Abstract
Exposure to inorganic arsenic induces skin cancer and abnormal pigmentation in susceptible humans. High-throughput gene transcription assays such as DNA microarrays allow for the identification of biological pathways affected by arsenic that lead to initiation and progression of skin cancer and abnormal pigmentation. The overall purpose of the reported research was to determine knowledge building insights on biomarker genes for arsenic toxicity to human epidermal cells by integrating a collection of gene lists annotated with biological information. The information sets included toxicogenomics gene-chemical interaction; enzymes encoded in the human genome; enriched biological information associated with genes; environmentally relevant gene sequence variation; and effects of non-synonymous single nucleotide polymorphisms (SNPs) on protein function. Molecular network construction for arsenic upregulated genes TNFSF18 (tumor necrosis factor [ligand] superfamily member 18) and IL1R2 (interleukin 1 Receptor, type 2) revealed subnetwork interconnections to E2F4, an oncogenic transcription factor, predominantly expressed at the onset of keratinocyte differentiation. Visual analytics integration of gene information sources helped identify RAC1, a GTP binding protein, and TFRC, an iron uptake protein as prioritized arsenic-perturbed protein targets for biological processes leading to skin hyperpigmentation. RAC1 regulates the formation of dendrites that transfer melanin from melanocytes to neighboring keratinocytes. Increased melanocyte dendricity is correlated with hyperpigmentation. TFRC is a key determinant of the amount and location of iron in the epidermis. Aberrant TFRC expression could impair cutaneous iron metabolism leading to abnormal pigmentation seen in some humans exposed to arsenicals. The reported findings contribute to insights on how arsenic could impair the function of genes and biological pathways in epidermal cells. Finally, we developed visual analytics resources to facilitate further exploration of the information and knowledge building insights on arsenic toxicity to human epidermal keratinocytes and melanocytes.
Keywords: arsenic, disulfide bond, environmental response genes, functional annotation, hyperpigmentation, iron uptake, keratinocyte, melanocyte, RAC1, SNPs, skin cancer, TFRC, toxicogenomics, vicinal cysteines Isokpehi et al
Introduction
Arsenic is a naturally occurring environmental toxicant that is widely distributed in the earth’s crust.1,2 The air, water, and soil environment can be contaminated with arsenic from industrial smelting of metals, power generation with coal, and applications of pesticides and herbicides.3 Human exposure occurs mainly through the accumulation of arsenic from soil into food sources including grains, vegetables, fish, and meats.4–6 Arsenic enters the human body by dermal contact, inhalation, or food consumption, but ingestion of contaminated drinking water is the most common route. The tissues most often affected by arsenic exposure are the skin, nasal passages, lungs, gastrointestinal tract, and liver.7
Human exposure to chronic low-dose arsenic is associated with an increased risk of epithelial cancers of the skin such as intraepidermal carcinomas (Bowen disease), squamous cell carcinomas (SCC), basal cell carcinomas (BCC),8 and Merkel cell carcinoma (MCC).9 In most cases where internal cancers are attributed to arsenic exposure, there are other evidences of cutaneous cancers due to the adverse effects of arsenic expressed in the form of arsenical keratosis, hyperpigmentation, and multiple cutaneous malignancies.10 Clinical manifestations of arsenic exposure include epithelial cancers of the bladder, liver, kidney, prostate, and lung; cardiovascular and skin diseases; as well as neuropathies of the central nervous system.7
The mechanisms of toxicity and carcinogenicity of arsenicals on various cell types have been proposed.11–15 These mechanisms include perturbation of signaling cascades, oxidative stress, and chromosomal aberrations.16 There could also be perturbation of the transcriptional activity and changes in the global gene expression.15,17,18 Exposure to arsenic trioxide leads to the alteration in mitochondrial integrity and the generation of reactive oxygen species (ROS),19 enhanced cell proliferation,20 and alteration of DNA methylation.21
Advances in genomics and other “-omics” technologies are providing massive numbers of datasets and the accompanying scientific publications that describe potential gene, protein, and biological processes as potential biomarkers of adverse health effects of environmental chemicals. In particular toxicogenomics, which is a scientific field that investigates how the entire genome response to environmental toxicant effects, is a recognized approach to discover potential biomarkers of adverse toxicity and exposure as well as to validate/quantify biomarker signatures. 22 The growth in data from toxicogenomics research has led to the development of bioinformatics databases such as the Comparative Toxicogenomics Database (CTD) for curating toxicogenomics relationships (chemical-gene, chemical-disease, and gene-disease) found in scientific publications.23 As of the December 7, 2011, the CTD contained 28,413 PubMed references, 352,925 chemical-gene relationships, 6,605 unique chemicals, 20,710 unique genes, and 334 unique organisms. These toxicogenomics relationships and data when combined with biological information on human genes from other bioinformatics databases can lead to knowledge building (discovering previously unknown relationships from datasets) on potential biomarkers. Significant over-representation (enrichment) of certain biological topics for a particular gene is an example of the biological information that can be obtained from a bioinformatics database such as ConceptGen.24 ConceptGen database currently consists of approximately 18,000 concepts, each with 5 or more assigned genes. Further, diverse bioinformatics tools are now available to reconstruct molecular interactions from predicted and experimentally validated datasets.25–29 The reconstruction of molecular pathways involving potential biomarker genes and proteins can also yield insights on how the normal cellular activities are altered in different chemical exposures or disease conditions. These diverse datasets from bioinformatics databases, therefore, present opportunities for discovery and inferences on the biological processes affected by arsenicals. In particular, we are interested in identifying genes for further research on the mechanisms of toxic action and cancer initiation of arsenic on skin keratinocytes.
We have used a visual analytics approach to integrate and make sense of data on arsenic-associated genes stored in two bioinformatics resources: Comparative Toxicogenomics Database and ConceptGen. According to Boulos et al,30 the goal of visual analytics is to facilitate the discourse between the user and the data by providing dynamic displays and versatile visual interaction opportunities with the data that can support analytical reasoning and the exploration of data from multiple user-customizable aspects. Interactive visual analytics approaches are increasingly required to assist human cognition and analytical reasoning to identify patterns in datasets of diverse sizes.6,30,31 The primary objectives of the reported research were to (1) compare the toxicogenomics relationships for genes in a molecular network involving two genes (IL1R2 and TNFSF18) that were upregulated in HaCaT keratinocytes exposed to chronic, low dose arsenic trioxide;18 and (2) identify arsenic-interacting genes enriched for the melanosome (melanin-pigment bearing organelles transferred from melanocytes to keratinocytes) biological information. These objectives are to help generate hypotheses for further research on the mechanisms of arsenic toxicity on skin keratinocytes, in particular, aberrant skin pigmentation and cancer initiation.
Our visual analytics protocols identified (1) five genes in the IL1R2 and TNFSF18 molecular network that were perturbed by an arsenical in at least one of the three keratinocyte cell lines (2) 106 genes were enriched for the skin pigmentation topics: melanocyte, melanogenesis, melanosome(s), melanocyte differentiation, melanosome membrane, and skin pigmentation and (3) nine genes were localized in the melanosome but also known to be perturbed by arsenic trioxide in other cell types. Molecular network construction on selected upregulated genes encoding membrane proteins revealed interconnections to E2F4, an oncogenic transcription factor, predominantly expressed at the onset of keratinocyte differentiation. Two melanosome localized proteins TFRC and RAC1 were also prioritized from the visual analytics process. The reported findings provide knowledge building insights on how arsenic could impair the function of genes and the biological pathways related to arsenic toxicity in keratinocytes and melanocytes.
