Abstract
Understanding protein function and interaction is central to the elucidation of biological processes. Systematic analysis of protein interactions have shown that the eukaryotic proteome is highly interconnected and that biological function frequently depends on the orchestrated action of many proteins. Perturbation of these functions or interactions can lead to various disease states and pharmacologic intervention can result in corrective therapies. The fact that proteins rarely act in isolation, but rather comprise complex machines that stably and/or transiently interact with many different partners at different times, demands the need for robust tools that allow comprehensive global analyses of these events. Here we describe a powerful protein fusion technology, the HaloTag platform, and how it enables the study of many facets of protein biology by offering a broad choice of applications. We review the development of the key aspects of the technology and it’s performance in both in vitro and in vivo applications. In particular, we focus on HaloTag’s multifunctional utility in protein imaging, protein isolation and display, and in the study of protein complexes and interactions. We demonstrate it’s potential to help elucidate important facets of proteomic biology across complex biological systems at the biochemical, cell-based and whole animal level.
Keywords: HaloTag technology, protein fusion tags, protein interactions, protein imaging, protein purification.
INTRODUCTION
Study of proteins and elucidation of their function is central to our understanding of biological processes at the cellular and whole organism level. Given that protein interactions are focal points in almost all biological processes, the study of protein interactions among themselves and with other molecules continues to expand and occupy a center stage in research. Contributing to the efforts for deciphering protein function is the explosion of new experimental tools for the study of genomics and proteomics. Among the many approaches in proteomics, one of the most common is tagging of proteins with protein fusion tags, in order to equip proteins with additional functionalities that allow specific protein manipulation. Many different protein tags have been developed and two classes in particular have profoundly affected how proteins are studied in vivo and in vitro: auto-fluorescent and affinity tags. Auto-fluorescently tagged proteins revolutionized the way protein function is studied in living cells [1-3]. These tags are useful not only for protein localization studies, but also for the study of dynamic processes, conformational changes and protein-protein interactions. Similarly, affinity fusion tags transformed the in vitro analysis of proteins. Affinity tags provide a selective and efficient tool for protein isolation and immobilization [4-7]. They may also provide some advantage of enhancing solubility and stability thereby permitting higher level of soluble protein to be expressed [8, 9]. The number of new affinity tags and applications for their use continues to expand [8, 10-13]. However, there are limitations to these technologies. With auto-fluorescent proteins we are limited with respect to the types of fluorophores and, in addition, these tags often do not provide ready options to isolate and immobilize proteins for in vitro studies. The use of an additional affinity tag (e.g. His tag) is then required for protein immobilization when using fluorescent proteins. Alternatively, affinity tags provide a very efficient method for in vitro protein studies, but they do not enable specific labeling and imaging of proteins in living cells. Thus, comprehensive analysis of protein function often requires use of multiple tags and therefore multiple genetic constructs and, importantly, multiple assumptions regarding the null effects of these various additions. Thus, using multiple tags is not only less efficient but it also complicates data comparison across these multiple platforms. The desire to combine the advantages of both of these technologies and overcome some of the limitations has driven the development of a new category of protein tags, one where a single tag could be readily reconfigured to meet different experimental needs. The approach utilizes a single protein tag, the function of which can be altered by attaching various chemical moieties (fluorescent labels, affinity handles, etc.). This approach benefits from the specificity and ease afforded by using one genetically encoded protein tag with the functional diversity and adaptability supplied by synthetic chemistry. Several such multi-functional tags have recently been developed such as HaloTag, Tetra-cysteine tag, and SNAP-tag [12, 14-20]. In this article we will focus on the HaloTag technology highlighting the principles of its performance, development, functional diversity, and multiple applications which enable a comprehensive analysis of protein biology.
