Abstract
Background
Crows and ravens (Passeriformes: Corvus) are large-brained birds with enhanced cognitive abilities relative to other birds. They are among the few non-hominid organisms on Earth to be considered intelligent and well-known examples exist of several crow species having evolved innovative strategies and even use of tools in their search for food. The 40 Corvus species have also been successful dispersers and are distributed on most continents and in remote archipelagos.
Results
This study presents the first molecular phylogeny including all species and a number of subspecies within the genus Corvus. We date the phylogeny and determine ancestral areas to investigate historical biogeographical patterns of the crows. Additionally, we use data on brain size and a large database on innovative behaviour and tool use to test whether brain size (i) explains innovative behaviour and success in applying tools when foraging and (ii) has some correlative role in the success of colonization of islands. Our results demonstrate that crows originated in the Palaearctic in the Miocene from where they dispersed to North America and the Caribbean, Africa and Australasia. We find that relative brain size alone does not explain tool use, innovative feeding strategies and dispersal success within crows.
Conclusions
Our study supports monophyly of the genus Corvus and further demonstrates the direction and timing of colonization from the area of origin in the Palaearctic to other continents and archipelagos. The Caribbean was probably colonized from North America, although some North American ancestor may have gone extinct, and the Pacific was colonized multiple times from Asia and Australia. We did not find a correlation between relative brain size, tool use, innovative feeding strategies and dispersal success. Hence, we propose that all crows and ravens have relatively large brains compared to other birds and thus the potential to be innovative if conditions and circumstances are right.
Keywords: Biogeography, Brain, Cognition, Crow, Systematics, Tool use
Background
Crows are large passerine birds that are considered intelligent because of their flexible behaviour, problem-solving abilities and social learning [1,2]. Several species show a number of fascinating innovations including tool use in their foraging, the best-known example being that of the New Caledonian crow (C. moneduloides) [3-5]. Such innovations in foraging are not only unique but are also expected to require increased cognitive abilities, which have been shown to be related to a brain size relatively larger than that of other birds [1,4,6]. Thus, the combination of opportunistic behaviour and intelligence should make corvids highly adaptable, competitive and potentially good colonizers of new environments [7,8].
The family Corvidae (crows, jays, magpies and allies) contains 117 species [9] distributed across most continents except Antarctica. Within the family, crows (genus Corvus) make up about one third of the species diversity (40 species) and they occur on all continents except South America and Antarctica as well as in remote archipelagos such as Hawaii, Micronesia and Melanesia [10]. The Corvidae is part of the core Corvoidea radiation that contains more than 750 species. Recent studies have argued that the core Corvoidea originated in an archipelago environment north of Australia in the late Oligocene/early Miocene and dispersed via the historically complex Indo-Pacific archipelagos to the rest of the world [11]. Thus, we may expect that some or all of the core Corvoidea’s member groups could have been preadapted for dispersal and colonization and in the case of Corvus, it can be expected that this combined with large brains and the increased associated cognitive abilities would make them ideal dispersers and colonizers across the planet [8].
Species-level systematics within Corvus has been based largely on morphological data [12] or very sparse sampling for molecular phylogenies e.g. [13-15] and even vocalizations have been used to infer phylogeny e.g. C. enca and C. mellori, [16,17]. A molecular phylogeny based on extensive taxon sampling is required to establish systematic relationships within Corvus so that questions pertaining to historical biogeography, brain size and the evolution of innovative foraging habits and tool use might be addressed. Additionally, a robust and densely sampled phylogeny will provide a framework for future work on plumage evolution and various aspects of macroecology and macroevolution.
In the present study, we present a molecular phylogeny including all extant crow species and a number of subspecies sometimes assigned species rank [10]. We use the phylogeny to assess systematic relationships and to elucidate historical biogeographical patterns by dating the phylogeny and estimating ancestral areas across the tree. Furthermore, taking into account the Corvus phylogeny, we test whether (i) brain size is correlated with the ability to disperse to and colonize islands and (ii) brain size correlates with innovative feeding behaviour and tool use within crows.
Methods
Taxon sampling and laboratory procedures
We sampled all forty extant species of Corvus[9] (Table 1). Where possible we included multiple individuals and, for widespread species, multiple subspecies (e.g. Corvus encaCorvus macrorhynchosCorvus coronoides and Corvus frugilegus). We also included some well-documented closely related genera to test for monophyly of Corvus: GarrulusPicaNucifraga[14]. Lanius was used to root the tree.
Table 1.
List of taxa included in the study
| Species | Origin of sample | Voucher number | GAPDH | ODC | ND3 | ND2 |
|---|---|---|---|---|---|---|
|
Corvus albicollis |
South Africa |
FMNH 447947 |
|
JQ024104 |
JQ024061 |
JQ023991 |
|
Corvus albus |
South Africa |
FMNH 443790 |
JQ023921 |
JQ024103 |
JQ024060 |
JQ023990 |
|
Corvus bennetti |
Australia |
ANWC 33292 |
JQ023900 |
JQ024082 |
JQ024019 |
JQ023945 |
|
Corvus bennetti |
Australia |
ANWC 52018 |
JQ023901 |
JQ024083 |
JQ024020 |
JQ023946 |
|
Corvus brachyrhynchos |
USA |
UWBM 86268 |
JQ023920 |
JQ024102 |
JQ024039 |
JQ023966 |
|
Corvus capensis* |
Abyssinia |
FMNH 370464 |
|
|
|
JQ023977 |
|
Corvus caurinus |
USA |
UWBM 58841 |
JQ023915 |
JQ024097 |
JQ024034 |
JQ023961 |
|
Corvus corax |
Denmark |
ZMUC 131662 |
JQ023891 |
JQ024073 |
|
JQ023935 |
|
Corvus corone cornix |
Denmark |
ZMUC 143486 |
JQ023894 |
JQ024076 |
JQ024013 |
JQ023939 |
|
Corvus corone corone |
Denmark |
ZMUC 138386 |
JQ023892 |
