SUMMARY
Combination of stem cell-based approaches with gene-editing technologies represents an attractive strategy for studying human disease and developing therapies. However, gene-editing methodologies described to date for human cells suffer from technical limitations including limited target gene size, low targeting efficiency at transcriptionally inactive loci, and off-target genetic effects that could hamper broad clinical application. To address these limitations, and as a proof of principle, we focused on homologous recombination-based gene correction of multiple mutations on lamin A (LMNA), which are associated with various degenerative diseases. We show that helper-dependent adenoviral vectors (HDAdVs) provide a highly efficient and safe method for correcting mutations in large genomic regions in human induced pluripotent stem cells and can also be effective in adult human mesenchymal stem cells. This type of approach could be used to generate genotype-matched cell lines for disease modeling and drug discovery and potentially also in therapeutics.
INTRODUCTION
The development of targeted gene-editing technologies in human induced pluripotent stem cells (hiPSCs) and human embryonic stem cells (hESCs) may not only open the opportunity for à la carte cell engineering but also offer the potential for the efficient treatment and/or cure of multiple human diseases. Recently, several technologies have been developed allowing for gene targeting in human embryonic stem cells (hESCs) and hiPSCs (Buecker et al., 2010; Händel and Cathomen, 2011; Hockemeyer et al., 2009; Howden et al., 2011; Irion et al., 2007; Khan et al., 2010; Lombardo et al., 2007; Mitsui et al., 2009; Nieminen et al., 2010; Sancho-Martinez et al., 2011; Song et al., 2010; Suzuki et al., 2008; Tenzen et al., 2010; Zou et al., 2009; Zwaka and Thomson, 2003). However, despite numerous efforts and recent advancements, major problems remain unresolved. First, the classical nonviral vectors and delivery approaches are associated with low efficiency of gene targeting in human pluripotent stem cells, especially for transcriptionally silenced genomic loci (Goulburn et al., 2011; Ruby and Zheng, 2009; Wang et al., 2011; Xue et al., 2009). Second, the current technologies introduce DNA double-strand breaks (DSBs) (Arnould et al., 2011; Händel and Cathomen, 2011; Miller et al., 2011). Accordingly, the potential for off-target effects leading to unexpected mutations and chromosomal aberrations might not be acceptable for therapeutic applications. Moreover, for genes bearing mutational “hot spots” (Worman et al., 2010), efficient tools targeting multiple sites spanning a large DNA sequence still need to be developed.
In the present work we sought to address some of these issues by taking advantage of a recently established LMNA-based iPSC model of disease (Liu et al., 2011). LMNA is a known transcriptionally inactive locus in pluripotent cells. Importantly, more than 400 different mutations of LMNA have been reported, with additional mutations continuously being discovered (Zaremba-Czogalla et al., 2011). Thus, LMNA represents an ideal candidate locus for the study of hot-spot gene correction of silenced genes by using single vectors targeting large genomic regions. LMNA encodes lamin A, an architectural component of nuclear lamina in somatic cells indispensable for nuclear integrity (Dechat et al., 2008). Mutations on lamin A are associated with disruption of the nuclear structure, aberrant cellular function, and disease development, defining a group of human degenerative diseases referred to as laminopathies (Burke et al., 2001; Worman et al., 2010; Zaremba-Czogalla et al., 2011). One of the best-known laminopathies is Hutchinson-Gilford progeria syndrome (HGPS or progeria), a premature aging disorder associated with the presence of a specific mutation of the LMNA gene. Such mutation results in a truncated mRNA encoding the mutant protein progerin (Burtner and Kennedy, 2010). Progerin plays a dominant role in the nucleus and its accumulation can result in a number of different nuclear defects, accelerated aging, and cellular senescence (Dechat et al., 2008; Scaffidi and Misteli, 2005).
Recently we and others have reported the generation of iPSCs (HGPS-iPSCs) from HGPS patient fibroblasts (Liu et al., 2011; Zhang et al., 2011). Here we report the establishment of an efficient and highly specific method for correction of a number of LMNA mutations. Indeed, the use of a single helper-dependent adenoviral vector (HDAdV) was sufficient for the correction of different mutations spanning a substantially large region of the LMNA gene. Targeted correction of LMNA led to the restored expression of wild-type lamin A and to the abolishment of progerin expression. Consequently, lack of progerin expression resulted in the correction of the disease-associated cellular phenotypes, including decelerated senescence and restoration of normal nuclear morphology. Furthermore, the use of HDAdV-based approaches demonstrated very high efficiencies in the absence of any observed genetic and epigenetic abnormality and led to indistinguishable cell genetic backgrounds before and after gene targeting. Thus, the method described here might represent a more efficient and safer alternative to other current technologies. Along the same line, HDAdV-based approaches can be used for the generation of genetically matched iPSCs serving as a more reliable control for disease modeling and drug discovery. Lastly, our methodologies demonstrate the use of HDAdV-based gene targeting as a reliable tool for the engineering of adult mesenchymal stem cells (MSCs).
