar and miRanda for the in silico prediction of binding sites in the Bach1 mRNA for let-7 miRNA and found two perfect seed region matches between let-7 miRNA and Bach1 3’-UTR (Table 1 and Suppl. Fig. 1). These sites are highly conserved in the human, mouse, rat, dog, chicken and pig. No perfect binding site predicted for let-7 miRNA in the coding region of Bach1 was identified.
Table 1.
Computational analysis of binding sites for let-7a, let-7b, let-7c, let-7e and miR-98 in the 3’-UTR of Bach1 mRNA
 |
3.2 let-7b, let-7c and miR-98 directly act on the 3’-UTR of Bach1 mRNA in Huh-7 cells
To determine whether let-7 miRNA directly binds the predicated sites of the Bach1 3’-UTR, we performed luciferase reporter assays. We chose three representative members in the let-7 miRNA family, namely let-7b, let-7c and miR-98, and over-expressed their levels by transfecting Huh-7 cells with let-7b, let-7c, miR-98 miRNA mimics. We also transfected Huh-7 cells with MMNC as controls. Two firefly luciferase reporter constructs, pLSV40-GL3/Bach1-WT with two perfect seed match sites for let-7 miRNA (Fig.1A), and pLSV40-GL3/Bach1-Mut with partial seed match for let-7b, let-7c and miR-98 that failed to meet the criteria for miRNA binding (Fig.1B), were used. let-7b, let-7c, or miR-98 mimic resulted in a significant decrease in Bach1 3’-UTR-dependent luciferase activity but not in mutant Bach1 3’-UTR-dependent luciferase activity, whereas MMNC had no effect on WT or mutant reporter luciferase activities (Fig.1C). These results demonstrated that let-7b, let-7c and miR-98 directly target Bach1 3’-UTR.
Figure 1. let-7 miRNA mimics repress wild type but not mutant reporter activities.
(A) The firefly luciferase reporter construct pLSV40-GL3/Bach1 containing two seed match sites for let-7 miRNA is schematically illustrated; (B) Four nucleotides in two seed match sites of the Bach1 3’-UTR (highlighted) were mutated, leading to a partial seed match at positions 2–8 of let-7b, let-7c and miR-98 that failed to meet the criteria for miRNA binding. Effects of let-7 miRNA on WT and mutant reporter activities were illustrated. Huh-7 cells were co-transfected with 0.4 µg/mL of pLSV40-GL3/Bach1-WT or pLSV40-GL3/Bach1-Mut, 0.4 µg/mL of pRL-TK (renilla) and with 50 nM let-7 mimic, or MMNC as negative control by Lipofectamine 2000 as indicated. 48 h after transfection, the luciferase reporter activities were measured using Dual Luciferase Assay System from Promega. Firefly luciferase activities were normalized to renilla luciferase activities and total protein, as indicated in Materials and Methods. Values for cells with 50 nM of MMNC transfection were set equal to 1. Data are presented as means ± SE, n=3; (C) let-7b, let-7c and miR-98 significantly inhibit WT but not mutant reporter activities (*differs from MMNC only, p=0.012, let-7b; p=0.016, let-7c; and p=0.012, miR-98).
To obtain additional evidence of direct interaction between let-7 miRNA and Bach1-3’-UTR, mutant let-7b with four nucleotides replaced in the seed region was created to re-establish perfect seed region matches between mutant let-7b and mutant Bach1 3’-UTR (Fig.2A). We predicted that mutant let-7b, containing two perfect seed region sites for mutant Bach1 3’-UTR, should restore its effect on mutant reporter activity. Huh-7 cells were co-transfected with pLSV40-GL3/Bach1-Mut, pRL-TK, and with increasing concentrations of let-7b or mutant let-7b mimic, and luciferase reporter activity was assayed. As expected, mutant let-7b resulted in a significant reduction in let-7b, let-7c mutant reporter luciferase activity, whereas let-7b did not affect mutant reporter luciferase activity (Fig.2B), further indicating the direct interaction between let-7 miRNA and the 3’-UTR of Bach1 mRNA.
Figure 2. Mutant let-7b mimic inhibits the luciferase activity of mutant Bach1 3’-UTR reporter.