JQ024074 |
JQ024010 |
JQ023936 |
|
Corvus coronoides coronoides |
Australia, QLD |
ANWC 32675 |
JQ023908 |
JQ024090 |
JQ024027 |
JQ023953 |
|
Corvus coronoides coronoides |
Australia, NSW |
ANWC 49539 |
JQ023911 |
JQ024093 |
JQ024030 |
JQ023956 |
|
Corvus coronoides coronoides |
Australia, NSW |
ANWC 29239 |
JQ023904 |
JQ024086 |
JQ024023 |
JQ023949 |
|
Corvus coronoides coronoides |
Australia, ACT |
ANWC 34200 |
JQ023910 |
JQ024092 |
JQ024029 |
JQ023955 |
|
Corvus coronoides perplexus |
Australia, WA |
ANWC 31774 |
JQ023906 |
JQ024088 |
JQ024025 |
JQ023951 |
|
Corvus coronoides perplexus |
Australia, WA |
ANWC 31706 |
JQ023905 |
JQ024087 |
JQ024024 |
JQ023950 |
|
Corvus coronoides perplexus |
Australia, WA |
ANWC 50365 |
JQ023902 |
JQ024084 |
JQ024021 |
JQ023947 |
|
Corvus coronoides perplexus |
Australia, WA |
ANWC 31869 |
JQ023907 |
JQ024089 |
JQ024026 |
JQ023952 |
|
Corvus coronoides perplexus |
Australia, WA |
ANWC 50476 |
JQ023912 |
JQ024094 |
JQ024031 |
JQ023957 |
|
Corvus crassirostris* |
Ethiopia |
NRM 551730 |
|
|
JQ024002 |
JQ023927 |
|
Corvus cryptoleucus |
USA |
UWBM 80762 |
JQ023917 |
JQ024099 |
JQ024036 |
JQ023963 |
|
Corvus dauuricus |
Mongolia |
UWBM 58041 |
JQ023913 |
JQ024095 |
JQ024032 |
JQ023958 |
|
Corvus edithae* |
Kenya |
FMNH 370461 |
|
|
JQ024056 |
JQ023986 |
|
Corvus enca celebensis* |
Sulawesi |
RMNH 60561 |
|
|
JQ024042 |
JQ023969 |
|
Corvus enca compilator* |
Borneo |
RMNH 60563 |
|
|
JQ024059 |
JQ023989 |
|
Corvus enca pusillus* |
Palawan |
RMNH 100023 |
|
|
JQ024043 |
JQ023970 |
|
Corvus florensis* |
Flores |
RMNH 85140 |
|
|
JQ024046 |
JQ023973 |
|
Corvus frugilegus frugilegus |
Denmark |
ZMUC 143511 |
|
|
JQ024011 |
JQ023937 |
|
Corvus frugilegus pastinator* |
China |
NRM 570731 |
|
|
JQ024068 |
JQ023999 |
|
Corvus fuscicapillus* |
New Guinea |
AMNH 300970 |
|
|
JQ024048 |
JQ023975 |
|
Corvus hawaiiensis* |
Hawaii |
AMNH 196263 |
|
|
|
JQ023982 |
|
Corvus imparatus* |
Mexico |
AMNH 706673 |
|
|
|
JQ023978 |
| Corvus insularis |
New Britain |
AM 0.60592 |
JQ023888 |
JQ024070 |
JQ024007 |
JQ023932 |
|
Corvus jamaicensis* |
Jamaica |
AMNH 155238 |
|
|
JQ024052 |
JQ023981 |
|
Corvus kubaryi* |
Micronesia |
NRM 570711 |
|
|
JQ024003 |
JQ023928 |
|
Corvus leucognaphalus* |
Hispaniola |
NRM 570710 |
|
|
JQ024004 |
JQ023929 |
|
Corvus macrorhynchos japonensis* |
Japan |
NRM 570732 |
|
|
JQ024069 |
JQ024000 |
|
Corvus macrorhynchos levaillantii* |
N. Siam |
NRM 570733 |
|
|
JQ024067 |
JQ023998 |
|
Corvus macrorhynchos mandschuricus |
Russia |
UWBM 47167 |
JQ023918 |
JQ024100 |
JQ024037 |
JQ023964 |
|
Corvus macrorhynchos philippinus* |
Philippines |
ZMUC 104586 |
|
|
JQ024054 |
JQ023984 |
|
Corvus meeki* |
Bougainville |
AMNH 221033 |
|
|
JQ024058 |
JQ023988 |
|
Corvus mellori |
Australia |
ANWC 45128 |
JQ023895 |
JQ024077 |
JQ024014 |
JQ023940 |
|
Corvus mellori |
Australia |
ANWC 52403 |
JQ023903 |
JQ024085 |
JQ024022 |
JQ023948 |
|
Corvus mellori |
Australia |
ANWC 34099 |
JQ023909 |
JQ024091 |
JQ024028 |
JQ023954 |
|
Corvus minutus* |
Cuba |
AMNH 501484 |
|
|
JQ024051 |
JQ023980 |
|
Corvus monedula |
Denmark |
ZMUC 143533 |
JQ023893 |
JQ024075 |
JQ024012 |
JQ023938 |
|
Corvus moneduloides* |
New Caledonia |
FMNH 268468 |
|
|
JQ024040 |
JQ023967 |
| Corvus nasicus* |
Cuba |
NRM 570734 |
|
|
JQ024066 |
JQ023997 |
|
Corvus orru |
Australia |
ANWC 32239 |
JQ023898 |
JQ024080 |
JQ024017 |
JQ023943 |
|
Corvus orru |
Australia |
ANWC 50885 |
JQ023899 |
JQ024081 |
JQ024018 |
JQ023944 |
|
Corvus ossifragus |
USA |
UWBM 86680 |
JQ023914 |
JQ024096 |
JQ024033 |
JQ023960 |
|
Corvus palmarum* |
Hispaniola |
FMNH 352731 |
|
|
JQ024050 |
JQ023979 |
|
Corvus palmarum |
Hispaniola |
AMNH DOT 16134 |
JQ023922 |
JQ024105 |
|
JQ023992 |
|
Corvus pectoralis* |
China |
AMNH 261595 |
|
|
JQ024053 |
JQ023983 |
|
Corvus pectoralis* |
China |
NRM 570709 |
|
|
JQ024005 |
JQ023930 |
|
Corvus rhipidurus* |
Niger |
FMNH370467 |
|
|
JQ024057 |
JQ023987 |
|
Corvus ruficollis* |
Iran |
FMNH284717 |
|
|
JQ024055 |
JQ023985 |
|
Corvus sinaloae |
Mexico |
UWBM 81200 |
JQ023916 |
JQ024098 |
JQ024035 |
JQ023962 |
|
Corvus splendens |
Singapore |
UWBM 83598 |
JQ023919 |
JQ024101 |
JQ024038 |
JQ023965 |
|
Corvus tasmanicus boreus |
Australia |
AM 0.70670 |
JQ023889 |
JQ024071 |
JQ024009 |
JQ023934 |
|
Corvus tasmanicus boreus |
Australia |
AM 0.70687 |
JQ023890 |
JQ024072 |
JQ024008 |
JQ023933 |
|
Corvus tasmanicus tasmanicus |
Australia |
ANWC 44920 |
JQ023896 |
JQ024078 |
JQ024015 |
JQ023941 |
|
Corvus tasmanicus tasmanicus |
Australia |
ANWC 45502 |
JQ023897 |
JQ024079 |
JQ024016 |
JQ023942 |
|
Corvus tristis* |
New Guinea |
NRM 543594 |
|
|
JQ024006 |
JQ023931 |
|
Corvus tristis* |
New Guinea |
RMNH 22732 |
|
|
JQ024049 |
JQ023976 |
|
Corvus typicus* |
Sulawesi |
RMNH 101686 |
|
|
JQ024045 |
JQ023972 |
|
Corvus unicolor* |
Sulawesi |
AMNH 673967 |
|
|
JQ024041 |
JQ023968 |
|
Corvus validus* |
Halmahera |
RMNH 140643 |
|
|
JQ024047 |
JQ023974 |
|
Corvus violaceus* |
Seram |
RMNH 140590 |
|
|
JQ024044 |
JQ023971 |
|
Corvus woodfordi |
Solomon Islands |
UWBM 63090 |
|
|
|
JQ023959 |
|
Corvus woodfordi |
Solomon Islands |
AMNH DOT6705 |
JQ023923 |
JQ024106 |
JQ024062 |
JQ023993 |
|
Outgroups |
|
|
|
|
|
|
|
Nucifraga caryocatactes |
Sweden |
ZMUC 138408 |
JQ023924 |
JQ024107 |
JQ024064 |
JQ023995 |
|
Garrulus garrulus |
Denmark |
ZMUC 136378 |
|
|
JQ024063 |
JQ023994 |
|
Pica pica |
Denmark |
ZMUC 144204 |
JQ023925 |
JQ024108 |
JQ024065 |
JQ023996 |
|
Lanius collaris |
Cameroon/Tanzania |
GenBank/ZMUC 138905 |
FJ357916 |
EU272112 |
JQ024001 |
JQ023926 |
|
Dicrurus bracteatus/hottentottus |
New Guinea/Philippines |
GenBank |
EF052813 |
EU272113 |
GQ145422 |
GQ145384 |
|
Sturnus vulgaris |
Sweden |
GenBank |
EF441231 |
EF441253 |
GU816823 |
DQ146346 |
|
Menura |
Australia |
GenBank |
EF441220 |
EF441242 |
AY542313 |
AY542313 |
|
Pitta angolensis |
Tanzania |
GenBank |
AY336596 |
DQ785940 |
GU816799 |
GU816827 |
| Acanthisitta | New Zealand | GenBank | EU726202 | EU726220 | AY325307 | AY325307 |
Acronyms are: AM, Australian Museum, Sydney, Australia; AMNH, American Museum of Natural History, USA; ANWC, Australian National Wildlife Collection, Canberra, Australia; FMNH, Field Museum of Natural History, Chicago, USA; NRM, Naturhistoriska Riksmuseet, Stockholm, Sweden; RMNH, Rijksmuseum van Natuurlijke Histoire, Leiden, Netherlands; UWBM, University of Washington, Burke Museum, Seattle, USA; ZMUC, Zoological Museum, University of Copenhagen, Denmark. Asterisks after taxon names indicate that sequences were obtained from toe-pads of old museum specimens.