RESULTS AND DISCUSSION
HDAdV-Based Gene Correction Shows High Efficiency in Transcriptionally Inactive Locus
Of the 12 exons of the LMNA gene, exons 2 to 12 cluster closely together. This region covers around 80% of the whole LMNA coding sequence and accumulates up to 246 of the published mutations in the LMNA gene (http://www.umd.be/LMNA/). With the aim to develop a single method amenable for the correction of multiple LMNA mutations, we engineered a HDAd-based gene-correction vector (LMNA-c-HDAdV) (Figure 1A). HDAdV has been proven to mediate efficient and precise gene editing in hESCs without the requirement for artificial DNA DSBs (Suzuki et al., 2008). Because of the complete removal of viral genes from the vector, HDAdVs show low cytotoxicity while displaying the major advantage of having a large cloning capacity (~37 kb). Thus, HDAdVs permit the insertion of long homologous DNA regions that are designed to facilitate targeted integration via homologous recombination (HR) (Suzuki et al., 2008).
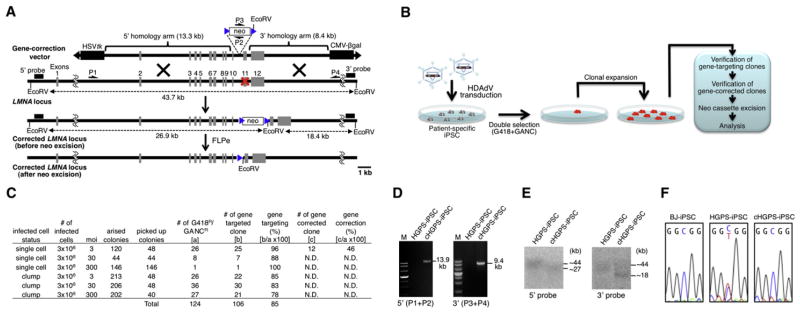
Figure 1. Correction of HGPS-Associated LMNA Mutation in iPSCs with LMNA-c-HDAdV.
(A) Schematic molecular representation of LMNA gene correction with LMNA-c-HDAdV. The primers for PCR are shown as arrows (P1, P2, P3, and P4). The probes for Southern analyses are shown as black bars (5′ probe and 3′ probe). HSVtk stands for herpes simplex virus thymidine kinase gene cassette used for negative selection; neo stands for neomycin-resistant gene cassette used for positive selection; CMV-β-gal indicates the β-gal expression cassette for determination of HDAdV titer; blue triangle, FRT site; red X, mutation sites in exon 11.
(B) Schematic representation of the gene-correction approach employed in iPSCs with laminopathy-associated mutation(s).
(C) Gene-targeting and gene-correction efficiencies at the LMNA locus in HGPS-iPSCs achieved by different infection conditions. N.D., not determined.
(D) PCR analyses of HGPS-iPSCs and cHGPS-iPSCs via 5′ primer pair (P1 and P2; 13.9 kb) or 3′ primer pair (P3 and P4; 9.4 kb). M, DNA ladder.
(E) Southern blot analyses of HGPS-iPSCs and cHGPS-iPSCs. The approximate molecular weights (kb) corresponding to the bands are indicated.
(F) Sequencing results of C1824T mutation site in exon 11 of LMNA in BJ-iPSCs (wild-type), HGPS-iPSCs, and cHGPS-iPSCs. All iPSCs employed represent high-passage iPSCs (>30).
See also Figure S1.
In view of these advantages, we first evaluated the ability of LMNA-c-HDAdV to correct the HGPS-associated LMNA mutation C1824T in exon 11 in HGPS-iPSCs. We initially tested various viral infection conditions as well as different multiplicities of infection (MOIs). The system here employed includes a negative selection step by ganciclovir (GANC) resistance. If the vector randomly integrates into the genome, the integrated DNA will still include an HSVtk cassette leading to cell death by GANC selection. On the other hand, if the vector integrates at the targeted site by HR, the HSVtk cassette is removed and the cell displays GANC resistance (Mansour et al., 1988). Thus, negative selection helps to minimize ectopic vector integration (Suzuki et al., 2008). Accordingly, after sequential positive (G418) and negative (GANC) selection (Mansour et al., 1988), double-resistant colonies were expanded and maintained in hESC medium (Figure 1B). Effective gene targeting at the LMNA loci in drug-resistant clones was verified by PCR and Southern blot (Figures 1C–1E; Figure S1A available online). We speculated that because of the transcriptional inactivation of the LMNA locus in HGPS-iPSCs (Liu et al., 2011; Zhang et al., 2011), gene-targeting efficiencies would be considerably low as previously described for other methodologies (Goulburn et al., 2011; Ruby and Zheng, 2009; Wang et al., 2011; Xue et al., 2009). Surprisingly, efficient gene targeting of LMNA (78%–100%) was consistently obtained even with the lowest MOI used (Figure 1C). These results indicate that high-targeting efficiency might saturate the system, allowing for reduced number of viral particles. We arbitrarily selected the iPSC colonies that were infected as single cells with the lowest viral titer (MOI = 3) for further analysis. Since most laminopathy-associated mutations are heterozygous, we expected gene-correction efficiencies to be 50% of the total frequency of gene-targeting events. Accordingly, DNA sequencing showed that mutations in 46% of the drug-resistant colonies were successfully corrected (Figures 1C and 1F).
