Abstract
Here we report the discovery of a highly conserved novel binding site located at the interface between the protease and helicase domains of the Hepatitis C Virus (HCV) NS3 protein. Using a chemical lead, identified by fragment screening and structure-guided design, we demonstrate that this site has a regulatory function on the protease activity via an allosteric mechanism. We propose that compounds binding at this allosteric site inhibit the function of the NS3 protein by stabilising an inactive conformation and thus represent a new class of direct acting antiviral agents.
Viruses are highly evolved pathogenic agents that have developed efficient strategies to program all essential functions in the limited genomic space available. The HCV genome encodes only 10 viral proteins, but their pleiotropic nature ensures the survival and propagation of progeny virus 1,2. The HCV polyprotein is processed by host and viral proteases into structural (E1, E2, C) and nonstructural (p7, NS2, NS3, NS4a, NS4b, NS5a, NS5b) proteins. The NS3 protein is a bi-functional enzyme with a serine protease domain at the N-terminus and an ATP dependent helicase domain at the C-terminus. The two functional domains of NS3 remain connected in vivo and profoundly influence each other’s enzymatic parameters 3-6. The protease performs the cis cleavage to release itself from the poly-protein and forms a non-covalent complex with NS4a, an essential peptide-cofactor for the trans cleavages that release the NS4b, NS5a and NS5b proteins. The NS4a stabilizes the protease and anchors the complex to the membrane of the endoplasmic reticulum (ER). The helicase domain binds to nucleic acid chains and, fueled by the hydrolysis of ATP, tracks along them in a 3′ to 5′ direction to displace annealed strands or bound proteins. The mechanism by which the NS3 protein switches between different activities is currently unknown, however, as with other viral proteins such as HIV integrase and reverse transcriptase, allosteric modulation may be an important factor 7-9.
The NS3 protein has been the focus of intense research as a drug discovery target, with two new protease inhibitor drugs approved in 2011 for the treatment of HCV infection 10. However, as with all antiviral drugs, the emergence of resistant strains will accentuate the need for new drugs with different and complementary modes of action. Both the NS3 protease and helicase domains have been used independently and extensively as models for the in vivo relevant full length enzyme 11-15. However a growing body of evidence has demonstrated that the juxtaposition of the domains in the full length protein has profound effects on the selectivity, catalytic rate and affinity for their respective substrates 1-4. Thus, the isolated helicase domain displays a preference for DNA substrates, whereas the presence of the protease in the full length protein greatly enhances the binding affinity and processivity of RNA substrates, as would be expected for an RNA virus 5. Similarly, poly-nucleotides, and poly-uracil in particular, stimulate protease activity in the full length context while not having any effect on the isolated protease domain 3. Comparable effects have been reported for the related flavivirus NS3 proteins from Dengue and West Nile virus, where despite the different genomic and structural arrangement, both functional domains also remain attached 16, 17. In these latter cases, the full-length and protease domain proteins show similar in vitro kinetics, whilst the ATPase and unwinding functions of the helicase are significantly influenced by the presence of the protease domain 16, 17.
Fragment screening has been successfully used to discover novel binding sites in a range of other systems, including HIV protease and integrase, glycogen phosphorylase and the oncogenic Ras protein 18-24. In this study we report the use of X-ray crystallography to identify fragments which bind at a conserved allosteric site on the full length NS3/4a protein and the structure based optimization of these inhibitors, which function by stabilizing a pre-existing autoinhibited conformational state of the protein. We show that this conformation is relevant in the virus lifecycle by eliciting resistance mutations in the subgenomic replicon system.
Results
X-ray crystallographic fragment screen
We performed a fragment-based screen using crystals of the HCV NS3/4a full length genotype 1b holo enzyme [Figure 1] 25. Crystal structures of the protein in complex with a series of fragment hits revealed the existence of a new binding site at the interface of the two domains [Figure 1, 2a,b]. The high sensitivity of fragment screening using X-ray crystallography allowed the detection of very weakly binding fragment hits, often in the mM range with acceptable ligand efficiencies (LE) (compounds 1 and 2, IC50 > 5mM (LE < 0.3) and ~ 500μM (LE = 0.3) respectively). The binding site is relatively hydrophobic, with residues Met485, Val524, Cys525, Gln526 and Val630 from the helicase domain and His57, Val78, Asp79, Asp81 and Arg155 from the protease domain making key van der Waals contacts with the fragments. The pocket is near, yet distinct from the protease active site, which is occupied by the C-terminus of the helicase domain in the crystal structure [Figure 1].
Figure 1.
