Abstract
Background
Role of KRAS, BRAF and PIK3CA mutations in pathogenesis of colorectal cancer (CRC) has been recently investigated worldwide. In this population-based study, we evaluated the incidence rates and distribution of such somatic mutations in genetically isolated population from Sardinia.
Methods
From April 2009 to July 2011, formalin-fixed paraffin-embedded tissues (N = 478) were prospectively collected from Sardinian CRC patients at clinics across the entire island. Genomic DNA was isolated from tissue sections and screened for mutations in KRAS, BRAF, and PIK3CA genes by automated DNA sequencing.
Results
Overall, KRAS tumour mutation rate was 30% (145/478 positive cases). Distribution of mutation carriers was surprisingly different within the island: 87/204 (43%) in North Sardinia vs. 58/274 (21%) in Middle-South Sardinia (p<0.001). Among 384 CRC cases whose DNA was available, only one (0.3%) patient carried a mutation in BRAF gene; PIK3CA was found mutated in 67 (17%) patients. A significant inverse distribution of PIK3CA mutation rates was observed within Sardinian population: 19/183 (10%) cases from northern vs. 48/201 (24%) cases from central-southern island (p<0.001). This heterogeneity in frequencies of KRAS/PIK3CA somatic mutations is consistent with already-reported discrepancies in distribution of germline mutations for other malignancies within Sardinian population. Preliminary clinical evaluation of 118 KRAS wild-type patients undergoing anti-EGFR-based treatment indicated lack of role for PIK3CA in predicting response to therapy.
Conclusions
Our findings support the hypothesis that differences in patients’ origins and related genetic backgrounds may contribute to even determine the incidence rate of somatic mutations in candidate cancer genes.
Keywords: Colorectal carcinoma, KRAS gene, BRAF gene, PIK3CA gene, Mutation analysis, Cancer genetic heterogeneity
Introduction
Colorectal carcinoma (CRC) develops through different pathways, all involving changes at the chromosomal or gene levels. It is now widely accepted that sporadic colorectal cancers frequently arise from preneoplastic lesions through the activation of oncogenes (KRAS and BRAF) as well as the inactivation of tumour suppressor genes (APC, p16, p53, and DCC) and mismatch repair genes, such as MLH1 and MSH2 and, to a lower extent, PMS2 and hMSH6[1]. In addition, activating mutations in BRAF, a member of the RAF gene family, which encode kinases that are regulated by members of the RAS protein family (HRAS, KRAS, and NRAS) and mediate cellular responses to growth signals, were found to be associated with microsatellite instability (MSI) cancers [2,3]. As stated above, KRAS is the member of the RAS gene family mostly mutated in CRC; unlike the BRAF mutations, the KRAS mutations have been found to be equally distributed in all tumours, regardless of their MSI status [4]. These findings acquire an important role from the pathogenetic point of view since the mutations that hit these two genes are reciprocally exclusive. On such a basis, the occurrence of an activating mutation at either one of the two genes may be linked to different molecular processes and, therefore, may generate at least three distinct tumour subtype: BRAFmut/MSI+, KRASmut/MSI+, and KRASmut/MSI- [2-4].
Mutation of KRAS is an established predictor of absence of response to epidermal growth factor receptor (EGFR)-targeted agents [5]. The utility of KRAS as a prognostic marker remains uncertain. A recent meta-analysis reported that KRAS mutation may act as a negative prognostic indicator in both a trans-stage and stage-specific setting [6], whereas other studies, such as the large PETACC-3 translational trial [7], reported the absence of any prognostic value. On this regard, survival from recurrence was markedly worse in BRAF-mutant tumours into the PETACC-3 trial and this observation was consistent with the previously reported poor prognosis of BRAF mutations in advanced (stage IV) CRCs [8]. Unlike KRAS mutations, BRAF mutations might not be predictive of lack of anti-EGFR therapy benefit [3]. Among these genes, nearly all mutations affect the kinase domains at codons 12 or 13 of KRAS and codon 600 of BRAF[3,9].
