Summary
Long-term memory and synaptic plasticity are thought to require the synthesis of new proteins at activated synapses. The CPEB family of RNA binding proteins, including Drosophila Orb2, has been implicated in this process. The precise mechanism by which these molecules regulate memory formation is however poorly understood. We used gene targeting and site-specific transgenesis to specifically modify the endogenous orb2 gene in order to investigate its role in long-term memory formation. We show that the Orb2A and Orb2B isoforms, while both essential, have distinct functions in memory formation. These two isoforms have common glutamine-rich and RNA-binding domains, yet Orb2A uniquely requires the former and Orb2B the latter. We further show that Orb2A induces Orb2 complexes in a manner dependent upon both its glutamine-rich region and neuronal activity. We propose that Orb2B acts as a conventional CPEB to regulate transport and/or translation of specific mRNAs, whereas Orb2A acts in an unconventional manner to form stable Orb2 complexes that are essential for memory to persist.
Highlights
► Orb2A and Orb2B function by distinct mechanisms in long-term memory ► Orb2A function requires its glutamine-rich domain but not its RNA-binding domain ► Orb2B function requires the RNA-binding domain but not its glutamine-rich domain ► Neuronal activation induces Orb2 heteromers dependent upon Orb2A’s glutamine-rich domain
RNA binding proteins at synapses may underlie plasticity, but their role is not well understood. Krüttner et al. show here that Orb2 isoforms affect memory by distinct mechanisms: through its RNA-binding domain or by forming heterocomplexes in a manner dependent on its glutamine-rich region and neuronal activity.
Introduction
Most behaviors can be modified through the process of learning and memory, allowing the individual to adapt its innate behavioral repertoire to the specific contingencies of the local environment. Depending on the duration, intensity and salience of the learning experience, memories can be either short or long lasting. These behavioral modifications are thought to reflect anatomical and functional changes at specific synapses. Long-term synaptic plasticity requires new protein synthesis both at the soma and locally at the synapse (Sutton and Schuman, 2006). To ensure that local protein synthesis is restricted to the relevant synapses, either through the local capture or translation of mRNAs only in specific synapses, a ”synaptic tag” has been postulated (Frey and Morris, 1997; Martin et al., 1997). Candidates for such a local protein synthesis regulator are members of the cytoplasmic polyadenylation element binding (CPEB) family. The founding members of this family mediate local protein synthesis in early development (Mendez and Richter, 2001), but some CPEB proteins are also thought to mediate protein synthesis in neurons (Alarcon et al., 2004; Atkins et al., 2004; Huang et al., 2002, 2003, 2006; Liu and Schwartz, 2003; Si et al., 2003a; Wells et al., 2001; Wu et al., 1998; Zearfoss et al., 2008; Miniaci et al., 2008; Si et al., 2003a).
CPEB proteins can be divided into two subfamilies. The CPEB-I subfamily includes the Xenopus CPEB1 and its Drosophila ortholog Orb1, both of which regulate mRNA translation during oogenesis (Mendez and Richter, 2001). CPEB1 and Orb1 bind cytoplasmic polyadenylation elements (CPEs) in the 3′UTR of dormant mRNAs, triggering their polyadenylation and translation (Fox et al., 1989; Hake et al., 1998). Members of the CPEB-II subfamily, including Drosophila Orb2, have been found to function in synaptic plasticity (mCPEB2–4) (Richter, 2001) or long-term memory formation (Drosophila Orb2) (Keleman et al., 2007; Majumdar et al., 2012). The mechanism by which these proteins might regulate protein synthesis is still unclear. Indeed, it has been suggested that neither polyadenylation nor CPEs are involved in translational regulation by CPEB-II proteins (Huang et al., 2006).
Almost all CPEBs exist in multiple variants generated by alternative mRNA splicing (Theis et al., 2003; Wang and Cooper, 2009). The orb2 locus potentially generates six distinct proteins, only two of which contain the well-conserved RNA-binding domain (RBD) in the C terminus that is characteristic of CPEB proteins. These two isoforms, Orb2A and Orb2B, also share a glutamine-rich domain (Q domain) in the N terminus similar to that found in some but not all CPEB proteins in other species (Hafer et al., 2011; Si et al., 2003a). Orb2A and Orb2B differ only in their N termini, which do not contain any conserved domains. In Drosophila, long-term memory mediated by Orb2 is critically dependent on the Q domain (Keleman et al., 2007). The corresponding Q domain in Aplysia CPEB is thought to maintain long-term synaptic facilitation, possibly due to its putative prion-like properties (Heinrich and Lindquist, 2011; Si et al., 2010; Si et al., 2003b).
In order to further understand the cellular and molecular contributions of Orb2 to learning and memory in Drosophila, we have conducted detailed genetic and biochemical analyses of the endogenous Orb2 protein. To ensure that the modified proteins are expressed at the appropriate level and in the appropriate spatial and temporal pattern, we have made all modifications directly in the orb2 locus. Our genetic and biochemical data support a model in which Orb2B acts as a conventional CPEB molecule by a mechanism dependent on its RBD. Orb2A appears to function in an unconventional mechanism that requires the Q domain but is independent of its RBD, possibly by seeding the formation of Orb2A:Orb2B complexes upon neuronal stimulation. We propose that these complexes mediate changes in mRNA translation at activated synapses, contributing to experience-dependent changes in synaptic function and animal behavior.
Results
Generation and Validation of orb2attP
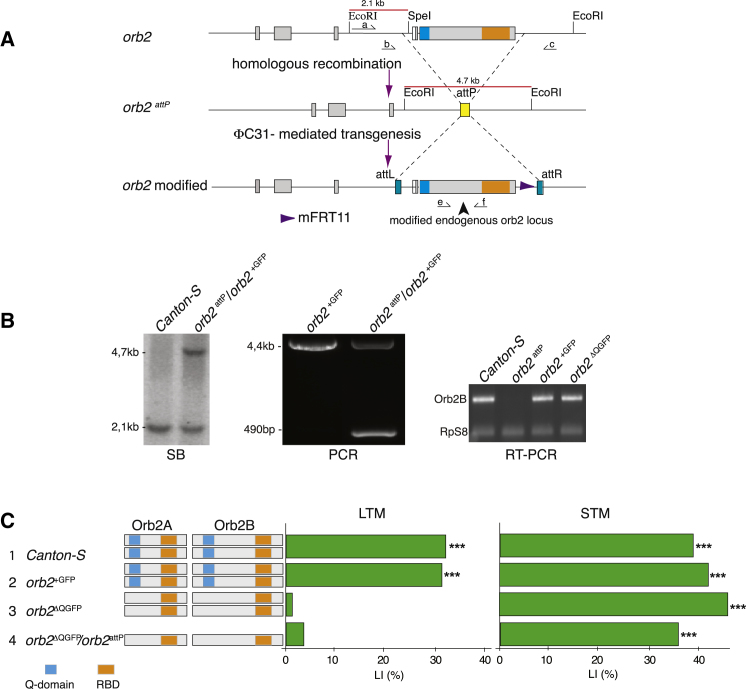
We generated by homologous recombination (Gong and Golic, 2003) an allele that allows rapid modification of the endogenous orb2 locus. This new allele, orb2attP, replaces most of the orb2 open reading frame (including sequences encoding the RBD and Q domains) with an attP recognition site. This attP site can be targeted by the site-specific recombinase phiC31 to insert any desired sequences directly into the orb2 locus (Bischof et al., 2007; Groth et al., 2004; Figure 1A).
Figure 1.
Generation of the orb2attP Allele
(A) Most of the orb2 open reading frame was replaced with the acceptor site, attP, for the phi-C31 recombinase by ends-out homologous recombination. Dashed lines indicate deleted region. orb2 modified alleles were generated by insertion into the orb2attP allele of the relevant donor construct bearing the donor site, attB, and the modified genomic orb2 fragment, by phi-C31 mediated transgenesis.
(B) orb2 modified alleles were verified molecularly by Southern blots (SB), PCR, and RT-PCR. SB (left panel) was performed using probe a indicated in (A) and EcoRI, SpeI restriction digest. PCR (middle panel) amplification was performed using primers b and c indicated in (A). Obtained products were of the predicted size. RT-PCR (right panel) fragments were amplified using primers e and f indicated in (A). Product sizes were as predicted and were verified by DNA sequencing. RpS8 was used as an internal control. orb2attP/orb2+GFP heterozygotes were used for SB and PCR. Adult rare homozygous escapers orb2attP were used for RT-PCR.
(C) LIs (green bars) of males carrying the indicated orb2 alleles (on the left) with the corresponding Orb2A and Orb2B protein organization (in the middle), tested in single-pair assays with mated females as trainers and testers for long-term memory (LTM) and short-term memory (STM). Control Canton-S flies, wild-type rescue allele (orb2+GFP), Q domain-deleted allele (Orb2ΔQGFP), and transheterozygote (Orb2ΔQGFP/orb2attP) were tested. p values are for H0: LI = 0, ∗∗p < 0.01, ∗∗∗p < 0.001 (permutation test).
