Abstract
Multiple sclerosis (MS) is a complex disease with underlying genetic and environmental factors. Although the contribution of alleles within the major histocompatibility complex (MHC) are known to exert strong effects on MS risk, much remains to be learned about the contributions of loci with more modest effects identified by genome-wide association studies (GWASs), as well as loci that remain undiscovered. We use a recently developed method to estimate the proportion of variance in disease liability explained by 475,806 single nucleotide polymorphisms (SNPs) genotyped in 1,854 MS cases and 5,164 controls. We reveal that ~30% of MS genetic liability is explained by SNPs in this dataset, the majority of which is accounted for by common variants. These results suggest that the unaccounted for proportion could be explained by variants that are in imperfect linkage disequilibrium with common GWAS SNPs, highlighting the potential importance of rare variants in the susceptibility to MS.
Multiple sclerosis (MS) is an inflammatory disease of the central nervous system, and is the most common neurological disorder affecting young adults1. Current evidence implicates roles for both environmental and genetic factors in the onset and progression of the disease2,3,4. The importance of genetic factors in MS was recognized early in the study of the disease, and is best illustrated by observations of strong familial clustering and a significantly increased risk in first-degree relatives5,6,7. Further support for the role of genes in MS comes from studies of monozygotic and dizygotic twins, which also indicate a strong genetic component; however, heritability estimates from these studies range from roughly 25% to 75%8,9,10,11. Alleles of the major histocompatibility complex (MHC) are so far known to make the single strongest contribution to MS susceptibility12. In addition, many loci of more modest effect have also recently been identified in genome-wide association studies (GWASs)13,14,15,16. While risk alleles at the MHC are thought to represent a significant proportion of MS genetic susceptibility13, the contribution of variants outside of the MHC, specifically those represented by single nucleotide polymorphisms (SNPs) genotyped by GWASs, has not been extensively explored. To investigate in more detail the role of common GWAS variants in MS susceptibility, we used publically available genotype data from the United Kingdom (UK) MS patient and control cohorts16 and a recently described approach that assesses contributions made by all genotyped SNPs, rather than solely risk loci that reach genome-wide significance17,18,19,20. From this analysis we show that approximately 30% of the genetic variation in liability to MS is directly explained by variants represented by current GWAS arrays.
Results