Methods
Data sources for knowledge building insights on arsenic toxicity to keratinocytes and melanocytes
The data sources for gene lists used in the research were (1) molecular interaction of arsenic-regulated genes IL1R2 and TNFSF18, (2) Comparative Toxicogenomics Database,23 (3) ConceptGen database,24 (4) Human Genome Organization Nomenclature Committee Gene Names,32 (5) SNPs3D (molecular functional effects of non-synonymous SNPs based on structure and sequence analysis),33 and (6) Environmental Genome Sequencing Project.34 The retrieval and processing of the gene lists are described in the following sections.
Prediction of molecular network of genes for arsenic-regulated genes IL1R2 and TNFSF18
Genes for interleukin 1 receptor, type II (IL1R2) and tumor necrosis factor (ligand) superfamily, member 18 (TNFSF18) encode membrane proteins that form ligand-receptor systems. We therefore assumed that the predicted molecular interactions for IL1R2 will have connections to ligands IL1B and IL1A,35 while TNFSF18 will have connection to its receptor TNFRSF18.36 We used the Michigan Molecular Interactions (MiMI) plugin for Cytoscape37 to retrieve molecular interactions as well as interaction attributes in the Michigan Molecular Interactions resource for query genes IL1B, IL1R2, IL1A, TNFSF18, TNFRSF18, and their nearest neighbors. The Cytoscape platform was used to display the biological networks with the biological entities (genes/proteins) represented as nodes and the biological interactions represented as edges between nodes.38,39
Construction of arsenical-gene interactions datasets
Arsenicals are defined in the Medical Subject Headings database (http://www.ncbi.nlm.nih.gov/mesh) as inorganic or organic compounds that contain arsenic, and the term is assigned an accession number of D001152. The Batch Query tool (http://ctdbase.org/tools/batchQuery.go) of the Comparative Toxicogenomics Database was used to download the arsenical-gene interaction datasets for (1) all the genes curated in the Comparative Toxicogenomics Database having at least an interaction with arsenicals and (2) all the genes in the predicted molecular network involving IL1R2 and TNFSF18. In both cases, the fields that constitute each record were Input (Gene Symbol or NCBI Entrez Gene ID), GeneSymbol (Approved Gene Symbol), GeneName (Approved Gene Name), GeneID (Entrez Gene ID), ChemicalName (NCBI MeSH Chemical Name), ChemicalID (NCBI MeSH Chemical ID), CasRN (Chemical Abstracts Service Registry Number), Organism (Scientific Name of Organism), OrganismID (NCBI Taxonomy ID), Interaction (Curated Interaction), InteractionActions (Effect of chemical on gene), and PubMedIDs (PubMed Identifier). The CTD chemical-gene interaction data were determined from peer-reviewed articles published and indexed in PubMed (www.pubmed.gov).
For the molecular interaction network, each record of the dataset was further annotated with the cell or organ type, sequence type (mRNA or protein), chemical classification (arsenical or non-arsenical) and the in vivo or in vitro experimental systems associated with the entry. The additional annotations (data fields) were determined after reading the abstract and full text of the PubMed indexed article. The sequence type category was included to identify proteins that may bind to arsenic via vicinal cysteines.
Construction of dataset on enriched biological information for arsenic-interacting genes
Enriched biological topics for genes curated to interact with arsenicals were programmatically retrieved from the ConceptGen data repository. Each gene can be enriched in 14 concept types: Gene Ontology (GO) biological process; GO molecular function; GO cellular component; Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway; Biocarta pathway; Protein ANalysis THrough Evolutionary Relationships (Panther) Pathways; Protein Domains (Pfam); Medical Subject Headings (MeSH); Online Mendelian Inheritance in Man (OMIM); Drug Bank Target Sets; microRNA Predicted Tagets; Gene Expression Omnibus (GEO) datasets, Michigan Molecular Interactions (MiMI) and Protein Interactions; Metabolites; and Cytoband (chromosomal locations). The dataset constructed consists of Gene Symbol, ConceptID, Concept Name and Concept Type.
Dataset of human genes encoding enzymes
Since enzymes are components of biological pathways, we were interested in determining the subset of CTD arsenic-interacting genes that are enzymes. The complete Human Genome Nomenclature Committee (HGNC) gene names dataset consisting of approved symbols of over 33,000 loci were downloaded from www.genenames.org.
Dataset on prioritized environmental response human genes
The gene symbols for 647 prioritized environmental response human genes were obtained from the National Institute of Environmental Health Sciences’ Environmental Genome Project (NIEHS EGP) at http://egp.gs.washington.edu/. These genes are involved in DNA repair, cell cycle regulation, apoptosis, and metabolism and are suspected to have a function in the susceptibility to environmental exposures in a panel of 95 individuals representing the ethnic diversity found in the United States.34,40 The putative functional effects of non-synonymous coding single nucleotide polymorphisms (ncSNPS) on the prioritized genes were extracted from NIEHS EGP website using a set of computer scripts. Two programs SIFT (Sorting Intolerant from Tolerant) and Polyphen (polymorphism phenotyping) were used to classify the ncSNPs. The SIFT classifies ncSNPs into tolerant or intolerant, while Polyphen classifies ncSNPs to benign, possibly damaging, and probably damaging. We were interested in genes with ncSNPs that change cysteine residues in the protein sequence. These ncSNPs may help to identify proteins that have vicinal cysteines, which are possible targets for arsenic binding.
Dataset of arsenic-interacting genes with candidate SNPs affecting disulfide bonds
The supplementary table from our previous publication on candidate single nucleotide polymorphisms for arsenic responsiveness in proteins was retrieved from the PubMed Central website for the article (http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2964045/bin/BBI-4-supplementary.xls).14 The set of genes that have breakage of disulfide bond as annotation of the impact of SNP on protein stability was selected for further analysis.
Visual analytics of datasets (gene lists)
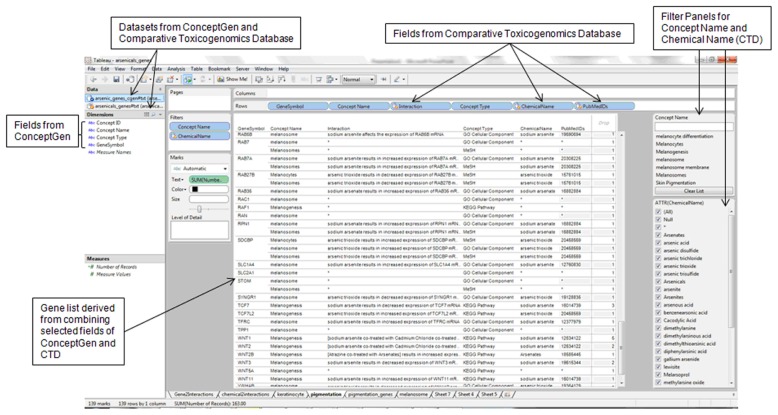
The GeneSymbol (Gene Symbol) field was present in all the datasets analyzed and, therefore, was used to link the datasets. A visual analytics software, Tableau Professional (http://tableausoftware.com/), was used to address the primary research objectives: (1) compare the toxicogenomics relationships for genes in a molecular network involving the two genes (IL1R2 and TNFSF18) that were upregulated in HaCaT keratinocytes exposed to chronic, low dose arsenic trioxide18 and (2) identify arsenic-interacting genes enriched for the melanosome topics (melanin-pigment bearing organelles transferred from melanocytes to keratinocytes). Additional objectives conducted to support the two primary objectives were to (1) identify the arsenic-interacting enzymes with melanosome localization that are perturbed by arsenic; (2) identify the environmental response human genes with potential vicinal cysteines for arsenic binding; and (3) design a visual analytic dashboard to enable integration and interactive exploration of data on effects of arsenic on genes with candidate single nucleotide polymorphisms affecting disulfide bonds. In each case, the data fields in the datasets were arranged on the visual analysis interface to enable interaction, integration, and presentation of the contents of the data fields. For example, in the integration of the CTD and Concept- Gen datasets (gene lists), the following data fields were designed to be listed in the following order: GeneSymbol (CTD), Concept Name (ConceptGen), Interaction (CTD), Concept Type (ConceptGen), ChemicalName (CTD), and PubMedIDs (CTD) (Fig. 1).