To validate our approach we first reintroduced into the orb2attP locus either wild-type sequences (orb2+GFP) or a modification designed to delete the Q domain (orb2ΔQGFP). In both cases, as in most of the modifications reported here, the targeted orb2 allele additionally carried sequences encoding a C-terminal GFP tag. The structure of these modified orb2 loci were confirmed by Southern blots, RT-PCR and sequencing (Figure 1B). As expected, the orb2attP mutants were homozygous lethal, whereas the orb2+GFP and orb2ΔQGFP alleles were viable (Keleman et al., 2007). The latter two alleles were tested for memory in a courtship conditioning paradigm, in which males that experience futile courtship of mated and therefore unreceptive females subsequently suppress their courtship toward other mated females (Gailey et al., 1982; Kamyshev et al., 1999; McBride et al., 1999; Siegel and Hall, 1979; Tompkins et al., 1983). Memory in this assay is quantified as a learning index (LI), which measures the extent of the courtship suppression (Experimental Procedures). Males homozygous for orb2+GFP had long-term, 24 hr memory comparable to that of control Canton-S males (2, orb2+GFP, LI = 31.7; 1, Canton-S, LI = 32.6), whereas orb2ΔQGFP and orb2ΔQGFP/orb2attP males had no long-term memory (3, orb2ΔQGFP, LI = 0.63; 4, orb2ΔQGFP/orb2attP, LI = 3.03) (Figure 1C; see Table S1 available online). These data are consistent with the lack of long-term memory previously reported for homozygous and hemizygous orb2ΔQ mutants (Keleman et al., 2007). Short-term, 1 hr memory of all tested genotypes was normal (Figure 1C; Table S2), as previously reported also for orb2ΔQ mutants (Keleman et al., 2007).
These results validate our general strategy for introducing targeted modifications at the orb2 locus and confirm that the C-terminal GFP tag does not impair Orb2 function. Accordingly, we also introduced the GFP tag for other modifications to the orb2 locus reported below, although for simplicity it is only indicated in allele or protein names when it is exploited in immunolabeling or biochemistry experiments.
Orb2A and Orb2B Have Distinct Expression Patterns and Developmental Roles
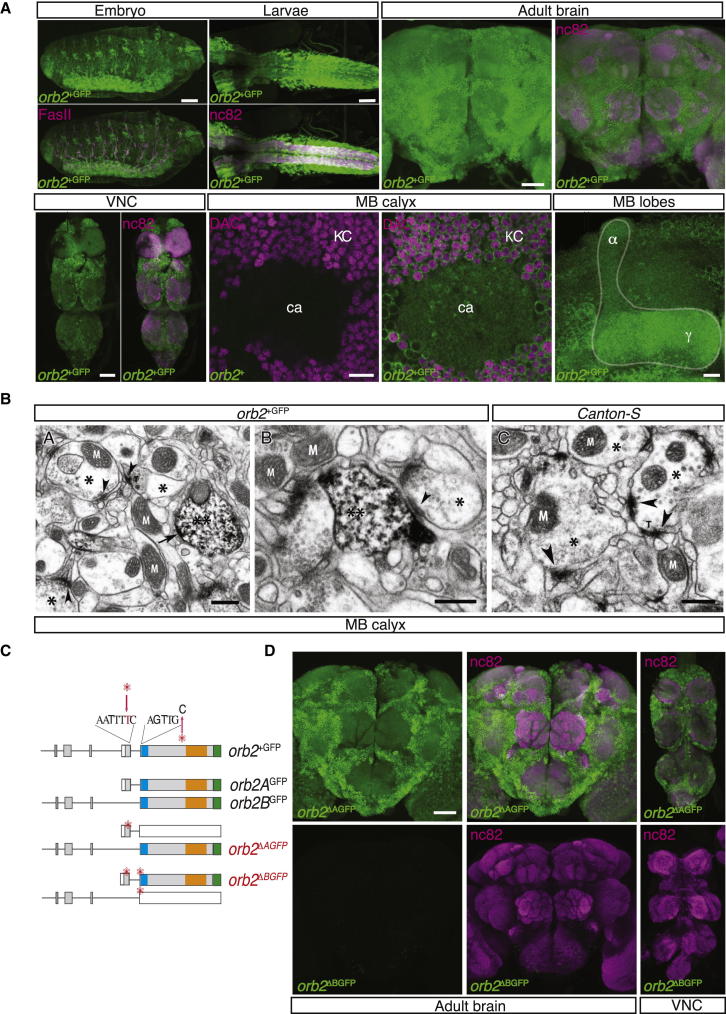
We used antibodies against the GFP tag on the endogenous Orb2 protein encoded by orb2+GFP to determine its expression pattern and subcellular localization. At the level of light microscopy, Orb2 appeared to be broadly expressed throughout the nervous system of embryo, larvae, and adult, including the ventral nerve cord (VNC) and the brain. In the adult brain Orb2 appeared to be widely expressed throughout various regions including the lobes, calyces, and soma of the mushroom bodies (MB), a center for olfactory memory formation in insect brains (Heisenberg, 2003; Figure 2A).
Figure 2.
Orb2 Is Enriched in the Nervous System and Localizes to Pre- and Postsynaptic Compartments
(A) Confocal projections of the Drosophila embryo, larvae, and adult brain of orb2+GFP animals (upper panels) stained with antibody to GFP (green) and counterstained with the general neuropil marker, antibody to either FasII or nc82 (red). Scale bar is 50 μm. Confocal projections of the adult Drosophila ventral nerve cord (VNC), mushroom body calyx (ca), lobes (α, γ), and cell bodies of the Kenyon cells (KC) (lower panels) of orb2+GFP animals stained with antibody to GFP (green) and counterstained with either general neuropil marker, antibody nc82 (red), or Kenyon cell body marker, antibody to Dachshund (DAC) (red). Scale bar is 50 μm except close up of the calyx and MB lobes, 10 μm.
(B) Immuno-EM of the Drosophila orb2+GFP and canton-S brains in the region of calyx. See also Figure S1.
(A) A GFP positive presynaptic cell (two asterisks) labeled by dark DAB precipitates is shown with an electron-dense active zone (arrow). Three GFP negative neurons indicated by a single asterisk display clear electron-dense active zones (arrowheads) with associated synaptic vesicles. Occasionally T bars (T), the presumptive docking site for vesicles, are also visible. (B) A GFP positive neuron (two asterisks) that is postsynaptic to a GFP negative neuron (asterisk) with an active zone (arrowhead) and synaptic vesicles is shown. The closely aligned pre- and postsynaptic membranes of the synaptic cleft are visible.
(C) A similar region of the MB calyx to those above, but from a control Canton-S animal, is shown. Arrowheads indicate the presynaptic specializations: electron-dense active zones, associated synaptic vesicles, and T bars. In all panels, mitochondria are labeled with M and scale bar is 500 nm.
(C) Schematic of the strategy to generate isoform specific orb2 alleles.
(D) Confocal projections of the Drosophila brain and the VNC of orb2ΔAGFP (upper panels) and orb2ΔBGFP (lower panels) mutant flies stained with the antibody to GFP (green) and counterstained with the general neuropil marker, antibody nc82 (red). Scale bar is 50 μm. See also Figures S2 and S3.
Previous studies in other species have variously placed CPEBs at either pre- or postsynaptic sites. Mouse CPEB3, for example, was reported to be present in postsynaptic densities, whereas Aplysia CPEB was shown to localize in presynaptic compartments (Huang et al., 2003, 2006; Liu and Schwartz, 2003; Wu et al., 1998). To examine the subcellular localization of Drosophila Orb2, we examined the calyx (input) region of the MB (see Experimental Procedures for details). Using immuno-electron microscopy, we detected Orb2 both in the presynaptic compartment of the extrinsic MB neurons, characterized by the presence of presynaptic specializations such as electron-dense active zones, synaptic vesicles and occasionally T bars, and the postsynaptic compartment likely to be the termini of the Kenyon cells (KCs) in the calyx, characterized by the presence of close membrane alignments with the presumptive presynaptic region (Figure 2B). Furthermore, consistent with the reported role for Orb2 during development (Hafer et al., 2011; Keleman et al., 2007), we observed morphological defects in the brains of rare adult escapers of orb2attPGFP null mutants (Figure S1).
To distinguish the expression of the Orb2A and Orb2B isoforms, we next generated alleles designed to tag one isoform while eliminating the other (Figure 2C). By inserting a single nucleotide in the exon specific to orb2A, we disrupted the Orb2A reading frame while leaving the GFP-tagged Orb2B reading frame intact. In a second allele, we additionally removed a single nucleotide in the first common exon, thereby restoring the reading frame of Orb2A, including the GFP tag, while now disrupting that of Orb2B. We refer to these two alleles as orb2ΔAGFP and orb2ΔBGFP, respectively. Homozygous orb2ΔBGFP mutants were lethal, whereas orb2ΔAGFP flies were viable and healthy, indicating that Orb2B but not Orb2A has an essential role in development. To examine the respective distributions of the GFP-tagged Orb2B and Orb2A proteins we used homozygous orb2ΔAGFP and adult escaper orb2ΔBGFP animals. The distribution of Orb2B was grossly similar to that observed for Orb2, but Orb2A was undetectable in our experiments (Figure 2D). However, Orb2A has been reported to be expressed in the Drosophila brain at very low levels using a GFP-tagged genomic rescue transgene (Majumdar et al., 2012), consistent with the genetic data presented below that reveal a functional requirement for Orb2A in long-term memory. We therefore conclude that Orb2A is indeed expressed in the adult brain, but either at very low levels, in very few cells, under specific conditions, or in a conformation in which the GFP tag is not readily accessible. Importantly, deletion of either isoform did not affect the various orb2 transcript levels, as revealed by quantitative PCR experiments (Figure S2; Table S3). We therefore attribute the distribution patterns, and the phenotypes reported below, to the specific modifications introduced to each isoform rather than any indirect result of altered transcription from the orb2 locus.