(A) Four nucleotides in the seed match sites of let-7b were mutated, resulting in formation of perfect seed region matches between mutant let-7b and mutant Bach1 3’-UTR; (B) Effect of wild type or mutant let-7b mimic on mutant Bach1 3’-UTR reporter activity. Huh-7 cells were co-transfected with 0.4 µg/mL of pLSV40-GL3/Bach1, 0.4 µg/mL of pRL-TK and with 0, 10, 20 or 50 nM wild type let-7b or mutant let-7b mimic for 48 h, firefly and renilla luciferase activities were measured using Dual Luciferase Assay System from Promega. Firefly luciferase activities were normalized to renilla luciferase activities and total protein, as indicated in Materials and Methods. Values for cells without miRNA transfection were set equal to 1. Data are presented as means ± SE, n=3. *differs from 0, p=0.017, 10 nM; p=0.004, 20 nM; and p=0.003, 50 nM).
3.3 let-7 miRNA directly down-regulates Bach1 gene expression at a post-transcriptional level in Huh-7 and HepG2 cells
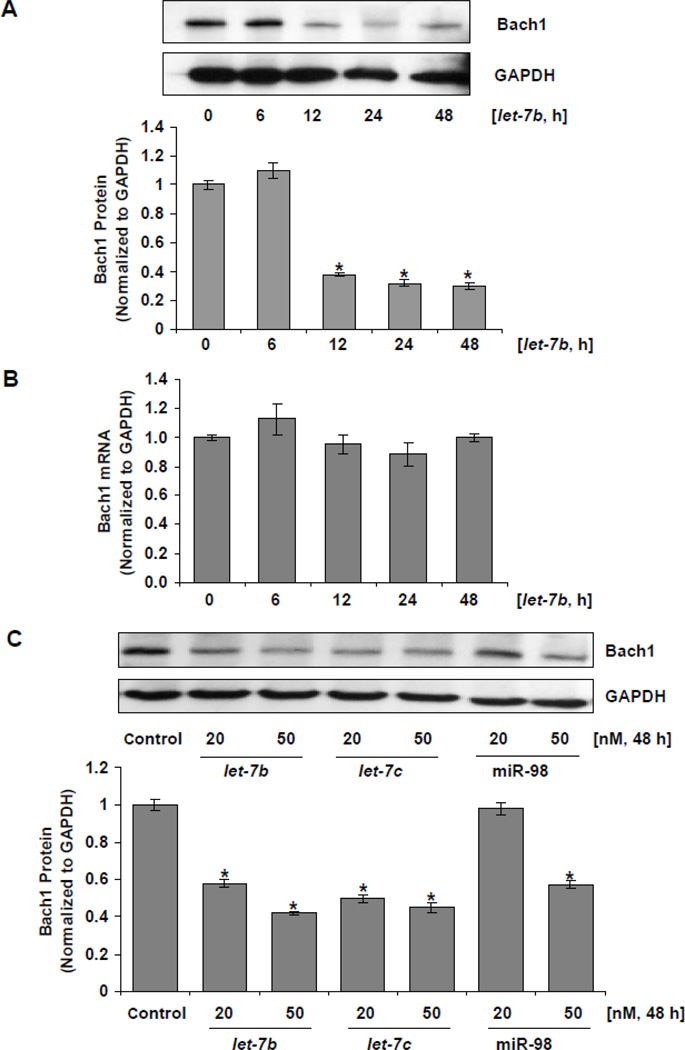
Having shown that let-7b, let-7c and miR-98 directly act on the 3’-UTR of Bach1 mRNA, we next determined whether let-7 miRNA regulates Bach1 gene expression in Huh-7 cells and hepG2 cells. The effects of let-7 miRNA mimics or inhibitors on Bach1 mRNA and protein levels were examined by real-time qRT-PCR and WB, respectively. We first determined time-course effect of let-7b mimic on Bach1 mRNA and protein levels, and then chose three representative members, including let-7b, let-7c and miR-98, and over-expressed their levels by transfecting Huh-7 cells with let-7b, let-7c, miR-98 miRNA mimics. let-7b, let-7c, or miR-98 mimic resulted in 50–70% reduction of Bach1 protein levels compared with MMNC (Figs.3A&C). No significant effect of these mimics on Bach1 mRNA levels was observed (Figs.3B&D), which is consistent with our previous observations.
Figure 3. let-7 miRNA mimics down-regulate Bach1 protein levels in human Huh-7 cells.