Gene Correction by LMNA-c-HDAdV Maintains Genetic and Epigenetic Integrity and Reverses Disease-Associated Phenotypes
In order to achieve genomic integrity in the corrected clones, we next excised the neomycin-resistant gene cassette, flanked by FRT sites, by transient expression of FLPe recombinase (Figure S1B). The neo-removed corrected HGPS-iPSC lines (hereafter referred to “cHGPS-iPSCs”) showed a normal karyotype, expressed pluripotent markers, and exhibited demethylation of the OCT4 promoter and pluripotency (Figures S1C–S1I; Movie S1). Genome-wide single nucleotide polymorphism (SNP) genotyping demonstrated that the genetic background of the cHGPS-iPSCs was the same as the parental HGPS fibroblasts, but distinct from H9 hESCs and the control BJ-iPSCs (Figure 2A; Table S1). By copy number variation (CNV) of the SNP genotyping data, using the method described in Laurent et al. (2011), there were no detected new duplications or deletions in the cHGPS-iPSCs compared to the HGPS-fibroblast cells (Figure 2B). Although this result may appear to be at variance with a recent report by Hussein et al. (2011), there were significant differences between the samples analyzed. Hussein et al. (2011) reported a higher frequency of CNVs in very early passage (<10 passages) when compared to later-passage iPSCs and postulated that these CNVs occurred during the process of reprogramming and were negatively selected in culture. Thus, because our gene targeting experiments started with HGPS-iPSCs at passage 30, our corrected iPSCs (>passage 30) clearly surpassed that early window of high-frequency CNV. Both SNP arrays used in the two studies assayed ~1 million SNPs, and the analyses required consistent calls for a minimum of ten adjacent markers to identify a CNV; therefore the controversies observed between Hussein’s and our own results were not due to technical but to passage number differences. DNA microarray analysis revealed very similar expression profiles between cHGPS-iPSCs and their parental HGPS-iPSCs (Figure 2C), indicating that the targeting procedure did not alter global gene expression. To examine whether the gene-targeting procedure interfered with the global epigenetic state, we performed genome-wide DNA methylation analysis. An average of 12 million single-end 80 bp sequencing reads was generated for each sample, yielding an average read depth of >50× for all CpG sites called, which is roughly twice as high as the published whole-genome bisulfite sequencing data (Lister et al., 2011). The methylation levels measured on the Watson strand and the Crick strand were highly consistent for the same CpG sites (Pearson correlation coefficient r = 0.965 for site with read depth ≥ 10), indicating that the methylation measurements were very accurate. As shown in Figure 2D, the cHGPS-iPSCs showed a highly similar methylation profile as its parental iPSCs, in contrast to hESCs and wild-type iPSCs. Thus, our gene correction approach effectively maintained genetic and epigenetic cell integrity.
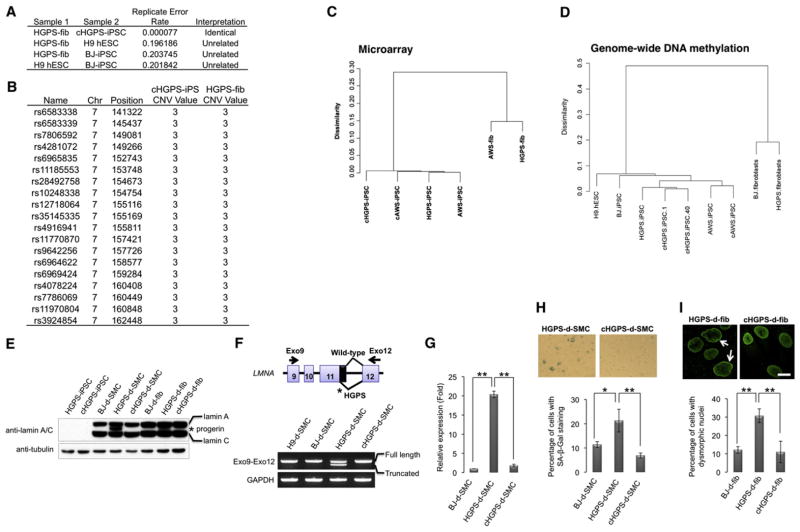
Figure 2. Abrogation of HGPS-Associated Phenotypes by Correction of LMNA in iPSCs.