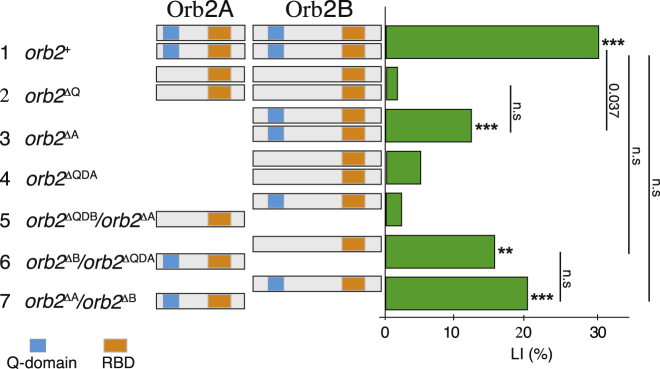
The Q Domain in Orb2A Is Both Required and Sufficient for Long-Term Memory
We used orb2 isoform-specific alleles to test the function of Orb2A and Orb2B in long-term memory. Viable orb2ΔA mutant males, expressing only the B isoform, were tested as homozygotes. These mutants had a normal short-term memory (Table S5D) and a strong detriment in long-term memory in comparison to the wild-type flies (3, orb2ΔA, LI = 12.69; 1, orb2+, LI = 30.31), almost as severe as mutants lacking the Q domain in both isoforms (2, orb2ΔQ, LI = 2.15), suggesting that Orb2A function is critically required for long-term memory (Figure 3; Table S4). However, these mutant flies were able to form residual but statistically significant memory likely to be mediated by Orb2B.
Figure 3.
The Q Domain in Orb2A Is Both Required and Sufficient for Long-Term Memory
LIs (green bars) of males carrying the indicated orb2 alleles (left) with the corresponding Orb2A and Orb2B protein organization (middle) tested in single-pair assays with mated females as trainers and testers for long-term memory. p values are for H0: LI = 0, ∗∗p < 0.01, ∗∗∗p < 0.001 and H0 LI = LI1 (bars) (permutation test). See also Figure S4 and Table S4.
To assess the role of the Q domain in Orb2 isoforms, we generated a specific deletion of this domain by reinserting into orb2attP a genomic fragment in which disruption of either Orb2A or Orb2B was combined with the deletion of the Q domain. orb2ΔQΔA males, expressing only Orb2B lacking its Q domain, had a normal short-term memory (Table S5D) but, like orb2ΔQ mutants, almost no long-term memory (4, orb2ΔQΔA, LI = 5.16; 2, orb2ΔQ, LI = 2.15) (Figure 3; Table S4), suggesting that the residual memory of the orb2ΔA mutants might be mediated by the Q domain of Orb2B.
Since the orb2ΔQΔB mutation was lethal when homozygous, we tested this allele in combination with the viable orb2ΔA allele. These flies, which lack the Q domain specifically in Orb2A, had a normal short-term memory (Table S5D) but no long-term memory (5, orb2ΔQΔB/orb2ΔA, LI = 2.86) (Figure 3; Table S4). This lack of memory shows that the Q domain in Orb2A is essential, and that of Orb2B insufficient, for long-term memory.
To test for the sufficiency of the Q domain in Orb2A, we tested the memory of the transheterozygotes in which the Q domain is present only in Orb2A. The learning index of these mutants was indistinguishable from control flies in which both isoforms are intact (6, orb2ΔB/orb2ΔQΔA, LI = 16.97; 7, orb2ΔB/orb2ΔA LI = 20.83) (Figure 3; Table S4). These results indicate that Orb2A has a specific role in long-term memory that requires the Q domain, which in Orb2B is both dispensable and insufficient.
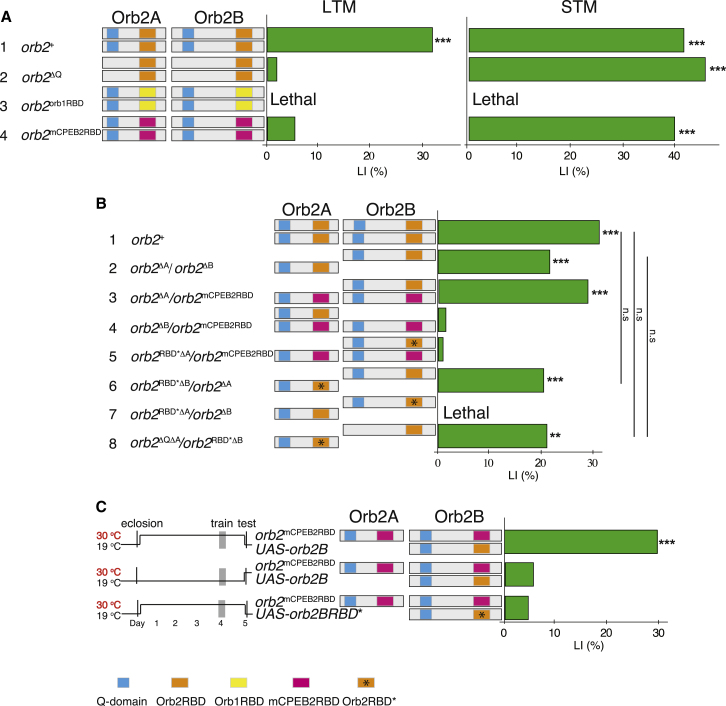
The RBD Is Essential for Function of Orb2B but Not of Orb2A in Long-Term Memory
To assess the role of the RBD in long-term memory, as a first step we chose to replace the Orb2 RBD with the RBDs of other CPEBs, reasoning that such chimeric proteins might retain activity toward conserved and common RNA targets but not Orb2-specific targets involved in long-term memory formation. A swap of the Orb2 RBD with the RBD of Orb1 (3, orb2orb1RBD) did not rescue viability, whereas the swap with the RBD of the mCPEB2 (4, orb2mCPEB2RBD) rendered flies viable and healthy (Figure 4A; Table S5A). This indicated that RNA binding properties of this domain are required during the development, and moreover suggested a potential conservation in RNA targets between the CPEB II family members at least in development. The conservation of RNA targets is consistent with the high homology in this region, ∼90% (Theis et al., 2003).
Figure 4.
The RBD Is Essential for Function of Orb2B but Not of Orb2A in Long-Term Memory
(A and B) LIs (green bars) of males carrying the indicated orb2 alleles (left) with the corresponding Orb2A and Orb2B protein organization (middle), tested in single-pair assays with mated females as trainers and testers for long-term memory (LTM) (A and B) and short-term memory (STM) (A). p values are for H0: LI = 0, ∗∗p < 0.01, ∗∗∗p < 0.001 and H0: LI = LI1 (bars) (permutation test). See also Table S5B.
(C) LIs (green bars) of 247-Gal4, orb2mCPEB2RBD/tub-Gal80ts, UAS-orb2B or UAS-orb2BRBD∗ males, cultured according to two temperature regimes (left) in tests for long-term memory. p values are for H0: LI = 0, ∗∗p < 0.01, ∗∗∗p < 0.001 (permutation test).
See also Table S5C.
Interestingly, orb2mCPEB2RBD mutants showed strong long-term memory impairment in comparison to the control flies (4, orb2mCPEB2RBD LI = 6.16; 1, orb2+, LI = 32.39) (Figure 4A; Table S5A). In contrast, short-term memory was normal (4, orb2mCPEB2RBD, LI = 40.0; 1, orb2+, LI = 42.34) (Figure 4A; Table S5A′), indicating that the long-term memory impairment is unlikely the result of developmental defects caused by the RBD swap. Most importantly, this allele provided us with the unique opportunity to assess the role of the RBD in Orb2B in long-term memory, independently of its role in development. In the orb2mCPEB2RBD background, expression of the wild-type Orb2B, but not Orb2A, fully rescued memory (3, orb2ΔA/orb2mCPEB2RBD, LI = 28.5; 4, orb2ΔB/orb2mCPEB2RBD, LI = 1.68) (Figure 4B; Table S5B), and this rescue was dependent on its RBD (5, orb2RBD∗ΔA/orb2mCPEB2RBD, LI = 1.04, see the paragraph below on the mutated RBD∗). We therefore conclude that the RBD in Orb2B has a specific function in long-term memory, likely in the adult.
To test if Orb2B has a specific role in memory in the adult, we manipulated Orb2B expression in a temporal fashion in a viable orb2mCPEB2RBD background using the tripartite UAS/Gal4/Gal80 expression system (McGuire et al., 2003). Expression in the adult MBs of the wild-type UAS-orb2B, but not UAS-orb2B∗ with the RBD mutated, was sufficient for full rescue of long-term memory (1, TubG80ts, orb2mCPEB2RBD, UAS-orb2B, MB247G4 (29°C), LI = 28.4; 3, TubG80ts, orb2mCPEB2RBD, UAS-orb2B∗, MB247G4 (29°C), LI = 5.89) (Figure 4C; Table S5C). This result shows that in addition to its role during development, Orb2B has a specific function in long-term memory in adult animals that requires its RBD.
If, as we propose, Orb2A does not require its RBD, and Orb2B does not require its Q domain, then we might expect complementation between the relevant orb2 alleles. To test this, we generated a series of Orb2 mutant alleles in which we mutated the key residues in the RBD predicted to be essential for binding to its RNA targets (Mendez et al., 2002). Transheterozygous orb2RBD∗ΔB/orb2ΔA flies, with the functional RBD only in Orb2B, are viable and have normal memory (6, orb2RBD∗ΔB/orb2ΔA, LI = 20.59; 2, orb2ΔA/orb2ΔB, LI = 20.83) (Figure 4B; Table S5B). By contrast, flies with the functional RBD only in Orb2A are lethal (7, orb2RBD∗ΔA/ orb2ΔB). Moreover, transheterozygotes which lack a functional RBD in Orb2A and the Q domain in Orb2B have memory at the level of both wild-type animals and animals with only one wild-type copy of each isoform (8, orb2ΔQΔA/orb2RBD∗ΔB, LI = 20.68; 2, orb2ΔA/orb2ΔB, LI = 20.83) (Figure 4B; Table S5B).