Figure 1.
Sample design for integration of gene information sources for knowledge building insights on biomarker genes of arsenic toxicity to keratinocytes and melanocytes.
Note: The design shows the integration of datasets on arsenic-interacting genes from the Comparative Toxicogenomics Database (http://ctdbase.org/) and enriched biological information from ConceptGen (http://conceptgen.ncibi.org/).
Results
Data sources for knowledge building insights
The data sources for developing knowledge building insights using a visual analytics approach are presented in Table 1. The starting data source was a list of 3,826 genes curated to interact with arsenicals in the Comparative Toxicogenomics Database. The ConceptGen data was available for 3,419 arsenic-interacting genes. Additionally, 647 environmental response genes from the Environmental Genome Project were retrieved as well as 79 genes with associated SNPs that target cysteine residues. Finally, 36,805 human gene names were obtained from the Human Genome Organization Nomenclature Committee (HGNC) website (www.genenames.org). Table 2 is a list of 14 genes that were found in all the data sources. We integrated the gene list with Gene Ontology (GO) Cellular Component annotation stored in ConceptGen to determine those found in skin cell types. The RAC1, Ras-related C3 botulinum toxin substrate 1, met our filtering criteria as encoding a protein localized to the melanosome. The RAC1-arsenic interactions and associated PubMed Identifiers of the source articles are presented in Table 3. Visual analytics workbook is available as a Supplemental File and can be viewed with the free Tableau Reader (http://www.tableausoftware.com/products/reader).
Table 1.
Data sources and data types for knowledge building on arsenic toxicity.
| Data sources and data types | Count |
|---|---|
| Comparative toxicogenomics database (http://ctdbase.org/) | |
| Arsenic interacting genes | 3,286 |
| Arsenicals | 29 |
| Arsenical-gene interactions | 7,724 |
| Arsenicals interaction actions | 207 |
| Organisms investigated for arsenical action | 20 |
| PubMed identifiers for articles on arsenic interacting genes | 591 |
| ConceptGen (http://conceptgen.ncibi.org/) | |
| Arsenic interacting genes in conceptGen | 3,419 |
| Concept names for arsenic interacting genes | 13,587 |
| Concept types for arsenic interacting genes: | |
| Biocarta pathway | 3,486 |
| Cytoband | 5,192 |
| Drug bank | 1,387 |
| Gene expression | 19,2432 |
| GO biological process | 70,403 |
| GO cellular component | 14,286 |
| GO molecular function | 25,266 |
| KEGG pathway | 6,198 |
| MeSH | 73,530 |
| Metabolite | 7,315 |
| miRBase | 12,8227 |
| OMIM | 252 |
| Panther pathway | 2,526 |
| pFAM | 6,083 |
| TransFac | 1,035 |
| Environmental genome project (http://egp.gs.washington.edu/) | 647 |
| Arsenical interacting genes with single nucleotide polymorphisms targeting cysteine residues (http://www.snps3D.org/) | 79 |
| Human gene names (http://www.genenames.org) | 36,805 |
Table 2.
Genes observed in all the five gene information sources for knowledge building insights on arsenic toxicity to keratinocytes and melanocytes.
| Gene symbol | Approved name | CTD | ConceptGen | EGP | HGNC | SNPs3D |
|---|---|---|---|---|---|---|
| ABCC1 | ATP-binding cassette, sub-family C (CFTR/MRP), member 1 | 28 | 309 | 1 | 1 | 13 |
| ATM | Ataxia telangiectasia mutated | 8 | 233 | 1 | 1 | 1 |
| BRCA1 | Breast cancer 1, early onset | 4 | 316 | 1 | 1 | 3 |
| BRCA2 | Breast cancer 2, early onset | 3 | 314 | 1 | 1 | 1 |
| CASP2 | Caspase 2, apoptosis-related cysteine peptidase | 4 | 72 | 1 | 1 | 1 |
| CDH1 | Cadherin 1, type 1, E-cadherin (epithelial) | 7 | 1267 | 1 | 1 | 1 |
| EEF1A1 | Eukaryotic translation elongation factor 1 alpha 1 | 3 | 140 | 1 | 1 | 1 |
| ERCC5 | Excision repair cross-complementing rodent repair deficiency, complementation group 5 | 3 | 119 | 1 | 1 | 1 |
| IGFBP7 | Insulin-like growth factor binding protein 7 | 2 | 106 | 1 | 1 | 1 |
| LTA | Lymphotoxin alpha (TNF superfamily, member 1) | 4 | 471 | 1 | 1 | 1 |
| MAPK9 | Mitogen-activated protein kinase 9 | 26 | 160 | 1 | 1 | 2 |
| MSH5 | Muts homolog 5 (E. coli) | 1 | 109 | 1 | 1 | 1 |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | 11 | 339 | 1 | 1 | 3 |
| XPC | Xeroderma pigmentosum, complementation group C | 4 | 120 | 1 | 1 | 1 |
Notes: CTD: Comparative Toxicogenomics Database (http://ctdbase.org/); ConceptGen: ConceptGen database at National Center for Integrative Biomedical Informatics (http://conceptgen.ncibi.org/); EGP: Environmental Genome Project Gene-Oriented Sequence Variation Discovery (http://egp. gs.washington.edu/); HGNC: Human Genome Organization Gene Nomenclature Committee (http://www.genenames.org/); SNPs3D: Molecular functional effects of non-synonymous SNPs based on structure and sequence analysis (http://www.snps3d.org/).
Table 3.
Interactions of arsenicals with RAC1 protein and mRNA.
| Interaction | PubMed IDs* |
|---|---|
| [Ethanol co-treated with sodium arsenite] results in increased localization of RAC1 protein | 18182400 |
| [Tetradecanoylphorbol acetate co-treated with arsenic trioxide] results in increased expression of RAC1 mRNA | 18922899 |
| [Tetradecanoylphorbol acetate co-treated with arsenic trioxide] results in increased expression of RAC1 protein | 18922899 |
| NSC 23766 inhibits the reaction [sodium arsenite results in increased activity of RAC1 protein] | 19349368 |
| Pertussis Toxin inhibits the reaction [sodium arsenite results in increased activity of RAC1 protein] | 19349368 |
| RAC1 protein affects the reaction [sodium arsenite results in increased expression of PECAM1 protein] | 19033667 |
| S1PR1 protein affects the reaction [sodium arsenite results in increased activity of RAC1 protein] | 19349368 |
| Sodium arsenate results in increased expression of RAC1 mRNA | 16882884 |
| Sodium arsenite results in increased activity of RAC1 protein | 19349368 |
| Sodium arsenite results in increased localization of and results in increased activity of RAC1 protein | 18182400 |
| Sodium arsenite results in increased localization of RAC1 protein | 18182400 |
Notes:
Abstract of article can be obtained from www.pubmed.gov. Additional information on arsenic-gene interactions can be obtained from http://ctdbase.org.
Genes in molecular network perturbed by arsenicals in keratinocyte cell lines
A 24-node molecular interaction network containing IL1R2 and TNFSF18 was predicted using the MiMI Cytoscape Plugin (Fig. 2). As expected, TNFSF18 interacted directly with its receptor, TNFRSF18. Further, IL1R2 interacted directly with its ligands, IL1A and IL1B. An oncogenic transcription factor, E2F4, served as connecting node for the subnetwork containing the interleukin genes and the subnetwork containing TNFSF18 and TNFRSF18. The molecular interaction map also predicted Necdin (NDN) as a connection between E2F4 and IL1A. TNF receptor associated factor 2 (TRAF2) was predicted as a connection between E2F4 and TNFSF18. The activating transcription factor-2 (ATF2), a sequence-specific DNA-binding protein, was predicted to interact with both E2F4 and IL1B.