(A) Replicate error analysis indicating that cHGPS-iPSCs and HGPS fibroblasts were essentially genetically identical, whereas HGPS fibroblasts, H9 hESCs, and BJ-iPSCs came from unrelated individuals.
(B) CNV calling via CNV Partition, identifying one region of duplication (CNV value = 3) at chr7:141322-162448, which was present in both the original HGPS fibroblasts and cHGPS-iPSCs.
(C) Hierarchical clustering on RMA-normalized probe sets intensity values for HGPS- and AWS-iPSCs before and after gene correction, and their original fibroblast cell lines.
(D) Hierarchical clustering on genome-wide DNA methylation profiles of the indicated cell lines.
(E) Immunoblotting analysis of lamin A/C and progerin expression in the indicated cell lines with lamin A/C antibody. Tubulin was used as loading control. d-SMC, iPSC-derived SMC; d-fib, iPSC-derived fibroblast.
(F) RT-PCR analysis of expression of full-length lamin A (top band) and truncated lamin A (progerin; bottom band) in hESC/iPSC-derived SMCs (passage 3) with the indicated primer pair. GAPDH was used as loading control.
(G) Quantitative RT-PCR analysis of progerin expression in BJ-iPSC-, HGPS-iPSC-, or cHGPS-iPSC-derived SMCs at passage 3. **p < 0.01.
(H) Senescence-associated (SA)-β-gal staining of SMCs derived from HGPS-iPSC or cHGPS-iPSC at passage 5. Correction of HGPS-iPSCs resulted in more than a 68% reduction on the derived SMCs undergoing senescence; *p < 0.05, **p < 0.01.
(I) Immunostaining of lamin A in iPSC-derived fibroblasts at passage 15. Arrows denote dysmorphic nuclei. Dysmorphic nuclei were quantified and represented in the lower panels. **p < 0.01. Data are shown as mean ± SD; n = 3. Scale bar represents 20 μm. All iPSCs employed represent high-passage iPSCs (>30).
To ascertain whether correction of the mutant LMNA in HGPS-iPSCs is able to prevent development of some of the disease-associated phenotypes, we differentiated the cHGPS-iPSCs into vascular smooth muscle cells (SMCs) and fibroblasts, two known HGPS-affected cell types (Liu et al., 2011; Zhang et al., 2011). Immunoblotting and RT-PCR analysis did not detect progerin mRNA or protein in cHGPS-iPSCs-derived cells (Figures 2E–2G). These results correlated with a significant rescue of the senescence phenotype. Senescence-associated (SA)-β-gal staining was reduced from 21.3% in SMCs differentiated from the noncorrected HGPS-iPSCs to 6.8% in cHGPS-iPSCs-derived SMCs, compared to 11.4% observed in BJ-iPSC-derived SMCs (Figure 2H). In addition, cHGPS-iPSCs-derived fibroblasts showed more than 60% reduction in the number of abnormal nuclei as compared to noncorrected counterparts (Figure 2I). Altogether, our results show that genetic correction of HGPS-iPSCs completely abrogated expression of progerin and resulted in expression of wild-type lamin A, thus leading to restored nuclear architecture and a normal cell senescence program.
LMNA-c-HDAdV Spans Large Genomic Regions and Could Be Used for the Correction of Multiple Mutations
Considering the large DNA-carrying capacity of our generated LMNA-c-HDAdV, we next decided to examine whether the same approach could be used to correct other LMNA mutations. We generated iPSCs from a patient with atypical Werner syndrome (AWS), a milder premature aging disease caused by a different LMNA mutation (A1733T in exon 11, E578V) (Csoka et al., 2004). AWS-iPSCs demonstrated acquisition of the typical iPSC characteristics including expression of pluripotency markers, ability to differentiate in vitro, formation of teratomas, and pluripotent gene expression and DNA methylation signatures (Figures 2C, 2D, 3A, and 3B; Figure S1G). In addition, AWS-iPSCs showed a normal karyotype and carried the disease-specific LMNA mutation (Figure 3F; Figure S1C). By employing the same approach used for the correction of HGPS-iPSCs, we obtained gene-targeting efficiencies ranging from 89% to 100% and specific gene correction of up to 35% (Figures 3C–3F; Figure S1J). The corrected AWS-iPSCs (cAWS-iPSCs) expressed pluripotent markers and showed a normal karyotype as well as a highly similar DNA methylation and gene expression profile as their parental AWS-iPSCs (Figures 2C and 2D; Figure S1C and S1E). These observations highlight the possibility of using a single LMNA-c-HDAdV vector for the correction of multiple mutations of the LMNA gene and thus for the treatment of different laminopathies.