Two nuclear gene regions, ornithine decarboxylase (ODC) introns 6 to 7 (chromosome 3), and glyceraldehyde-3-phosphodehydrogenase (GAPDH) intron-11 (chromosome 1), and two mitochondrial markers NADH dehydrogenase subunit 2 (ND2) and subunit 3 (ND3) were sequenced and used to estimate phylogenetic relationships. Primer pairs used for amplification were: ND2: Lmet [18]/H6312 [19]; ND3: ND3-L10755/ND3-H11151 [20]; ODC: OD6/OD8 [21], G3P13/G3P14b [22]. For the old museum specimens we only sequenced the mitochondrial genes. Corresponding laboratory procedures for study skins are detailed in Irestedt et al. [23]. Additional internal primers were designed for this study, ND3-corvR1: GTCAAATAGTAGAAACAGGATTGC; ND3-CorvF1: TTTTCAATTCGATTCTTCCTAGT; ND2-CorvR1: CTTGAACTAGAAAGTATTTGGTTGC; ND2-CorvF2:CCCCTAATCTCAAAATCTCACCA; ND2-CorvR2: CCTTGTAGGACTTCTGGGAATC; ND2-CorvF3: CTAGGACTAGTGCCATTTCACTT; ND2-CorvR3: AGATAGAGGAGAAGGCCATAATT; ND2-CorvF4: CTGAATAGGACTAAACCAAACACAA; ND2-CorvR4: AGTGTTAGTAGGAGGATTGTGCT; ND2-CorvF5: CCACACTAATAACTGCATGAACAAA; ND2-CorvR5: TGTGGGGTGGAAGTGTGATTGT; ND2-CorvF6: TCACTACTGGGCCTCTTCTTCTA. Purified PCR products were cycle-sequenced using the Big Dye terminator chemistry (ABI, Applied Biosystems) in both directions with the same primers used for PCR amplification and run on an automated AB 3100 DNA sequencer. Sequences were assembled with SeqMan II (DNASTAR). Positions where the nucleotide could not be determined with certainty were coded with the appropriate IUPAC code. GenBank accession numbers are provided in Table 1.
Alignment and phylogenetic analyses
Sequence alignment was performed using MegAlign. The concatenated alignment consisted of 2346 base pairs (bp) and the lengths of the individual alignments were GAPDH: 299 bp, ODC intron-6 and 7: 611 bp, NADH dehydrogenase subunit 2: 1041 and NADH dehydrogenase subunit 3: 395 bp. Coding genes (ND2 and ND3) were checked for the presence of stop codons or insertion/deletion events that would have disrupted the reading frame. We used Bayesian inference [24,25], as implemented in MrBayes 3.1.2 [26,27] to estimate phylogenetic relationships. The most appropriate substitution models were determined with MrModeltest 2.0 [28], using the Akaike information criterion [29,30]. Bayesian analyses for the concatenated data set were performed allowing the different parameters (base frequencies, rate matrix or transition/transversion ratio, shape parameter, proportion of invariable sites) to vary between the six partitions (GAPDH, ODC, 1st, 2nd, 3 rd codon positions for mtDNA and tRNA), i.e. mixed-models analyses [27,28]. Two independent runs initiated from random starting trees were performed for each data set, and in all MrBayes analyses, the Markov Chain Monte Carlo (MCMC) was run using Metropolis-coupling, with one cold and three heated chains, for 10 million (individual analyses) to 20 million (combined analysis) iterations with trees sampled every 100 iterations. The number of iterations discarded before the chains had reached their apparent target distributions (i.e. the length of the “burn-in” period) was graphically estimated using AWTY [31,32] by monitoring the change in cumulative split frequencies, and by the loglikelihood values and posterior probabilities for splits and model parameters. We used GARLI 0.95 [33] to perform maximum likelihood analyses on the concatenated data set. Five independent analyses of 50 million generations were performed. Nodal support was evaluated with 100 nonparametric bootstrap pseudoreplications.
Dating analyses
To estimate the relative divergence times within Corvus, we used beast v.1.6 [34-36] and assigned the best fitting model, as estimated by mrmodeltest 2.0 [28], to each of the four partitions. We assumed a Yule Speciation Process for the tree prior and an uncorrelated lognormal distribution for the molecular clock model [35,37]. We used default prior distributions for all other parameters and ran MC3 chains for 50 million generations. The program Tracer [38] was used to assess convergence diagnostics. To obtain absolute date estimates we calibrated the tree using secondary calibration points derived from Barker et al. [39] who used various approaches to date the all Passeriformes tree. Thus we used the age of Acanthisittidae versus other passerines at 76 ± 8 My SD (age within 95% confidence intervals = 62.8–89.2 My) and the split between Menura noveahollandiae and all other oscines 63 ± 2 My SD (confidence intervals = 59.7–66.3 My). In order to apply these calibration points, some additional taxa were included in the dating analyses (see Table 1). We also compared our age estimates with the classic mitochondrial 2% rule [40].
Biogeographical analysis
We used Bayes-lagrange[41] to assess ancestral patterns within Corvus. In a Maximum-Likelihood biogeographical analysis [42,43] as implemented in the software lagrange [[42]], ancestral areas are optimized onto internal nodes. lagrange enables maximum likelihood estimation of the ancestral states (range inheritance scenarios) at speciation events by modelling transitions between discrete states (biogeographical ranges) along phylogenetic branches as a function of time. With the Bayes-lagrange approach it is possible to optimize on multiple trees whereby topological uncertainty is taken into account. We sampled 2000 trees (by thinning the chain stochastically) from the MCMC BEAST output, and ran lagrange on all of them. The frequency of ancestral areas for clades was then recorded and plotted as marginal distributions on the majority-rule consensus tree derived from the MCMC. The major advantage of the Bayes-Lagrange method is that the marginal distributions for the alternative ancestral areas at each node in the tree are the product of both the phylogenetic uncertainty in the rest of the tree and the uncertainty in the biogeographical reconstruction of the node of interest.