Orb2 A and Orb2B Form Heteromeric Complexes Mediated by the Q Domain
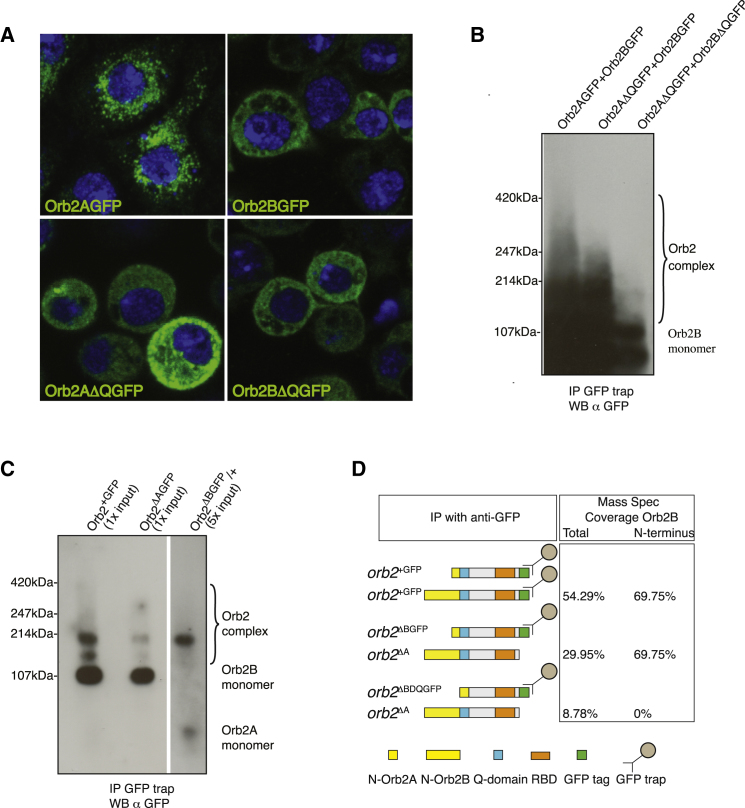
One possible explanation for this interallelic complementation between orb2A and orb2B alleles could be that the proteins they encode form a functional complex. We examined this possibility by light microscopy and biochemistry. Due to the small size of Drosophila neurons, we used expression studies in the Drosophila S2 cell line. S2 cells do not express Orb2 (our unpublished deep seq. data); therefore, we had a clean background in which to test for aggregation and the potential role of the Orb2 Q domain in this process. When individually expressed, Orb2A and Orb2B, have distinct localizations. While both isoforms are localized to the cytoplasm, Orb2A has a granular appearance whereas Orb2B is diffuse. Interestingly, loss of the Q domain in Orb2A (Orb2AΔQGFP) led to a loss of the granular appearance whereas deletion of this domain in Orb2B (Orb2BΔQGFP) did not cause any detectable change (Figure 5A). This observation was extended by IP experiments. In immunoprecipitates from S2 cells transfected with both Orb2AGFP and Orb2BGFP, we observed large Orb2 complexes ranging between 100–400 kDa. Deletion of the Q domain in Orb2A eliminated higher molecular weight multimers above 250 kDa, and deletion of the Q domain both in A and B isoforms eliminated them completely, leaving only the Orb2 monomer band at ∼100 kDa (Figure 5B). These experiments suggest that Orb2 can form multimers in S2 cells that are dependent on the Q domain of both isoforms.
Figure 5.
Orb2A and Orb2B Form Multimeric Complexes Mediated by the Q Domain
(A) Representative confocal images of the Drosophila S2 cells expressing either Orb2AGFP or Orb2BGFP wild-type (upper panels) or with the Q domain deleted (lower panels) stained with the antibody to GFP (green) and counterstained with DAPI (blue).
(B) Immunoprecipitation (IP), SDD-AGE, and western blot (WB) analysis of cell extracts from the Drosophila S2 cells coexpressing Orb2AGFP and Orb2BGFP isoforms either wild-type or with the Q domain deleted.
(C) Immunoprecipitation (IP), SDD-AGE, and western blot (WB) analysis of the orb2+GFP, orb2ΔAGFP, and orb2ΔBGFP/+ head extracts. Note that Orb2A is detectable only with the 5× protein input (12.5 mg).
(D) Mass spectroscopy (MS) analysis of the immunoprecipitates from head extracts of the animals carrying the indicated orb2 alleles (left). In the middle, schematics of the Orb2 protein organization are shown. On the right, % coverage of the total and the unique Orb2B peptides identified by MS after immunoprecipitation of either wild-type or Q domain-deleted Orb2AGFP.
To examine multimerization of both isoforms in vivo, we analyzed immunoprecipitates from fly brains. In orb2+GFP brains, we found Orb2 present both in monomers and oligomers (∼100 and 200 kDa), while in immunoprecipitates from orb2ΔAGFP brains we found Orb2B mostly in a lower molecular weight band of ∼100 kDa. Since deletion of Orb2B is lethal, to analyze multimerization properties of Orb2A we immunoprecipitated Orb2A from the brains of heterozygous animals (orb2ΔBGFP/+). We observed Orb2A almost exclusively in a high molecular weight band of ∼200 kDa. Consistent with Orb2B being expressed at higher levels than Orb2A, Orb2A could not be detected from the same amount of input material as for Orb2B (Figure 5C). In summary, Orb2A preferentially exists in multimeric complexes, whereas Orb2B has a lower propensity to aggregate but may be induced to aggregate in the presence of Orb2A.
In order to test whether Orb2A and Orb2B are present in the same complex, we turned to mass spectrometry (MS), which can readily distinguish between the two isoforms. As we were unable to detect the 9 amino acids specific to Orb2A, we looked for Orb2B-specific peptides when Orb2A was immunoprecipitated. The presence of Orb2B in such immunoprecipitates would indicate that Orb2A is able to pull down Orb2B, and that these two proteins are present in one complex. We precipitated Orb2A from orb2ΔBGFP/+ and orb2ΔBΔQGFP/+ transheterozygous animals. Orb2B-specific peptides were found only from orb2ΔBGFP/+ but not from orb2ΔBΔQGFP/+ brains (Figure 5D). These results show that both Orb2 isoforms are present in the Drosophila brain in one complex, provided Orb2A has an intact Q domain.
Neuronal Activity Induces Orb2 Multimers through the Q Domain of Orb2A
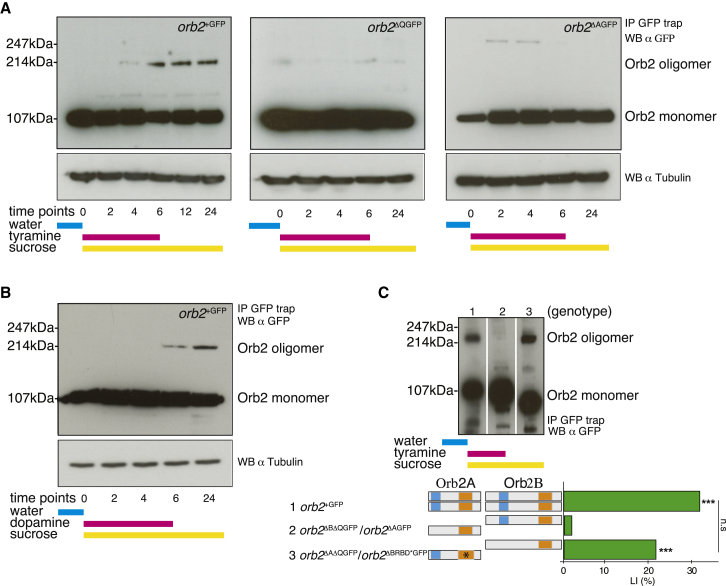
Both dopamine and octopamine have been shown to mediate memory formation in olfactory and courtship learning paradigms (Keleman et al., 2012; Schwaerzel et al., 2003; Tempel et al., 1984). We fed adult flies carrying wild-type orb2+GFP with either dopamine or tyramine (a neurotransmitter and precursor of octopamine) to stimulate broadly neuromodulatory pathways in the brain, and monitored Orb2 multimers at specific time points postfeeding. Orb2 in brain extracts from flies fed with either tyramine or dopamine exists both as monomers (∼100 kDa) and oligomers (∼200 kDa). The oligomer band appears between 4–6 hr postfeeding and lasts for at least 20 more hours (Figures 6A and 6B). This result parallels our previous finding that memory in orb2ΔQ mutants does not last beyond 6 hr (Keleman et al., 2007). In control animals fed with sucrose only (point 0), the oligomer band was absent. The amount of extract we used for these experiments should only monitor the Orb2B isoform. Therefore, we interpret our results as demonstrating that Orb2B the mono- to oligomeric state upon neuronal stimulation.
Figure 6.
Neuronal Stimulation Induces Orb2 Multimers through the Q Domain of Orb2A
(A and B) Adult flies of indicated genotypes, after being starved for 16 hr, were fed with either tyramine or dopamine and sucrose for 6 hr and then continued on sucrose only. At the indicated time points, head extracts were analyzed by IP and WB for Orb2 multimers. Tubulin was used as a loading control.
(C) Adult flies of indicated genotypes, after being starved for 16 hr, were fed with tyramine and sucrose for 6 hr. Head extracts were analyzed by IP and WB for Orb2 oligomers after 24 hr (upper panel). LIs (green bars) of males carrying the indicated orb2 alleles (left) with the corresponding Orb2A and Orb2B protein organization (middle), tested in single-pair assays with mated females as trainers and testers for long-term memory (lower panel). p values are for H0, LI = 0, ∗∗∗p < 0.001 and H0, LI = LI1 (bar) (permutation test).