Crystal structure of the full length NS3/4a protein
Ribbon diagram of the crystal structure of the full length NS3/4A protein, with the novel allosteric site represented as a red surface. The protease domain is coloured in grey, the NS4a cofactor in pink, helicase domain in blue with the C-terminus of the helicase highlighted in orange. This color scheme is maintained in all figures. The location of the ATP and RNA binding sites are shown for orientation. The inset represents a magnified view of the protease-helicase interface and the newly discovered allosteric pocket. The six C-terminal residues of the helicase domain (Asp626-Thr631) bind in the active site of the protease domain and form backbone hydrogen-bonding interactions which result in the formation of an anti-parallel β-sheet. These interactions stabilise the protein in a closed conformation. The catalytic triad (Ser139, His57, Asp81) and other key residues are labeled.
Figure 2.
Co-crystal structures of fragment hits
a and b. Compounds 1 and 2 were identified by fragment screening using crystals of the full length NS3/4a protein. The crystal structures show the compounds bound in the allosteric pocket, with binding predominantly dominated by van der Waals interactions. The pink mesh represents the electron density map (Fo-Fc omit map contoured at 3.9 σ) for each of the ligands. Compound 2 was one of the most potent fragments identified during the screening phase, with an IC50 ~ 500 μM (LE ~ 0.30) against FL NS3/4a, and no inhibition of the protease domain alone.
Structure based lead optimisation
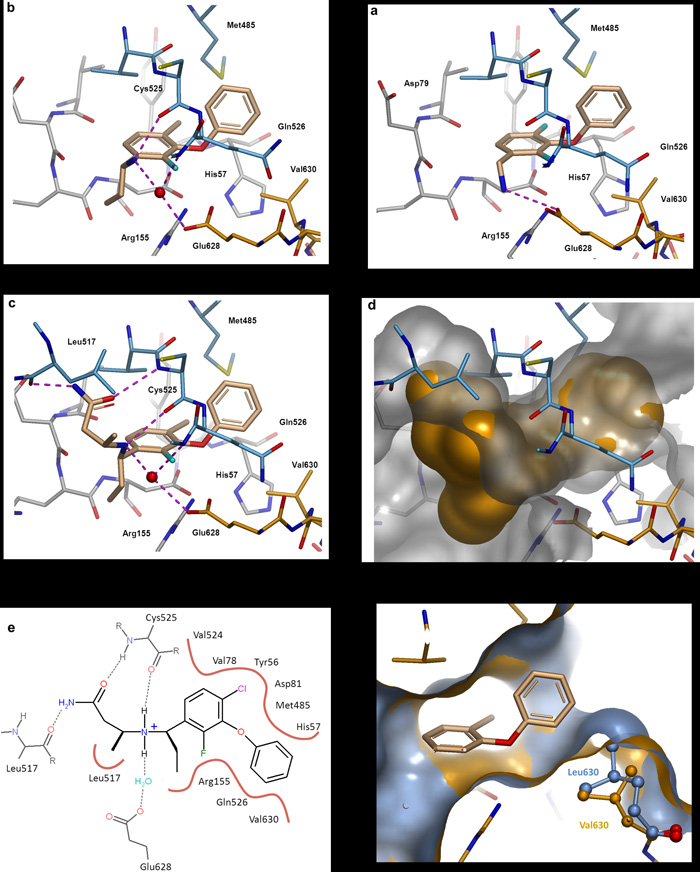
We used structure guided optimisation to elaborate the low affinity fragments into tight binding chemical leads in order to investigate the functional relevance of this site. Affinity against the NS3/4a protein improved by over four orders of magnitude, from ~ 500μM to < 0.01μM, as illustrated by compounds 2 and 6 [Table 1]. Analogue screening and structure guided optimisation of compound 2 initially focused on efficiently filling the hydrophobic core of the binding site and stabilising the bound conformation [compound 3, see Figures 3a]. The aminomethyl group was moved to the meta position in order to allow the positively charged NH3 group to interact with the acidic side chain of Glu628, leading to an increase in binding affinity and a significant improvement in LE (IC50 = 20μM, LE = 0.38). In order to maximize van der Waals interactions with the core of the binding site, the 4-fluoro of compound 3 was replaced with a larger chlorine atom. An ethyl group was introduced at the benzylic position to restrict the rotation of the aminomethyl side chain, which now interacts with Glu628 via a water molecule and also forms a hydrogen bond to the backbone carbonyl of Cys525 [Figure 3b]. Compound 4 represents a conformationally constrained and efficient ligand (IC50 ~ 1.3μM, LE = 0.42) with well-defined growing vectors. Subsequent optimisation centered on the identification of further specific hydrogen bonding interactions to the protein. The primary amine of compound 4 was used as a synthetic handle which provided an excellent vector for probing the pocket formed by Tyr516, Leu517, Val524 and Cys525. This led to the design of compound 5, which makes key interactions with residues from both the protease and helicase domains, including the C-terminus of the latter and shows good shape complementarity with the site [Figure 3c-e]. 5 was the first compound identified to bind at this site with sub 100nM affinity (LE = 0.38), and was subsequently used as a tool molecule to study the functional role of the site. Additional affinity was achieved by growing into the pocket formed by residues Asp79, Thr519 and Leu522. Introduction of an ethylamino side-chain (compound 6) led to a further improvement in potency (Kd = 0.022μM; IC50 < 0.01μM; LE ~ 0.39). Compound 7, the enantiomer of 5 (identical physical properties, but structurally the mirror image) was tested against the full length enzyme and showed a 400-fold reduction in binding affinity. Analysis of the protein-ligand crystal structure of compound 5 indicated that the key binding interactions were stereospecific, and were inaccessible to compound 7, thus explaining the loss in potency.