In addition to the RAS/RAF/MEK pathway, the phosphatidylinositol 3-kinase (PI3K)/AKT/mTOR signaling cascade does participate in regulating cell proliferation and survival, apoptosis, and migration [10]. Activation of the PI3K/AKT/mTOR pathway is frequently mediated by mutations in the p110α subunit of PI3K, PIK3CA, with most mutations (>80%) occurring either in exon 9, which codes for the helical domain, or exon 20, which codes for the kinase domain [10].
In population-based studies, the prevalence of KRAS, BRAF, and PIK3CA mutations ranges from 30% to 40% for KRAS mutations, from 5% to 15% for BRAF mutations, and from 10% to 15% for PIK3CA mutations [11]. Somatic mutations of PIK3CA may coexist with either KRAS or BRAF mutations within the same tumor [12], but KRAS and BRAF mutations appear to be mutually exclusive [13].
In Sardinia, which has experienced little immigration due to its remote location and whose population has inherited many of the same genetic traits, the contribution of somatic mutations in these three genes to the CRC pathogenesis has not been estimated yet. Colorectal cancer represents the second principal death-causing malignancy in Sardinia, with an incidence (standardized rate, 104 per 100.000 inhabitants per year; Sardinian population includes about one million and half inhabitants) quite comparable with that observed in Western countries [14].
As previously demonstrated by our group for other malignancies (mainly, breast cancer and malignant melanoma), the geographical distribution of germline sequence variants across the island seems to be significantly heterogeneous, suggesting that the genetic background may influence the occurrence of cancer gene mutations [15-17]. In this study, we assessed the prevalence and distribution of KRAS, BRAF, and PIK3CA mutations at somatic level among 478 consecutively-collected CRC patients from Sardinian population.
Materials and methods
Samples
Four hundred and seventy-eight patients with histologically-proven diagnosis of colorectal carcinoma (CRC) and regularly participating to the follow-up programs at the Institutions across Sardinia island were included into the study. To avoid any bias, CRC patients were consecutively collected from April 2009 to September 2011; they were included regardless of age at diagnosis and disease characteristics. No CRC case from our series was associated with clinically relevant colorectal polyposis. Sardinian origin was ascertained in all cases through genealogical studies; for all patients, place of birth of their parents and grandparents was assessed in order to assign their geographical origin within the island. Clinical and pathological features for the assessment of the disease stage at diagnosis as well as of the onset age and tumour anatomical location were confirmed by medical records and/or pathology reports. Disease stage classification was assigned according to the American Joint Committee on Cancer guidelines [18].
Formalin-fixed paraffin embedded tissue samples from CRC patients were obtained from the archives of the Institutes and Services of Pathology participating to the study. Tissue samples were estimated to contain at least 70% neoplastic cells by light microscopy.
All patients were informed about the aims of this study and, before the tissue sample was collected, gave a written informed consent. The study was reviewed and approved by the ethical review board of the University of Sassari.
Mutation analysis
All tumour tissues were collected and processed at the laboratory of the Institute of Biomolecular Chemistry of Sassari; genomic DNA was isolated from tissue sections using a standard protocol and DNA quality assessed for each specimen. In particular, paraffin was removed from formalin-fixed paraffin-embedded (FFPE) samples by treatment with Bio-Clear (Bio-optica, Milan, Italy) and DNA was purified using the QIAamp DNA FFPE Tissue kit (QIAGEN Inc., Valencia, CA, USA).