For this study, we used genome-wide genotype data for 475,806 autosomal SNPs collected from 1,854 MS cases and 5,164 controls sampled from the UK16. After assessing the relatedness between individuals, and thus accounting for effects of population structure, we first estimated the proportion of variance explained by all autosomal SNPs simultaneously. This analysis revealed that 30.7% (standard error (SE) = 2.05%) of the variance in liability to MS is accounted for by SNPs in this dataset.
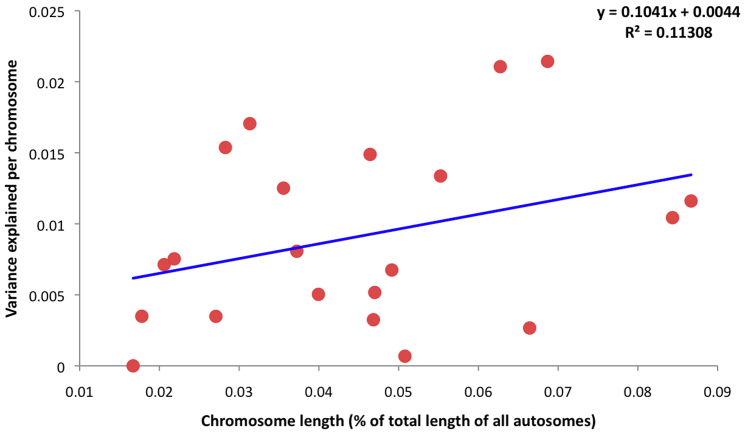
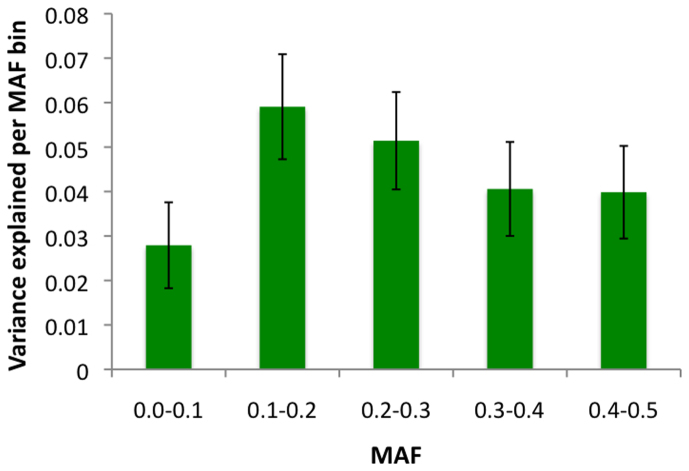
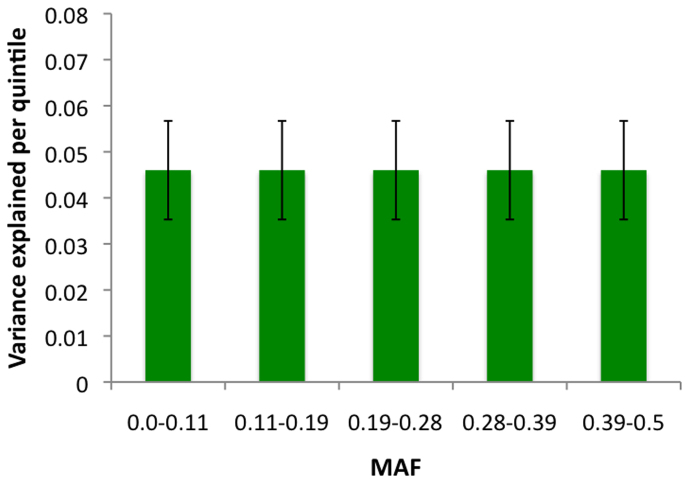
We next partitioned SNPs by autosome and recalculated the proportion of variance explained by variants found on each chromosome (Table 1); estimated values ranged from ~0–8% per chromosome. Not surprisingly, given the known contribution of the MHC, which is located on chromosome 6, SNPs on this chromosome account for 8.11% of the variance (SE = 0.72%). By calculating the proportion of the genome represented by each chromosome (not including the length of sex chromosomes), we tested for a correlation between the variance explained by each chromosome relative to its size, excluding chromosome 6 (Figure 1). Although it was evident that several of the smaller chromosomes contributed less to the overall variance than several of the larger chromosomes, the overall trend was not significant (r = 0.336, P = 0.136). To assess the contribution made by common versus rare variants, we also binned SNPs based on minor allele frequency (MAF; Figure 2). From this, we observed that common variants (MAF > 0.1; ~4–6%), which are most abundantly sampled on GWAS arrays, make a greater contribution than rare variants (MAF < 0.1; ~2.8%). However, because of the unequal number of SNPs in each bin, we also binned SNPs by quintile (Figure 3). Based on this analysis, we found that all quintiles displayed an equivalent variance, highlighting that no particular frequency of MAF makes a larger or smaller contribution to MS, and that all should be captured and tested.
Table 1. Proportion of variance in MS liability explained per chromosome.
| chr | Variance Explained | Standard Error |
|---|---|---|
| 1 | 0.011606 | 0.006417 |
| 2 | 0.010433 | 0.006207 |
| 3 | 0.021433 | 0.006129 |
| 4 | 0.002666 | 0.005454 |
| 5 | 0.021062 | 0.005955 |
| 6 | 0.081112 | 0.007155 |
| 7 | 0.013365 | 0.005453 |
| 8 | 0.000678 | 0.004836 |
| 9 | 0.006747 | 0.004896 |
| 10 | 0.005168 | 0.004938 |
| 11 | 0.003246 | 0.004827 |
| 12 | 0.014884 | 0.005266 |
| 13 | 0.005035 | 0.004257 |
| 14 | 0.008067 | 0.004431 |
| 15 | 0.01251 | 0.004326 |
| 16 | 0.01705 | 0.004983 |
| 17 | 0.015371 | 0.004533 |
| 18 | 0.003484 | 0.004116 |
| 19 | 0.007125 | 0.003979 |
| 20 | 0.007533 | 0.004086 |
| 21 | 0 | 0.002963 |
| 22 | 0.003493 | 0.003107 |
Figure 1. Contribution of GWAS SNPs and chromosome length.