Table 1.
In vitro activity profile of compounds binding at the allosteric binding site
| Cmpd | Structure | Kd ITC (μM) |
FL IC50 (μM) |
LE | PD IC50 (μM) |
EC50 (μM) |
CC50 (μM) |
|---|---|---|---|---|---|---|---|
| 1 |

|
ND | > 5000 | < 0.30 | Inactive | ND | ND |
| 2 |
|
ND | 55% I ± 3 @ 500 μM |
0.30 | Inactive | ND | ND |
| 3 |

|
29 | 20 ± 4 | 0.38 | 54 ± 4% I @ 1000 μM |
Inactive | 40 |
| 4 |

|
ND | 1.3 ± 0.1 | 0.42 | Inactive | Inactive | 11 |
| 5 |

|
0.062 | 0.10 ± 0.03 | 0.38 | Inactive | 0.4±0.2 | >10 |
| 6 |

|
0.022 | 68% I ± 6 @ 0.01 |
~ 0.39 | Inactive | 0.0083± 0.001 |
4.5 |
| 7 |

|
24 | 59% I ± 5 @ 300 |
< 0.25 | ND | Inactive | >30 |
Kd refers to the binding affinity of the ligand to the full length protein measured by isothermal titration calorimetry (ITC); IC50’s reflect the inhibition of protease activity of the full length (FL) and protease domain (PD) proteins respectively. Assay conditions for FL and PD are optimised for maximum signal and differ from each other (see methods for details). LE (ligand efficiency) is defined as the binding affinity in kcal per heavy atom, in this instance the IC50 value from the full length protease assay have been used rather than the Kd to allow comparison of all of the compounds. EC50 is the cell based replicon activity of the compounds and CC50 reflects the cytotoxicity index. Compounds were assayed independently 3 or more times, and the reported value is either the geometric or arithmetic mean ± the standard deviation (SD). All ITC data is n = 1.
Figure 3.
Structure guided optimisation
Protein-ligand co-crystal structures of compounds 3 to 5.
a. Compound 3. Flanking fluorine substituents were introduced to stabilise the bound conformation of 2 and also improve hydrophobic interactions. The aminomethyl group was moved to the meta position allowing the positively charged NH3 group to interact with the acidic side chain of Glu628.
b. Compound 4. The 4-fluoro was replaced with a larger chlorine atom to maximize van der Waals interactions. An ethyl group was introduced at the benzylic position to restrict the rotation of the aminomethyl side chain, which now interacts with Glu628 via a water molecule and also forms a hydrogen bond to the backbone carbonyl of Cys525.
c. The X-ray crystal structure of 4 was used to design a small set of compounds to probe the pocket formed by Tyr516, Leu517, Val524 and Cys525. This resulted in the identification of compound 5 which makes key interactions with residues from the protease domain (Arg155, His57) and the helicase domain (Leu517, Cys525, Gln526, Met485) including the C-terminus (Glu628, Val630). H-bonds = dashed lines, conserved water molecules = red spheres.
d. Surface representation of compound 5 and the suface of the allosteric pocket.
e. 2D representation of compound 5 bound in the allosteric site. H-bonds = dashed lines, hydrophobic contacts = solid lines.
f. Surface representation of the wild type protein (gold) complexed with compound 5, overlaid with the crystal structure of the Val630Leu mutant protein (grey). The larger protein side-chain constricts the allosteric binding site.
Cell based sub-genomic replicon assay
Compounds 5, 6 and 7 were tested for functional activity in the cell based genotype 1b sub-genomic replicon, an in vitro system which mimics viral replication and is commonly used as an antiviral activity screen 26, 27. The results showed a good correlation between the enzyme and cell based assays, where both compounds 5 and 6 displayed reproducible antiviral activity with no cytotoxicity at the concentrations tested. In contrast compound 7 showed no anti-viral effect as predicted from the enzyme data [Table 1]. In addition, in vitro functional assays also showed that compounds binding at this novel site inhibited the full length protein but crucially not the isolated protease domain, consistent with the observed binding mode. These results suggest that compounds of this type inhibit the protease activity of NS3/4a via an allosteric mechanism. Furthermore, to investigate whether these compounds inhibit both the cis and trans cleavage activities, we explored whether the accumulation of unprocessed polyprotein could be detected in the replicon system upon addition of compound 6. Using western blot analysis and antibodies against NS5b, we were able to detect faint bands that corresponded to the un-cleaved proteins, but as the processing of the polyprotein is a constantly occurring event in the cell culture, the signal to noise was tenuous, precluding unambiguous interpretation of the results [Supplementary figure 1].