The coding sequence and splice junctions of exons 2 and 3 in KRAS gene (where all pathogenetic mutations occur [9]), exon 15 in BRAF gene (nearly all oncogenic mutations have been detected at the kinase domain in exon 15 [19,20]), and exons 9 and 20 in PIK3CA gene (they represent the mostly mutated domains of this gene [10,21]) were screened for mutations by direct automated sequencing. Briefly, polymerase chain reaction (PCR) was performed on 25–50 ng of isolated genomic DNA in a 9700 Thermal cycler (Applied Biosystems, Foster City, CA, USA); all PCR-amplified products were directly sequenced using an automated fluorescence-based cycle sequencer (ABIPRISM 3100, Applied Bio-systems, Foster City, CA), as previously described by our group [17]. Primer sequences were as follow: KRAS exon 2 forward, TGTGTGACATGTTCTAATATAGTCACAT - exon 2 reverse, GGTCCTGCACCAGTAATATGC - exon 3 forward, GACTGTGTTTCTCCCTTCT - exon 3 reverse, TGGCAAATACACAAAGAAAG; PIK3CA exon 9 forward, GGGAAAAA TATGACAAAGAAAGC - exon 9 reverse, CTGAGATCAGCCAAATTCAGTT - exon 20 forward, CTCAATGATGCTTGGCTCTG - exon 20 reverse, TGGAATCCAGAGTGAGC TTTC; BRAF exon 15 forward, TCATAATGCTTGCTCTGATAGGA - exon 15 reverse, GGCCAAAAATTTAATCAGTGGA. Protocols for PCR-based assays were designed and optimized in our laboratory; they will be available upon request. Screening for BRAF and PIK3CA genes was incomplete in a fifth of patients (94/478; 20%) due to the low amount of available tumour tissue samples.
Statistical analysis
Statistical analysis for the presence of KRAS, BRAF, or PIK3CA mutations versus different variables (sex, age at diagnosis, anatomical site of primary CRC, disease stage, geographical origin of patients) was performed by Pearson’s Chi-Square test. The odds ratio (OR) and 95% confidence interval (CI) values were calculated by logistic regression analysis. The exact coefficient for sample proportion analysis was performed to determine all significant parameters (below 0.05 level). All analyses were performed using the statistical package SPSS/7.5 per Windows.
Results
Paraffin-embedded tumour tissues from a total of 478 patients with advanced colorectal carcinoma (CRC) originating from different geographical areas within Sardinia island were screened for mutations in candidate genes. Considering the primary tumour, left colon was the most frequent anatomical location (left colon, 192 [40%]; right-transverse colon, 172 [36%]; rectum, 114 [24%]) (Table 1). The median age was 64 years (range, 31–87 years), with a preponderance of males (291 men; 61%). At the time of diagnosis, minority of patients presented with localized disease (AJCC stage II, 173 [36%] versus AJCC advanced stages III and IV, 160 [34%] and 145 [30%], respectively) (Table 1).
Table 1.
Distribution of mutations according to the characteristics of CRC patients
|
Characteristic |
|
KRASmut |
|
PI3Kmut |
|---|---|---|---|---|
| No. (N = 478) | No. (N = 384) | |||
|
Sex | ||||
| Male |
293 |
84 (29%) |
231 |
38* (16%) |
| Female |
185 |
61 (33%) |
153 |
29 (19%) |
|
Tumor site | ||||
| Right-transverse colon |
172 |
54 (31%) |
138 |
25* (18%) |
| Left colon |
192 |
59 (31%) |
151 |
23 (15%) |
| Rectum |
114 |
32 (28%) |
95 |
19 (20%) |
|
Disease stage | ||||
| Stage II |
173 |
44 (25%) |
143 |
22 (15%) |
| Stage III |
160 |
56 (35%) |
128 |
25 (20%) |
| Stage IV |
145 |
45 (31%) |
113 |
20* (18%) |
|
Tumor grading | ||||
| Well differentiated |
56 |
17 (30%) |
44 |
6 (14%) |
| Moderately differentiated |
383 |
118 (31%) |
309 |
54 (17%) |
| Poorly differentiated |
39 |
10 (26%) |
31 |
7* (23%) |
|
Age, years | ||||
| < 50 |
50 |
16 (32%) |
45 |
5 (11%) |
| 50-59 |
116 |
38 (33%) |
102 |
14 (14%) |
| 60-69 |
178 |
56 (31%) |
151 |
30* (20%) |
| 70+ | 134 | 35 (26%) | 86 | 18 (21%) |
* 1 patient also mutated in BRAF.