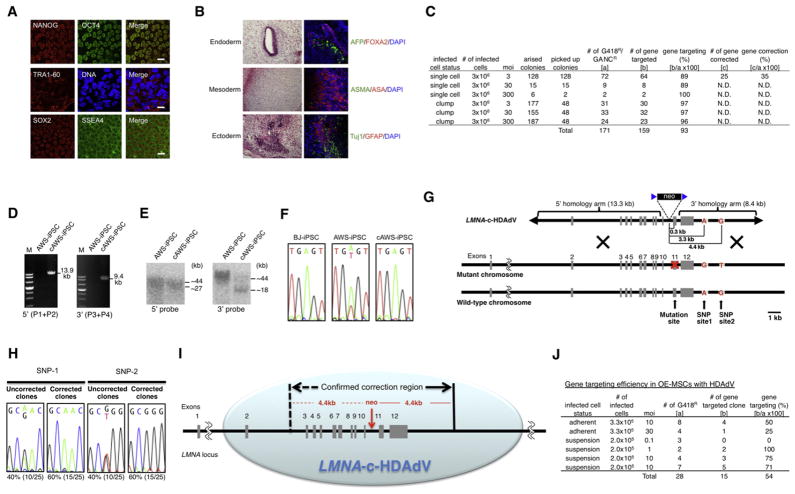
Figure 3. Editing the LMNA Locus in iPSCs and MSCs via LMNA-c-HDAdV.
(A) Immunostaining of different pluripotency markers in AWS-iPSCs. Nuclei were stained with Hoechst 33342. Scale bars represent 20 μm.
(B) H&E staining (left) and immunofluorescence (right) of three germ layer-specific markers (endoderm: AFP, FOXA2; mesoderm: ASMA, ASA; ectoderm: Tuj1, GFAP) in teratomas derived from AWS-iPSCs. Nuclei were stained with DAPI.
(C) Gene-targeting and gene-correction efficiencies at the LMNA locus in AWS-iPSCs achieved by different infection conditions. N.D., not determined.
(D) PCR analyses of AWS-iPSCs and gene-corrected AWS-iPSCs (cAWS-iPSCs) via 5′ primer pair (P1 and P2; 13.9 kb) or 3′ primer pair (P3 and P4; 9.4 kb). M, DNA ladder.
(E) Southern blot analyses of AWS-iPSCs and cAWS-iPSCs. The approximate molecular weights (kb) corresponding to the bands are indicated.
(F) Sequencing results of A1733T mutation site in exon 11 in BJ-iPSCs (wild-type), AWS-iPSCs, and cAWS-iPSCs.
(G) Schematic molecular representation of mutation- and SNP-correction with LMNA-c-HDAdV in AWS-iPSCs. The mutation site (A1733T) and SNP sites 1 and 2 are located 0.3, 3.3, and 4.4 kb downstream of the neo-insertion site, respectively, on the same chromosome as the AWS mutation.
(H) Sequencing results of SNP sites 1 and 2 downstream of exon 12 in cAWS-iPSCs with and without SNP correction.
(I) Schematic illustration of the putative gene editing capacity of LMNA-c-HDAdV at LMNA locus.
(J) Gene-targeting efficiency in OE-MSCs with LMNA-c-HDAdV achieved by different infection conditions. All iPSCs employed represent high-passage iPSCs (>30).
See also Figure S1.
We next sought to determine the maximal region that LMNA-c-HDAdV covers. To this end, we sequenced the whole LMNA locus in HGPS- and AWS-iPSCs looking for specific SNP sites across the LMNA locus that are not present in the vector. Two AWS-specific SNP sites located downstream of exon 12 were found on the same chromosome as the AWS mutation in the noncorrected cells and were used as a read-out for our analysis (Figure 3G). These two SNP sites are located 3.3 and 4.4 kb downstream of the neo-insertion site, respectively. By examining the presence of these two SNPs in the corrected AWS-iPSCs, we found that both of them were effectively replaced by the wild-type LMNA-c-HDAdV sequence (60% of the gene-corrected clones) (Figure 3H), indicating that the majority of HR events occur outside of the second SNP site. Since exons 3–12 are also located within the 4.4 kb region from neo-insertion site (Figure 3I), our results suggest that LMNA-c-HDAdV can at least replace the whole exon 3–12 region. Thus, our method could potentially be used to correct more than 200 different mutations described in the LMNA locus.