Human Huh-7 cells were transfected with 0–50 nM of let-7 miRNA mimics, or MMNC as control by Lipofectamine 2000 for indicated times. Cells were harvested, and total RNA and proteins were extracted. The levels of Bach1 mRNA and protein were assessed by real-time qRT-PCR and WB, respectively, as described in Materials and Methods. The amounts of Bach1 mRNA and protein were normalized to GAPDH, which did not vary with transfection. Values for cells at time 0 or with a MMNC transfection were set equal to 1. Data are presented as means ± SE, n=3. * differs from time 0 or negative control, P<0.05. (A) WB analysis of time-course effects of let-7b mimic on Bach1 protein levels; (B) Time-course effects of let-7b mimic on Bach1 mRNA levels; (C) WB analysis of effects of let-7b, let-7c and miR-98 mimics on Bach1 protein levels; (D) Effects of let-7b, let-7c and miR-98 mimics on Bach1 mRNA levels.
Next, we transfected Huh-7 cells with a combination of let-7a, let-7c and let-7e inhibitors and assessed effects of these inhibitors on Bach1 mRNA and protein levels by real-time qRT-PCR and WB, respectively. We chose to inhibit let-7a, let-7c and let-7e levels since these members of the let-7 miRNA family are moderately abundantly expressed in Huh-7 cells and are the most abundant let-7 miRNAs in the let-7 miRNA family members expressed in Huh-7 cells (Suppl. Fig. 2), and inhibition of these miRNAs were expected to largely decrease let-7 miRNA levels in Huh-7 cells (Suppl. Fig.3B). let-7a, let-7c and let-7e inhibitors (20 nM each) significantly up-regulated Bach1 protein levels in comparison to negative controls in Huh-7 cells (Figs.4A&B) without affecting Bach1 mRNA levels (Fig.4C).
Figure 4. let-7 miRNA inhibitors up-regulate Bach1 protein levels in human Huh-7 cells.
Huh-7 cells were transfected with let-7 miRNA inhibitors which contain a combination let-7a, let-7c and let-7e inhibitors, or MINC as control by Lipofectamine 2000. 48 h after transfection, cells were harvested and total RNA and proteins were extracted. Bach1 mRNA and protein levels were assayed by real-time qRT-PCR and WB, respectively. The amounts of Bach1 mRNA and protein were normalized to GAPDH, which did not vary with transfection. Values for cells with a mock transfection (0) were set equal to 1. Data are presented as means ± SE, n=3. (A) Up-regulation of Bach1 protein levels by let-7 miRNA inhibitors; (B) Densimetric analysis of Bach1 and GAPDH band intensities (*p=0.009, let-7 inhibitors vs MINC); (C) Lack of effect of let-7 miRNA inhibitors on Bach1 mRNA levels.
3.4 let-7 miRNA up-regulates HMOX1 gene expression and protects cells from oxidative injury in Huh-7 cells
We asked whether down-regulation of Bach1, a well-documented transcriptional repressor of the HMOX1 gene, increases HMOX1 gene expression. We determined the effects of let-7b, let-7c and miR-98 mimics on HMOX1 mRNA levels by real-time qRT-PCR. Huh-7 cells were transfected with let-7b, let-7c or miR-98 mimic or MMNC as control. As anticipated, these let-7 miRNA mimics significantly up-regulated HMOX1 mRNA levels by 3–4-fold in comparison with non-specific MMNC (Fig.5A). The up-regulation of HMOX1 by these miRNAs was correlated with down-regulation of Bach1 gene expression. Furthermore, a combination of let-7a, let-7c and let-7e inhibitors (20 nM each) significantly inhibited let-7 miRNA-mediated up-regulation of HMOX1 gene expression in Huh-7 cells (Fig.5B).
Figure 5. Effects of let-7 miRNA mimics and inhibitors on expression of the HMOX1 gene in human Huh-7 cells.
Huh-7 cells were transfected with let-7 miRNA mimics, MMNC, let-7 miRNA inhibitors (a combination of let-7a, let-7c and let-7e inhibitors) or MINC by Lipofectamine 2000 for 48 h. Cells were harvested, and total RNA was then extracted. The levels of HMOX1 mRNA were assessed by real-time qRT-PCR. The amounts of HMOX1 mRNA were normalized to GAPDH which did not vary with transfection. Values for cells with mimic or inhibitor negative control transfection were set equal to 1. Data are presented as means ± SE, n=3. (A) let-7 miRNA mimics up-regulate HMOX1 mRNA levels (*differs from MMNC only, p=0.004, let-7b; p=0.002, let-7c; and p=0.001, miR-98); (B) let-7 miRNA inhibitors down-regulate HMOX1 mRNA levels (*p=0.005, let-7 inhibitors vs MINC).