The proportion of variance in MS liability explained by SNPs partitioned by autosome (based on data from Table 1, excluding chr 6) relative to chromosome size, which was determined by dividing the length of each autosome by the sum of the lengths of all autosomes.
Figure 2. Contribution of GWAS SNPs partitioned by minor allele frequency.
The total proportion of variance explained and standard errors for SNPs in each of five MAF bins. The number of SNPs included in each bin varied slightly (0.0–0.1%, n = 76046; 0.1–0.2%, n = 112435; 0.2–0.3%, n = 97482; 0.3–0.4%, n = 89704; 0.4–0.5%, n = 86625).
Figure 3. Contribution of GWAS SNPs partitioned by quintile.
The total proportion of variance explained and standard errors for all SNPs tested after binning by quintile. The number of SNPs included in each quintile are as follows: 0.0–0.11%, n = 93079; 0.11–0.19%, n = 93074; 0.19–0.28%, n = 93076; 0.28–0.39%, n = 93089; 0.39–0.5%, n = 93116).
Lastly, we carried out an association analysis using only the UK GWAS data. We identified 15 associated autosomal SNPs in this cohort outside of the MHC with P values <1×10−5. These SNPs, their positions (hg18; NCBI Build 36.1), and the nearest RefSeq gene to each are listed in Table 2. Using association analysis data, we also examined the contribution made by all associated SNPs to the observed variance after binning by P value, including those SNPs within the MHC (Table 3).
Table 2. Top SNPs from association analysis using UK GWAS data.
| SNP | Chr | Position | Gene | P value |
|---|---|---|---|---|
| rs6662618 | 1 | 92707999 | GFI1 | 1.95E-06 |
| rs11809572 | 1 | 101122894 | EXTL2 | 9.34E-06 |
| rs16849327 | 3 | 104970212 | ZPLD1, ALCAM | 7.17E-06 |
| rs16869665 | 4 | 20095328 | SLIT2 | 3.14E-06 |
| rs2214543 | 7 | 10763417 | NDUFA4 | 8.31E-06 |
| rs11984075 | 7 | 37403379 | ELMO1 | 6.40E-07 |
| rs10749170 | 10 | 116302100 | ABLIM1 | 5.67E-06 |
| rs10502249 | 11 | 122009461 | UBASH3B | 6.38E-06 |
| rs11069349 | 13 | 98572648 | DOCK9 | 1.83E-06 |
| rs727263 | 13 | 98802109 | UBAC2 | 3.26E-06 |
| rs7325747 | 13 | 98827933 | UBAC2 | 4.36E-06 |
| rs9303323 | 17 | 37341634 | TTC25 | 5.30E-06 |
| rs12952314 | 17 | 37398449 | DNAJC7 | 8.18E-06 |
| rs7209012 | 17 | 37414849 | DNAJC7 | 9.42E-07 |
| rs335516 | 18 | 28048065 | MEP1B | 5.99E-06 |
Table 3. Contribution of associated SNPs from UK GWAS dataset to MS liability after binning by P value.
| Bin: P value | # of SNPs | Variance Explained | Standard Error |
|---|---|---|---|
| 1.00E-03 | 1195 | 0.176747 | 0.007402 |
| 1.00E-04 | 429 | 0.108225 | 0.010376 |
| 1.00E-05 | 298 | 0.069538 | 0.009827 |
| 1.00E-06 | 244 | 0.044657 | 0.008199 |
| 1.00E-10 | 149 | 0.035719 | 0.007789 |
Discussion
Using available data from a large UK case-control cohort16, we have conducted a comprehensive assessment of the contribution of genome-wide SNPs on the variance in liability to MS. The power of the approach used here is that contributions of genotypes at all available loci across the genome (in this case, 475,806), rather than only a set of identified MS risk loci, can be accounted for using this method. Thus, from our analysis, we conclude that approximately 30% of MS heritability is explained by variants on current GWAS arrays, including SNPs on chromosome 6, which alone account for ~8% and reflect the major contribution of the MHC. The role of the MHC in MS has long been known; specifically, HLA-DRB1*1501 confers a 2-fold increase in risk13. However, the underlying genetic architecture of MS is presumed to be polygenic, involving a large number of loci with smaller effects22,23. Our findings lend support to this notion, as we observed that the genetic contributions of SNPs on autosomes other than chromosome 6 were at least in part correlated to autosome length. However, this relationship was not significant, and not as convincing as that illustrated previously for other polygenic disorders17,21. This might hint at the possibility that some unidentified MS risk loci have slightly larger effects than others, which has been discussed recently23. Additionally, our study was smaller than that of Yang et al.17 and Lee et al.21, and thus would be comparatively underpowered.