Generation of resistant mutations
To corroborate the functional role for the novel site we applied long term selective pressure on the replicon with increasing concentrations (10-30 × EC50) of compound 5 for 39 days and allowed resistance mutations to emerge. The emergence of antiviral resistance is a key factor in establishing the mechanism by which compounds exert their antiviral activity and is thus the gold standard for the demonstration of mode of action 28, 29. Sequencing of the resistant replicons identified mutations in the helicase domain at positions Met485 and Val630 individually, both of which map to the allosteric site and would be expected to hinder binding of compound 5 [Figure 3c]. Resistant replicons showed a 30-fold decreased susceptibility to compound 5, but not telaprevir, an active site protease inhibitor used as a control [Supplementary figure 2]. Incorporation of site directed mutations Met485Val and Val630Leu into the wild-type enzyme resulted in 40-50-fold weaker IC50 values, in good agreement with the cell based data. Finally, the crystal structure of the Val630Leu mutant protein in complex with compound 5 showed the binding pocket to be smaller, due to the larger leucine side-chain [Figure 3f].
Cross genotypic analysis of the allosteric site
We examined the sequences of known HCV variants to determine whether the site was likely to be present across all genotypes 30. Given the high replication and mutation rates of the virus, we hypothesized that the degree of conservation of the sequence could give an indication of the functional significance of any given site. The analysis was performed on the residues that line the allosteric site and showed a high degree of conservation ranging from 75-96% sequence identity [Supplementary table 1]. This suggests that the allosteric site is conserved across all genotypes, consistent with a functional role. To test this hypothesis, we produced and characterized equivalent protein constructs to the genotype 1b protein, with sequences corresponding to the 1a, 2a, 3a, 4, 5 and 6 genotypes. Compound 6 demonstrated comparable inhibition against genotypes 1a, 1b, 3a, 5 and 6, a 10-fold loss in activity against genotype 4 and a significant reduction against genotype 2a, as anticipated from the sequence comparison [Supplementary table 2]. Specifically, modeling of the allosteric pocket using the genotype 4 sequence suggested that an Leu517Phe substitution would constrict the site and hinder the compound from binding optimally. An analogous substitution in genotype 2a, together with a Val630Met substitution, prevents compounds of this type from binding effectively, providing an explanation for the reduced affinity.
Biophysical characterization of conformational states
We investigated the molecular basis by which compounds binding at the novel site exert their allosteric effect by analyzing the HCV NS3/4a crystal structure. The structure of the protein shows the six C-terminal residues of the helicase domain (Asp626-Thr631) bound in the active site of the protease domain forming key backbone hydrogen-bonding interactions, as previously reported [Figure 1] 25. For the protease to perform its catalytic role, the C-terminus is required to vacate the active site to allow access to the substrates. This necessitates a concomitant movement of the functional domains, which would result in an open conformation [Figure 5]. Though the crystal structure of the full length NS3/4a protein in an open conformation has not been reported, its presence in solution is implied by the catalytic activity of the protease. Sedimentation velocity analysis of the NS3/4a showed that in solution, and under conditions used for functional analysis, the protein populates two different monomeric species, consistent with open and closed conformations [Supplementary figure 3]. Our data agree with a recent publication, in which sedimentation velocity and functional RNA unwinding assays on different full length NS3 constructs suggested equivalent extended and compact conformations 31. We also performed size exclusion chromatography and dynamic light scattering experiments, which albeit not having the resolution to be conclusive, were consistent with the presence of two different conformational species in solution, adding further support for the presence of an alternative conformational state in solution [Supplementary figure 4]. Finally, analysis of all our data collected by isothermal titration calorimetry (ITC) reflected a trend consistent with NS3/4a conformational flexibility being reduced upon binding of these compounds [Supplementary table 4 and figure 5].
Discussion
In this study we identify a novel allosteric pocket in the HCV NS3/4a protein located at the interface between the protease and helicase domains. We propose that an equilibrium exists between an open and closed conformation of the full length HCV NS3/4a protein and that chemical leads which bind at the allosteric pocket can shift this equilibrium and inhibit protease activity. Specifically, we suggest that the interactions observed in the apo crystal structure are consistent with the NS3/4a protein populating an ‘auto-inhibited’ form in solution, and that compound 5 stabilizes the auto-inhibited (closed) conformation by binding at the allosteric site [Figure 5]. This places the compound in close proximity to the C-terminal tail and results in direct interactions including a water-mediated salt bridge to Glu628 and van der Waals contact with Val630 [Figure 3c]. We propose that the stabilisation of the auto-inhibited state by small molecules binding at the allosteric site inhibit protease activity via a non-competitive mechanism. Furthermore we speculate that binding at this allosteric site may also affect helicase activity, a function which has been shown to require an open conformation of the NS3 protein 31. An analogous report of an equilibrium between alternative extended and compact conformations adopted by the related dengue virus NS3 protein further underscores the potential biological significance of inter-domain interactions and allosteric modulation 31,32. The successful coupling of the two apparently unrelated functional activities within a single polypeptide chain requires sufficient flexibility for the protein to adopt its optimal catalytic conformation, whilst restricting the number of energetically unfavorable conformational species 32. The co-evolution of this strategy across the Flaviviridae family argues for its importance.