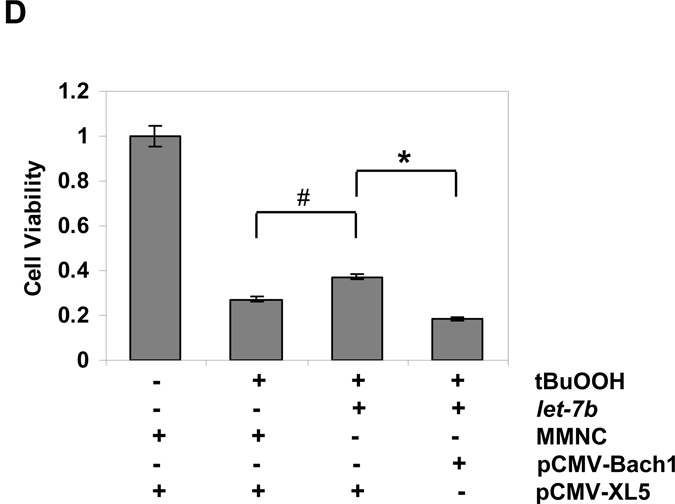
In our last set of experiments, we determined whether over-expression of let-7 miRNA could protect cells from oxidative injury. tBuOOH is widely used model oxidant for experiments to increase intracellular ROS levels and to produce oxidative injury, and therefore was used to increase cellular ROS and produce oxidative injury in Huh-7 cells. Huh-7 cells were transfected with let-7b mimic or MMNC as control for 48 h. Cells with let-7b mimic transfection showed significant protection from oxidant-mediated cytotoxicity compared with cells transfected with MMNC (Figs.6A & B). Over-expression of let-7b significantly blocked increased cellular ROS induced by tBuOOH in Huh-7 cells transfected with let-7b mimic but not MMNC (Suppl. Fig. 5). And then we asked if the protection against oxidative injury by let-7b is through the pathway to down-regulate Bach1 and up-regulate HMOX1. To this end, we performed experiments to over-express Bach1 by using an expression vector containing the coding region of the human Bach1 gene [30] and determine the effects of over-expression of Bach1 on the let-7b-mediated protection against tBuOOH induced oxidative stress. Transfection of Bach1 expression vector resulted in a marked increase in Bach1 protein levels. let-7b mimic significantly protected cells from tBuOOH induced oxidative injury, whereas the protection was abrogated by over-expression of Bach1 in Huh-7 cells (Figs. 6C&D).
Figure 6. Over-expression of let-7b protects cells from oxidative injury in Huh-7 cells.
Huh-7 cells were transfected for 48 h with 50 nM of let-7b or MMNC by Lipofectamine 2000. Cells were then incubated with the indicated concentrations of tBuOOH for 4 or 6 h. CellTiter-Glo® Reagent was added for CellTiter-Glo luminescent cell viability assay on a Synergy HT microplate reader with integration time set for 0.25 to 1 second. Decreases in luminescence were taken as an index of cellular cytotoxicity. Data represent means ± SE of triplicate determinations. Data are presented as means ± SE, n=3. * differs from MMNC only, P<0.05. Susceptibility of Huh-7 cells transfected with MMNC and let-7b to tBuOOH induced oxidative injury was compared. (A) 4 h after tBuOOH treatment (*differs from MMNC, p= 0.03); (B) 6 h after tBuOOH treatment (*differs from MMNC, p=0.03); (C) WB analysis of over-expression of Bach1 by pCMV-Bach1; (D) The protection of cells by let-7b against oxidative injury was ablated following over-expression of Bach1. Huh-7 cells were transfected with 0.4 µg/mL of pCMV-Bach1 or pCMV-XL5, a control vector without Bach1 cDNA insertion for 24 h, and then transfected with 50 nM of let-7b or MMNC for 48 h. Cells were then incubated with 400 µM of tBuOOH for 6 h. Cell viability assays were performed as described above. Data are presented as means ± SE, n=3. #p=0.006 and *p=0.001.
4. DISCUSSION
The major novel findings of this study are summarized as follows: 1) The predicted binding sites in the 3’-UTR of Bach1 for let-7 miRNA are functional as demonstrated by multiple lines of evidence, including (a) two conserved 7-mer sites in the 3’-UTR of Bach1 that perfectly match the “seed region” (positions 2–8) of let-7 miRNA in initial in silico analysis (Table 1 and Suppl. Fig.1); (b) the expected effects of these miRNAs on luciferase activities of the luciferase reporter constructs harboring the wild type and mutant Bach1 3’-UTRs (Figs.1&2); (c) down-regulation of Bach1 protein levels by over-expression of let-7b, let-7c and miR-98 (Figs.3A&C); and (d) up-regulation of Bach1 protein levels by let-7 miRNA inhibitors (Figs.4A&B); 2) Over-expression of let-7 miRNA attenuates oxidative injury via enhancement of the HMOX1 gene expression in human hepatocytes (Figs.5&6, Suppl.Fig.5). These results suggest that over-expression or up-regulation of let-7b, let-7c and miR-98 represents a novel therapeutic approach for protecting human hepatocytes from oxidant injury as may occur in hepatitis, and other forms of liver disease.