We assigned species distributions to one or more of nine geographical areas for the Bayes-lagrange analysis basing these on evidence of historical relationships of tectonic plates and terranes in the Indo-Pacific [44,45]: Nearctic, Caribbean, Palaearctic, Africa, Indomalaya (including the Philippines), Wallacea, Australo-papua and the Pacific. The analysis was carried out using the maxareas (= 2) option in lagrange. However, we also ran additional analyses exploring the importance of changing the maxareas (setting maxareas = 3 and 4).
Brain size, tool use and innovation
Data on brain size, which are considered a good proxy for cognition and intelligence [46], and body mass, were taken from Mlikovsky [47] and Iwanuik & Nelson [48]. Although the data are drawn from two sources, the data have been converted to reflect inner brain case volumes and are therefore directly comparable. These two datasets together include brain size data for 29 Corvus species [47,48]. By comparing the brain sizes for those species that are represented in both datasets it is clear that most discrepancies between the two datasets are explained by the size of the bird individuals measured. Therefore we believe that the measurements from the two datasets can be analysed combined. For a few species information on body mass was lacking in which case we used data from the CRC Handbook [49]. We compared the data on body mass used in our analyses [47-49] with body mass data provided in Handbook of Birds of the World [10] and found the data to be in agreement. After ln transforming the data we regressed brain size against body mass for 30 out of 40 species of Corvus. We also ran separate analyses based on the two datasets from Mlikovsky [47] and Iwanuik & Nelson [48] to account for potential differences in measuring body mass and brain size. We compiled data on tool use and innovative foraging behaviour from studies by Lefebvre et al. [4], Overington et al. [50] and Bentley-Condit & Smith [51]. Additionally, we searched the Handbook of Australian, New Zealand and Antarctic birds [52] for data on Australian crows. Altogether we found that 10 out of 40 Corvus species have a documented record of using tools and that 17 out 40 species use innovative foraging strategies. We note that opinions differ on what it means to be a “real” tool user and that some species are only known to use tools in captivity (Corvus frugilegus). However, this only underscores the high plasticity of this behaviour among Corvus species.
To investigate whether tool use, innovative foraging strategy and colonization of islands was associated with relative larger brain size across Corvus species, we ran a phylogenetic generalized least squares model (PGLS) in a phylogenetic framework using R version 2.10.1 [53] and the caper R package [54,55] as well as the ape package [56]. This statistical approach fits a linear model, taking into account phylogenetic non-independence. We tested the correlation of ln transformed brain size and ln transformed body mass as explanatory variables, with potential effect of tool use, innovation or island/continent distribution.
Results
Molecular phylogenetics and dating
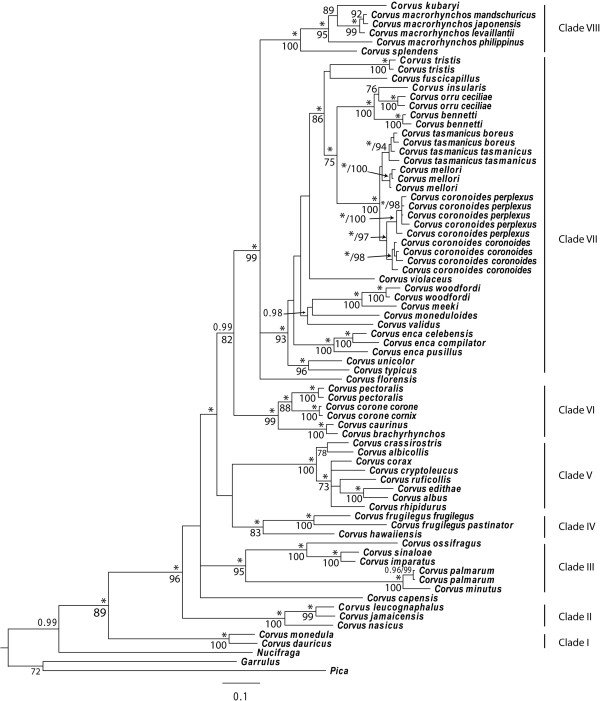
Model based analyses performed on the concatenated dataset (six partitions: GAPDH, ODC, 1st, 2nd, 3 rd codon positions for mtDNA and tRNA; maximum likelihood (ML): –ln 16528.5601, Bayesian inference (BI) harmonic mean: –ln 15872.64) yielded a 50% majority-rule consensus tree (BI) that was topologically congruent with the Maximum Likelihood tree (Figure 1), (for well-supported nodes receiving posterior probabilities >0.95 or bootstrap values >70%). Scores of the best likelihood trees were within 0.05 likelihood units of the best tree recovered in each of the other four garli runs, suggesting that the five runs had converged.
Figure 1.
The 50% majority-rule consensus tree of the Corvus obtained from the Bayesian analysis of the combined dataset (GAPDH, ODC, ND2 and ND3). Above the branch is the posterior probability (only values above 0.95 are shown, asterisks indicate 1.00 posterior probabilities). Below the branch is the maximum likelihood bootstrap value (only values above 70% are shown) from 100 pseudoreplicates. Clades I-VIII are discussed in the text.
We find that the genus Corvus is monophyletic and furthermore recovered eight well-supported sub-clades that contain members more or less restricted to biogeographical regions. The basal members (Clades I-III) are distributed across the Holarctic region, the Caribbean and Africa. The Caribbean members are found in two well-supported separate clades (Clades II and III) but resolution between the clades and Corvus capensis remain unresolved. Clade IV consists of the Eurasian Corvus frugilegus and Corvus hawaiiensis. The western and eastern subspecies of C. frugilegus represent a deep split in concordance with a previous study on this species complex [15]. Clade V consists of all the African Corvus species (except C. capensis) and the Holarctic C. corax. Clade VI consists of Holarctic species that are separated in two distinct well-supported clades, one clade of Neararctic species (C. caurinus and C. brachyrhynchos) and one clade of Palaearctic species (C. corone and C. pectoralis). Clade VII contains all the Australo-Papuan and Wallacean taxa except the Wallacean Corvus florensis. The latter species remains unresolved relative to Clades VII and VIII. Within Clade VII we also find some Pacific taxa. One subclade within Clade VII contains all Australian taxa and we also recover a well-supported Australo-Papuan clade. The relationships of the Wallacean species, however, remain unresolved at the base of Clade VII except that there is good support for a sister relationship between C. unicolor and C. typicus. Clade VIII includes the widespread C. macrorhynchos, the Indo-Malayan C. splendens and the Micronesian C. kubaryi.
Our BEAST dating analysis supports a mid-Miocene origin of Corvus dating to around 17.5 Mya (age within 95% HPD confidence intervals = 14.05–21.19 My). Our chronogram was consistent with the “2% rule” (uncorrected pairwise distances) for the rate of mitochondrial DNA sequence divergence per million years for young nodes (Pliocene to present) but suggested somewhat younger diversification times than those that BEAST determined as of Miocene age, which could be expected due to saturation in the mitochondrial genes [57]. According to the 2% rule the origin of Corvus dates to about 11 Mya. Knowledge of a Corvus fossil from North America dating back to the late Miocene [58] does not add much further insight because the author was unable to assign a systematic position. However, assuming that the fossil is closely related to the other North American taxa it supports our age estimates based on secondary calibration points.