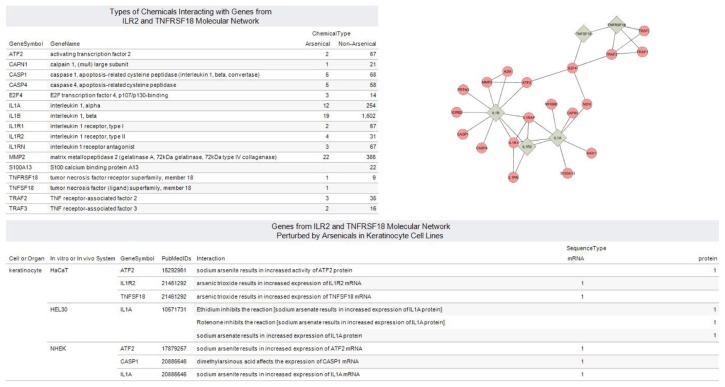
Figure 2.
Integrated view of chemical-gene interactions for molecular network with arsenic-upregulated genes TNFSF18 and IL1R2.
Notes: Interaction Map for IL1B, IL1R2, IL1A, TNFRSF18 and TNFSF18 reveals E2F4 Transcription Factor as link between subnetworks. E2F4 controls cell cycle and acts on tumor suppressor proteins. The expression of five genes ATF2, CASP1, IL1A, IL1R2 and TNFSF18 were shown to be perturbed by arsenicals in keratinocyte cell lines: immortalize human keratinocytes (HaCaT), murine keratinocyte cell line (HEL30) and normal human epidermal keratinocyte (NHEK). Visual Analytics resource for exploring and downloading views is available at http://public.tableausoftware.com/views/tnfsf18_il1r2_ctd/molnet_ctd.
The clustering coefficient for the network of 24 genes was 0.128, which is a factor of the connectivity of a node and the connectivity of the neighborhood to which this node is connected. The average number of neighbors was 2.833. Another parameter examined was the connection degree, which measures the number of partners directly connected to a particular node. The connection degrees for IL1A, IL1B, IL1R2, TNFRSF18 and TNFSF18 were 9, 7, 4, 4, and 3 respectively.
The dataset in the CTD for the predicted molecular network consisted of 2,792 chemical-gene interactions (as of August 10, 2011) for 16 of the 24 genes. Figure 2 is a dashboard view consisting of 4 views generated from the dataset. These interactions, which consist of 676 chemicals investigated in 22 organisms, were divided into arsenical and non-arsenical groups. A set of 16 interactions were not annotated with organism information. All genes except S100 A13 (S100 calcium binding protein A13) had at least a curated interaction with an arsenical. The expression of five genes ATF2, CASP1, IL1A, IL1R2, and TNFSF18 were shown to be perturbed by arsenicals in the following keratinocyte cell lines: immortalize human keratinocytes (HaCaT), murine keratinocyte cell line (HEL30), and normal human epidermal keratinocyte (NHEK). Based on the CTD data, the protein activity of ATF2 and IL1A genes were perturbed by arsenic in HaCaT and HEL30 respectively. In addition, we identified 9 genes (ATF2, CAPN1, CASP1, CASP4, E2F4, IL1A, IL1B, IL1R2, and MMP2) where their corresponding proteins have been studied for interaction with arsenic (Fig. 2).
Genes enriched for skin pigmentation topics
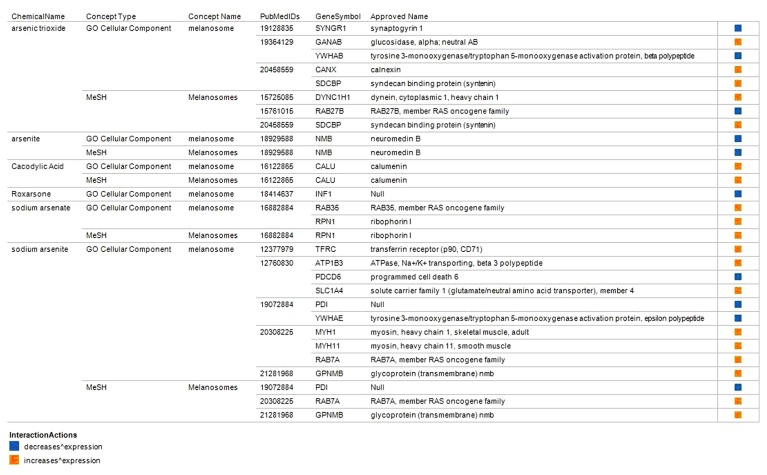
A total of 106 arsenic-interacting genes were enriched for the following skin pigmentation topics in Concept- Gen: melanocyte, melanogenesis, melanosome, melanosomes, melanocyte differentiation, melanosome membrane, and skin pigmentation. We also identified a subset of 22 genes annotated to encode products that are localized to the melanosome but their activity known to be perturbed by arsenic trioxide and other arsenicals in other cell types (Fig. 3). These gene-arsenic interactions were documented in 14 PubMed indexed articles (PubMed Identifiers shown in Fig. 3). We further prioritized these 22 genes based on the cell type used in the investigation with particular interest on skin cell types. The gene encoding an iron uptake protein, transferrin receptor (TFRC), was among the genes enriched for melanosome topic. The expression of TFRC was increased by sodium arsenite in normal human epidermal keratinocytes (NHEK).13
Figure 3.
Selected arsenic-interacting genes enriched for melanosome topics.
Notes: A total of 106 arsenic-interacting genes were enriched for the following skin pigmentation topics in ConceptGen. A subset of 22 genes is shown that were annotated to encode products that localize to the melanosome but their expression is known to be perturbed by arsenic trioxide and other arsenicals in other cell types. The changes in expression is limited to increase or decrease expression.
Arsenic-perturbed genes encoding enzymes localized in the melanosome
Integration of ConceptGen and HGNC datasets identified 8 enzymes localized in the melanosome and were also observed to be perturbed by arsenic in other cell types (Fig. 4). The enzymes were carbonic anhydrase II (CA2), cathepsin B (CTSB), cathepsin D (CTSD), glucosidase, alpha; neutral AB (GANAB), matrix metallopeptidase 1 (interstitial collagenase) (MMP1) and protein disulfide isomerase family A, member 4 (PDIA4). The gene lists integration revealed in arsenic trioxide and sodium arsenite had a decreased expression of MMP1 mRNA in leukemia and keratinocytes cell lines respectively.13,41 Further, the PDAI4 mRNA had an increased expression from sodium arsenite in a leukemia cell line but a decreased expression by tetraarsenic tetrasulfide in human vascular endothelial cells.42,43 Interaction of the two cathepsins (CTSB, CTSD) with arsenicals led to an increased protein or transcription levels.
Figure 4.
Enzymes localized in the melanosome and observed to be perturbed by arsenic in other cell types.
Notes: The enzymes lists were obtained by integrating datasets from ConceptGen and the Human Genome Organization (HUGO) Nomenclature Committee’s gene names website (http://www.genenames.org/). CA2: carbonic anhydrase II, CTSB: cathepsin B, CTSD: cathepsin D, GANAB: glucosidase, alpha; neutral AB, matrix; MMP1: metallopeptidase 1 (interstitial collagenase) and PDIA4: protein disulfide isomerase family A, member 4.