OVERVIEW OF THE HALOTAG TECHNOLOGY
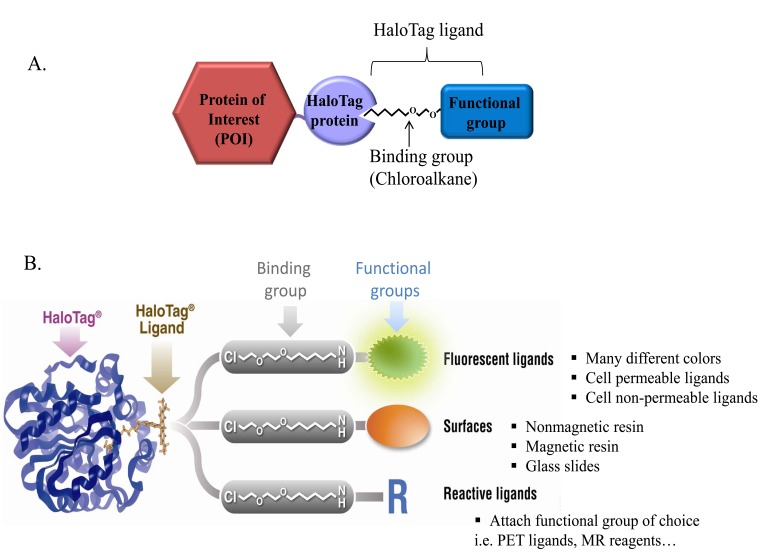
HaloTag technology consists of two elements, the HaloTag protein, a protein fusion tag which can be genetically fused to any protein of interest (POI), and a variety of organic molecules, HaloTag ligands, which irreversibly bind to HaloTag protein. To achieve efficient and specific binding of HaloTag protein with several different ligands we designed the ligands so that they all comprise two elements; the constant reactive group and a variable functional/reporter group. The reactive group consists of a chloroalkane, the substrate necessary for covalent attachment to the HaloTag protein. As this reaction is conserved, the reactive group remains constant for all ligands. The remaining part of the ligand, the functional group, encompasses many different entities including different fluorescent dyes, affinity handles (e.g. biotin, etc.) or solid supports (e.g. resins). This diversity of the HaloTag ligands bestow different functionalities onto HaloTag fusion protein allowing for numerous means to study proteins, ranging from protein imaging to protein capture and display. In summary, one genetic protein fusion construct can be used in various biochemical, cell-based and even whole animal protein analysis studies (Fig. 1).
Fig. (1). Multiple functionalities of the HaloTagTM Technology.
A. Schematic of the HaloTag technology consisting of the HaloTag protein and a selection of synthetic HaloTag ligands carrying different functional groups. HaloTag protein can be easily fused to any protein of interest (POI). HaloTag ligands specifically and covalently bind to the HaloTag fusions.
B. Schematic representation of HaloTag-TMR ligand binding to the active site in the binding tunnel of HaloTag protein. HaloTag ligands with different functional groups such as surfaces, fluorescent dyes or reactive groups are shown; the constant binding group (chloroalkane) is highlighted by the grey shaded rectangle.The ligands, depending on functional group impart multiple functions to a HaloTag fusion protein including imaging, immobilization and others. Thus one genetic construct can be used in various in vitro and in vivo assays.
DEVELOPMENT OF HALOTAG PROTEIN AND LIGANDS
Several criteria were considered during the development of HaloTag to ensure optimal performance and broad utility of HaloTag-POI fusions. Important considerations included: the reversible or irreversible nature of attachment of chemical functionalities (ligands) to the protein, high specificity of ligand binding, ability to perform reactions in vitro, in living cells and ideally also in whole animals. In addition, we focused on particular attributes of HaloTag ligands that differentiate them from other systems, such as the highly efficient reactivity with protein even at low protein concentration within timescales compatible with biological processes. Many different chemical functionalities (HaloTag ligands) have now been created by us and others, without affecting the specificity and kinetics of binding, ensuring that the ligands are not toxic to the cell and, when appropriate, cell permeable. In order to enable broad utility and easy adoption, most of these ligands are commercially available, easy-to-use, and do not require any special knowledge of chemistry synthesis.
The considerations described above played a critical role in our selection of the system that was used for development of HaloTag. We chose the bacterial haloalkane dehalgenase, because it exhibits features that allowed us to develop a tag with all the desired attributes. A brief summary of the development and optimization is described below; a more complete description of the evolution of HaloTag can be found in work by Los and Encell [20, 21].
The bacterial dehalogenase catalyzes a reaction that forms a covalent transition state intermediate with the chloroalkane substrate. This covalent bond is subsequently hydrolyzed, releasing the substrate and regenerating the active site [20, 22]. It was previously shown that mutating a histidine residue in the active site of the dehalogenase from Xanthobacter prevented hydrolysis of the covalent bond, leading to formation of a irreversible bond with the substrate [23]. By applying an analogous mutation to the Rhodococcus dehalogenase we showed that an irreversible (covalent) bond can readily be formed with chloroalkane derivatives. The protein carrying this mutation was then called HaloTag and the chloroalkane derivatives, HaloTag ligands [20]. Together they allowed for development of a system where HaloTag ligands carrying different functional groups could be permanently associated with the protein thereby imparting many different functionalities onto the protein.