See also Figure S4.
To evaluate the impact of the Q domain deletion on oligomerization we fed orb2ΔQGFP mutant flies with tyramine and followed Orb2 protein as above. In orb2ΔQGFP mutant brains, although Orb2 protein was expressed at the same level as in the wild-type orb2+GFP animals, only an Orb2 monomer was observed (Figure 6A), implying an acute role for the Q domain in Orb2 oligomerization. This result parallels a complete lack of long-term memory in orb2ΔQ mutant flies.
To investigate if Orb2A regulates oligomerization of Orb2B, we fed animals lacking the Orb2A isoform with tyramine. As above, we did not detect Orb2B oligomers, suggesting that Orb2A is crucial for oligomerization (Figure 6A). Finally, to test the role of Orb2A’s Q domain in Orb2 oligomer formation, we analyzed transheterozygous animals in which the Q domain present only in Orb2A and RBD only in Orb2B, able to form long-term memory (3, orb2ΔQΔAGFP/orb2RBD∗ΔBGFP, LI = 20.68; 1, orb2+GFP, LI = 32.39) (Table S5B). As predicted, in brain extracts from these animals, Orb2 multimers were detected as in the wild-type brains. In contrast, in brain extracts of animals in which the Q domain was lacking specifically in Orb2A and present only in Orb2B, which are unable to form long-term memory, Orb2 oligomers were not detected (2, orb2ΔQΔBGFP/orb2ΔAGFP, LI = 2.86) (Table S4; Figure 6C). We conclude that Orb2 oligomers are induced by neuronal activity in Orb2A-dependent manner. The Q domain of Orb2A is both essential and sufficient, whereas that of Orb2B is dispensable and insufficient, for Orb2 oligomers formation. These results suggest that Orb2 complexes are essential for memory persistence.
Discussion
Local translation of mRNAs in both pre- and postsynaptic compartments is thought to be important for the synaptic modifications that underlie long-lasting memories (Frey and Morris, 1997; Kang and Schuman, 1996; Martin et al., 1997). The CPEB family of proteins regulate local translation (Alarcon et al., 2004; Huang et al., 2006; Si et al., 2003a; Wells et al., 2001; Wu et al., 1998; Zearfoss et al., 2008), and the Drosophila CPEB protein Orb2 is acutely required for long-term memory (Keleman et al., 2007; Majumdar et al., 2012). However, the detailed molecular mechanism of CPEB function in synaptic plasticity and memory formation remains elusive.
We have shown here that the two Orb2 isoforms, Orb2A and Orb2B, both contribute to long-term memory formation, albeit by distinct mechanisms. The two isoforms share the same RNA-binding and Q domains, yet each uniquely requires only one of these domains for its function in long-term memory formation. Specifically, the Q domain is essential in Orb2A but not Orb2B, whereas the RNA-binding domain is required in Orb2B but not Orb2A. Moreover, we found that Orb2A lacking its RNA-binding domain is able to fully complement Orb2B lacking its Q domain. Such interallelic complementation often reflects the formation of the heteromeric complexes between the encoded proteins (Garen and Garen, 1963; Zhang et al., 2006), and indeed we observed that Orb2A and Orb2B are present in the same protein complexes in vitro and in vivo, and that formation of these heteromeric Orb2A:Orb2B complexes acutely depends on Orb2A and its Q domain. Moreover, these complexes are induced within 6 hr after feeding with biogenic amines (thought to provide learning signals relevant for memory formation; Schwaerzel et al., 2003), corresponding to the time course of memory decay in Orb2ΔQ mutants (Keleman et al., 2007). We therefore propose that Orb2A:Orb2B heteromeric complexes are induced at specific synapses by the relevant learning signals and required for memory persistence beyond 6 hr.
Dopamine is thought to provide a reinforcement signal in Drosophila courtship learning (Tempel et al., 1984). For short-term memory this dopamine signal is provided by neurons that innervate the gamma lobe of the mushroom body (Keleman et al., 2012), and for long-term memory Orb2 is required in intrinsic gamma lobe neurons (Kenyon cells; Keleman et al., 2007). Gamma lobe synapses are thus a likely site of Orb2 complex formation and the structural and functional modifications that underlie courtship learning in Drosophila. Orb2 also functions in long-term memory in an appetitive learning paradigm (Majumdar et al., 2012), which likely maps to a distinct class of mushroom body neuron (Waddell, 2010). Indeed, specific long-term memories may be stored at various sites in the fly brain, extending even beyond the mushroom body (Chen et al., 2012; Davis, 2011). The broad distribution of Orb2 throughout the nervous system suggests that it may contribute generally to long-term synaptic plasticity and memory formation, regardless of where these memories are stored.
Why might Orb2A have such a critical role in Orb2 complex formation and long-term memory, when most of its residues are shared with the evidently more abundant Orb2B, including the Q and RNA-binding domains? The efficacy of complex formation of proteins containing Q domains is thought to be determined by the length of the preceding N-terminal sequences (Shorter and Lindquist, 2004). In this regard it is interesting to note that Orb2A has an N-terminal extension of 9 amino acids, compared to the 162 N-terminal residues of Orb2B. Additionally, a single point mutation in the unique Orb2A N-terminal extension decreases Orb2 multimer formation in the Drosophila brain and impairs long-term memory retention beyond 48 hr (Majumdar et al., 2012). Thus, both the size and sequence of Orb2A’s unique N-terminal extension might endow it with a greater propensity to aggregate than Orb2B, and thereby nucleate heteromeric Orb2 complexes through the Q domain of Orb2A.
It has been suggested that the activation of Orb2 and other CPEB proteins occurs via the prion-like properties of their Q domains (Heinrich and Lindquist, 2011; Krishnan and Lindquist, 2005; Majumdar et al., 2012; Si et al., 2003a, 2010). Such Q domains occur however in a wide range of proteins with diverse biochemical functions, in which they are generally thought to mediate homo- and heterotypic interactions (Michelitsch and Weissman, 2000). In some of these proteins, for example, the Q domains serve as polar zippers in the assembly of large multimeric complexes (Perutz et al., 1994). Whatever the means by which the Q domain of Orb2 contributes to complex formation, our data suggest that this is restricted to Orb2A, as Orb2B does not require its Q domain to interact with Orb2A and function in long-term memory formation. This mechanism is likely to be conserved among CPEB proteins, as the Q domain of Orb2A can be replaced with the analogous domain from CPEBs of Aplysia and mouse, but not with the prion domain of ScUre2 (Figure S4).
Our data support and extend a model (Majumdar et al., 2012) in which the two Orb2 isoforms form heteromeric complexes that are essential for long-term memory formation. We further propose that, upon neuronal stimulation, Orb2A, which may be present in more limiting amounts, restricted locations, or under specific circumstances, provides the spatial and temporal specificity for heteromeric complex formation and, we infer, synaptic plasticity. Orb2B, in contrast, appears to be more broadly and highly expressed and may mediate a more general function of Orb2 in development (Cziko et al., 2009; Hafer et al., 2011; Richter, 2007; Shieh and Bonini, 2011). Orb2 has been reported to be present in the messenger RNPs, as we have also observed here specifically for Orb2B (Figure S3), and is thought to control mRNA transport and translational repression (Cziko et al., 2009; Mendez and Richter, 2001). During learning, Orb2A might interact with Orb2B-containing RNPs at the relevant synapses, releasing the associated mRNAs from translational repression or possibly even converting Orb2B from a translation repressor to an activator.
CPEB molecules are conserved across a wide range of species and most of them exist in multiple isoforms generated through alternative splicing, often varying only in their N terminus (Theis et al., 2003; Wang and Cooper, 2009). This is the case for mCPEB3, for example, the Q domain of which is able to substitute for the Q domain of Orb2A both biochemically and behaviorally (Figure S4). It is tempting to speculate that the model we propose here is not unique for Drosophila Orb2 but might also extend to other members of the CPEB family. Moreover, because Orb2A functions in long-term memory without its RNA-binding domain, it is possible that proteins lacking an RNA-binding domain, and hence not even recognized as canonical CPEB molecules, might function in a fashion analogous to Orb2A in Drosophila and other species.
Experimental Procedures
Generation and Verification of the orb2attP and Subsequent Modified orb2 Alleles
orb2attP was generated by ends-out homologous recombination (Gong and Golic, 2003), using homology arms of 3.3 kb and 3.7 kb flanking the A isoform specific and common exons of Orb2 (CG5735). In the initial recombinant, this region was replaced with an attP followed by a white+ marker flanked by mFRT11 recognition sites for the mFLP5 recombinase (Hadjieconomou et al., 2011). Removal of the white+ marker using hs-mFLP5 generated the final orb2attP allele, in which the A and common exons were replaced by an attP site (Groth et al., 2004) and a single mFRT11 site. The targeted allele was verified by genomic PCR and DNA sequencing across the entire homology region. Southern blot and RT-PCR confirmed the intended modifications (Figure 1B). Modified orb2 alleles were generated by cloning of the genomic fragments with the relevant modification first into vector containing donor attB site for subsequent reinsertion by phiC31 (Bischof et al., 2007) mediated transgenesis into orb2attP allele (details in the Supplemental Experimental Procedures). The intended modifications were verified by PCR amplification and DNA sequencing across the modified region.