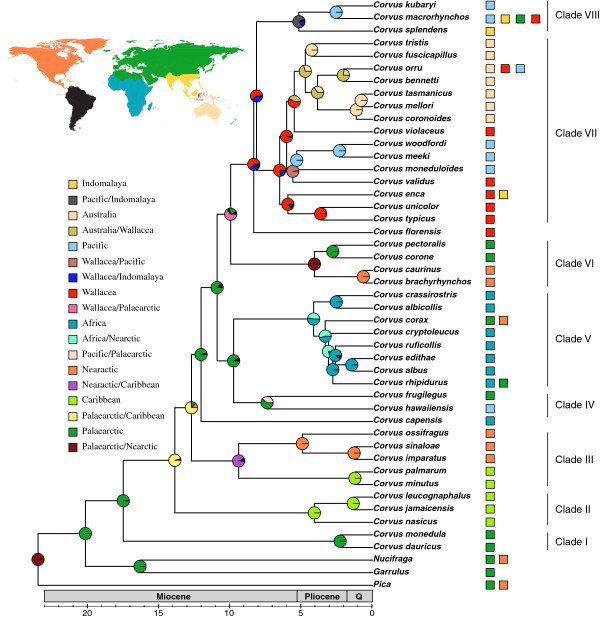
Biogeographical analysis
The Bayes-lagrange analysis (Figure 2) finds the origin of Corvus and its closest relatives (Pica, Nucifraga, Garrulus) to be within the Holarctic region. The origin of Corvus, however, is Palaearctic, although some deep branches lead to taxa distributed in North America and the Caribbean. Colonization of Africa took place in the Pliocene, and colonization of Wallacea took place in the late Miocene and led to further colonization of Australo-Papua around 5 Mya. We find evidence for four colonization events of the Pacific from Asia and Australia. The Caribbean was colonized twice. One Caribbean clade is sister to a clade of North American taxa (Clade III) indicative of colonization from there. However, the ancestral area analysis postulates a Palaearctic/Caribbean origin for the other Caribbean clade (II).
Figure 2.
A summary of the BAYES-LAGRANGE ancestral area analysis for the genus Corvus. The tree is a chronogram (pruned to include one individual per species) based on the BEAST dating analysis of a combined data set of mitochondrial (ND2 and ND3) and nuclear (GAPDH and ODC) DNA sequences. Pie charts at internal nodes indicate the probability of a given area of origin. The inset map indicates the regions demarcated for the ancestral area analyses and colours to the right of the taxon names indicate present distributions (Nearctic, Palaeearctic, Caribbean, Africa, Indomalaya, Wallacea, Australo-Papua and Pacific) and thus coding for the ancestral area analyses. Black parts of the pie charts indicate a mixture of other areas.
Brain size and tool use
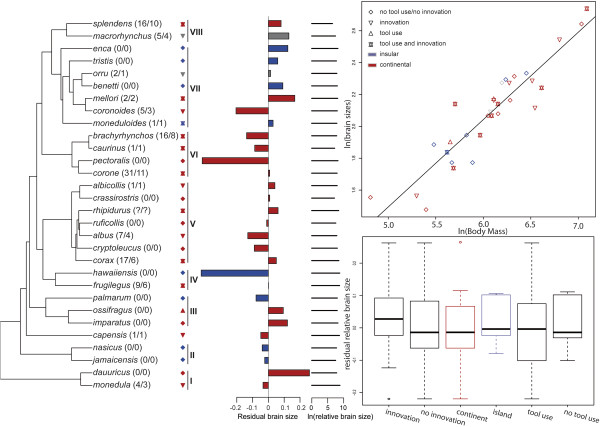
We find a highly significant correlation between body mass and brain size in Corvus (P < 0.001 Figure 3) both for the combined dataset and when analyzing the individual datasets from Mlikovsky [47] and Iwanuik & Nelson [48]. This suggests that there are no significant differences in relative brain size between large and small Corvus species. Our analysis (Phylogenetic Generalized Least Squares) taking covariance between taxa into account, finds no correlation between brain size, tool use (P = 0.67) and innovative behaviour (P = 0.69), and no correlation between brain size and the ability to colonize islands (P = 0.46) (Figure 3C). Seemingly, all members of Corvus have the same relative brain size and species of all sizes have innovative foraging strategies/use tools and have been able to colonize islands.
Figure 3.
A) Phylogeny showing the taxa used in the comparative brain size analyses. Numbers in parentheses indicate the number of innovations followed by the diversity of innovations. Symbols indicate whether the taxon applies tools (upwards pointing triangle), innovative strategies (downwards pointing triangle) or both (star combining the two triangles) in its search for food. Distributions are indicated for islands (blue), continents (red) or both (grey). Island taxa are indicated in blue, continental taxa in red and combinations in grey. Residual brain size and relative brain size for the taxa are indicated to the right of the phylogeny B) Linear regression between brain and body mass. C) Box-plot displaying the difference (median, 25% and 75% percentiles and sample minimum and maximum) in relative brain size between Corvus species that use tools/no tools, Corvus species that apply innovation/no innovation and Corvus species that occur on islands/continents. Relative brain size represents residual values obtained from a linear regression between ln-transformed brain size and ln-transformed body mass.
Discussion
Systematics and biogeography
The early history of the classification of the family Corvidae has been summarized by Goodwin [12] but is restricted to morphology. We present the first complete molecular species level phylogeny for the crows and ravens (Corvus spp) including several subspecies of widespread species (Figure 1). Most notably, we demonstrate that what is currently classified as the Australian Raven C. coronoides, comprises two clades, one in the west (C. c. perplexus) and one in the east (C. c. coronoides). They are geographically isolated from each other only by approximately 100 km of apparently unsuitable habitat across the continent’s south coast at the Great Australian Bight. We propose that these two taxa be elevated to species rank. In contrast, populations of C. tasmanicus, geographically isolated from each other by >500 km, were not reciprocally monophyletic. Currently recognized as two subspecies, their isolation and divergence is presumably very recent. In accordance with a previous study including samples from throughout the Palaearctic, we show that C. frugilegus may also represent two distinct species, one in the western Palaearctic and one in the eastern Palaearctic [15]. C. macrorhynchos is found to be paraphyletic such that the Philippine C. macrorhynchos philippinus is sister to a clade comprising all other C. macrorhyncos, which occur in East Asia, and C. kubaryi. However, denser taxon sampling for C. macrorhynchos and population sampling for other widespread species (e.g. Corvus enca and Corvus orru) is needed to properly revise taxonomic issues at species and subspecies levels.
Our dating analysis suggests that the radiation of Corvus began in the mid-Miocene and our ancestral area analysis indicates a Palaearctic origin (Figure 2). This is consistent with the two most basal members, Corvus monedula and Corvus dauuricus (Clade I) being Eurasian and with the closest extant relatives of Corvus distributed in Eurasia and North America (Pica and Nucifraga). This results in a signature of a Holarctic origin for Corvus and its closest relatives. We infer that two clades independently colonized the Caribbean islands in the late Miocene (Clades II and III). One Caribbean clade (II) has a long branch that leads to C. leucognaphalusC. jamaicensis and C. nasicus and our analyses suggest a Palaearctic/Caribbean origin. This could be interpreted as evidence for long distance ocean dispersal similar to that inferred in other passerine bird groups that have crossed the Atlantic e.g. Turdus[59]. Two alternative interpretations of the Caribbean having been colonized from North America are possible (i) extinction of a North American ancestor, (ii) an ancestral form was widely distributed in the Holarctic (like Corvus corax) and gave rise to independent colonizations to the Caribbean followed by isolation of the North American population and a second colonization to the Caribbean (Clade III). The other Caribbean species (C. palmarum and C. minutus; Clade III) are sister to three North American species (C. ossifragusC. sinaloae and C. imparatus). This provides evidence for colonization by C. palmarum and C. minutus of the Caribbean from North America.