The NS4a protein is known to play a key role in the function and localization of the HCV NS3 protein. It has been previously demonstrated that NS4a intercalates into the NS3 protease β-sheet prior to the cis cleavage, and that the amphipathic helix α0 of the NS3 protein associates with the ER membrane and positions the complex for the subsequent insertion of the N-terminus of NS4a into the membrane 33-36. Based on this data a model was proposed, which places the protease active site facing the ER membrane and requires the partial immersion of the hydrophilic helicase domain into the membrane during the cis cleavage 35. The subsequent rotation of the helicase domain away from the protease domain would then populate a functionally active, extended conformation, analogous to the model proposed here and to that recently reported for the dengue NS3 protein 31, 32, 35. The stabilisation of the NS3/4a auto-inhibited conformation via binding of allosteric inhibitors, could thus not only inhibit the further processing of the HCV polyprotein by the protease, but also prevent the formation of the replicase complex.
The discovery of an allosteric site on the HCV NS3/4a protein offers a new avenue to develop novel anti-viral agents as well as an opportunity to further probe the regulatory mechanisms employed by viral enzymes. This discovery is an example of how chemical leads, this time found using fragment-based screening methods, can reveal novel mechanisms that modulate the function of complex biological systems. The chemical probes reported herein, may provide invaluable tools by which the mechanism of membrane insertion and the specific conformation of the NS3/4a protein at different stages of the HCV lifecycle may be ascertained.
Methods
Full details of the protocols followed can be found in the supplementary information. Brief descriptions of the main methods are described below.
Cloning, expression and purification of HCV NS3/4a
The full length genotype 1b NS3/4a construct comprises residues NS4a21-32NS33-631 25 and cloned into a pET 28 vector via NdeI and XhoI restriction sites. The plasmid was freshly transformed into BL21 (DE3) or Rosetta 2 (DE3) E. coli cells and grown at 37°C in Terrific Broth supplemented with 50ug/ml of kanamycin and 100μM ZnCl2 or ZnSO4, and induced with IPTG after lowering the temperature to 18°C. Cells were suspended in lysis buffer at 4°C, lysed by sonication and clarified by centrifugation. The protein was captured by IMAC chromatography (Qiagen) and further purified using ion exchange chromatography (GE Healthcare) and final polish was done using a gel filtration column(GE Healthcare). Fractions were analysed by SDS PAGE (Novex) pooled and concentrated to a final concentration of ~6mg/ml. Analysis by mass spectrometry showed the mass of the purified wildtype protein to be 70663D. The expected mass was 70794D indicating the loss of the N-terminal methionine, which was verified by N-terminal sequencing. Further details and a representative gel illustrating the purity level achieved is shown in supplementary material (figure 5).
Crystallisation and X-ray fragment screen
The protein was ultracentrifuged prior to crystallization and then used to in a 1:1 ratio with 0.2 M 2-(N-morpholino)ethanesulfonic acid (MES)-NaOH pH 6.6, 14-20% w/v polyethylene glycol (PEG) 6000, 10% w/v 2-methyl-2,4-pentandiol (MPD) in hanging drop crystallisation at 20°C. Seeding was used to increase the number of diffraction-grade crystals obtained, which formed over 24-48 hours.
176 fragments were screened by X-ray crystallography in cocktails of four. Each compound stock solution was made up in 100% DMSO and added to the cocktail crystal soaking solution (0.225M MES 6.6, 22.5% PEG 6000, 11.25% MPD) to give a final nominal concentration of 4 × 50mM compound. The pH of cocktail soaking solutions was adjusted to pH6.6 +/− 0.5 pH units. Crystals were soaked in the cocktail solutions for up to 10 days, prior to freezing directly from the cocktail solution using cryo loop and liquid nitrogen. Individual compounds were soaked at 5-50mM using stock solutions of 0.1-1.0M compound in 100% d6 DMSO. X-ray data were collected at 100K on the macromolecular beamlines ID23 and ID29-1 at ESRF and I03 at Diamond, using 0.5 or 1.0 degree oscillations, and a total oscillation range of 70-90 degrees, depending on the orientation of the crystal within the cryo loop. Of the 176 fragments screened a total of 16 were observed to bind at the allosteric site.