Generation of the Fly Strains
To generate fly strains carrying modified orb2 alleles, donor constructs containing genomic fragments with the specific modification were injected into the embryos from a cross between orb2attP flies and phiC31 integrase-expressing flies, ZH11 (Bischof et al., 2007). DNA injection resulted in a site directed integration of the attB containing constructs into orb2attP. mHSFLP5 (Hadjieconomou et al., 2011) was used to excise the w+ marker.
Southern Blot
A probe (a) as indicated in Figure 1A, was generated by PCR using the primer SB1 and SB2 (Supplemental Experimental Procedures). Fifteen micrograms genomic DNA was digested using EcoRI/SpeI. DNA was run on a 0.5% agarose gel at 60V at 4°C over night. The gel was blotted in 20× SSC over night. After cross-linking, the membrane was incubated in hybridization solution (ULTRAhyb Ultrasensitive Hybridization Buffer, Ambion, AM8670) before incubation with the labeled probe (Prime-It Random Primer Labeling Kit, Strategene, 300385) for 16 hr.
RT-PCR
Total RNA was extracted using Trizol and reverse transcribed using random primers. Twenty-five cycles were used for amplification using primers e and f as indicated in Figure 1A (Supplemental Experimental Procedures). RpS8, was amplified with primers HH142 and HH143 (Supplemental Experimental Procedures) and used as an internal control.
Behavioral Tests
All orb2 alleles were backcrossed for five generations into a Canton-S background before being used in behavioral assays. Flies were raised on semi defined medium at 25°C in a 12 hr dark-light cycle. Virgin males were collected at eclosion and aged individually for 5 days before training. Canton-S premated females were aged for 4 days in groups of 50–100 with Canton-S males collected at the same time. All assays were performed at circadian time 6:00–10:00 on at least 3 independent days.
Courtship Conditioning
Males were assayed for courtship conditioning as described (Siwicki and Ladewski, 2003). For training, individual males were placed in food chambers either with (trained) or without (naive) a single premated female. After training (or sham training), each male was recovered, transferred to a fresh food vial and kept in isolation until testing. For long-term memory, males were trained for 6–7 hr and tested after 24 hr. For short-term memory, the training period was 1 hr and the test was performed after 30 min. Tests were performed in a 10 mm diameter courtship chamber and videotaped for 10 min (JVC handycam, 30 GB HD). Videos were scored manually and blind to the genotype for CI, which is the percentage of time each male spent courting during the test. Courtship index (CI) was used to calculate the learning Index (LI): CInaive-CI trained/CInaive × 100. Values are mean ± SEM. LIs were analyzed using a MATLAB script by permutation test (Kamyshev et al., 1999). Briefly, the entire set of courtship indices for both the naive and trained flies were pooled and then randomly assorted into simulated naive and trained sets of the same size as in the original data. A LIp was calculated for each of 100,000 randomly permuted data sets, and p values were estimated as the fraction for which LIp > LI (to test H0, LI = 0) or | LIp | > | LI - LI0 | (to test H0, LI = LI0). p values are for H0: LI = LI1 (permutation test) and ∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001 for H0, LI = 0 (permutation test).
Feeding with Biogenic Amines
Three days old adult flies were starved on water (at 18°C in the dark) prior to feeding with either 10 mM tyramine or 5 mM dopamine supplemented with 2% sucrose. At the indicated time points the heads were harvested and used for IPs and WBs as described below.
Biochemistry
Immunoprecipitation and Western Blot
Adult heads of the indicated genotype were lysed in homogenization buffer (PBS, 150 mM NaCl, 0.1 mM CaCl2, 3 mM MgCl2, 5% Glycerol, 1 mM DTT, 0.1% Triton X-100, 0.1% NP-40, Protease inhibitor cocktail from Roche, EDTA free). The lysate was cleared by centrifugation prior to incubation with Chromotek GFP-trap beads (according to the manufacturer protocol). The proteins were transferred to a PVDF membrane (Millipore) overnight in the cold room at 35 mv. Membrane was blocked in 5% milk prior to incubation for 1 hr with primary antibody. After three washes in PBST (PBS + 0.05% Tween20) membrane was incubated for 1 hr in secondary antibody. The membrane was developed using SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific).
Antibodies used: anti-GFP (Abcam 6556 rabbit polyclonal, 1:2,000), anti-tubulin (mouse monoclonal, Sigma, 1:25,000), ECL anti-mouse IgG, Horseradish Peroxidase linked F (ab′)2 fragment (from donkey) (GE healthcare, 1:10,000), ECL anti-Rabbit IgG, Horseradish Peroxidase linked whole antibody (from donkey) (GE healthcare, 1:10,000).
SDD-AGE
SDD-AGE was performed as described in Halfmann and Lindquist (2008). IP samples were loaded on horizontal 3% TAE agarose gel containing 0.1% SDS and run for 7 hr at 50 V in TAE buffer. Gel was than blotted over night onto nitrocellulose membrane using 1× TBS buffer containing 0.1% SDS.
Immunohistochemistry
Adult Brains, VNC, and Larval CNS
Immunohistochemistry for adult brains, VNC, and larval CNS was performed as described (Yu et al., 2010). Fly brain and VNC were dissected (between 5 to 8 days after eclosion) in PBS and fixed using 4% paraformaldehyde in PBST (PBS with 0.3% Triton X-100) for 20 min at 24°C. After washing in PBST, the tissue was blocked in 5% normal goat serum in PBST for at least 2 hr. The primary antibody and secondary antibody were incubated for 48 hr at 4°C. The brains were washed with PBST 3 × 10 min and then overnight at 4°C between the primary and secondary antibody incubations. After the secondary antibody incubation, samples were washed 3 × 10 min and overnight at 4°C before mounting in Vectashield (VectorLabs).
Antibodies used: rabbit polyclonal anti-GFP (1:5,000, Torri Pines); mouse nc82 (1:50, Hybridoma Bank); mouse anti-DAC2-3 (1:200, Hybridoma Bank); rabbit anti-eIF4e (Nakamura et al., 2004) (1:5,000); rabbit anti-Trailer-hitch (Tral) (Boag et al., 2005) (1:5,000); secondary Alexa-488, -568 antibodies (1:1,000, Invitrogen).
Embryos
Immunohistochemistry for embryos was as described (Patel et al., 1987). Embryos were collected and incubated in 50% bleach for 3 min and rinsed into a sieve using tap water. Next, they were transferred to the eppendorf tubes containing 500 μl heptane and 450 μl PBS. For fixation 50 μl formaldehyde was added for 20 min at RT. Lower phase was removed first, and the heptane was replaced by fresh heptane and ice-cold methanol. Then embryos were agitated strongly for 1 min to remove their vitelline membrane. After that, 3 × 5 min washes in methanol were performed followed by three washes in PBST to remove residual methanol. Next, the embryos were blocked for 1 hr in 5% normal goat serum prior to antibody incubation. Antibody incubation was done either for 1hr at RT or O/N at 4°C. Antibodies used: rabbit polyclonal anti-GFP (1:5,000, Torri Pines), mouse anti-FasII (1:50, Hybridoma Bank, 1D4), secondary Alexa-488, -568 antibodies (1:1,000, Invitrogen).
Confocal Microscopy
Tissues were scanned using a Zeiss LSM 510 with a Zeiss Multi Immersion Plan NeoFluar 25×/0.8 objective (as described; Yu et al., 2010). On average 8 brains or 5 VNCs were imaged for each genotype. Scanning parameters were set to image the central brain or the entire ventral nerve cord within 30 min. Images were taken at 512 × 512 pixels and 180 slices at 1.2 μm interval. A macro plug-in was used to automate the scanning process. Images were processed in ImageJ (NIH) to obtain maximum intensity Z projections.
Immuno-EM on Adult Brains
The heads of 3 days old orb2+GFP and Canton-S male flies were fixed in 4% paraformaldehyde, 0.1% glutaraldehyde, 0.07 M phosphate buffer (pH 7.3) for 3 hr at 4°C. Frontal vibratome sections (80 μm) were collected from each head from the anterior to the posterior region and the last two sections were processed for immuno-EM. Fifteen heads were used per genotype. Sections were incubated with rabbit anti-GFP (Molecular Probes, dilution 1:200) for 44 hr at 4°C and avidin-biotinylated-peroxidase complexes (Vectastain Elite Kit Vector, Burlingame, CA) were formed as described (Yasuyama et al., 2002). Sections were post fixed in 0.1% osmium tetroxide in 0.1M cacodylate buffer (pH 7.4) in 0.08% potassium ferrocyanide for 1 hr at 4°C, dehydrated in ethanol and embedded in LX112 resin (Ladd Research, Williston, VT) following standard procedures. Semithin sections were collected (0.8 μm thickness) using a Leica EM UC6 microtome (Leica, Vienna, Austria), stained with toluidine blue and examined under the light microscope to determine the location in the brain. Ultrathin serial sections (110 nm) were collected on EM specimen grids when the region of the mushroom body calyx were visible, post-stained with uranyl acetate and lead citrate and imaged with a FEI Tecnai 12 transmission electron microscope, operated at 120 kV and equipped with an Eagle 4kx4k camera (FEI, Eindhoven, The Netherlands). At least three brains were analyzed per genotype.
Mass Spectrometry
Tryptic Digestion
For reduction 2 μl DTT of 1mg/ml (dissolved in 100 mM ammonium bicarbonate) stock solution were added to the samples. Reduction was performed for 30 min at 56°C. Alkylation was performed with 2 μl of 40 mM (in 100 mM ammonium bicarbonate) stock solution for 30 min at room temperature in the dark. Afterward the samples were digested with 400 ng of trypsin (Gold, Promega) for 16 hr. The digestion was stopped with 10 μl of 10% TFA.