Also notable, we observed that the majority of variation represented by GWAS SNPs was explained by common variants with MAFs over 0.1%, perhaps not surprisingly given that these outnumbered rare variants. This highlights both, the utility of GWAS arrays, which have placed much emphasis on the inclusion of common SNPs, and the fact that the use of larger sample sizes in GWAS should increase power and yield discoveries of additional risk loci, a point that has recently been noted in the context of schizophrenia21. Importantly though, this observation does not delimit the potentially significant role of rare variants in MS. For example, rare variants in CYP27B1, a gene essential to vitamin D synthesis, have been reported at low frequencies in MS patients, but not in controls (odds ratio = 4.7)24. Rare variants in the TYK2 gene have also more recently been shown to influence MS risk25. Furthermore, we found that even after including the effects of over 400,000 SNPs in this cohort, most of the variance in MS liability remains unaccounted for. As has been discussed previously in the context of the “missing heritability” of complex diseases, one of the more likely explanations for this is that GWAS SNPs are in imperfect linkage disequilibrium (LD) with disease-causing variants26. Again, this points to the possible importance of rare variants, as allele frequency differences between causative alleles and genotyped SNPs impact LD, and may also implicate a potential role for structural variants (e.g., large deletions or duplications), which are also only partially represented by neighboring SNPs, especially those that are multi-allelic and in regions of the genome characterized by segmental duplication27. Imputation based methods to increase the number of common variants tested can also be applied to datasets such as the one used here, but it has recently been observed in schizophrenia that the application of imputation methods only yielded an approximate 2% increase in heritability estimates21.
In conclusion, we estimate that approximately 30% of genetic variation in liability to MS is captured by considering all genotyped SNPs simultaneously. The remaining missing heritability most likely reflects imperfect LD between causal variants and the genotyped SNPs.
Methods
Genotypes for UK MS cases and controls were obtained from GWAS data recently generated by the International Multiple Sclerosis Genetics Consortium and the Wellcome Trust Case Control Consortium 216. Estimates of the proportion of variance explained were calculated using the Genome-wide Complex Trait Analysis (GCTA) tool (http://gump.qimr.edu.au/gcta/)17,18,19,20,21,28. Genetic relatedness between individuals was conducted by principal component analysis using the GCTA tool; for this step, the threshold used to identify and remove related individuals was set to a pairwise genetic relationship value of >0.025 (no individuals met this criteria). The top 20 eigenvectors from this analysis were then used as covariates in a restricted maximum likelihood analysis, again conducted within the GCTA tool; this was used to estimate the proportion of the variance explained by SNPs at the genome-wide level, and after partitioning SNP data by autosomes, MAFs, and quintiles. Assembly statistics for GRCh37 (hg19) were used to calculate autosome lengths (autosome length/total length of all autosomes). Association analysis of GWAS SNPs was conducted using PLINK (http://pngu.mgh.harvard.edu/purcell/plink/)29.
Author Contributions
S.V.R., C.T.W., and G.D. conceived of analysis and analyzed the data. C.T.W. and S.V.R. wrote the manuscript, which was critically revised for important intellectual content by F.B., G.D., and G.G. The study was supervised by S.V.R.