The fact that dehalogenase is monomeric, relatively small (33 kDa) and that neither eukaryotic cells nor most bacteria (e.g. E. coli) possess the haloalkane dehalogenase or its ligands provide excellent advantages. There is no endogenous equivalent in any of the standardly used experimental biological systems which would compete for binding of Halo Tag ligands and interfere with its specificity and efficiency. However, certain important performance attributes were still lacking in the original HaloTag variant. For example, the slow kinetics of substrate binding of HaloTag posed a significant limitation. In an effort to improve the properties of HaloTag, it was subjected to several rounds of site specific and random mutagenesis. The result was a HaloTag exhibiting a rapid substrate binding rate, comparable to that of tetrameric streptavidin with biotin [20 ,21]. This protein engineering process also improved the thermostability and solubility properties of HaloTag. The resulting HaloTag protein displayed rapid, specific and covalent binding to the HaloTag ligands and allowed for expression of highly soluble fusion proteins in the absence of any endogenous competitors in essentially any biological system of interest.
As noted above, all HaloTag ligands comprise the same chloroalkane based binding group, onto which a variety of functional groups are appended using standard chemical synthesis. A series of fluorescent HaloTag ligands were developed by coupling TMR, Oregon Green, Coumarin, Alexa Fluor® 488, Alexa Fluor® 660 and R110 dyes with the reactive binding group. The resulting ligands readily permeate into mammalian cells, with exception of the Alexa Fluor 488 substrate, which is an important cell impermeable variant. Importantly, at the appropriate concentrations these ligands show no cytotoxicity. Thus, with these fluorescent dyes it is now possible to make any HaloTag-POI fusion permanently fluorescent and, in addition, by selecting different dyes it is possible to label the HaloTag fusion with different fluorescent dye in different experiments or even at different time points within the same experiment.
To enable applications beyond fluorescent labeling, such as protein capture and display, HaloTag ligand surfaces were generated. Significant efforts were dedicated towards development of surfaces with very low nonspecific binding and efficient specific binding of HaloTag fusion proteins. Currently three formats of surfaces, magnetic resin, non-magnetic resin and glass slides enable HaloTag mediated protein immobilization and display. Further development of new HaloTag surfaces, fluorescent ligands and ligands with other functionalities should allow creation of additional applications of HaloTag. The availability of HaloTag reactive intermediate substrates (reactive ligands), consisting essentially of the binding group carrying a reactive group, enables creation of custom reagents by adding any functional group of interest following standard chemical synthesis. This substrate has allowed the development of novel applications by other users [24-29]. Several key applications of HaloTag are described below.
KEY APPLICATIONS OF HALOTAG TECHNOLOGY
a. Protein Imaging
Selective protein visualization through creation of genetic fusions is a powerful analytical method commonly used to elucidate protein function in complex biological environments such as cells or organisms. Unlike GFP, HaloTag and HaloTag-POI fusions are per se not fluorescent, but rather they only become fluorescent upon the addition of a fluorescent HaloTag ligand. This is an important distinction as it gives spatial and temporal control of labeling. The process of fluorescent labeling is simple and rapid requiring only the addition of the fluorescent HaloTag ligand to the media for a brief 15-30 min incubation followed by a few quick washes. Wash steps can be avoided by using an optimized concentration of the ligand such as HaloTag R110Direct ligand. This ligand is simply added to the cell media at the time of plating of the cells. The labeling on demand aspect of HaloTag fusions allows for development of unique applications such as pulse chase and differential spatial labeling of proteins. Traditionally, metabolic pulse-chase with isotopically labeled amino acids has been used to monitor stability and synthesis of specific proteins in cells [30]. The low temporal resolution of this method limits the use of this approach especially when studying proteins with relatively long half-lives. The ability to rapidly label proteins at a particular time point in combination with availability of different fluorescent ligands makes HaloTag particularly suitable for pulse chase based analysis of protein turnover [31]. In a typical experiment a HaloTag-protein fusion is labeled with one of the available fluorescent ligands (e.g. green), followed by removal of this ligand by media exchange and addition of a ligand with a different color (e.g. red). The second ligand is present in the media throughout the duration of the experiment and will specifically label all newly synthesized protein in red, while the old protein remains green. As a result of this process the two pools of protein can be distinguished allowing analysis of degradation and synthesis. Importantly, it is the covalent binding property of HaloTag ligands that imparts long-term fluorescent labeling and prevents dissociation of the first bound ligand from the labeled protein while subsequent labeling is occurring with the second added ligand. The rapid binding kinetics is also important since it allows labeling of cellular proteins in minutes and ensures complete labeling of low abundant proteins, which is critical during the short pulse labeling phase. Without these performance attributes of HaloTag, the complete labeling of the first protein pool would be compromised.
Dual labeling using different fluorescent ligands can also be used if studying membrane bound versus intracellular protein populations. In this approach the different color ligands also differ in regards to permeability. By combining a cell permeable ligand in one color with a non-permeable ligand of different color we can differentially label surface exposed proteins where HaloTag is exposed on the cellular surface, from the intracellular pool of the same protein. This affords analysis of protein trafficking, endocytosis and study of membrane protein biology [32].