NanoLC-MS
The nano HPLC system used in all experiments was an UltiMate 3000 Dual Gradient HPLC system (Dionex, Amsterdam, The Netherlands), equipped with a Proxeon nanospray source (Proxeon, Odense, Denmark), coupled to an LTQ Velos Orbitrap mass spectrometer (Thermo Fisher Scientific). Instrument was operated in data-dependent mode using a full scan in the ICR cell followed by MS/MS scans of the twelve most abundant ions in the linear ion trap. MS/MS spectra were acquired in the multistage activation mode. Precursor ions selected for fragmentation were put on a dynamic exclusion list for 90 s. Monoisotopic precursor selection was enabled.
Analysis of MS Data
For peptide identification, all MS/MS spectra were searched using Mascot 2.2.04 (Matrix Science, London, UK) against the flybase database (49,832 sequences; 31,566,328 residues). The following search parameters were used: beta-methylthiolation on cysteine was set as a fixed modification; oxidation on methionine was set as variable modification. Monoisotopic masses were searched within unrestricted protein masses for tryptic peptides. The peptide mass tolerance was set to ± 5 ppm and fragment mass tolerance to ± 0.5 Da. The maximal number of missed cleavages was set to 3. Results were imported to Scaffold 3.3.2 software with minimum two peptides per protein resulting a FDR rate from 0%.
Inducible S2 Cell Lines Expressing orb2 Alleles
Cloning Strategy
Orb2AGFP was PCR amplified from the Drosophila cDNA with the primers CP156 and CP155, Orb2BGFP with the primers CP154 and CP155, Orb2ADQGFP with the primers CP157 and CP155, and Orb2BDQGFP with the primers CP154 and CP158 and CP155 and CP159. PCR products were used in overlap PCR using primers CP154 and CP155. Final PCR products were recombined with pDONR221 vector (Invitrogen) and resulting entry clones were recombined with pMK/AWG to generate expression vectors. pMK/AWG was generated by subcloning the recombination cassette of pAWG (The Drosophila Gateway Vector Collection, T. Murphy) EcoRV/NheI with EcoRV/SpeI into the backbone of pMK33/pMtHy (Koelle et al., 1991) thus resulting in stable and inducible expression vectors.
Generation of the Stable S2 Cell Lines Expressing orb2 Alleles
S2 cells were grown as semi-adhering cultures at 27°C in water jacketed incubator with 5% CO2 in liquid Schneider’s Drosophila Medium (Invitrogen, 11720-034) supplemented with 10% FCS (fetal calf serum) and 1× PenStrep (Invitrogen, 15070) without agitation. Transfected cell lines were grown in the medium supplemented with hygromycin B (Roche, 10-843-555-001) at 300 μg/ml.
For transfection, confluent S2 cell cultures were split 1:3 to a fresh medium and grown overnight. Cultures were washed with 1× PBS and resuspended to 2 million cells/ml in fresh medium before being transfected with 800 ng appropriate plasmid using Effectene Transfection Reagent (QIAGEN, 301425) according to manufacturer’s protocol. Transfected cells were subsequently grown for 72 hr and then changed to the selection medium containing hygromycin B by spinning 5 min at 1,000 g and resuspending the cell pellet in a fresh culture medium containing the antibiotic. For expression of Orb2 protein S2 cells were induced O/N with 0.25 mM CuSO4 (final concentration).
Acknowledgments
We are very grateful to Mark T. Palfreyman for critical comments on the manuscript. We thank Pawel Pasierbek for help with the confocal imaging, Maria Novatchkova for bioinformatic analysis, and Jos Onderwater and Anja de Jong for technical assistance with the immuno-EM experiments. eIF4E and Tral antibodies were gift from Akira Nakamura. Basic research at the IMP is funded in part by Boehringer Ingelheim GmbH. This work was additionally supported by Austrian Science Fund, FWF (S.K.) to K.K., Vienna Science and Technology Fund, WWTF (B.S.) to K.K., and FW7 EU grant, GENCODYS.
Published: October 17, 2012
Footnotes
Supplemental Information includes four figures, six tables, and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.neuron.2012.08.028.
Contributor Information
Barry J. Dickson, Email: dickson@imp.ac.at.
Krystyna Keleman, Email: keleman@imp.ac.at.
Supplemental Information
References
- Alarcon J.M., Hodgman R., Theis M., Huang Y.S., Kandel E.R., Richter J.D. Selective modulation of some forms of schaffer collateral-CA1 synaptic plasticity in mice with a disruption of the CPEB-1 gene. Learn. Mem. 2004;11:318–327. doi: 10.1101/lm.72704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins C.M., Nozaki N., Shigeri Y., Soderling T.R. Cytoplasmic polyadenylation element binding protein-dependent protein synthesis is regulated by calcium/calmodulin-dependent protein kinase II. J. Neurosci. 2004;24:5193–5201. doi: 10.1523/JNEUROSCI.0854-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J., Maeda R.K., Hediger M., Karch F., Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boag P.R., Nakamura A., Blackwell T.K. A conserved RNA-protein complex component involved in physiological germline apoptosis regulation in C. elegans. Development. 2005;132:4975–4986. doi: 10.1242/dev.02060. [DOI] [PubMed] [Google Scholar]
- Chen C.C., Wu J.K., Lin H.W., Pai T.P., Fu T.F., Wu C.L., Tully T., Chiang A.S. Visualizing long-term memory formation in two neurons of the Drosophila brain. Science. 2012;335:678–685. doi: 10.1126/science.1212735. [DOI] [PubMed] [Google Scholar]
- Cziko A.M., McCann C.T., Howlett I.C., Barbee S.A., Duncan R.P., Luedemann R., Zarnescu D., Zinsmaier K.E., Parker R.R., Ramaswami M. Genetic modifiers of dFMR1 encode RNA granule components in Drosophila. Genetics. 2009;182:1051–1060. doi: 10.1534/genetics.109.103234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R.L. Traces of Drosophila memory. Neuron. 2011;70:8–19. doi: 10.1016/j.neuron.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox C.A., Sheets M.D., Wickens M.P. Poly(A) addition during maturation of frog oocytes: distinct nuclear and cytoplasmic activities and regulation by the sequence UUUUUAU. Genes Dev. 1989;3(12B):2151–2162. doi: 10.1101/gad.3.12b.2151. [DOI] [PubMed] [Google Scholar]
- Frey U., Morris R.G. Synaptic tagging and long-term potentiation. Nature. 1997;385:533–536. doi: 10.1038/385533a0. [DOI] [PubMed] [Google Scholar]
- Gailey D.A., Jackson F.R., Siegel R.W. Male courtship in Drosophila: the conditioned response to immature males and its genetic control. Genetics. 1982;102:771–782. doi: 10.1093/genetics/102.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garen A., Garen S. Complementation in vivo between structural mutants of alkaline phosphatase from E. coli. J. Mol. Biol. 1963;7:13–22. doi: 10.1016/s0022-2836(63)80015-7. [DOI] [PubMed] [Google Scholar]
- Gong W.J., Golic K.G. Ends-out, or replacement, gene targeting in Drosophila. Proc. Natl. Acad. Sci. USA. 2003;100:2556–2561. doi: 10.1073/pnas.0535280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A.C., Fish M., Nusse R., Calos M.P. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjieconomou D., Rotkopf S., Alexandre C., Bell D.M., Dickson B.J., Salecker I. Flybow: genetic multicolor cell labeling for neural circuit analysis in Drosophila melanogaster. Nat. Methods. 2011;8:260–266. doi: 10.1038/nmeth.1567. [DOI] [PubMed] [Google Scholar]
- Hafer N., Xu S., Bhat K.M., Schedl P. The Drosophila CPEB protein Orb2 has a novel expression pattern and is important for asymmetric cell division and nervous system function. Genetics. 2011;189:907–921. doi: 10.1534/genetics.110.123646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake L.E., Mendez R., Richter J.D. Specificity of RNA binding by CPEB: requirement for RNA recognition motifs and a novel zinc finger. Mol. Cell. Biol. 1998;18:685–693. doi: 10.1128/mcb.18.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R., Lindquist S. Screening for amyloid aggregation by semi-denaturing detergent-agarose gel electrophoresis. J. Vis. Exp. 2008 doi: 10.3791/838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich S.U., Lindquist S. Protein-only mechanism induces self-perpetuating changes in the activity of neuronal Aplysia cytoplasmic polyadenylation element binding protein (CPEB) Proc. Natl. Acad. Sci. USA. 2011;108:2999–3004. doi: 10.1073/pnas.1019368108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisenberg M. Mushroom body memoir: from maps to models. Nat. Rev. Neurosci. 2003;4:266–275. doi: 10.1038/nrn1074. [DOI] [PubMed] [Google Scholar]
- Huang Y.S., Jung M.Y., Sarkissian M., Richter J.D. N-methyl-D-aspartate receptor signaling results in Aurora kinase-catalyzed CPEB phosphorylation and alpha CaMKII mRNA polyadenylation at synapses. EMBO J. 2002;21:2139–2148. doi: 10.1093/emboj/21.9.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.S., Carson J.H., Barbarese E., Richter J.D. Facilitation of dendritic mRNA transport by CPEB. Genes Dev. 2003;17:638–653. doi: 10.1101/gad.1053003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.S., Kan M.C., Lin C.L., Richter J.D. CPEB3 and CPEB4 in neurons: analysis of RNA-binding specificity and translational control of AMPA receptor GluR2 mRNA. EMBO J. 2006;25:4865–4876. doi: 10.1038/sj.emboj.7601322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamyshev N.G., Iliadi K.G., Bragina J.V. Drosophila conditioned courtship: two ways of testing memory. Learn. Mem. 1999;6:1–20. [PMC free article] [PubMed] [Google Scholar]