Targeting of LMNA Locus with LMNA-c-HDAdV Can Be Achieved in Adult Stem Cells
Finally, we evaluated the possibility of using HDAd-based vectors for gene editing in human adult stem cells. We focused on mesenchymal stem cells (MSCs) because of their current and extensive use in regenerative medicine (Salem and Thiemermann, 2010). LMNA mutations mainly affect mesoderm-derived tissues, such as muscle and adipocytes. Thus, LMNA correction on MSCs could represent an attractive alternative for therapeutic purposes (Zaremba-Czogalla et al., 2011). Because of their high cell-cycle kinetics which enables effective clonal expansion, we specifically focused on olfactory ectomesenchymal stem cells (OE-MSCs) (Delorme et al., 2010). Furthermore, OE-MSCs are easily accessible by noninvasive biopsies and therefore represent an attractive cell population for clinical applications (Delorme et al., 2010). As shown in Figure 3J and Figures S1K and S1L, LMNA-c-HDAdV demonstrated gene-editing efficiency of 54% in wild-type OE-MSCs, without disruption of the normal lamin A/C expression. Although OE-MSCs from patients with laminopathies are not available at this stage, our study suggests in particular the possibility that LMNA-c-HDAdV could be used to genetically modify laminopathy-specific MSCs in the future, and more generally, the feasibility of targeting adult stem cells for gene correction.
In summary, we report the successful correction of different laminopathy-associated LMNA mutations in patient-specific iPSCs with a single gene correction vector. Moreover, HDAdV-based approaches efficiently target specific genomic loci on adult stem cells, a scenario not previously addressed. Our results demonstrate that the long homology arms used in HDAdVs have the capability to edit the targeted genomic locus without off-target effects and/or introduction of additional mutations, thus presenting an advantageous alternative to the use of other gene-editing technologies (Hockemeyer et al., 2009). Although bacterial artificial chromosome (BAC)-mediated vectors also used long homology arms, their ability and efficiency to target the genome was restricted to transcriptionally active loci (Howden et al., 2011; Song et al., 2010). Here we show that LMNA-c-HDAdV can specifically edit transcriptionally inactive loci with high efficiencies. Thus, the HDAdV represents a robust and versatile tool that could be applied toward the correction of multiple monogenic diseases. Finally, this approach could serve to generate appropriate genotype-matched iPSC lines in disease modeling and drug discovery studies.
EXPERIMENTAL PROCEDURES
Construction and Preparation of LMNA-c-HDAdV
LMNA-c-HDAdV was generated with a BAC clone, containing the human LMNA loci (RP11-54H19, BACPAC resources), that were modified with BAC recombineering (Datsenko and Wanner, 2000). In brief, the FRT-PGK-EM7-neo-bpA-FRT fragment was recombined into intron 10 of LMNA in the BAC clone. A total of 23.2 kb of LMNA homology, including the marker cassette, was subcloned into the HDAdV plasmid pAMHDAdGT8-4 (kindly provided by Dr. Kohnosuke Mitani). The generated LMNA-c-HDAdV was linearized by PmeI and then transfected into 116 cells (kindly provided by Dr. Philip Ng) in the presence of helper virus AdHPBGF35 (kindly provided by Dr. André M. Lieber) (Shayakhmetov et al., 2004). Crude virus extracts were serially amplified in 116 cells and then purified according to the method previously described (Palmer and Ng, 2004). β-gal-transducing units (btu) were determined in 293 cells to define infectious vector titers.
Immunofluorescence Analysis
4% formaldehyde in PBS was used to fix cells at room temperature (RT) for 20–30 min. After fixation, cells were exposed to 0.4% Triton X-100 in PBS for 5 min at RT. Cells were blocked with 10% FBS in PBS for 30 min and incubated with primary antibody for 1 hr at RT or overnight at 4°C. Washing was conducted with PBS followed by incubation with a corresponding secondary antibody for 1 hr at RT. Hoechst 33342 or DAPI was used to stain nuclei.
Teratoma Analysis
To test pluripotency in vivo, iPSC lines were injected into NOD-SCID IL2Rgammanull mice (Jackson Laboratories) and teratoma formation was assessed. In short, ~106 iPSCs in ~50 μl of hESC medium were injected into the kidney capsule or testis of anesthetized mice. Teratoma formation was monitored and animals were sacrificed ~6–12 weeks after injection. Harvested teratomas were processed and analyzed by immunostaining and hematoxylin-eosin staining. All murine experiments were conducted with approval of The Salk Institute Institutional Animal Care and Use Committee (IACUC).
Statistical Analysis
Results are presented as mean ± SD. Student’s t test was applied to all comparisons. Statistical significance was defined as p < 0.05.
Supplementary Material
Acknowledgments
We would like to thank K. Mitani, P. Ng, and A. Lieber for kindly providing experimental materials, J. Karlseder, L. Gerace, M. Hetzer, and C. Rodriguez Esteban for helpful discussions, C. Lynch for running SNP genotyping arrays, M.C. Llach for karyotyping, M. Marti for teratoma analysis, Y. Xia and S.-L. Yang for technical help, and M. Schwarz for administrative help. G.-H.L. was partially supported by a CIRM grant (TG2-01158), and J.Q. was partially supported by an AFAR/Ellison Medical Foundation postdoctoral fellowship. E.N. was partially supported by a F.M. Kirby Foundation postdoctoral fellowship. L.C.L. is supported by NIH K12HD001259-11. The work by L.C.L. and J.F.L. was supported by CIRM TR1-01250. This study was supported by grants from DA025779 (K.Z.), the G. Harold and Leila Y. Mathers Charitable Foundation, Sanofi-Aventis, Ellison Medical Foundation, Helmsley Foundation, MICINN, and Fundacion Cellex (J.C.I.B.).