References
- Noseworthy J. H., Lucchinetti C., Rodriguez M. & Weinshenker B. G. Multiple Sclerosis. N. Engl. J. Med. 343, 938–952 (2000). [DOI] [PubMed] [Google Scholar]
- Dyment D. A., Ebers G. C. & Sadovnick A. D. Genetics of multiple sclerosis. Lancet Neurol. 3, 104–110 (2004). [DOI] [PubMed] [Google Scholar]
- Ebers G. C. Environmental factors and multiple sclerosis. Lancet Neurol. 7, 268–277 (2008). [DOI] [PubMed] [Google Scholar]
- Ramagopalan S. V., Deluca G. C., Degenhardt A. & Ebers G. C. The genetics of clinical outcome in multiple sclerosis. J. Neuroimmunol. 201, 183–199 (2008). [DOI] [PubMed] [Google Scholar]
- Sadovnick A. D., Baird P. A. & Ward R. H. Multiple sclerosis: updated risks for relatives. Am. J. Med. Gen. 29, 533–541 (1988). [DOI] [PubMed] [Google Scholar]
- Robertson N. P., Fraser M., Deans J., Clayton D., Walker N. & Compston D. A. Age-adjusted recurrence risks for relatives of patients with multiple sclerosis. Brain 119, 449–455 (1996). [DOI] [PubMed] [Google Scholar]
- Ebers G. C., Sadovnick A. D., Dyment D. A., Yee I. M., Willer C. J. & Risch N. Parent-of-origin effect in multiple sclerosis: observations in half-siblings. Lancet 363, 1773–1774 (2004). [DOI] [PubMed] [Google Scholar]
- Mumford C. J., Wood N. W., Kellar-Wood H., Thorpe J. W., Miller D. H. & Compston D. A. The British Isles survey of multiple sclerosis in twins. Neurology 44, 11–15 (1994). [DOI] [PubMed] [Google Scholar]
- Willer C. J. et al. Twin concordance and sibling recurrence rates in multiple sclerosis. Proc. Natl. Acad. Sci. U S A. 100, 12877–12882 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T., Skytthe A., Stenager E., Petersen H. C., Kyvik K. O & Brønnum-Hansen, H. Risk for multiple sclerosis in dizygotic and monozygotic twins. Mult. Scler. 11, 500–503 (2005). [DOI] [PubMed] [Google Scholar]
- Hawkes C. H. & Macgregor A. J. Twin studies and the heritability of MS: a conclusion. Mult. Scler. 15, 661–667 (2009). [DOI] [PubMed] [Google Scholar]
- Lincoln M. R. et al. A predominant role for the HLA class II region in the association of the MHC region with multiple sclerosis. Nat. Genet. 37, 1108–1112 (2005). [DOI] [PubMed] [Google Scholar]
- Hafler D. A. et al. Risk alleles for multiple sclerosis identified by a genomewide study. N. Engl. J. Med. 357, 851–862 (2007). [DOI] [PubMed] [Google Scholar]
- De Jager P. L. et al. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat. Genet. 41, 776–782 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patsopoulos N. A. et al. Genome-wide meta-analysis identifies novel multiple sclerosis susceptibility loci. Ann. Neurol. 70, 897–912 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawcer S. et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 476, 214–219 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42, 565–269 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H. et al. Estimating Missing Heritability for Disease from Genome-wide Association Studies. Am. J. Hum. Genet. 88, 294–305 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat. Genet. 43, 519–525 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 44, 369–375 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H. et al. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. .Nat Genet. 44, 247–250 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawcer S. The complex genetics of multiple sclerosis: pitfalls and prospects. Brain 131, 3118–3131 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush W. S. et al. Evidence for polygenic susceptibility to multiple sclerosis–the shape of things to come. Am. J. Hum. Genet. 86, 621–625 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramagopalan S. V. et al. Rare variants in the CYP27B1 gene are associated with multiple sclerosis. Ann. Neurol. 70, 881–886 (2011). [DOI] [PubMed] [Google Scholar]
- Dyment D. A. et al. Exome sequencing identifies a novel, multiple sclerosis susceptibility variant in the TYK2 gene. .Neurology (in press). [DOI] [PMC free article] [PubMed]
- Manolio T. A. et al. Finding the missing heritability of complex diseases. Nature 461, 747–753 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell C. D. et al. Population-genetic properties of differentiated human copy-number polymorphisms. Am. J. Hum. Genet. 88, 317–332 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. et al. GCTA: a tool for Genome-wide Complex Trait Analysis. Am. J. Hum. Genet. 88, 76–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]