Other imaging applications such as super-resolution imaging and single molecule imaging approaches have also adopted HaloTag technology [24, 33-36]. In one of these studies the observation was made that the relatively hydrophobic nature of HaloTag ligand appeared to support more efficient permeation of the super-resolution dye (ATTO655) when coupled to the HaloTag reactive ligand using the N-hydroxyl succinimidyl chemistry [33]. Combination of pho-toactivatable fluorescent proteins and HaloTag-ATTO655 allowed for multi-color super-resolution imaging of live cells and submicroscopic resolution of receptor organization in the plasma membrane. HaloTag imaging can also provide the means of reliably measuring intracellular pH by precise targeting of 5(and 6-) carboxyseminaphthorhodafluor (SNARF-1) to sub-cellular compartments. This was accomplished by coupling SNARF-1 to the HaloTag reactive ligand and by generating HaloTag fusions with proteins which reside in specific sub-cellular locations such as nucleus or cytoplasm [37]. The localization of HaloTag protein fusions is dictated by the fusion partner allowing targeted positioning of HaloTag protein to different cellular compartments. Subsequently, cells are treated with the HaloTag ligand carrying the pH sensor dye (SNARF) which binds to HaloTag protein localized to a particular location in a cell. Depending on the pH of the environment the fluorescent signal of SNARF changes allowing discernible measurement of pH in different locations.
The simplicity and flexibility of the technology makes this system amenable for development of other imaging applications and recently, in vivo animal imaging using HaloTag has been reported [26, 28, 38]. In these studies in vivo imaging in animal models expressing HaloTag fusions on the surface of tumorigenic cells were performed allowing detection of growing tumors upon injection into animals using a variety of different fluorescent or PET HaloTag ligands.
b. Protein Purification, Isolation of Protein Complexes and Display of Proteins
As noted earlier HaloTag is a powerful tool for purification of proteins, isolation of cellular protein complexes (protein-protein and protein-DNA), and for protein capture and display. To utilize HaloTag for direct protein purification, a chromatographic matrix carrying the chloroalkane ligand (HaloLink resin) was created. The covalent nature of the HaloTag-fusion protein capture combined with rapid binding kinetics overcomes the equilibrium-based limitations associated with traditional non-covalent affinity purification and enables efficient capture of target proteins even at very low abundance. Furthermore, it allows for extensive and stringent washing without loss of bound protein due to diffusion. While the covalent association clearly has its advantages it also creates a challenge in eluting the protein of interest. As the covalent bond between HaloTag and chloroalkane cannot be readily reversed, traditional approaches to elute proteins from the resin cannot be utilized. Instead the protein of interest can be released by specific protease (TEV) cleavage, as the TEV protease recognition site is present between the HaloTag and the fusion partner. Upon cleavage, HaloTag stays bound to the resin while the fusion partner is released, yielding highly pure protein free of tag. It is important to note that a few amino acids of the linker segment connecting the two proteins remain associated with the protein after the cleavage. Depending on the N or C orientation of the HaloTag, 6 or 13 amino acids of the linker sequence, respectively, remain with the purified protein.
Successful purification of functional proteins generally requires efficient expression of recombinant proteins in soluble form. This is especially a challenge when heterologous or large proteins are expressed in E. coli, one of the most frequently used expression hosts due to easy manipulation, rapid growth and low cost [39-41]. One approach to overcome the limitations of low expression of soluble protein is to optimize expression conditions such as temperature, growth media, induction conditions etc. To simplify the screening of these variables, fluorescent tags such as GFP have been utilized [42, 43]. HaloTag offers an excellent alternative not only because it is an excellent purification tool, but also because it allows for easy fluorescent detection and quantitation of expressed proteins. Furthermore, unlike GFP, the HaloTag fluorescent signal survives boiling and SDS-PAGE gel electrophoresis and therefore allows for gel based resolution and quantification using the fluorescent signal. Thus, HaloTag can be used for rapid screening and purification. Since HaloTag protein labeled with fluorescent HaloTag ligand can no longer bind to the resin, the labeling reaction is performed on small aliquots of the sample. The utility and superior performance of this system for purification of proteins from E. coli has been demonstrated [44]. In the same report the authors also note that HaloTag improves the solubility of the fusion proteins.