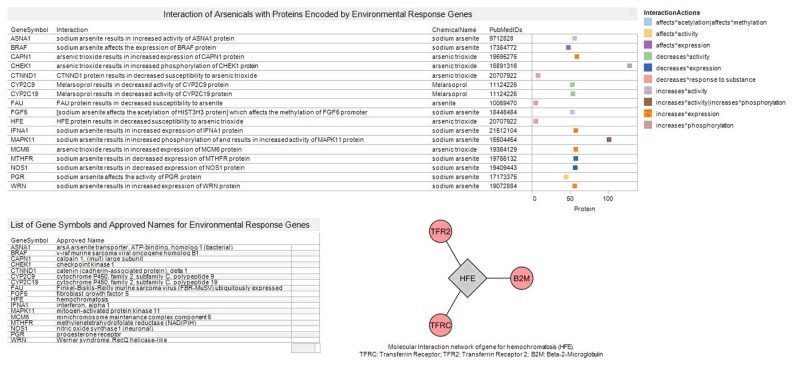
Environmental response genes with potential vicinal cysteines for arsenic binding
The visual analytics pipeline retrieved 17 genes encoding protein isoforms that could have vicinal cysteines liable to arsenic binding. The gene for hemachromatosis (iron overload) was present in this list of genes. Molecular interaction network construction using MiMI cytoscape plug-in revealed the HFE interacts with Transferrin Receptor (TFRC), Transferrin Receptor 2 (TFR2) and Beta-2-Microglobulin (B2M) (Fig. 5).
Figure 5.
Integration of gene information sources for selected environmental response genes that are potential arsenic-binding proteins.
Notes: The list of arsenic response genes was obtained from the Environmental Genome Project (http://egp.gs.washington.edu/) and the approved names from the Human Genome Organization Nomenclature Committee (HGNC) website (www.genenames.org). Arsenic-protein interaction relationships were obtained from Comparative Toxicogenomics (CTD http://ctdbase.org/). Molecular interaction of HFE gene was obtained from Michigan Molecular Interaction (MiMI). The color-coded boxes indicate the type of interaction with arsenic. The position of each box corresponds to the position of the word “protein” in the interaction text from CTD.
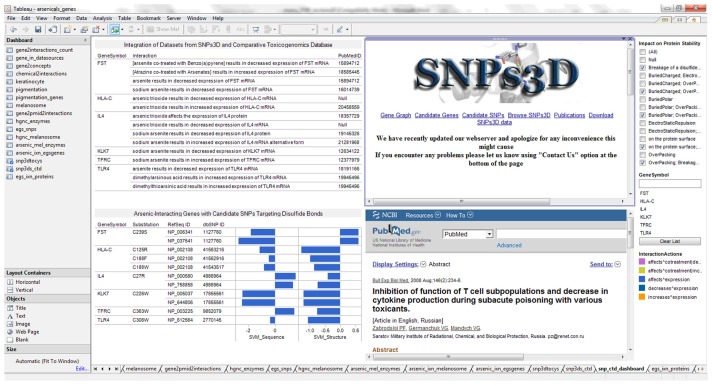
Arsenic-gene interaction for skin keratinocyte relevant genes with candidate SNPs targeting disulfide bonds
A total of six genes (FST, HLA-C, IL4, KLK7, TFRC, and TLR4) were identified as having single nucleotide polymorphisms that cause breakage of disulfide bond. The visual analytics dashboard was designed to facilitate knowledge building insights on arsenic toxicity (Fig. 6). The dashboard integrates data streams from the Comparative Toxicogenomics Databases (genechemical interactions and curated literature), SNPs3D (effects of SNPs on sequence and structure of proteins predicted by Support Vector Machines [SVM]). Further, web pages for SNPs3D and PubMed were integrated to enable online searching for additional information for knowledge building. Interestingly, the transferrin receptor (TFRC) was also identified as having vicinal cysteines for arsenic sensing based on SNP targeting.
Figure 6.
Visual analytics resource for exploring arsenic-interacting genes with candidate single nucleotide polymorphisms affecting disulfide bonds.
Notes: The dashboard view enables user to search SNPs3D (www.snps3d.org) and PubMed (www.pubmed.gov) for genes with SNP associated breakage of disulfide bonds. Additional impacts on protein stability and genes available in as supplementary file of previous publication: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2964045/bin/BBI-4-supplementary.xls.
Discussion
We report, for the first time, a unique visual analytics approach to integrate and interactively explore the gene information sources to determine knowledge building insights on arsenic-induced hyperpigmentation and skin cancer. The overall purpose of the reported research was to determine knowledge building insights on biomarker genes for arsenic toxicity to human epidermal cells by integrating a collection of gene lists annotated with biological information. The information sets included toxicogenomics gene-chemical interaction, enzymes encoded in the human genome, enriched biological information associated with genes, environmentally relevant sequence variation, and effects of non-synonymous Single Nucleotide Polymorphisms on protein function.
Two datasets on arsenic-gene interactions form the basis of the reported research. First, we constructed a dataset of chemical-gene interactions for a network of 16 genes that included two genes upregulated in HaCaT keratinocytes exposed to chronic (22 passages), low-dose (0.5 mg/L) arsenic trioxide.18 Additionally, we enriched the dataset with data on cell/organ type and in vitro/in vivo experimental system for each chemical-gene interaction (Fig. 2). Second, the arsenic-gene interaction dataset of 3,826 genes curated for interaction with arsenic was integrated with other datasets for knowledge building insights on arsenic-gene interaction in the melanosome. The human epidermal melanosomes are pigment (melanin)-containing large elliptical or spheroidal organelles (approximately 500 nm in diameter) formed in the melanocytes, donated to neighboring keratinocytes by passing through dendrites of melanocytes.44–46 Taken together, the two starting datasets, the additional annotations, the data sources, and the visual analytics approach enabled a set of research objectives to be accomplished.
We have previously reported that the genes for cytokines TNFSF18 and IL1R2 are perturbed when human skin keratinocytes (HaCaT) was chronically exposed to low dose of arsenic trioxide.18 TNFSF18 is a ligand for receptor TNFRSF18 and it modulates T-lymphocyte survival in peripheral tissues playing a vital role in resistance to infection and cancers. TNFSF18 is found in extracellular space and is integral to membranes47 and belongs to the TNF ligand superfamily that contains uniform structural motif and the TNF homology domain (THD), which binds to cysteine-rich domains (CRDs) of TNF receptors.48 TNF can exert many of its effects by binding to cell membrane receptors. The members of the TNF receptor superfamily possess an identical characteristic of an extra cellular domain containing two to six repeats of cysteine rich motifs.49 The sequence homology between the CRDs and the DNA-binding “zinc-fingers” may be used to speculate intracellular protein phosphorylation by protein kinase C (PKC).50 IL1R2, also referred to as IL1RB, encodes membrane bound proteins known to be crucially involved in immune response and also functions as a decoy receptor for inflammatory interleukin 1 (IL-1).51 Over-expression of IL1R2 has been reported in human uroepithelial cell line (HUC-1) chronically exposed to arsenite, and our results concur with this observation using HaCaT cell line.52 The cell membrane modulates protein function through localization with the substrate, activator, or downstream target and activation of the protein by a conformational switch.53 Membrane localized proteins could bind with arsenic at the cell surface and lead to subsequent changes in cellular biological pathways.