Relationships of an African species, C. capensis, were difficult to ascertain but it seems to represent a single Miocene colonization of Africa from the Palaearctic. C. capensis does not seem closely related to members of Clade V, which includes all other taxa that have colonized Africa and radiated within the continent. Clade VII of Indo-Pacific species has sequentially colonized Southeast Asia, Wallacea, Australo-Papua and the Pacific islands. However, the Hawaiian crow (C. hawaiiensis) is not a member of this clade, instead it clusters with the Palaearctic C. frugilegus (Clade IV) and so we infer it to have colonized Hawaii from East Asia. This is unexpected because the Hawaiian biota generally evolved through colonization from America whereas that of the rest of the Pacific was mostly colonized from Asia and Australo-Papua [60]. For the Corvus radiation, Asia has been the main source area for colonization of the Pacific as opposed to Australo-Papua, which is only the source area for one out of four Pacific lineages. Overall, the ancestral area analysis provides a rather clear pattern of separate colonizations of all continents except South America from the Palaearctic/(Nearctic).
Brain size, tool use and colonization
Tool use is rare in the animal kingdom and is considered restricted to primates [61], Cetaceans [62], and some birds (e.g. Psittaciformes and Passeriformes) [1]. By far the most well-known tool using bird is the New Caledonian crow (Corvus moneduloides) and several studies have demonstrated this island endemic crow’s ingenious abilities to use sticks to probe for larvae [5,63]. It is well established in the literature that cognitive abilities correlate with larger relative brain size, and that the family Corvidae have unusually large brains compared to other birds [64,65]. Particularly, the New Caledonian crow’s ability to use tools has been explained by its extraordinary large brain [6]. However, in the study by Cnotka et al. [6] phylogenetic relationships among crows were unknown and they were therefore unable to make appropriate phylogenetic corrections.
Several members of the genus Corvus use a variety of natural tools or advanced innovative strategies when foraging [summarized in 4]. Until now, innovative feeding techniques have been reported for 17 of the 40 species of Corvus and tool use for 10 of the 40 Corvus species ( Additional file 1: Table S1). There is only one tool using species (Corvus ossifragus) that is not known to have foraging innovations and it should be noted that tool-use is usually seen as species typical and often hard-wired as evidenced by experiments on young woodpecker finches [66] and young New Caledonia crows [67], both of which “know” about tools from the start without having to learn about them.
Our analyses on body mass and brain size demonstrate that there is a significant correlation between the two variables, meaning that all crows have the same large relative brain size (similar body mass/brain size ratio). However, our comparative analyses on brain size and innovative feeding/tool use strategies within Corvus, corrected for phylogenetic relationships, reveal no correlation between the variables. This could be interpreted in two ways. Either brain size has little to do with innovative foraging strategies/tool use and thus other factors are more important in determining whether or not crows use tools or innovative feeding strategies. Alternatively, all crow species have large brains relative to other birds and thus have the potential to use tools or other innovative feeding strategies. Given that a number of studies have already demonstrated a link between cognitive abilities and brain size in both birds [3,4] and mammals [68,69] and that it is well-established that corvids have larger brains than many other birds [1,4], it seems the most likely hypothesis that all crows have the potential to develop innovative foraging strategies and to use tools in their search for food and there could be many reasons why this potential is only realised in some species across the Corvus tree. However, it has also been argued that total brain size may not be the ideal proxy for cognition and that measures should be taken to explore particular brain regions to explain innovation and tool-use [70]. We do not consider this study the final word on the topic, but merely a first attempt to combine phylogeny with functional traits associated with cognition and innovation in crows.
The most persistent hypothesis of large brains and corresponding enhanced cognition is that they evolved as an adaptation to handle novel or altered environmental conditions [71]. Island environments may prove particularly challenging as they, depending on the size and nature of the island, may provide fewer available niches, inferior access to food and new unknown dangers. On the other hand, a new island colonizer, could also find itself in an environment free of closely related competitors and free of inhibitors leading to occupancy of a wider range of habitats – ecological release [72,73]. A combination of these two extreme scenarios, however, could to some extent counteract each other, which may explain the lack of correlation between relative brain size and island colonisations in crows and ravens.
Conclusion
The analyses based on molecular sequence data from all recognized crow and raven species (genus: Corvus) demonstrate that the genus is monophyletic and that it originated in the Palaearctic in the Miocene. From the centre of origin crows dispersed to North America and the Caribbean, to Africa and to Australasia, with several independent colonizations of remote Pacific islands. Our analysis comparing brain size and colonization of islands within Corvus found no correlation and we therefore conclude that colonization of islands by crows cannot be explained by brain size. We did not find a correlation between brain size, tool use and innovative foraging strategies as otherwise suggested by other studies e.g. [6,8]. Thus, there seems no reason to believe that brain size alone has any influence on tool use, innovative foraging stragegies and colonization ability within the crow lineages. Rather it would appear that large brains had already evolved in the ancestor of crows, leading to a generally high cognitive ability to deal with new challenges for crows and other corvid lineages.
Competing interests
The authors have no competing interests.
Authors’ contributions
KAJ designed the study, carried out the lab work, performed the phylogenetic analyses, and drafted the manuscripts. PHF carried out additional comparative analyses. MI assisted with lab work. All authors read, commented upon and approved the manuscript.
Supplementary Material
Table S1. List of taxa used in the comparative analyses Distribution is either insular or continental. Body mass (D) according to Dunning body mass (I) according to Iwanuik & Nelson and body mass (M) according to Mlikovsky. Brain size according to Mlikovsky and Iwanuik & Nelson. Feeding innovation according to Lefebvre et al. Bentley-Condit & Smith and Higgins et al. Feeding innovations according to Lefebvre et al. Bentley-Condit & Smith and Overington et al. [4,7,47-51].
Contributor Information
Knud A Jønsson, Email: kajonsson@snm.ku.dk.
Pierre-Henri Fabre, Email: phfabre@snm.ku.dk.
Martin Irestedt, Email: martin.irestedt@nrm.se.
Acknowledgements
We thank a number of institutions for supplying samples for this study: AM, Australian Museum, Sydney, Australia; AMNH, American Museum of Natural History, USA; ANWC, Australian National Wildlife Collection, Canberra, Australia; FMNH, Field Museum of Natural History, Chicago, USA; NRM, Naturhistoriska Riksmuseet, Stockholm, Sweden; RMNH, Rijksmuseum van Natuurlijke Histoire, Leiden, Netherlands; UWBM, University of Washington, Burke Museum, Seattle, USA; ZMUC, Zoological Museum, University of Copenhagen, Denmark. KAJ and PHF acknowledge the Danish National Research Foundation for support to the Center for Macroecology, Evolution, and Climate. We also thank Leo Joseph, Jon Fjeldså and two anonymous reviewers for comments on the manuscript.