X-ray diffraction data were processed using autoPROC with either mosflm or XDS as the indexing and integration program and then reduced further using the CCP4 suite of programs (see supplementary table 2) 37-40. The crystals belong to spacegroup P212121 with unit cell dimensions a=91.2Å, b=110.7Å, c=142.3Å, and with two copies in the asymmetric unit. The ligand complexes were solved using 1CU1 as a search model 25 rebuilt using COOT and refined using refmac and autoBUSTER 41-43. Ligand placement was performed using AutoSolve 44. The pdb accession codes for the complex structures are 4b6e, 4b6f, 4b76, 4b71,4b73 and 4b74 for compounds 1-6 in the wildtype enzyme respectively, and 4b75 for compound 5 in the Val630Leu mutant enzyme.
NS3/4a protease Assays
The protease activity of the full length NS3/4a and the protease domain were measured using a FRET-based assay utilizing a peptide substrate derived from the NS4A/B cleavage site and labelled at one end with a quencher (QXL520) and at the other with a fluorophore (5-FAMsp) (Anaspec). The reaction was followed by monitoring the change in fluorescence on a Gemini plate reader (Molecular Devices) for 30 minutes at room temperature. Initial rates were calculated from the progress curves using SoftMax Pro (Molecular Devices). The IC50 value was then calculated from replicate curves using Prism GraphPad software. Full details can be found in the supplementary methods.
Isothermal Titration Calorimetry
Isothermal Titration Calorimetry (ITC) experiments were performed on a MicroCal VP-ITC at 25°C. Protein was dialysed overnight and the dialysate was used to dilute protein to between 8-20μM and compound to the final working concentration. Compound stocks were prepared in 100% DMSO. The final experimental DMSO concentration was typically between 1 and 3%. All ITC experiments were set up with protein in the sample cell and compound in the injection syringe. Data were fit to a single site binding model using Origin 7.0 software. The stoichiometry parameter was fixed at 1 in cases where the Kd value was greater than the protein concentration. A typical titration curve of compound 5 against the WT NS3/4a protein is shown in supplementary figure 6.
Compound Supply
Compound 1 was purchased from Bionet (LC/MS purity > 98%; M− = 132). Compound 2 was purchased from Fluorochem (HPLC purity > 90%; [M+H-NH3]+ = 183). Compounds 3-6 were synthesized in house as described in the Supplementary Methods.
Replicon Assays
About 4 × 10−3 huh-7 cells persistently infected with the HCV subgenomic replicon construct pFKI3889luc-ubi-neo/NS3-3′/ET were plated/well in a 96 well tissue culture plate and allowed to attach overnight in DMEM medium supplemented with 10% FBS 1% NEAA, and 250 μg/ml gentamicin. The medium was replaced with fresh medium as described above lacking gentamicin. Semilog dilutions of the compound in medium were added to triplicate wells to give a 0.1% DMSO final concentration. Plates were incubated at 37°C in 5% CO2 and air for 72 h. The antiviral effect of the compound, EC50 values were determined by measuring the luciferase activity of the cells by adding an equal volume of 2x Steady-Glo reagent to the wells and incubating at room temperature for 5 minutes (Promega). A volume of 100 μl/well was transferred to a white bottom 96 well plate to read in a luminometer as per supplier protocol (Promega).
Sedimentation Velocity Analysis
Sedimentation Velocity Experiments were done at CAMIS, New Hampshire, USA (C. May, T. Laue) and the protocol followed is described in the supplementary methods.
Supplementary Material
Acknowledgements
We gratefully acknowledge the contributions of M. Vinkovic, H. Angove, F. Holding, E. Chiarparin and M.L.Verdonk. We thank C. Cartwright for producing figures 4 and supplementary table 1. X-ray data were collected at the European Synchrotron Radiation Facility and at the Diamond Lightsource. Sedimentation velocity experiments were performed at CAMIS, New Hampshire, USA.
Figure 4.
Proposed mode of action of allosteric inhibitors
A. Closed conformation - product of the cis-cleavage between the NS3 and NS4a proteins. The C-terminus of the helicase domain occupies the protease active site and stabilises the protein in an auto-inhibited conformation.
B. Open conformation - required for proteolytic activity and also reportedly required for helicase activity 31. This conformation allows substrates to access the protease active site and is inhibited by the peptidomimetic active site inhibitors such as telaprevir and boceprevir.
C. Closed conformation – Compounds binding at the protease-helicase interface stabilise the auto-inhibited conformation of the protein and block catalytic function via an allosteric mechanism.
Colour key: red oval - allosteric inhibitor; orange line - C-terminus of the helicase domain; brown line - flexible linker between the protease and helicase domains.
This research program was supported by grant 087738 from the Wellcome Trust.
Footnotes
Competing Financial Interests
All authors are employees of Astex Pharmaceuticals and own shares in the company.
Additional information
Supplemenatry information and chemical compound information is available in the online version of the paper at http://www.nature.com/naturechemicalbiology/.