There is an increasing need and desire to express and purify mammalian derived proteins directly from cultured mammalian cells retaining the physiological relevance and providing a more compatible environment for producing properly folded and functional mammalian proteins. Mammalian cells possess the proper machinery to produce mature proteins through proteolytic processing and are able to carry out relevant post–translational modification (PTM) which many proteins require for biological activity [45-47]. Therefore, achieving satisfactory purification of proteins from mammalian cells is of high interest and importance. Unfortunately, many of the traditional affinity tags that have been developed, fail to adequately purify proteins from mammalian cells. This is often due to inefficient binding of the tagged proteins to their purification matrices at lower protein expression levels typically observed in mammalian cells, as well as the nonspecific binding of endogenous proteins with similar affinities resulting in lower yields and purity of recovered proteins [48-50]. These limitations are reduced significantly when HaloTag is used for purification of proteins from mammalian cells. This is due to the performance attributes of HaloTag described above; the high binding rate, the covalent binding and TEV specific release all work in concert to improve protein recovery, purity and yield. Absence of proteins analogous to HaloTag in mammalian cells also contributes to improved purity of proteins purified via HaloTag. Moreover, HaloTag fluorescent detection can be used for easy evaluation of optimization of expression and parameters such as transfection conditions, culturing conditions, expression clone selection, etc. [51, 52].
Protein interactions, protein complexes and networks remain the focal point of current studies in cellular biology. Significant effort has been focused on analysis of global protein network analysis and genome mapping. Trapping both transient and often weak protein interactions presents a significant challenge as dissociation occurs before complexes can be adequately captured. The same properties which benefit HaloTag-based purification also make HaloTag especially well suited for capture of protein complexes directly from mammalian cells. Rapid and highly specific capture of HaloTag eliminates the need for long binding incubations and extensive washing thereby increasing the chance of recovery of transient interactions. With this approach a number of known transiently interacting partners of RNAP II were identified, in addition to three previously uncharacterized interacting proteins [53]. A similar approach was also used to capture cross-linked protein-DNA complexes providing an antibody free alternative to ChIP analysis [54].
Importantly, the same HaloTag fusion used for isolation of various protein complexes can also be used for covalent capture and surface display and further confirmation, validation or discovery of new protein interactions. The system was uniquely designed for allowing specific capture of the proteins onto glass slides (HaloLink protein arrays system) and other surfaces directly from lysates, thus alleviating the need for tedious protein purification, a major bottleneck in fabrication of current protein arrays.
The advantage of this system is that unlike other covalent immobilization techniques, where binding of protein is through multiple attachment sites and leads to random, often improper orientation, immobilization using HaloTag is a single point attachment through its active site ligand and therefore provides uniform orientation of the POI. Single point, oriented attachment increases capacity, effectiveness and reproducibility of the system preventing surface induced protein unfolding and improving protein activity and native conformation [55]. The need for protein specific optimization of capture conditions is also eliminated as the immobilization relies on the same chemistry, namely attachment of HaloTag to the HaloTag ligand-modified surface. This simplifies the process, and significantly improves the uniformity of the arrays and reproducibility of the assays. The rapid preparation of HaloTag-fusion arrays and excellent performance opens-up the possibility for screening hundreds or thousands of different protein interactions [56, 57].
c. Protein Degradation
The various applications and analyses described above are invaluable for studies directed at the elucidation of protein function. Such studies are often complemented by approaches that attempt to achieve specific disruption/ elimination of the activity of proteins in an effort to understand the underlying mechanisms governing a particular biological system. Historically, methods such as genetic knock-out technology, mutagenesis, antisense RNA and RNA interference (RNAi) have been used to specifically alter or eliminate a function of a particular protein. However, these methods are all indirect effectors of the POI and suffer from genetic compensation, low time resolution and can be difficult to use in live animals. HaloTag provides a new analytical tool for specific and direct protein depletion or inactivation of protein function at the protein level [27, 29, 58, 59]. Studies showed that binding of HaloTag ligands carrying a hydrophobic moiety to HaloTag fusion proteins destines these proteins for effective proteosomal degradation. This novel hydrophobic tagging technology of HaloTag fusions efficiently induced degradation of proteins in cell culture systems, as well as directly in animals, potentially making this system ideal for validating potential drug targets in disease models [29, 58].
Clearly, one major effort of biological research is to understand protein function and the interactions which regulate the majority of cellular processes. Development of new tools enabling a more comprehensive analysis of proteins is essential for furthering our understanding of this complex field. Here we presented a powerful platform, based on HaloTag technology, which offers both the reality and future promise of overcoming many of the limitations of current protein analysis approaches.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflicts of interest.
ACKNOWLEDGEMENT
Declared none.