The molecular interaction map for IL1B, IL1R2, IL1A, TNFRSF18, and TNFSF18 revealed the E2F4 transcription factor as link (hub) between subnetworks. E2F4 controls cell cycle and function in the suppression of proliferation-associated genes, and its gene mutation and increased expression may be associated with human cancer.54 E2F4 is predominantly expressed at the onset of keratinocyte differentiation. 55 Further, E2F4 is a promoter of proliferation of human intestinal epithelial crypt cells and colorectal cancer cells.56 The known predominant expression of E2F4 in the onset of keratinocyte differentiation raises the possibility that arsenic perturbation of protein- protein interactions that include E2F4 could alter keratinocyte differentiation.57
From the dataset of 3,829 arsenic-interacting genes from CTD, 106 genes were identified to be enriched for skin pigmentation topics in the ConceptGen enriched biological information (Fig. 3 and Supplementary File 2). Based on our interest in understanding abnormal pigmentation observed in arsenic toxicity, we prioritized two genes, RAC1, a small guanosine triphosphate (GTP)-binding protein, and transferrin receptor (TFRC), an iron uptake receptor, for further discussion Isokpehi et al in this article. According to the Gene Ontology (GO) Cellular Component annotation, RAC1 and TFRC are localized to the melanosome.24 RAC1 was observed in all the data sources (Fig. 2), while TFRC had been previously identified as possessing vicinal cysteines that are potential binding sites for arsenic.14
The curated molecular interactions in Comparative Toxicogenomics Database indicate that arsenicals increase RAC1 activity, localization, or expression (Table 2).
RAC1 is involved in the signaling process, and actin polymerization is required for the formation of dendrites, which transport the melanosomes from the melanocytic cells to approximately 36 neighboring keratinocytes.58–60 Specifically, RAC1 mediates formation of lamellipodia (protrusions at the edge of the cell important for cell migration) and melanocyte dendricity.61,62 The transfer of melanosomes to keratinocytes is a mechanism for photoprotection of the skin from ultraviolet (UV) radiation and, thus, the prevention of skin cancer.63 Melanocyte dendricity is critical for skin pigmentation.64 Thus arsenic-induced “increase in localization or activity of the RAC1 protein” could result in an increase in melanocyte dendricity leading to hyperpigmentation.
The visual analytics–facilitated integration of multiple gene information sources revealed that the transferrin receptor gene (TFRC) was annotated in the ConceptGen database to be enriched for the melanosome topic. Further, the Comparative Toxicogenomics Database records that in the normal human epidermal keratinocytes (NHEK), sodium arsenite resulted in an increased expression of TFRC mRNA13 (Fig. 3). We previously showed that TFRC contain vicinal cysteines using inferences from SNP-induced effects of breakage of disulfide bond.13 Vicinal cysteines in proteins are potential locations for arsenic binding that can affect protein function.14,65,66 Further, TFRC is a key determinant of the amount and location of iron in the epidermis, and there is a correlation between the expression of TFRC and iron uptake.67 While 20% to 25% of the absorbed iron is normally eliminated from the body by epidermal desquamation,68 an aberrant TFRC expression could impair cutaneous iron metabolism leading to the abnormal pigmentation seen in some humans exposed to arsenicals. The environmentally relevant HFE protein associated with hemochromatosis (characterized by excessive dietary iron) resulted in a decreased susceptibility to arsenic trioxide in tumor cell lines. HFE competes with transferrin for binding to the TFRC, which inhibits cellular uptake of transferrin (Fig. 5).69 Reduced iron import (low TFRC and high HFE) was a signature for favorable prognosis (P < 0.005) in breast cancer.70 Changes in iron content of keratinocytes could also be relevant in the susceptibility to arsenic-induced skin cancer.
Conclusion
Exposure to inorganic arsenic induces skin cancer and abnormal pigmentation in susceptible humans. High-throughput gene transcription assays such as DNA microarrays allow for the identification of biological pathways affected by arsenic. Investigation of arsenic-induced aberrations in gene expression can help predict initiation and progression of skin cancer and abnormal pigmentation. Molecular network construction for the arsenic upregulated genes TNFSF18 and IL1R2 revealed subnetwork interconnections to E2F4, an oncogenic transcription factor, predominantly expressed at the onset of keratinocyte differentiation. Visual analytics integration of gene information sources helped identify RAC1, a GTP binding protein, and TFRC, an iron uptake protein as prioritized arsenic-perturbed protein targets for biological processes leading to skin hyperpigmentation. RAC1 regulates the formation of dendrites that transfer melanin from the melanocytes to neighboring keratinocytes. An increased melanocyte dendricity is correlated with hyperpigmentation. TFRC is a key determinant of the amount and location of iron in the epidermis. Aberrant TFRC expression could impair cutaneous iron metabolism leading to abnormal pigmentation seen in some humans exposed to arsenicals. The reported findings contribute to new hypotheses and insights on how arsenic could impair the function of genes and biological pathways related to arsenic toxicity in keratinocytes and melanocytes. Finally, we developed visual analytics resources to facilitate further exploration of the information and the discovery of previously unknown relationships from datasets.
Acknowledgements
Dr. Steve F. Jennings of the University of Arkansas at Little Rock for facilitating the collaboration between Jackson State University and the University of Arkansas at Little Rock. National Institutes of Health: Research Centers in Minority Institutions (RCMI)—Center for Environmental Health at Jackson State University (NIH-NCRR 2G12RR013459); Mississippi IDeA Network for Biomedical Research Excellence (NIH-NCRR-P20RR016476 and NIH-NIGMS- 8P20GM103476); Arkansas IDeA Network for Biomedical Research Excellence (NIH-NCRRP20RR016460); Bioinformatics Programs in Minority Institutions (1T36GM095335); National Center for Integrative Biomedical Informatics (U54DA021519). National Science Foundation: Mississippi NSF-EPS-CoR Grant Awards (EPS-0903787); Undergraduate Research and Mentoring Program (DBI-0958179) and Visual Analytics in Biology Curriculum Network (DBI-1062057). U.S. Department of Homeland Security Science & Technology Directorate (2009-ST-062-000014; 2011-ST-062-000048); Disclaimer: The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the funding agencies.
Footnotes
Author Contributions
Conceived and designed the experiments: RDI, UKU, MNA, ANM, ORA. Analysed the data: RDI, UKU, MNA, ANM, MOJ, KF, MAB, RAH. Wrote the first draft of the manuscript: RDI, UKU, MAN, ANM. Contributed to the writing of the manuscript: RDI, UKU, MNA, ANM, MOJ, KF, ORA. Agree with manuscript results and conclusions: RDI, UKU, MNA, ANM, MOJ, KF, MAB, RAH, ORA. Made critical revisions and approved final version: RDI, UKU, MNA, ANM, MOJ, KF, MAB, RAH, ORA. All authors reviewed and approved of the final manuscript.
Competing Interests
Authors disclose no potential conflicts of interest.
Disclosures and Ethics
As a requirement of publication author(s) have provided to the publisher signed confirmation of compliance with legal and ethical obligations including but not limited to the following: authorship and contributorship, conflicts of interest, privacy and confidentiality and (where applicable) protection of human and animal research subjects. The authors have read and confirmed their agreement with the ICMJE authorship and conflict of interest criteria. The authors have also confirmed that this article is unique and not under consideration or published in any other publication, and that they have permission from rights holders to reproduce any copyrighted material. Any disclosures are made in this section. The external blind peer reviewers report no conflicts of interest.