References
- Emery NJ, Clayton NS. The mentality of crows: Convergent evolution of intelligence in Corvids and Apes. Science. 2004;306:1903–1907. doi: 10.1126/science.1098410. [DOI] [PubMed] [Google Scholar]
- Cornell HN, Marzluff JM, Pecoraro S. Social learning spreads knowledge about dangerous humans among American crows. Proc R Soc London B Biol Sci. 2012;279:499–508. doi: 10.1098/rspb.2011.0957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre L, Whittle P, Lascaris E, Finkelstein A. Feeding innovations and forebrain size in birds. Anim Behav. 1997;53:549–560. doi: 10.1006/anbe.1996.0330. [DOI] [Google Scholar]
- Lefebvre L, Nicolakakis N, Boire D. Tools and brains in birds. Behaviour. 2002;139:939–973. doi: 10.1163/156853902320387918. [DOI] [Google Scholar]
- Rutz C, Bluff LA, Reed N, Toscianko J, Newton J, Inger R, Kacelnik A, Bearhop S. The ecological significance of tool use in New Caledonian crows. Science. 2010;329:1523–1526. doi: 10.1126/science.1192053. [DOI] [PubMed] [Google Scholar]
- Cnotka J, Güntürkün O, Rehkämper G, Gray RD, Hunt GR. Extraordinary large brains in tool-using New Caledonian crows (Corvus moneduloides) Neurosci Lett. 2008;433:241–245. doi: 10.1016/j.neulet.2008.01.026. [DOI] [PubMed] [Google Scholar]
- Sol D, Timmermans S, Lefebvre L. Behavioural flexibility and invasion success in birds. Animal Behav. 2002;63:495–502. doi: 10.1006/anbe.2001.1953. [DOI] [Google Scholar]
- Sol D, Duncan RP, Blackburn TM, Cassey P. Lefebvre L Big brains, enhanced cognition, and response of birds to novel environments. Proc Natl Acad Sci USA. 2005;102:5460–5465. doi: 10.1073/pnas.0408145102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson EC. The Howard and Moore complete checklist of the birds of the world. 3. Princeton University Press, Princeton; 2003. [Google Scholar]
- Dos Anjos L. In: Handbook of the Birds of the World Bush-shrikes to Old World Sparrows. vol 14. Hoyo J, Elliot A, Christie DA, editor. Lynx Edicions, Barcelona; 2009. Family Corvidae (Crows) pp. 494–641. [Google Scholar]
- Jønsson KA, Fabre PH, Ricklefs RE, Fjeldså J. Major global radiation of corvoid birds originated in the proto-Papuan archipelago. Proc Natl Acad Sci USA. 2011;108:2328–2333. doi: 10.1073/pnas.1018956108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin D. Crows of the world. British Museum (Natural History), London, UK; 1976. [Google Scholar]
- Omland KE, Tarr CL, Boarman WI, Marzluff JM, Fleischer RC. Cryptic genetic variation and paraphyly in ravens. Proc R Soc London B Biol Sci. 2000;267:2475–2482. doi: 10.1098/rspb.2000.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson PGP, Jansén A-L, Johansson US, Ekman J. Inter-generic relationships of the crows, jays, magpies and allied groups (Aves: Corvidae) based on nucleotide sequence data. J Avian Biol. 2005;36:222–234. doi: 10.1111/j.0908-8857.2001.03409.x. [DOI] [Google Scholar]
- Haring E, Gamauf A, Kryukov A. Phylogeographic patterns in widespread corvid birds. Mol Phylogenet Evol. 2007;45:840–862. doi: 10.1016/j.ympev.2007.06.016. [DOI] [PubMed] [Google Scholar]
- Martens J, Böhner J, Hammerschmidt K. Calls of the Jungle Crow (Corvus macrorhynchos s.l.) as a taxonomic character. J Ornithology. 2000;141:275–284. [Google Scholar]
- Rowley I. A fourth species of Australian corvid. Emu. 1967;66:191–210. [Google Scholar]
- Hackett SJ. Molecular phylogenetics and biogeography of tanagers in the genus Ramphocelus (Aves) Mol Phylogenet Evol. 1996;5:368–382. doi: 10.1006/mpev.1996.0032. [DOI] [PubMed] [Google Scholar]
- Cicero C, Johnson NK. Higher level phylogeny of vireos (Aves: Vireonidae) based on sequences of multiple mtDNA genes. Mol Phylogenet Evol. 2001;20:27–40. doi: 10.1006/mpev.2001.0944. [DOI] [PubMed] [Google Scholar]
- Chesser RT. Molecular systematics of the rhinocryptid genus Pteroptochos. Condor. 1999;101:439–446. doi: 10.2307/1370012. [DOI] [Google Scholar]
- Allen ES, Omland KE. Novel intron phylogeny (ODC) supports plumage convergence in orioles (Icterus) Auk. 2003;120:961–969. [Google Scholar]
- Fjeldså J, Zuccon D, Irestedt M, Johansson US, Ericson PGP. Sapayoa aenigma: a New World representative of Old World suboscines. Proc R Soc London B Biol Sci Suppl. 2003;270:238–241. doi: 10.1098/rsbl.2003.0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irestedt M, Ohlson JI, Zuccon D, Källersjö M, Ericson PGP. Nuclear DNA from old collections of avian study skins reveals the evolutionary history of the Old World suboscines (Aves, Passeriformes) Zool Script. 2006;35:567–580. doi: 10.1111/j.1463-6409.2006.00249.x. [DOI] [Google Scholar]
- Holder MT, Lewis PO. Phylogeny estimation: traditional and Bayesian approaches. Nat Rev Genet. 2003;4:275–284. doi: 10.1038/nrg1044. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MrBayes: a program for the Bayesian inference of phylogeny Version 3.1.2. 2003. Available from: http://mrbayes.scs.fsu.edu/index.php.
- Huelsenbeck JP, Ronquist F, Hall B. MrBayes: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Nylander JAA. MrModeltest2 Available from: http://www.abc.se/~nylander/; 2004.
- Akaike H. In: Second International Symposium on Information Theory. Petrov BN, Csaki F, editor. Hungary: Akademiai Kiado, Budapest; 1973. Information theory as an extension of the maximum likelihood principle; pp. 276–281. [Google Scholar]
- Posada D, Buckley TR. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol. 2004;53:793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- Wilgenbusch JC, Warren DL, Swofford DL. AWTY: a system for graphical exploration of MCMC convergence in Bayesian phylogenetic inference. 2004. Available from: http://ceb.csit.fsu.edu/awt. [DOI] [PubMed]
- Nylander JAA, Wilgenbusch JC, Warren DL, Swofford DL. AWTY (are we there yet): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics. 2008;24:581–583. doi: 10.1093/bioinformatics/btm388. [DOI] [PubMed] [Google Scholar]
- Zwickl DJ. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD thesis, The University of Texas at Austin; 2006. [Google Scholar]
- Drummond AJ, Nicholls GK, Rodrigo AG, Solomon W. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics. 2002;161:1307–1320. doi: 10.1093/genetics/161.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST v1.4.7. 2007. Available from: http://beast.bio.ed.ac.uk/ [DOI] [PMC free article] [PubMed]
- Ho SYW. Calibrating molecular estimates of substitution rates and divergence times in birds. J Avian Biol. 2007;38:409–414. [Google Scholar]
- Rambaut A, Drummond AJ. Tracer v.1.4. 2007. Available from: http://beast.bio.ed.ac.uk/Tracer.