REFERENCES
- 1.Zhang J, Campbell RE, Ting AY, Tsien RY. Creating new fluorescent probes for cell biology. Nat Rev Mol Cell Biol. 2002;3:906–18. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 2.Lippincott-Schwartz J, Patterson GH. Development and use of fluorescent protein markers in living cells. Science. 2003;300:87–91. doi: 10.1126/science.1082520. [DOI] [PubMed] [Google Scholar]
- 3.Miyawaki A, Sawano A, Kogure T. Lighting up cells: labeling proteins with fluorophores. Nat Cell Biol . 2003;6:S1–S7. [PubMed] [Google Scholar]
- 4.Porath J, Carlsson J, Olsson I, Belfrage G. Metal chelate affinity chromatography, a new approach to protein fractionation. Nature. 1975;258:598–9. doi: 10.1038/258598a0. [DOI] [PubMed] [Google Scholar]
- 5.Loennerdal B, Keen CL. Metal chelate affinity chromatography of proteins. J Appl Biochem. 1982;4:203–8. [Google Scholar]
- 6.Smith DB, Johnson KS. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;7:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 7.Smyth DR, Mrozkiewcz MK, McGrath WJ, Listwan P, Kobe B. Crystal structures of fusion proteins with large-affinity tags. Protein Sci. 2003;12:1313–22. doi: 10.1110/ps.0243403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kapust RB, Waugh DS. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci. 1999;8:1668–74. doi: 10.1110/ps.8.8.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braun P, Hu Y, Shen B, et al. Proteome-scale purification of human proteins from bacteria. Proc Natl Acad Sci USA. 2002;99:2654–9. doi: 10.1073/pnas.042684199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Terpe K. Overview of tag protein fusions: from molecular and biochemical fundamentals to commercial systems. Appl Microbiol Biotechnol. 2003;60:523–33. doi: 10.1007/s00253-002-1158-6. [DOI] [PubMed] [Google Scholar]
- 11.Lichty JJ, Malecki JL, Agnew DH, Michelson-Horowitz DJ, Tan S. Comparison of affinity tags for protein purification. Protein Expr Purif. 2005;41:98–105. doi: 10.1016/j.pep.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 12.Gronemeyer T, Godin G, Johnsson K. Adding value to fusion proteins through covalent labeling. Curr Opin Biotechnol. 2005;16:453–8. doi: 10.1016/j.copbio.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 13.Chapman S, Poarka KJ, Robers AG. New tools for in vivo fluorescence tagging. Curr Opin Plant Biol. 2005;8:565–73. doi: 10.1016/j.pbi.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 14.Marks KM, Nolan GP. Chemic labeling strategies for cell biology. Nat Methods. 2006;3:591–6. doi: 10.1038/nmeth906. [DOI] [PubMed] [Google Scholar]
- 15.Chen I, Ting A. Site-specific labeling of proteins with small molecules in live cells. Curr Opin Biotechnol. 2005;16:35–40. doi: 10.1016/j.copbio.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 16.Gronemeyer T, Godin G, Johnsson K. Adding value to fusion proteins through covalent labeling. Curr Opin Biotechnol. 2005;16:453–8. doi: 10.1016/j.copbio.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 17.Miller LW, Cornish VW. Selective chemical labeling of proteins in living cells. Curr Opin Chem Biol. 2005;9:1–6. doi: 10.1016/j.cbpa.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 18.Griffin BA, Adams SR, Tsien RY. Specific covalent labeling of recombinant protein molecules inside live cells. Science. 1998;281:269–72. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- 19.Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat Biotechnol. 2003;21:86–9. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 20.Los GV, Encell LP, McDougall MG, et al. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol. 2008;3(6 ):73–82. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 21.Encell LP, Ohana RF. Kris Zimmerman Development of a Dehalogenase-Based protein fusion tag capable of rapid, selective and covalent attachment to customizable ligands. Curr Chem Genom. 2012;6:55–71. doi: 10.2174/1875397301206010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janssen DB. Evolving haloalkane dehalogenase. Curr Opin Chem Biol. 2004;8:150–9. doi: 10.1016/j.cbpa.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 23.Pries F, Kingma GH, Jeronimus-Stratingh CM, Bruins AP, Janssen DB. Histidine 289 is essential for hydrolysis of the alkyl-enzyme intermediate of haloalkane dehalogenase. J Biol Chem. 1995;270:10405–11. doi: 10.1074/jbc.270.18.10405. [DOI] [PubMed] [Google Scholar]