miRNAs have emerged as a class of important modulators of gene expression at the post-transcriptional level, acting mainly through translational regulation and mRNA destabilization [32–36]. In mammals, miRNAs are predicted to regulate more than 60% of all protein-coding genes[37]. The numbers of miRNAs continue to grow, and additional candidate genes regulated by miRNAs continue to be identified. The role for miRNA in regulating HMOX1, either directly or indirectly, has remained largely unknown. HMOX1 contains a relatively short 3’-UTR of 603 nucleotides (nt). Therefore, there are limited numbers of candidate miRNAs that are predicted to bind this gene and modulate its expression directly. A recent study demonstrated that miRNA-217 and miR-377 in combination decrease HMOX1 protein expression and enzymatic activity by directly interacting with the 3’-UTR of the HMOX1 mRNA, and knockdown of both miRNA-217 and miR-377 by their antagonists leads to a 2-fold increase in HMOX1 protein expression in primary human umbilical vein cells (HUVEC)[38]. Bach1, an important repressor of expression of HMOX1 and other heme-sensitive genes, has a much longer of 3’-UTR (3313 nt) which provides greater opportunity for putative miRNAs binding. We recently reported that miR-196, which has two putative good match binding sites for the 3’-UTR of Bach1 mRNA, translationally represses the expression of this protein, and up-regulates HMOX1 in human hepatoma cells [12]. Targeting Bach1 by miRNAs has emerged as an effective approach to enhance the expression of the HMOX1 gene in the liver. In this work, computational analysis predicted binding sites for let-7a, b, c, d, e, f, g, i and miR-98 in the 3’-UTR of Bach1 (Table 1 and Suppl. Fig.1). We over-expressed levels of the let-7 miRNA by using let-7b, let-7c and miR-98 mimics, or inhibited the let-7 miRNA levels by let-7a, let-7c and let-7e inhibitors, to validate or rule out a role for these miRNAs in regulation of Bach1 and HMOX1. This selection was based on the TargetScan context scores and miRNAs abundance profile relative to the total pool of miRNAs expressed in human liver cells (Suppl. Fig. 2). We demonstrated that let-7 miRNA bind to the 3’-UTR of Bach1 and directly down-regulate expression of this protein at a post-transcriptional level. Recent work has reported that let-7g suppresses Bach1 expression of both mRNA and protein levels in breast cancer lines [39], a system which is different from that in the present work. Our results in human hepatoma Huh-7 and HepG2 cells provide strong evidence that the let-7b, c and miR-98 down-regulate Bach1 gene expression at the protein level (Figs. 3&4, Suppl. Fig. 4). Since no perfect binding site predicted for let-7 miRNA in the coding region and 3’-UTR of HMOX1 exists, we do not expect a direct effect of let-7 miRNA on HMOX1. Rather we propose that enhancement of HMOX1 by let-7 miRNA is achieved through direct interaction of let-7 miRNA with Bach1, a well-recognized transcriptional repressor to directly regulate HMOX1 gene expression.
There is increasing evidence in recent studies that several members of the let-7 miRNA family are involved in human liver development, with let-7a, let-7b, and let-7c expressed at least 2-fold higher in adult compared to embryonic liver [21], and in liver disorders such as hepatocellular carcinoma [22, 23], hepatic fibrosis [24] and hepatitis [25]. In these and other liver disorders oxidative stress is known to accelerate the development and progression of disease, and antioxidant therapy has thus been proposed as treatment approach [40–43]. Over-expression of some of the let-7 miRNA family members was found to inhibit the proliferation of human hepatocellular carcinoma by targeting tumor-related genes [23], and to display anti-fibrotic effects, attributed to interaction with the mRNA of TGFβ-R1. However, there is also the possibility that these let-7 miRNA members could act indirectly, for example, by increasing expression of antioxidant proteins like HMOX1, a key cytoprotective enzyme, intimately and critically involved in protecting cells against oxidative stress and other types of stresses. This hypothesis was confirmed by our study. Over-expression of let-7b, let-7c and miR-98 led to enhancement of HMOX1 levels and protection of human liver cells against oxidative injury caused by tBuOOH. This is noteworthy since the liver is subjected to oxidative stress due to a high rate of oxidative metabolism, and oxidative stress has emerged as a key player in the development of diverse liver diseases, including alcoholic and non-alcoholic steatohepatitis, iron and copper overload, and chronic hepatitis C, etc. Therefore, over-expression of let-7 miRNA family members may represent a novel therapeutic approach for protecting hepatocytes from oxidative stress of diverse underlying cause.