References
- 1.Reed KE, Rice CM. Overview of hepatitis c virus genome structure, polyprotein processing and protein properties. Curr. Top. Microbiol. Immunol. 2000;242:55–84. doi: 10.1007/978-3-642-59605-6_4. [DOI] [PubMed] [Google Scholar]
- 2.Bartenschlager R, Lohmann V. Replication of hepatitis C virus. J. Gen. Virol. 2000;81:1631–1648. doi: 10.1099/0022-1317-81-7-1631. [DOI] [PubMed] [Google Scholar]
- 3.Morgenstern KA, et al. Polynucleotide Modulation of the Protease, Nucleoside Triphosphatase, and Helicase Activities of the Hepatitis C Virus NS3-4A Complex Isolated from Transfected COS Cells. J. Virol. 1997;71:3767–3775. doi: 10.1128/jvi.71.5.3767-3775.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beran RK, Pyle AM. Hepatitis C Viral NS3-4a protease Activity is Enhanced by the NS3 Helicase. J. Biol. Chem. 2008;283:29929–29937. doi: 10.1074/jbc.M804065200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beran RK, Serebrov V, Pyle AM. The Serine Protease domain of Hepatitis C Viral NS3 Activates RNA Helicase Activity by Promoting the Binding of RNA Substrate. J. Biol. Chem. 2007;282:34913–34920. doi: 10.1074/jbc.M707165200. [DOI] [PubMed] [Google Scholar]
- 6.Rajagopal V, Gurjar M, Levin MK, Patel SS. The Protease Domain Increases the Translocation Stepping Efficiency of the Hepatitis C Virus NS3-4A Helicase. J Biol. Chem. 2010;285:17821–17832. doi: 10.1074/jbc.M110.114785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hopkins AL, et al. Complexes of HIV-1 reverse transcriptase with inhibitors of the HEPT series reveal conformational changes relevant to the design of potent non-nucleoside inhibitors. J. Med.Chem. 1996;39:1589–1600. doi: 10.1021/jm960056x. [DOI] [PubMed] [Google Scholar]
- 8.Kessl JJ, et al. An Allosteric Mechanism for Inhibiting HIV-1 Integrase with a Small Molecule. Mol Pharmacol. 2009;76:824–832. doi: 10.1124/mol.109.058883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker MA. New approaches for inhibiting HIV integrase: a journey beyond the active site. Curr Opin Investig Drugs. 2009;10:129–36. [PubMed] [Google Scholar]
- 10.Chen KX, Njoroge FG. A review of HCV protease inhibitors. Curr Opin Investig Drugs. 2009;10:821–37. [PubMed] [Google Scholar]
- 11.Lamarre D, et al. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature. 2003;426:186–189. doi: 10.1038/nature02099. [DOI] [PubMed] [Google Scholar]
- 12.Thibeault D, et al. Use of the Fused NS4a Peptide-NS3 Protease Domain to Study the Importance of the Helicase Domain for Protease Inhibitor Binding to Hepatitis C Virus NS3-4a. Biochemistry. 2009;48:744–753. doi: 10.1021/bi801931e. [DOI] [PubMed] [Google Scholar]
- 13.Belon CA, Frick DN. Helicase inhibitors as specifically targeted antiviral therapy for hepatitis C. Future Virology. 2009;4:277–293. doi: 10.2217/fvl.09.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frick DN. The Hepatitis C Virus NS3 Protein: A Model RNA Helicase and Potential Drug Target. Curr Issues Mol. Biol. 2007;9:1–20. [PMC free article] [PubMed] [Google Scholar]
- 15.Raney KD, Sharma SD, Moustafa IM, Cameron CE. Hepatitis C Virus Non-structural Protein 3 (HCV NS3): A Multifunctional Antiviral Target. J. Biol Chem. 2010;285:22725–22731. doi: 10.1074/jbc.R110.125294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chappell KJ, et al. West Nile Virus NS2B/NS3 protease as an antiviral target. Curr Med Chem. 2008;15:2771–2784. doi: 10.2174/092986708786242804. [DOI] [PubMed] [Google Scholar]
- 17.Chernov AV, et al. The Two-component NS2B-NS3 Proteinase Represses DNA Unwinding Activity of the West Nile Virus NS3 Helicase. J. Biol. Chem. 2008;283:17270–17278. doi: 10.1074/jbc.M801719200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray CW, Blundell T. Structural biology in fragment-based drug design. Current Opinion in Structural Biology. 2010;20:497–507. doi: 10.1016/j.sbi.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Murray CW, Rees DC. The rise of fragment-based drug discovery. Nature chemistry. 2009;1:187–192. doi: 10.1038/nchem.217. [DOI] [PubMed] [Google Scholar]
- 20.Rhodes DI, et al. Structural basis for a new mechanism of inhibition of HIV-1 integrase identified by fragment screening and structure-based design. Antiviral Chemistry & Chemotherapy. 2011;21:155–168. doi: 10.3851/IMP1716. [DOI] [PubMed] [Google Scholar]