- Kang H., Schuman E.M. A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science. 1996;273:1402–1406. doi: 10.1126/science.273.5280.1402. [DOI] [PubMed] [Google Scholar]
- Keleman K., Krüttner S., Alenius M., Dickson B.J. Function of the Drosophila CPEB protein Orb2 in long-term courtship memory. Nat. Neurosci. 2007;10:1587–1593. doi: 10.1038/nn1996. [DOI] [PubMed] [Google Scholar]
- Keleman K., Vrontou E., Krüttner S., Yu J.Y., Kurtovic-Kozaric A., Dickson B.J. Dopamine neurons modulate pheromone responses in Drosophila courtship learning. Nature. 2012;489:145–149. doi: 10.1038/nature11345. [DOI] [PubMed] [Google Scholar]
- Koelle M.R., Talbot W.S., Segraves W.A., Bender M.T., Cherbas P., Hogness D.S. The Drosophila EcR gene encodes an ecdysone receptor, a new member of the steroid receptor superfamily. Cell. 1991;67:59–77. doi: 10.1016/0092-8674(91)90572-g. [DOI] [PubMed] [Google Scholar]
- Krishnan R., Lindquist S.L. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature. 2005;435:765–772. doi: 10.1038/nature03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Schwartz J.H. The cytoplasmic polyadenylation element binding protein and polyadenylation of messenger RNA in Aplysia neurons. Brain Res. 2003;959:68–76. doi: 10.1016/s0006-8993(02)03729-0. [DOI] [PubMed] [Google Scholar]
- Majumdar A., Cesario W.C., White-Grindley E., Jiang H., Ren F., Khan M.R., Li L., Choi E.M., Kannan K., Guo F. Critical role of amyloid-like oligomers of Drosophila Orb2 in the persistence of memory. Cell. 2012;148:515–529. doi: 10.1016/j.cell.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Martin K.C., Casadio A., Zhu H., Yaping E., Rose J.C., Chen M., Bailey C.H., Kandel E.R. Synapse-specific, long-term facilitation of aplysia sensory to motor synapses: a function for local protein synthesis in memory storage. Cell. 1997;91:927–938. doi: 10.1016/s0092-8674(00)80484-5. [DOI] [PubMed] [Google Scholar]
- McBride S.M., Giuliani G., Choi C., Krause P., Correale D., Watson K., Baker G., Siwicki K.K. Mushroom body ablation impairs short-term memory and long-term memory of courtship conditioning in Drosophila melanogaster. Neuron. 1999;24:967–977. doi: 10.1016/s0896-6273(00)81043-0. [DOI] [PubMed] [Google Scholar]
- McGuire S.E., Le P.T., Osborn A.J., Matsumoto K., Davis R.L. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- Mendez R., Richter J.D. Translational control by CPEB: a means to the end. Nat. Rev. Mol. Cell Biol. 2001;2:521–529. doi: 10.1038/35080081. [DOI] [PubMed] [Google Scholar]
- Mendez R., Barnard D., Richter J.D. Differential mRNA translation and meiotic progression require Cdc2-mediated CPEB destruction. EMBO J. 2002;21:1833–1844. doi: 10.1093/emboj/21.7.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelitsch M.D., Weissman J.S. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc. Natl. Acad. Sci. USA. 2000;97:11910–11915. doi: 10.1073/pnas.97.22.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miniaci M.C., Kim J.H., Puthanveettil S.V., Si K., Zhu H., Kandel E.R., Bailey C.H. Sustained CPEB-dependent local protein synthesis is required to stabilize synaptic growth for persistence of long-term facilitation in Aplysia. Neuron. 2008;59:1024–1036. doi: 10.1016/j.neuron.2008.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A., Sato K., Hanyu-Nakamura K. Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev. Cell. 2004;6:69–78. doi: 10.1016/s1534-5807(03)00400-3. [DOI] [PubMed] [Google Scholar]
- Patel N.H., Snow P.M., Goodman C.S. Characterization and cloning of fasciclin III: a glycoprotein expressed on a subset of neurons and axon pathways in Drosophila. Cell. 1987;48:975–988. doi: 10.1016/0092-8674(87)90706-9. [DOI] [PubMed] [Google Scholar]
- Perutz M.F., Johnson T., Suzuki M., Finch J.T. Glutamine repeats as polar zippers: their possible role in inherited neurodegenerative diseases. Proc. Natl. Acad. Sci. USA. 1994;91:5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter J.D. Think globally, translate locally: what mitotic spindles and neuronal synapses have in common. Proc. Natl. Acad. Sci. USA. 2001;98:7069–7071. doi: 10.1073/pnas.111146498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter J.D. CPEB: a life in translation. Trends Biochem. Sci. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Schwaerzel M., Monastirioti M., Scholz H., Friggi-Grelin F., Birman S., Heisenberg M. Dopamine and octopamine differentiate between aversive and appetitive olfactory memories in Drosophila. J. Neurosci. 2003;23:10495–10502. doi: 10.1523/JNEUROSCI.23-33-10495.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh S.Y., Bonini N.M. Genes and pathways affected by CAG-repeat RNA-based toxicity in Drosophila. Hum. Mol. Genet. 2011;20:4810–4821. doi: 10.1093/hmg/ddr420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J., Lindquist S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science. 2004;304:1793–1797. doi: 10.1126/science.1098007. [DOI] [PubMed] [Google Scholar]
- Si K., Giustetto M., Etkin A., Hsu R., Janisiewicz A.M., Miniaci M.C., Kim J.H., Zhu H., Kandel E.R. A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in aplysia. Cell. 2003;115:893–904. doi: 10.1016/s0092-8674(03)01021-3. [DOI] [PubMed] [Google Scholar]
- Si K., Lindquist S., Kandel E.R. A neuronal isoform of the aplysia CPEB has prion-like properties. Cell. 2003;115:879–891. doi: 10.1016/s0092-8674(03)01020-1. [DOI] [PubMed] [Google Scholar]
- Si K., Choi Y.B., White-Grindley E., Majumdar A., Kandel E.R. Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell. 2010;140:421–435. doi: 10.1016/j.cell.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Siegel R.W., Hall J.C. Conditioned responses in courtship behavior of normal and mutant Drosophila. Proc. Natl. Acad. Sci. USA. 1979;76:3430–3434. doi: 10.1073/pnas.76.7.3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siwicki K.K., Ladewski L. Associative learning and memory in Drosophila: beyond olfactory conditioning. Behav. Processes. 2003;64:225–238. doi: 10.1016/s0376-6357(03)00137-2. [DOI] [PubMed] [Google Scholar]
- Sutton M.A., Schuman E.M. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Tempel B.L., Livingstone M.S., Quinn W.G. Mutations in the dopa decarboxylase gene affect learning in Drosophila. Proc. Natl. Acad. Sci. USA. 1984;81:3577–3581. doi: 10.1073/pnas.81.11.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis M., Si K., Kandel E.R. Two previously undescribed members of the mouse CPEB family of genes and their inducible expression in the principal cell layers of the hippocampus. Proc. Natl. Acad. Sci. USA. 2003;100:9602–9607. doi: 10.1073/pnas.1133424100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompkins L., Siegel R.W., Gailey D.A., Hall J.C. Conditioned courtship in Drosophila and its mediation by association of chemical cues. Behav. Genet. 1983;13:565–578. doi: 10.1007/BF01076402. [DOI] [PubMed] [Google Scholar]
- Waddell S. Dopamine reveals neural circuit mechanisms of fly memory. Trends Neurosci. 2010;33:457–464. doi: 10.1016/j.tins.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.P., Cooper N.G. Characterization of the transcripts and protein isoforms for cytoplasmic polyadenylation element binding protein-3 (CPEB3) in the mouse retina. BMC Mol. Biol. 2009;10:109. doi: 10.1186/1471-2199-10-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells D.G., Dong X., Quinlan E.M., Huang Y.S., Bear M.F., Richter J.D., Fallon J.R. A role for the cytoplasmic polyadenylation element in NMDA receptor-regulated mRNA translation in neurons. J. Neurosci. 2001;21:9541–9548. doi: 10.1523/JNEUROSCI.21-24-09541.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Wells D., Tay J., Mendis D., Abbott M.A., Barnitt A., Quinlan E., Heynen A., Fallon J.R., Richter J.D. CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of alpha-CaMKII mRNA at synapses. Neuron. 1998;21:1129–1139. doi: 10.1016/s0896-6273(00)80630-3. [DOI] [PubMed] [Google Scholar]
- Yasuyama K., Meinertzhagen I.A., Schürmann F.W. Synaptic organization of the mushroom body calyx in Drosophila melanogaster. J. Comp. Neurol. 2002;445:211–226. doi: 10.1002/cne.10155. [DOI] [PubMed] [Google Scholar]
- Yu J.Y., Kanai M.I., Demir E., Jefferis G.S., Dickson B.J. Cellular organization of the neural circuit that drives Drosophila courtship behavior. Curr. Biol. 2010;20:1602–1614. doi: 10.1016/j.cub.2010.08.025. [DOI] [PubMed] [Google Scholar]
- Zearfoss N.R., Alarcon J.M., Trifilieff P., Kandel E., Richter J.D. A molecular circuit composed of CPEB-1 and c-Jun controls growth hormone-mediated synaptic plasticity in the mouse hippocampus. J. Neurosci. 2008;28:8502–8509. doi: 10.1523/JNEUROSCI.1756-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Billington C.J., Jr., Pan D., Neufeld T.P. Drosophila target of rapamycin kinase functions as a multimer. Genetics. 2006;172:355–362. doi: 10.1534/genetics.105.051979. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.