- 24.Lee HL, Lord SJ, Iwanaga S, et al. Superresolution imaging of targeted proteins in fixed and living cells using photoactivatable organic fluorophores. J Am Chem Soc. 2010;132 (43 ):15099–101. doi: 10.1021/ja1044192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, So MK, Loening AM, Yao H, Gambhir SS, Rao J. HaloTag protein-mediated site specific conjugation of bioluminescent proteins to quantum dots. Angew Chem Int Ed. 2006;45:4936–40. doi: 10.1002/anie.200601197. [DOI] [PubMed] [Google Scholar]
- 26.Strauch RC, Mastarone DJ, Sukerkar PA, Song Y, Ipsaro JJ, Meade TJ. Reporter protein-targeted probes for magnetic resonance imaging. J Am Chem Soc. 2011;133 (41 ):16346–9. doi: 10.1021/ja206134b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takemoto K, Matsuda T, McDougall M, Klaubert DH, Hasegawa AGV. Chromophore-assisted light inactivation of HaloTag fusion proteins labeled with eosin in living cells. ACS Chem Biol. 2011;6(5 ):401–6. doi: 10.1021/cb100431e. [DOI] [PubMed] [Google Scholar]
- 28.Hong H, Benink HA, Zhang Y, Yang Y, Uyeda TH, Engle JW. HaloTag: a novel reporter gene for positron emission tomography. Am J Transl Res. 2011;3(4 ):392–403. [PMC free article] [PubMed] [Google Scholar]
- 29.Neklesa TK, Tae HS, Schneekloth AR, Stulberg MJ, Corson TW, Sundberg TB. Small-molecule hydrophobic tagging-induced degradation of HaloTag fusion proteins. Nat Chem Biol. 2011;7:538–43. doi: 10.1038/nchembio.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dickson RC, Mendenhall MD, editors. 2nd. Totova NJ: Humana Press; 2004. Signal transduction protocols . [Google Scholar]
- 31.He Y, Xu Y Zhang, et al. Identification of a lysosomal pathway that modulates glucocorticoid signaling and the inflammatory. Sci Signal. 2011;4:180–8. doi: 10.1126/scisignal.2001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Svendsen S, Zimprich C, McDougall MG, Klaubert DH, Los GV. Spatial separation and bidirectional trafficking of proteins using a multi-functional reporter. BMC Cell Biol. 2008;9:7–31. doi: 10.1186/1471-2121-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilmes S, Staufenbiel M, Lisse D, et al. Triple-color super-resolution imaging of live cells: resolving submicroscopic receptor organization in the plasma membrane. Angew Chem Int Ed Eng. 2012;51(20 ):4868–71. doi: 10.1002/anie.201200853. [DOI] [PubMed] [Google Scholar]
- 34.Zhou ZP, Shimizu Y, Tadakuma H, Taguchi H, Ito K, Ueda T. Single molecule imaging of the trans-translation entry process via anchoring of the tagged ribosome. J Biochem. 2011;149 (5 ):609–18. doi: 10.1093/jb/mvr010. [DOI] [PubMed] [Google Scholar]
- 35.Kardona JR, Reck-Petersona SL, alea RD. Regulation of the processivity and intracellular localization of Saccharomyces cerevisiae dynein by dynactin. Proc Natl Acad Sci USA. 2009;106:5669–74. doi: 10.1073/pnas.0900976106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schroder J, Benink H, Dyba M, Los GV. In vivo labeling method using a genetic construct for nanoscale. Biophys J. 2009;96 (Issue 1 ):L1–L3. doi: 10.1016/j.bpj.2008.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benink HA, McDougall MG, Klaubert DH, Los GV. Direct pH measurements by using subcellular targeting of 5(and 6-) carboxyseminaphthorhodafluor in mammalian cells. BioTech. 2009;47:69–74. doi: 10.2144/000113220. [DOI] [PubMed] [Google Scholar]
- 38.Kosaka N, Ogawa M, Choyke PL, Karassina N, Corona C, McDougall M. In vivo stable tumor-specific painting in various colors using Dehalogenase-Based Protein-Tag fluorescent ligands. Boiconjug Chem. 2009;20:1367–74. doi: 10.1021/bc9001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baneyx F. Recombinant protein expression in Escherichia coli. Curr Opin Biotechnol. 1999;10:411–22. doi: 10.1016/s0958-1669(99)00003-8. [DOI] [PubMed] [Google Scholar]
- 40.Edwards AM, Arrowsmith CH, Christendat D, et al. Protein production: feeding the crystallographers and NMR spectroscopists. Nat Struct Biol. 2000; 7(Suppl ):970–2. doi: 10.1038/80751. [DOI] [PubMed] [Google Scholar]
- 41.Braun P, Hu Y, Shen B, et al. Proteome-scale purification of human proteins from bacteria. Proc Natl Acad Sci USA. 2002;99:2654–9. doi: 10.1073/pnas.042684199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waldo GS. Improving protein folding efficiency by directed evolution using the GFP folding reporter. Methods Mol Biol. 2003;230:343–59. doi: 10.1385/1-59259-396-8:343. [DOI] [PubMed] [Google Scholar]