In conclusion, the present study has established that let-7 miRNA directly interacts with the 3’-UTR of Bach1 mRNA and translationally represses the expression of this protein, leading to up-regulation of the HMOX1 gene. Over-expression of the let-7 miRNA family members may represent a potential novel therapeutic approach to liver disease characterized by increased oxidative stress. Our findings thus add an important class of miRNAs that influence expression of HMOX1, a key cytoprotective enzyme. Our studies also raise important new questions such as interactions between oxidative stress and miRNAs that need to be explored further. Studies are underway in our laboratory, designed to systematically identify precise miRNAs, including those unknown miRNA sequences that associate with oxidative stress, and to determine miRNA-mRNA interactions that regulate cellular gene expression related to oxidative stress in cell cultures and human liver tissues by using the latest high-throughput sequencing of RNAs isolated by cross-linking immunoprecipitation cross-linking immunoprecipitation (CLIP) technology (HITS-CLIP).
Supplementary Material
Highlights.
let-7 miRNA directly binds to the 3’-UTR of Bach1 and negatively regulates Bach1 gene expression at a post-transcriptional level.
let-7 miRNA thereby up-regulates the HMOX1 gene expression.
Over-expression of let-7 miRNA attenuates oxidative injury in human hepatocytes.
Acknowledgements
We thank Dr. Bryan R. Cullen (Duke University, Durham, NC) for providing pLSV40-Rluc, pLSV40-GL3 and pLSV40-GL3/Bach1 reporter vectors and Dr. Rolf Renne (University of Florida, Gainesville, FL) for the generous gift of luciferase reporter construct pGL3-Bach1.
Financial Support:
This work was supported by National Institutes of Health (NIH) grant R01-DK38825 to HLB, and Carolinas Medical Center Grants CRGC 08-35 and CRGC 10-010 to WH.
Abbreviations
- BCA
bicinchoninic acid
- bZip
basic leucine zipper
- CLIP
cross-linking immunoprecipitation
- CO
carbon monoxide
- DMEM
Dulbecco’s modified Eagle medium
- FBS
fetal bovine serum
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HITS
high-throughput sequencing
- HMOX1
heme oxygenase1
- MARE
Maf recognition element
- miRNA
microRNA
- MINC
microRNA inhibitor negative control
- MMNC
microRNA mimic negative control
- NQO1
NAD(P)H:quinone oxidoreductase
- nt
nucleotide(s)
- PAGE
polyacrylamide gel electrophoresis
- PVDF
polyvinylidene fluoride
- qRT-PCR
quantitative reverse transcription-polymerase chain reaction
- ROS
reactive oxygen species
- SDS
sodium dodecyl sulfate
- tBuOOH
tert-butyl hydroperoxide
- UTR
untranslated region
- WB
Western blot
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Elbirt KK, Bonkovsky HL. Heme oxygenase: recent advances in understanding its regulation and role. Proc Assoc Am Physicians. 1999;111:438–447. [PubMed] [Google Scholar]
- 2.Bonkovsky HL, Elbirt KK. Heme Oxygenase: Its Regulation and Role. In: Cutler RG, Rodriguez H, editors. Oxidative Stress and Aging, World Scientific. River Edge, NJ: 2002. pp. 699–706. [Google Scholar]
- 3.Brugger J, Schick MA, Brock RW, Baumann A, Muellenbach RM, Roewer N, Wunder C. Carbon monoxide has antioxidative properties in the liver involving p38 MAP kinase pathway in a murine model of systemic inflammation. Microcirculation. 2010;17:504–513. doi: 10.1111/j.1549-8719.2010.00044.x. [DOI] [PubMed] [Google Scholar]
- 4.Wu TW, Carey D, Wu J, Sugiyama H. The cytoprotective effects of bilirubin and biliverdin on rat hepatocytes and human erythrocytes and the impact of albumin. Biochemistry and cell biology. 1991;69:828–834. doi: 10.1139/o91-123. [DOI] [PubMed] [Google Scholar]