References
- 1.Hall M, Chen Y, Ahsan H, et al. Blood arsenic as a biomarker of arsenic exposure: results from a prospective study. Toxicology. 2006;225:225–33. doi: 10.1016/j.tox.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 2.Tchounwou PB, Patlolla AK, Centeno JA. Carcinogenic and systemic health effects associated with arsenic exposure—a critical review. Toxicol Pathol. 2003;31:575–88. doi: 10.1080/01926230390242007. [DOI] [PubMed] [Google Scholar]
- 3.Garelick H, Jones H, Dybowska A, Valsami-Jones E. Arsenic pollution sources. Rev Environ Contam Toxicol. 2008;197:17–60. doi: 10.1007/978-0-387-79284-2_2. [DOI] [PubMed] [Google Scholar]
- 4.Jackson BP, Taylor VF, Punshon T, Cottingham KL. Arsenic concentration and speciation in infant formulas and first foods. Pure Appl Chem. 2012;84:215–23. doi: 10.1351/PAC-CON-11-09-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson BP, Taylor VF, Karagas MR, Punshon T, Cottingham KL. Arsenic, organic foods, and brown rice syrup. Environ Health Perspect. 2012;120:623–6. doi: 10.1289/ehp.1104619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson MO, Cohly HH, Isokpehi RD, Awofolu OR. The case for visual analytics of arsenic concentrations in foods. Int J Environ Res Public Health. 2010;7:1970–83. doi: 10.3390/ijerph7051970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pimparkar BD, Bhave A. Arsenicosis: review of recent advances. J Assoc Physicians India. 2010;58:617–24. 629. [PubMed] [Google Scholar]
- 8.Yu HS, Chen GS, Sheu HM, et al. Alterations of skin-associated lymphoid tissue in the carcinogenesis of arsenical skin cancer. Proc Natl Sci Counc Repub China B. 1992;16:17–22. [PubMed] [Google Scholar]
- 9.Ho SY, Tsai YC, Lee MC, Guo HR. Merkel cell carcinoma in patients with long-term ingestion of arsenic. J Occup Health. 2005;47:188–92. doi: 10.1539/joh.47.188. [DOI] [PubMed] [Google Scholar]
- 10.Maloney ME. Arsenic in dermatology. Dermatol Surg. 1996;22:301–4. doi: 10.1111/j.1524-4725.1996.tb00322.x. [DOI] [PubMed] [Google Scholar]
- 11.Bae DS, Hanneman WH, Yang RS, Campain JA. Characterization of gene expression changes associated with MNNG, arsenic, or metal mixture treatment in human keratinocytes: application of cDNA microarray technology. Environ Health Perspect. 2002;110(Suppl 6):931–41. doi: 10.1289/ehp.02110s6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bae DS, Handa RJ, Yang RS, Campain JA. Gene expression patterns as potential molecular biomarkers for malignant transformation in human keratinocytes treated with MNNG, arsenic, or a metal mixture. Toxicoll Sci. 2003;74:32–42. doi: 10.1093/toxsci/kfg124. [DOI] [PubMed] [Google Scholar]
- 13.Hamadeh HK, Trouba KJ, Amin RP, Afshari CA, Germolec D. Coordination of altered DNA repair and damage pathways in arsenite-exposed keratinocytes. Toxicol Sci. 2002;69:306–16. doi: 10.1093/toxsci/69.2.306. [DOI] [PubMed] [Google Scholar]
- 14.Isokpehi RD, Cohly HH, Anyanwu MN, et al. Candidate single nucleotide polymorphism markers for arsenic responsiveness of protein targets. Bioinform Biol Insights. 2010;4:99–111. doi: 10.4137/BBI.S5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rea MA, Gregg JP, Qin Q, Phillips MA, Rice RH. Global alteration of gene expression in human keratinocytes by inorganic arsenic. Carcinogenesis. 2003;24:747–56. doi: 10.1093/carcin/bgg010. [DOI] [PubMed] [Google Scholar]
- 16.Kundu M, Ghosh P, Mitra S, et al. Precancerous and non-cancer disease endpoints of chronic arsenic exposure: the level of chromosomal damage and XRCC3 T241M polymorphism. Mutat Res. 2011;706:7–12. doi: 10.1016/j.mrfmmm.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das S, Pan D, Bera AK, et al. Sodium arsenite mediated immuno-disruption through alteration of transcription profile of cytokines in chicken splenocytes under in vitro system. Mol Biol Rep. 2011;38:171–6. doi: 10.1007/s11033-010-0091-5. [DOI] [PubMed] [Google Scholar]
- 18.Udensi UK, Cohly HH, Graham-Evans BE, et al. aberrantly expressed genes in HaCaT keratinocytes chronically exposed to arsenic trioxide. Biomark Insights. 2011;6:7–16. doi: 10.4137/BMI.S6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raghu KG, Cherian OL. Characterization of cytotoxicity induced by arsenic trioxide (a potent anti-APL drug) in rat cardiac myocytes. J Trace Elem Med Biol. 2009;23:61–8. doi: 10.1016/j.jtemb.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 20.Chowdhury R, Chatterjee R, Giri AK, Mandal C, Chaudhuri K. Arsenic-induced cell proliferation is associated with enhanced ROS generation, Erk signaling and CyclinA expression. Toxicology Lett. 2010;198:263–71. doi: 10.1016/j.toxlet.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 21.Ren X, McHale CM, Skibola CF, et al. An emerging role for epigenetic dys-regulation in arsenic toxicity and carcinogenesis. Environ Health Perspect. 2011;119:11–9. doi: 10.1289/ehp.1002114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Afshari CA, Hamadeh HK, Bushel PR. The evolution of bioinformatics in toxicology: advancing toxicogenomics. Toxicol Sci. 2011;120(Suppl 1):S225–37. doi: 10.1093/toxsci/kfq373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis AP, King BL, Mockus S, et al. The Comparative toxicogenomics database: update 2011. Nucleic Acids Res. 2011;39:D1067–72. doi: 10.1093/nar/gkq813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sartor MA, Mahavisno V, Keshamouni VG, et al. ConceptGen: a gene set enrichment and gene set relation mapping tool. Bioinformatics. 2010;26:456–63. doi: 10.1093/bioinformatics/btp683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chautard E, Fatoux-Ardore M, Ballut L, Thierry-Mieg N, Ricard-Blum S. MatrixDB, the extracellular matrix interaction database. Nucleic Acids Res. 2011;39:D235–40. doi: 10.1093/nar/gkq830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Croft D, O’Kelly G, Wu G, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39:D691–7. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stark C, Breitkreutz BJ, Reguly T, et al. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–9. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tarcea VG, Weymouth T, Ade A, et al. Michigan molecular interactions r2: from interacting proteins to pathways. Nucleic Acids Res. 2009;37:D642–6. doi: 10.1093/nar/gkn722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turner B, Razick S, Turinsky AL, et al. iRefWeb: interactive analysis of consolidated protein interaction data and their supporting evidence. Database (Oxford) 2010;2010:baq023. doi: 10.1093/database/baq023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamel Boulos MN, Viangteeravat T, Anyanwu MN, Ra NV, Kuscu E. Web GIS in practice IX: a demonstration of geospatial visual analytics using Microsoft Live Labs Pivot technology and WHO mortality data. Int J Health Georgrs. 2011;10:19. doi: 10.1186/1476-072X-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gribov A, Sill M, Luck S, et al. SEURAT: visual analytics for the integrated analysis of microarray data. BMC Med Genomics. 2010;3:21. doi: 10.1186/1755-8794-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Povey S, Lovering R, Bruford E, et al. The HUGO Gene Nomenclature Committee (HGNC) Hum Genet. 2001;109:678–80. doi: 10.1007/s00439-001-0615-0. [DOI] [PubMed] [Google Scholar]
- 33.Yue P, Melamud E, Moult J. SNPs3D: candidate gene and SNP selection for association studies. BMC Bioinformatics. 2006;7:166. doi: 10.1186/1471-2105-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rieder MJ, Livingston RJ, Stanaway IB, Nickerson DA. The environmental genome project: reference polymorphisms for drug metabolism genes and genome-wide association studies. Drug Metab Rev. 2008;40:241–61. doi: 10.1080/03602530801952880. [DOI] [PubMed] [Google Scholar]