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, one figure, one table, and one movie and can be found with this article online at doi:10.1016/j.stem.2011.04.019.
ACCESSION NUMBERS
Microarray data have been deposited in NCBI-GEO with the accession number GSE28607.
References
- Arnould S, Delenda C, Grizot S, Desseaux C, Pâques F, Silva GH, Smith J. The I-CreI meganuclease and its engineered derivatives: applications from cell modification to gene therapy. Protein Eng Des Sel. 2011;24:27–31. doi: 10.1093/protein/gzq083. [DOI] [PubMed] [Google Scholar]
- Buecker C, Chen HH, Polo JM, Daheron L, Bu L, Barakat TS, Okwieka P, Porter A, Gribnau J, Hochedlinger K, Geijsen N. A murine ESC-like state facilitates transgenesis and homologous recombination in human pluripotent stem cells. Cell Stem Cell. 2010;6:535–546. doi: 10.1016/j.stem.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke B, Mounkes LC, Stewart CL. The nuclear envelope in muscular dystrophy and cardiovascular diseases. Traffic. 2001;2:675–683. doi: 10.1034/j.1600-0854.2001.21001.x. [DOI] [PubMed] [Google Scholar]
- Burtner CR, Kennedy BK. Progeria syndromes and ageing: What is the connection? Nat Rev Mol Cell Biol. 2010;11:567–578. doi: 10.1038/nrm2944. [DOI] [PubMed] [Google Scholar]
- Csoka AB, Cao H, Sammak PJ, Constantinescu D, Schatten GP, Hegele RA. Novel lamin A/C gene (LMNA) mutations in atypical proge-roid syndromes. J Med Genet. 2004;41:304–308. doi: 10.1136/jmg.2003.015651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, Goldman RD. Nuclear lamins: Major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008;22:832–853. doi: 10.1101/gad.1652708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme B, Nivet E, Gaillard J, Häupl T, Ringe J, Devèze A, Magnan J, Sohier J, Khrestchatisky M, Roman FS, et al. The human nose harbors a niche of olfactory ectomesenchymal stem cells displaying neurogenic and osteogenic properties. Stem Cells Dev. 2010;19:853–866. doi: 10.1089/scd.2009.0267. [DOI] [PubMed] [Google Scholar]
- Goulburn AL, Alden D, Davis RP, Micallef SJ, Ng ES, Yu QC, Lim SM, Soh CL, Elliott DA, Hatzistavrou T, et al. A targeted NKX2.1 human embryonic stem cell reporter line enables identification of human basal forebrain derivatives. Stem Cells. 2011;29:462–473. doi: 10.1002/stem.587. [DOI] [PubMed] [Google Scholar]
- Händel EM, Cathomen T. Zinc-finger nuclease based genome surgery: It’s all about specificity. Curr Gene Ther. 2011;11:28–37. doi: 10.2174/156652311794520120. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden SE, Gore A, Li Z, Fung HL, Nisler BS, Nie J, Chen G, McIntosh BE, Gulbranson DR, Diol NR, et al. Genetic correction and analysis of induced pluripotent stem cells from a patient with gyrate atrophy. Proc Natl Acad Sci USA. 2011;108:6537–6542. doi: 10.1073/pnas.1103388108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein SM, Batada NN, Vuoristo S, Ching RW, Autio R, Närvä E, Ng S, Sourour M, Hämäläinen R, Olsson C, et al. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- Irion S, Luche H, Gadue P, Fehling HJ, Kennedy M, Keller G. Identification and targeting of the ROSA26 locus in human embryonic stem cells. Nat Biotechnol. 2007;25:1477–1482. doi: 10.1038/nbt1362. [DOI] [PubMed] [Google Scholar]
- Khan IF, Hirata RK, Wang PR, Li Y, Kho J, Nelson A, Huo Y, Zavaljevski M, Ware C, Russell DW. Engineering of human pluripotent stem cells by AAV-mediated gene targeting. Mol Ther. 2010;18:1192–1199. doi: 10.1038/mt.2010.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent LC, Ulitsky I, Slavin I, Tran H, Schork A, Morey R, Lynch C, Harness JV, Lee S, Barrero MJ, et al. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ES and iPS cells during reprogramming and time in culture. Cell Stem Cell. 2011;8:106–118. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O’Malley R, Castanon R, Klugman S, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GH, Barkho BZ, Ruiz S, Diep D, Qu J, Yang SL, Panopoulos AD, Suzuki K, Kurian L, Walsh C, et al. Recapitulation of premature ageing with iPSCs from Hutchinson-Gilford progeria syndrome. Nature. 2011;472:221–225. doi: 10.1038/nature09879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, Ando D, Urnov FD, Galli C, Gregory PD, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- Mansour SL, Thomas KR, Capecchi MR. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: A general strategy for targeting mutations to non-selectable genes. Nature. 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- Mitsui K, Suzuki K, Aizawa E, Kawase E, Suemori H, Nakatsuji N, Mitani K. Gene targeting in human pluripotent stem cells with adeno-associated virus vectors. Biochem Biophys Res Commun. 2009;388:711–717. doi: 10.1016/j.bbrc.2009.08.075. [DOI] [PubMed] [Google Scholar]