- 5.Zhu Z, Wilson AT, Mathahs MM, Wen F, Brown KE, Luxon BA, Schmidt WN. Heme oxygenase-1 suppresses hepatitis C virus replication and increases resistance of hepatocytes to oxidant injury. Hepatology. 2008;48:1430–1439. doi: 10.1002/hep.22491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oyake T, Itoh K, Motohashi H, Hayashi N, Hoshino H, Nishizawa M, Yamamoto M, Igarashi K. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol Cell Biol. 1996;16:6083–6095. doi: 10.1128/mcb.16.11.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ogawa K, Sun J, Taketani S, Nakajima O, Nishitani C, Sassa S, Hayashi N, Yamamoto M, Shibahara S, Fujita H, Igarashi K. Heme mediates derepression of Maf recognition element through direct binding to transcription repressor Bach1. EMBO J. 2001;20:2835–2843. doi: 10.1093/emboj/20.11.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kitamuro T, Takahashi K, Ogawa K, Udono-Fujimori R, Takeda K, Furuyama K, Nakayama M, Sun J, Fujita H, Hida W, Hattori T, Shirato K, Igarashi K, Shibahara S. Bach1 functions as a hypoxia-inducible repressor for the heme oxygenase-1 gene in human cells. J Biol Chem. 2003;278:9125–9133. doi: 10.1074/jbc.M209939200. [DOI] [PubMed] [Google Scholar]
- 9.Shan Y, Lambrecht RW, Ghaziani T, Donohue SE, Bonkovsky HL. Role of Bach-1 in regulation of heme oxygenase-1 in human liver cells: insights from studies with small interfering RNAS. J Biol Chem. 2004;279:51769–51774. doi: 10.1074/jbc.M409463200. [DOI] [PubMed] [Google Scholar]
- 10.Shan Y, Lambrecht RW, Donohue SE, Bonkovsky HL. Role of Bach1 and Nrf2 in up-regulation of the heme oxygenase-1 gene by cobalt protoporphyrin. FASEB J. 2006;20:2651–2653. doi: 10.1096/fj.06-6346fje. [DOI] [PubMed] [Google Scholar]
- 11.Hou W, Shan Y, Zheng J, Lambrecht RW, Donohue SE, Bonkovsky HL. Zinc mesoporphyrin induces rapid and marked degradation of the transcription factor Bach1 and up-regulates HO-1. Biochim Biophys Acta. 2008;1779:195–203. doi: 10.1016/j.bbagrm.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hou W, Tian Q, Zheng J, Bonkovsky HL. MicroRNA-196 represses Bach1 protein and hepatitis C virus gene expression in human hepatoma cells expressing hepatitis C viral proteins. Hepatology. 2010;51:1494–1504. doi: 10.1002/hep.23401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dhakshinamoorthy S, Jain AK, Bloom DA, Jaiswal AK. Bach1 competes with Nrf2 leading to negative regulation of the antioxidant response element (ARE)-mediated NAD(P)H:quinone oxidoreductase 1 gene expression and induction in response to antioxidants. J Biol Chem. 2005;280:16891–16900. doi: 10.1074/jbc.M500166200. [DOI] [PubMed] [Google Scholar]
- 14.Shan Y, Zheng J, Lambrecht RW, Bonkovsky HL. Reciprocal effects of micro-RNA-122 on expression of heme oxygenase-1 and hepatitis C virus genes in human hepatocytes. Gastroenterology. 2007;133:1166–1174. doi: 10.1053/j.gastro.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 16.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 17.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 18.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, Song E. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 19.Wulczyn FG, Smirnova L, Rybak A, Brandt C, Kwidzinski E, Ninnemann O, Strehle M, Seiler A, Schumacher S, Nitsch R. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J. 2007;21:415–426. doi: 10.1096/fj.06-6130com. [DOI] [PubMed] [Google Scholar]
- 20.Zhao C, Sun G, Li S, Lang MF, Yang S, Li W, Shi Y. MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:1876–1881. doi: 10.1073/pnas.0908750107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tzur G, Israel A, Levy A, Benjamin H, Meiri E, Shufaro Y, Meir K, Khvalevsky E, Spector Y, Rojansky N, Bentwich Z, Reubinoff BE, Galun E. Comprehensive gene and microRNA expression profiling reveals a role for microRNAs in human liver development. PloS one. 2009;4:e7511. doi: 10.1371/journal.pone.0007511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimizu S, Takehara T, Hikita H, Kodama T, Miyagi T, Hosui A, Tatsumi T, Ishida H, Noda T, Nagano H, Doki Y, Mori M, Hayashi N. The let-7 family of microRNAs inhibits Bcl-xL expression and potentiates sorafenib-induced apoptosis in human hepatocellular carcinoma. Journal of hepatology. 2010;52:698–704. doi: 10.1016/j.jhep.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 23.Lan FF, Wang H, Chen YC, Chan CY, Ng SS, Li K, Xie D, He ML, Lin MC, Kung HF. Hsa-let-7g inhibits proliferation of hepatocellular carcinoma cells by downregulation of c-Myc and upregulation of p16(INK4A) International journal of cancer. 2011;128:319–331. doi: 10.1002/ijc.25336. [DOI] [PubMed] [Google Scholar]