- 21.Perryman AL, et al. Fragment-based Screen against HIV Protease. Chem. Biol.Drug Des. 2010;75:257–268. doi: 10.1111/j.1747-0285.2009.00943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krimm I, Lancelin J-M, Praly J-P. Binding Evaluation of Fragment -Based Scaffolds for Probing Allosteric Enzymes. J.Med.Chem. 2012;55:1287–1295. doi: 10.1021/jm201439b. [DOI] [PubMed] [Google Scholar]
- 23.Maurer T, et al. Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc. Natl. Acad. Sci. USA. 2012;109:5299–5304. doi: 10.1073/pnas.1116510109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pommier Y, Marchand C. Interfacial inhibitors: targeting macromolecular complexes. Nature Reviews Drug Disc. 2012;11:25–36. doi: 10.1038/nrd3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao N, Reichert P, Taremi SS, Prosise WW, Weber PC. Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease–helicase. Structure. 1999;7:1353–1363. doi: 10.1016/s0969-2126(00)80025-8. [DOI] [PubMed] [Google Scholar]
- 26.Lohmann V, et al. Replication of subgenomic Hepatitis C virus RNA in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 27.Bartenschlager R. Hepatitis C replicons: potential role for drug development. Nature Rev. Drug Discov. 2002;1:911–916. doi: 10.1038/nrd942. [DOI] [PubMed] [Google Scholar]
- 28.De Clercq E. The design of drugs for HIV and HCV. Nature Reviews Drug Discovery. 2007;6:1001–1018. doi: 10.1038/nrd2424. [DOI] [PubMed] [Google Scholar]
- 29.Richman DD. The impact of drug resistance on the effectiveness of chemotherapy for chronic hepatitis B. Hepatology. 2000;32:866–867. doi: 10.1053/jhep.2000.18194. [DOI] [PubMed] [Google Scholar]
- 30.Kuiken C, Yusim K, Boykin L, Richardson R. The Los Alamos HCV Sequence Database. Bioinformatics. 2005;21:379–84. doi: 10.1093/bioinformatics/bth485. [DOI] [PubMed] [Google Scholar]
- 31.Ding SC, Kohlway AS, Pyle AM. Unmasking the Active Helicase Conformation of Nonstructural Protein 3 from Hepatitis C Virus. J. Virol. 2011;85:4343–4353. doi: 10.1128/JVI.02130-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo D, et al. Flexibility between the Protease and Helicase Domains of the Dengue Virus NS3 Protein Conferred by the Linker Region and Its Functional Implications. J. Biol. Chem. 2010;285:18817–18827. doi: 10.1074/jbc.M109.090936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang W, et al. Conserved C-Terminal Threonine of Hepatitis C Virus NS3 Regulates Autoproteolysis and Prevents Product Inhibition. J Virol. 2004;78:700–709. doi: 10.1128/JVI.78.2.700-709.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woelk B, et al. Subcellular localization, stability, and trans-cleavage competence of the hepatitis C virus NS3-NS4A complex expressed in tetracycline-regulated cell lines. J Virol. 2000;74:2293–2304. doi: 10.1128/jvi.74.5.2293-2304.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brass V, et al. Structural determinants for membrane association and dynamic organization of the hepatitis C virus NS3-4A complex. Proc. Natl. Acad. Sci. USA. 2008;105:14545–214550. doi: 10.1073/pnas.0807298105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horner SM, Park HS, Gale M., Jr Control of Innate Immune Signaling and Membrane Targeting by the Hepatitis C Virus NS3/4A Protease are Governed by the NS3 Helix α0. J. Virol. 2012;86:3112–3120. doi: 10.1128/JVI.06727-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vonrhein C, et al. Data processing and analysis with the autoPROC toolbox. Acta Cryst. 2011;D67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leslie AGW, Powell HR. Processing Diffraction Data with Mosflm. Evolving Methods for Macromolecular Crystallography. 2007;245:41–51. [Google Scholar]
- 39.Kabsch W. Integration, scaling, spacegroup-assignment and post-refinement. Acta Cryst. 2010;D66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 41.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 43.Bricogne G, et al. BUSTER version 1.11.2. Global Phasing Ltd; Cambridge, United Kingdom: 2011. [Google Scholar]
- 44.Mooij WTM, et al. Automated Protein-Ligand Crystallography for Structure-Based Drug Design. ChemMedChem. 2006;1:827–38. doi: 10.1002/cmdc.200600074. [DOI] [PubMed] [Google Scholar]
- 45.Schiering N, et al. A macrocyclic HCV NS3/4a protease inhibitor interacts with protease and helicase residues in the complex with its full-length target. Proc. Natl. Acad. Sci. USA. 2011;108:21052–21056. doi: 10.1073/pnas.1110534108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.