- Nieminen M, Tuuri T, Savilahti H. Genetic recombination pathways and their application for genome modification of human embryonic stem cells. Exp Cell Res. 2010;316:2578–2586. doi: 10.1016/j.yexcr.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Palmer DJ, Ng P. Physical and infectious titers of helper-dependent adenoviral vectors: a method of direct comparison to the adenovirus reference material. Mol Ther. 2004;10:792–798. doi: 10.1016/j.ymthe.2004.06.1013. [DOI] [PubMed] [Google Scholar]
- Ruby KM, Zheng B. Gene targeting in a HUES line of human embryonic stem cells via electroporation. Stem Cells. 2009;27:1496–1506. doi: 10.1002/stem.73. [DOI] [PubMed] [Google Scholar]
- Salem HK, Thiemermann C. Mesenchymal stromal cells: Current understanding and clinical status. Stem Cells. 2010;28:585–596. doi: 10.1002/stem.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho-Martinez I, Li M, Izpisua Belmonte JC. Disease correction the iPSC way: Advances in iPSC-based therapy. Clin Pharmacol Ther. 2011;89:746–749. doi: 10.1038/clpt.2010.341. [DOI] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nat Med. 2005;11:440–445. doi: 10.1038/nm1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shayakhmetov DM, Li ZY, Gaggar A, Gharwan H, Ternovoi V, Sandig V, Lieber A. Genome size and structure determine efficiency of postinternalization steps and gene transfer of capsid-modified adenovirus vectors in a cell-type-specific manner. J Virol. 2004;78:10009–10022. doi: 10.1128/JVI.78.18.10009-10022.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Chung SK, Xu Y. Modeling disease in human ESCs using an efficient BAC-based homologous recombination system. Cell Stem Cell. 2010;6:80–89. doi: 10.1016/j.stem.2009.11.016. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Mitsui K, Aizawa E, Hasegawa K, Kawase E, Yamagishi T, Shimizu Y, Suemori H, Nakatsuji N, Mitani K. Highly efficient transient gene expression and gene targeting in primate embryonic stem cells with helper-dependent adenoviral vectors. Proc Natl Acad Sci USA. 2008;105:13781–13786. doi: 10.1073/pnas.0806976105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenzen T, Zembowicz F, Cowan CA. Genome modification in human embryonic stem cells. J Cell Physiol. 2010;222:278–281. doi: 10.1002/jcp.21948. [DOI] [PubMed] [Google Scholar]
- Wang P, Rodriguez RT, Wang J, Ghodasara A, Kim SK. Targeting SOX17 in human embryonic stem cells creates unique strategies for isolating and analyzing developing endoderm. Cell Stem Cell. 2011;8:335–346. doi: 10.1016/j.stem.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman HJ, Ostlund C, Wang Y. Diseases of the nuclear envelope. Cold Spring Harb Perspect Biol. 2010;2:a000760. doi: 10.1101/cshperspect.a000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue H, Wu S, Papadeas ST, Spusta S, Swistowska AM, MacArthur CC, Mattson MP, Maragakis NJ, Capecchi MR, Rao MS, et al. A targeted neuroglial reporter line generated by homologous recombination in human embryonic stem cells. Stem Cells. 2009;27:1836–1846. doi: 10.1002/stem.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaremba-Czogalla M, Dubinska-Magiera M, Rzepecki R. Laminopathies: the molecular background of the disease and the prospects for its treatment. Cell Mol Biol Lett. 2011;16:114–148. doi: 10.2478/s11658-010-0038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Lian Q, Zhu G, Zhou F, Sui L, Tan C, Mutalif RA, Navasankari R, Zhang Y, Tse HF, et al. A human iPSC model of Hutchinson Gilford Progeria reveals vascular smooth muscle and mesenchymal stem cell defects. Cell Stem Cell. 2011;8:31–45. doi: 10.1016/j.stem.2010.12.002. [DOI] [PubMed] [Google Scholar]
- Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK, Chen G, Ye Z, Park IH, Daley GQ, et al. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaka TP, Thomson JA. Homologous recombination in human embryonic stem cells. Nat Biotechnol. 2003;21:319–321. doi: 10.1038/nbt788. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.