- Barker FK, Cibois A, Schikler P, Feinstein J, Cracraft J. Phylogeny and diversification of the largest avian radiation. Proc Natl Acad Sci USA. 2004;101:11040–11045. doi: 10.1073/pnas.0401892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir JT, Schluter D. Calibrating the avian molecular clock. Mol Ecol. 2008;17:2321–2328. doi: 10.1111/j.1365-294X.2008.03742.x. [DOI] [PubMed] [Google Scholar]
- Smith SA, Donoghue MJ. Combining historical biogeography with niche modeling in the Caprifolium clade of Lonicera (Caprifoliaceae, Dipsacales) Syst Biol. 2010;59:322–341. doi: 10.1093/sysbio/syq011. [DOI] [PubMed] [Google Scholar]
- Ree RH, Moore BR, Webb CO, Donoghue MJ. A likelihood framework for inferring the evolution of geographic range on phylogenetic trees. Evolution. 2005;59:2299–2311. [PubMed] [Google Scholar]
- Ree RH, Smith SA. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst Biol. 2008;57:4–14. doi: 10.1080/10635150701883881. [DOI] [PubMed] [Google Scholar]
- Audley-Charles MG. Tectonics of the New Guinea area. Annu Rev Earth Planet Sci. 1991;19:17–41. doi: 10.1146/annurev.ea.19.050191.000313. [DOI] [Google Scholar]
- Hall R. In: Biogeography and geological Evolution of SE Asia. Hall R, Holloway JD, editor. Backhuys Publishers, Leiden, the Netherlands; 1998. Biogeographic implications of the Tertiary palaeogeographic evolution of Sulawesi and Borneo; pp. 133–163. [Google Scholar]
- Lefebvre L. Taxonomic counts of cognition in the wild. Biol Lett. 2010;7:631–633. doi: 10.1098/rsbl.2010.0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlikovsky J. Brain size and forearmen magnum area in crows and allies (Aves: Corvidae) Acta Soc Zool Bohem. 2003;67:203–211. [Google Scholar]
- Iwaniuk AN, Nelson JE. Developmental differences are correlated with relative brain size in birds: a comparative analysis. Can J Zool. 2003;81:1913–1928. doi: 10.1139/z03-190. [DOI] [Google Scholar]
- Dunning JB. The CRC handbook of avian body masses. CRC Press, Boca Raton, FL, USA; 1993. [Google Scholar]
- Overington SE, Morand-Ferron J, Boogert NJ, Lefebvre L. Technical innovations drive the relationship between innovativeness and residual brain size in birds. Animal Behav. 2009;78:1001–1010. doi: 10.1016/j.anbehav.2009.06.033. [DOI] [Google Scholar]
- Bentley-Condit VK, Smith EO. Animal tool use: current definitions and an updated comprehensive catalog. Behaviour. 2010;147:185–221. doi: 10.1163/000579509X12512865686555. [DOI] [Google Scholar]
- Higgins PJ, Peter JM, Cowling SJ. Handbook of the Australian, New Zealand and Antarctic birds. Vol 7. Oxford University Press, Melbourne, Australia; 2006. [Google Scholar]
- R Development Core Team: R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. 2009. Available from: http://www.R-project.org.
- Orme D, Freckleton R, Thomas G, Petzoldt T, Fritz SE, Isaac N. 2011. http://cran.r-project.org/web/packages/caper/index.html.
- Freckleton RP, Harvey PH, Pagel M. Phylogenetic analysis and comparative data: A test and review of evidence. Am Nat. 2002;160:712–726. doi: 10.1086/343873. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Arbogast BS, Edwards SV, Wakely J, Beerli P, Slowinski JB. Estimating divergence times from molecular data on phylogenetic and population genetic timescales. Annu Rev Ecol Syst. 2002;33:707–740. doi: 10.1146/annurev.ecolsys.33.010802.150500. [DOI] [Google Scholar]
- Bickart KJ. In: Ornithological Monographs, No. 44, Recent Advances in the Study of Neogene Fossil Birds. Bickart KJ, Chandler RM, editor. University of California, USA; 1990. Part I: The Birds of the Late Miocene-Early Pliocene Big Sandy Formation, Mohave County, Arizona; pp. 1–72. [Google Scholar]
- Voelker G, Rohwer S, Outlaw DC, Bowie RCK. Repeated trans-Atlantic dispersal catalysed a global songbird radiation. Global Ecol Biogeogr. 2009;18:41–49. doi: 10.1111/j.1466-8238.2008.00423.x. [DOI] [Google Scholar]
- Gillespie RG, Claridge EM, Goodacre SL. Biogeography of the fauna of French Polynesia: diversification within and between a series of hot spot archipelagos. Phil Trans R Soc B. 2008;363:3335–3346. doi: 10.1098/rstb.2008.0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasello M, Call J. Primate Cognition. Oxford University Press, New York; 1997. [Google Scholar]
- Marino L. Convergence of complex cognitive abilities in cetaceans and primates. Brain Behav Evol. 2002;59:21–32. doi: 10.1159/000063731. [DOI] [PubMed] [Google Scholar]
- Hunt GR. Manufacture and use of hook-tools by new Caledonian crows. Nature. 1996;379:249–251. doi: 10.1038/379249a0. [DOI] [Google Scholar]
- Harvey PH, Krebs JR. Comparing brains. Science. 1990;249:140–146. doi: 10.1126/science.2196673. [DOI] [PubMed] [Google Scholar]
- Lefebvre L, Sol D. Brains, lifestyles and cognition: are there general trends? Brain Behav Evol. 2008;72:135–144. doi: 10.1159/000151473. [DOI] [PubMed] [Google Scholar]
- Tebbich S, Taborsky M, Fessl B, Blomqvist D. Do woodpecker finches acquire tool-use by social learning? Proc R Soc Lond B. 2001;268:2189–2193. doi: 10.1098/rspb.2001.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenward B, Weir AAS, Rutz C, Kacelnik A. Tool manufacture by naive juvenile crows. Nature. 2005;433:121. doi: 10.1038/433121a. [DOI] [PubMed] [Google Scholar]
- Baron G, Stephan H, Frahm HD. Comparative Neurobiology in Chiroptera. vol. I–III. Birkhäuser Verlag, Basel, Switzerland; 1996. [Google Scholar]
- Stephan H, Baron G. Frahm HD: Insectivora. SpringerVerlag, NewYork; 1991. [Google Scholar]
- Healy SD, Rowe C. A critique of comparative studies of brain size. Proc R Soc Lond B. 2007;274:453–464. doi: 10.1098/rspb.2006.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allman J. Evolving Brains. Scientific American Library, New York; 2000. [Google Scholar]
- MacArthur RH, Recher H, Cody ML. On the relation between habitat selection and species diversity. Am Nat. 1966;100:603–609. doi: 10.1086/282454. [DOI] [Google Scholar]
- Losos JB, De Queiroz K. Evolutionary consequences of ecological release in Caribbean Anolis lizards. Biol J Linn Soc. 1997;61:459–483. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of taxa used in the comparative analyses Distribution is either insular or continental. Body mass (D) according to Dunning body mass (I) according to Iwanuik & Nelson and body mass (M) according to Mlikovsky. Brain size according to Mlikovsky and Iwanuik & Nelson. Feeding innovation according to Lefebvre et al. Bentley-Condit & Smith and Higgins et al. Feeding innovations according to Lefebvre et al. Bentley-Condit & Smith and Overington et al. [4,7,47-51].