- 35.Mantovani A, Muzio M, Ghezzi P, Colotta C, Introna M. Regulation of inhibitory pathways of the interleukin-1 system. Ann N Y Acad Sci. 1998;840:338–51. doi: 10.1111/j.1749-6632.1998.tb09573.x. [DOI] [PubMed] [Google Scholar]
- 36.Krausz LT, Bianchini R, Ronchetti S, et al. GITR-GITRL system, a novel player in shock and inflammation. Scientific World Journal. 2007;7:533–66. doi: 10.1100/tsw.2007.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao J, Ade AS, Tarcea VG, et al. Integrating and annotating the interactome using the MiMI plugin for cytoscape. Bioinformatics. 2009;25:137–8. doi: 10.1093/bioinformatics/btn501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cline MS, Smoot M, Cerami E, et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2:2366–82. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–2. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guengerich FP. The Environmental Genome Project: functional analysis of polymorphisms. Environ Health Perspect. 1998;106:365–8. doi: 10.1289/ehp.98106365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chou WC, Chen HY, Yu SL, et al. Arsenic suppresses gene expression in promyelocytic leukemia cells partly through Sp1 oxidation. Blood. 2005;106:304–10. doi: 10.1182/blood-2005-01-0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujiwara Y, Honda A, Satoh M. DNA microarray gene expression analysis of human vascular endothelial cells exposed to arsenite. J Toxicol Sci. 2010;35:275–8. doi: 10.2131/jts.35.275. [DOI] [PubMed] [Google Scholar]
- 43.Zhang QY, Mao JH, Liu P, et al. A systems biology understanding of the synergistic effects of arsenic sulfide and Imatinib in BCR/ABL-associated leukemia. Proc Natl Acad of Sci U S A. 2009;106:3378–83. doi: 10.1073/pnas.0813142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kondo T, Hearing VJ. Update on the regulation of mammalian melanocyte function and skin pigmentation. Expert Rev Dermatol. 2011;6:97–108. doi: 10.1586/edm.10.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wasmeier C, Hume AN, Bolasco G, Seabra MC. Melanosomes at a glance. J Cell Sci. 2008;121:3995–9. doi: 10.1242/jcs.040667. [DOI] [PubMed] [Google Scholar]
- 46.Scott G, Leopardi S, Printup S, Madden BC. Filopodia are conduits for melanosome transfer to keratinocytes. J Cell Sci. 2002;115:1441–51. doi: 10.1242/jcs.115.7.1441. [DOI] [PubMed] [Google Scholar]
- 47.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–5. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bodmer JL, Schneider P, Tschopp J. The molecular architecture of the TNF superfamily. Trends Biochem Sci. 2002;27:19–26. doi: 10.1016/s0968-0004(01)01995-8. [DOI] [PubMed] [Google Scholar]
- 49.Idriss HT, Naismith JH. TNF alpha and the TNF receptor superfamily: structure-function relationship(s) Microsc Res Tech. 2000;50:184–95. doi: 10.1002/1097-0029(20000801)50:3<184::AID-JEMT2>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 50.Hommel U, Zurini M, Luyten M. Solution structure of a cysteine rich domain of rat protein kinase C. Nat Struct Biol. 1994;1:383–7. doi: 10.1038/nsb0694-383. [DOI] [PubMed] [Google Scholar]
- 51.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–5. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang SY, Su PF, Lee TC. Ectopic expression of interleukin-1 receptor type II enhances cell migration through activation of the pre-interleukin 1alpha pathway. Cytokine. 2009;45:32–8. doi: 10.1016/j.cyto.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 53.Johnson JE, Cornell RB. Amphitropic proteins: regulation by reversible membrane interactions (review) Mol Membr Biol. 1999;16:217–35. doi: 10.1080/096876899294544. [DOI] [PubMed] [Google Scholar]
- 54.Souza RF, Yin J, Smolinski KN, et al. Frequent mutation of the E2F-4 cell cycle gene in primary human gastrointestinal tumors. Cancer Res. 1997;57:2350–3. [PubMed] [Google Scholar]
- 55.Paramio JM, Segrelles C, Casanova ML, Jorcano JL. Opposite functions for E2F1 and E2F4 in human epidermal keratinocyte differentiation. J Biol Chem. 2000;275:41219–26. doi: 10.1074/jbc.M004973200. [DOI] [PubMed] [Google Scholar]
- 56.Garneau H, Paquin MC, Carrier JC, Rivard N. E2F4 expression is required for cell cycle progression of normal intestinal crypt cells and colorectal cancer cells. J Cell Physiol. 2009;221:350–8. doi: 10.1002/jcp.21859. [DOI] [PubMed] [Google Scholar]
- 57.Paramio JM, Lain S, Segrelles C, Lane EB, Jorcano JL. Differential expression and functionally co-operative roles for the retinoblastoma family of proteins in epidermal differentiation. Oncogene. 1998;17:949–57. doi: 10.1038/sj.onc.1202031. [DOI] [PubMed] [Google Scholar]
- 58.Kim MY, Choi TY, Kim JH, et al. MKK6 increases the melanocyte dendricity through the regulation of Rho family GTPases. J Dermatol Sci. 2010;60:114–9. doi: 10.1016/j.jdermsci.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 59.Carlson JA, Linette GP, Aplin A, Ng B, Slominski A. Melanocyte receptors: clinical implications and therapeutic relevance. Dermatol Clin. 2007;25:541–5ix. doi: 10.1016/j.det.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scott GA, Cassidy L. Rac1 mediates dendrite formation in response to melanocyte stimulating hormone and ultraviolet light in a murine melanoma model. J Invest Dermatol. 1998;111:243–50. doi: 10.1046/j.1523-1747.1998.00276.x. [DOI] [PubMed] [Google Scholar]
- 61.Jung E, Hwang W, Kim S, et al. Depigmenting action of platycodin D depends on the cAMP/Rho-dependent signalling pathway. Exp Dermatol. 2011;20:986–91. doi: 10.1111/j.1600-0625.2011.01379.x. [DOI] [PubMed] [Google Scholar]
- 62.Koivisto L, Hakkinen L, Matsumoto K, et al. Glycogen synthase kinase-3 regulates cytoskeleton and translocation of Rac1 in long cellular extensions of human keratinocytes. Exp Cell Res. 2004;293:68–80. doi: 10.1016/j.yexcr.2003.09.026. [DOI] [PubMed] [Google Scholar]
- 63.Scott GA, Arioka M, Jacobs SE. Lysophosphatidylcholine mediates melanocyte dendricity through PKC zeta activation. J Invest Dermatol. 2007;127:668–75. doi: 10.1038/sj.jid.5700567. [DOI] [PubMed] [Google Scholar]
- 64.Costin GE, Hearing VJ. Human skin pigmentation: melanocytes modulate skin color in response to stress. FASEB J. 2007;21:976–94. doi: 10.1096/fj.06-6649rev. [DOI] [PubMed] [Google Scholar]
- 65.Zhang X, Yang F, Shim JY, et al. Identification of arsenic-binding proteins in human breast cancer cells. Cancer Lett. 2007;255:95–106. doi: 10.1016/j.canlet.2007.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li YM, Broome JD. Arsenic targets tubulins to induce apoptosis in myeloid leukemia cells. Cancer Res. 1999;59:776–80. [PubMed] [Google Scholar]
- 67.Milstone LM, Adams BD, Zhou J, Bruegel Sanchez VL, Shofner J. Stratum-specific expression of human transferrin receptor increases iron in mouse epidermis. J Invest Dermatol. 2006;126:648–52. doi: 10.1038/sj.jid.5700127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jacob RA, Sandstead HH, Munoz JM, Klevay LM, Milne DB. Whole body surface loss of trace metals in normal males. Am J Clin Nutr. 1981;34:1379–83. doi: 10.1093/ajcn/34.7.1379. [DOI] [PubMed] [Google Scholar]
- 69.Pantopoulos K. Function of the hemochromatosis protein HFE: Lessons from animal models. World J Gastroenterol. 2008;14:6893–901. doi: 10.3748/wjg.14.6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miller LD, Coffman LG, Chou JW, et al. An iron regulatory gene signature predicts outcome in breast cancer. Cancer Res. 2011;71:6728–37. doi: 10.1158/0008-5472.CAN-11-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]