- 24.Venugopal SK, Jiang J, Kim TH, Li Y, Wang SS, Torok NJ, Wu J, Zern MA. Liver fibrosis causes downregulation of miRNA-150 and miRNA-194 in hepatic stellate cells, and their overexpression causes decreased stellate cell activation. American journal of physiology. 2010;298:G101–G106. doi: 10.1152/ajpgi.00220.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Lu Y, Toh ST, Sung WK, Tan P, Chow P, Chung AY, Jooi LL, Lee CG. Lethal-7 is down-regulated by the hepatitis B virus x protein and targets signal transducer and activator of transcription 3. Journal of hepatology. 2010;53:57–66. doi: 10.1016/j.jhep.2009.12.043. [DOI] [PubMed] [Google Scholar]
- 26.Hou W, Tian Q, Zheng J, Bonkovsky HL. Zinc mesoporphyrin induces rapid proteasomal degradation of hepatitis C nonstructural 5A protein in human hepatoma cells. Gastroenterology. 2010;138:1909–1919. doi: 10.1053/j.gastro.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi JT, Braich R, Manoharan M, Soutschek J, Ohler U, Cullen BR. A viral microRNA functions as an orthologue of cellular miR-155. Nature. 2007;450:1096–1099. doi: 10.1038/nature05992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics (Oxford, England) 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 29.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hintze KJ, Katoh Y, Igarashi K, Theil EC. Bach1 repression of ferritin and thioredoxin reductase1 is heme-sensitive in cells and in vitro and coordinates expression with heme oxygenase1, beta-globin, and NADP(H) quinone (oxido) reductase1. J Biol Chem. 2007;282:34365–34371. doi: 10.1074/jbc.M700254200. [DOI] [PubMed] [Google Scholar]
- 31.Hou WH, Rossi L, Shan Y, Zheng JY, Lambrecht RW, Bonkovsky HL. Iron increases HMOX1 and decreases hepatitis C viral expression in HCV-expressing cells. World J Gastroenterol. 2009;15:4499–4510. doi: 10.3748/wjg.15.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bushati N, Cohen SM. microRNA functions. Annual review of cell and developmental biology. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 34.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nature reviews. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 36.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science (New York, N.Y. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 37.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome research. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beckman JD, Chen C, Nguyen J, Thayanithy V, Subramanian S, Steer CJ, Vercellotti GM. Regulation of heme oxygenase-1 protein expression by miR-377 in combination with miR-217. J Biol Chem. 2011;286:3194–3202. doi: 10.1074/jbc.M110.148726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yun J, Frankenberger CA, Kuo WL, Boelens MC, Eves EM, Cheng N, Liang H, Li WH, Ishwaran H, Minn AJ, Rosner MR. Signalling pathway for RKIP and Let-7 regulates and predicts metastatic breast cancer. EMBO J. 2011;30:4500–4514. doi: 10.1038/emboj.2011.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi J, Ou JH. Mechanisms of liver injury. III. Oxidative stress in the pathogenesis of hepatitis C virus. Am J Physiol Gastrointest Liver Physiol. 2006;290:G847–G851. doi: 10.1152/ajpgi.00522.2005. [DOI] [PubMed] [Google Scholar]
- 41.Okuda M, Li K, Beard MR, Showalter LA, Scholle F, Lemon SM, Weinman SA. Mitochondrial injury, oxidative stress, and antioxidant gene expression are induced by hepatitis C virus core protein. Gastroenterology. 2002;122:366–375. doi: 10.1053/gast.2002.30983. [DOI] [PubMed] [Google Scholar]
- 42.Medina J, Moreno-Otero R. Pathophysiological basis for antioxidant therapy in chronic liver disease. Drugs. 2005;65:2445–2461. doi: 10.2165/00003495-200565170-00003. [DOI] [PubMed] [Google Scholar]
- 43.Webb C, Twedt D. Oxidative stress and liver disease. The Veterinary clinics of North America. 2008;38:125–135. doi: 10.1016/j.cvsm.2007.10.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.