- 43.Omoya K, Kato Z, Matsukuma E, et al. Systematic optimization of active protein expression using GFP as a folding reporter. Protein Expr Purif. 2004;36:327–32. doi: 10.1016/j.pep.2004.04.023. [DOI] [PubMed] [Google Scholar]
- 44.Friedman OR, Encell LP, Zhao K, et al. Halo Tag7: a genetically engineered Tag that enhances bacterial expression of soluble proteins and improves protein purification. Protein Expr Purif. 2009;68(1 ):110–20. doi: 10.1016/j.pep.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 45.Cazalla D, Sanford JR, Caceres JF. A rapid and efficient protocol to purify biologically active recombinant proteins from mammalian cells. Protein Expr Purif. 2005;42:54–8. doi: 10.1016/j.pep.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 46.Geisse S, Gram H, Kleuser B, Kocher HP. Eukaryotic expression systems: a comparison. Protein Expr Purif. 1996;8:271–82. doi: 10.1006/prep.1996.0101. [DOI] [PubMed] [Google Scholar]
- 47.Wurm FM. Production of recombinant protein therapeutics in cultivated mammalian cells. Nat Biotechnol. 2004;22:1393–8. doi: 10.1038/nbt1026. [DOI] [PubMed] [Google Scholar]
- 48.Pham PL, Perret S, Doan HC, et al. Large-scale transient transfection of serum-free suspension-growing HEK293 EBNA1 cells: peptone additives improve cell growth and transfection efficiency. Biotechnol Bioeng. 2003;84:332–42. doi: 10.1002/bit.10774. [DOI] [PubMed] [Google Scholar]
- 49.Cass B, Pham PL, Kamen A, Durocher Y. Purification of recombinant proteins from mammalian cell culture using a generic double-affinity chromatography scheme. Protein Expr Purif. 2005;40:77–85. doi: 10.1016/j.pep.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 50.Lichty JJ, Malecki JL, Agnew HD, Michelson-Horowitz DJ, Tan S. Comparison of affinity tags for protein purification. Protein Exp Purif. 2005;41:98–105. doi: 10.1016/j.pep.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 51.Ohana RF, Hurst R, Vidugiriene J, Slater MR, Wood KV, Urh M. HaloTag-based purification of functional human kinases from mammalian cells. Protein Expr Purif. 2011;76(2 ):54–64. doi: 10.1016/j.pep.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 52.Chumanov RS, Kuhn PA, Xu Wei, Burgess RR. Expression and purification of full-lengthmouseCARM1 from transiently transfected HEK293T cells using HaloTag technology. Protein Exp Purif. 2011;760:145–53. doi: 10.1016/j.pep.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Daniels DL, Méndez J, Mosley AL, et al. Examining the complexity of human RNA polymerase complexes using HaloTag technology coupled to label free quantitative proteomics. J Proteome Res. 2012;11 (2 ):564–75. doi: 10.1021/pr200459c. [DOI] [PubMed] [Google Scholar]
- 54.Hartzell DD, Trinklein ND, Mendez J, et al. A functional analysis of the CREB signaling pathway using HaloCHIP-chip and high throughput reporter assays. BMC Genom. 2009;10:497–512. doi: 10.1186/1471-2164-10-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nath N, Hurst R, Hook B, et al. Improving protein array performance: focus on washing and storage conditions. J Proteome Res. 2008;10:4475–82. doi: 10.1021/pr800323j. [DOI] [PubMed] [Google Scholar]
- 56.Hurst R, Hook B, Slater MR, Hartnett J, Storts DR, Nath N. Protein-protein interaction studies on protein arrays: effect of detection strategies on signal-to-background ratios. Anal Biochem. 2009;392(1 ):45–53. doi: 10.1016/j.ab.2009.05.028. [DOI] [PubMed] [Google Scholar]
- 57.Peterson SN, Kwon K. Application of a novel HaloTag to characterize protein-protein and protein-DNA interactions. Curr Chem Genom. 2012;6:8–17. doi: 10.2174/1875397301206010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tae HS, Sundberg TB, Neklesa TK, et al. Identification of hydrophobic tags for the degradation of stabilized proteins. Chem Bio Chem. 2012;13:538–41. doi: 10.1002/cbic.201100793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee HD, Lord SJ, Iwanaga S, et al. Superresolution imaging of targeted proteins in fixed and living cells using photoactivatable organic fluorophores. J Am Chem Soc. 2010;132 (43 ):15099–101. doi: 10.1021/ja1044192. [DOI] [PMC free article] [PubMed] [Google Scholar]