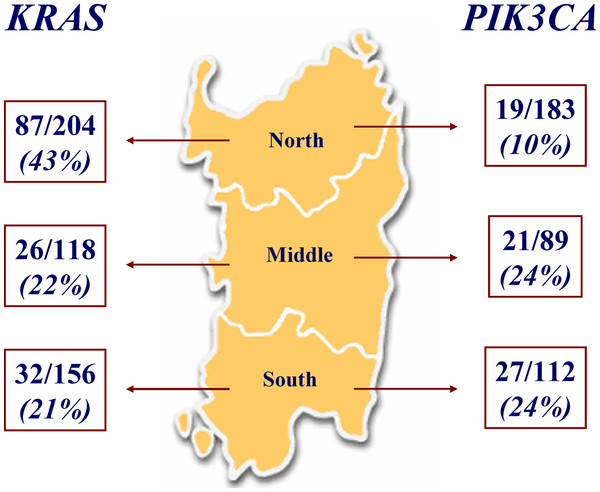
The full coding sequences and intron-exon junctions of the KRAS gene were sequenced in the entire series of 478 CRC patients; KRAS mutations were detected in 145 (30%) primary tumours (one patient had two mutations, G12D and Q61L). In terms of the gene positions of the identified KRAS mutations, 73% (N = 106) of them affected codon 12 and 20% (N = 29) affected codon 13, whereas the remaining 7% (N = 11) affected other codons (mainly, codon 61) (Table 2). All KRAS mutations detected in the present study have been previously reported in the Human Gene Mutation Database (HGMD) [22] and in the Catalogue Of Somatic Mutations In Cancer (COSMIC) [23]. Considering the patients’ origin within the Sardinia island, distribution of mutations was significantly heterogeneous: 87/204 (43%) mutated cases in North Sardinia versus 58/274 (21%) in Middle-South Sardinia [p<0.001 (OR, 2.82; 95% CI, 2.75-2.89); p normalized scientific notation = 6.8 x 10-4 (Figure 1). Such discrepancies did not result from incorrect standard sequencing as confirmed by an independent duplicate analysis.
Table 2.
Somatic mutations inKRASgene
| Mutation | No. | % |
|---|---|---|
|
Codon 12 | ||
|
G12A |
13 |
9 |
|
G12C |
11 |
8 |
|
G12D* |
41 |
28 |
|
G12R |
4 |
3 |
|
G12S |
5 |
3 |
|
G12V |
32 |
22 |
|
Total |
106 |
73 |
|
Codon 13 | ||
|
E49K |
1 |
0.5 |
|
Codon 49 | ||
|
G13C |
4 |
3 |
|
G13D |
23 |
16 |
|
G13S/V |
2 |
1 |
|
Total |
29 |
20 |
|
Codon 59 | ||
|
Q61R |
2 |
1 |
|
Q61L* |
7 |
5 |
|
Total |
9 |
6 |
|
Codon 61 | ||
| A59E | 1 | 0.5 |
Percentages are referred to mutation frequencies among the 145 positive cases.
* 1 patient with two mutations.
Figure 1.
Frequencies ofKRASandPIK3CAmutations across Sardinia. The three geographical regions within the island are indicated.
Among available DNA samples, 384 primary tumours were also evaluated for occurrence of BRAF (in exon 15) and PIK3CA (in exons 9 and 20) mutations. Overall, mutations were detected in 1 (0.3%) patient for BRAF gene and 67 (17.4%) cases for PIK3CA gene.
The single BRAF-mutated patient presented: a) the substitution of valine by a glutamic acid at position 600 (V600E), which has been demonstrated to account for majority (about 90%) of the BRAF mutations identified [19,20,24]; and b) a concomitant PIK3CA mutation, whereas no KRAS mutation coexisted. For PIK3CA mutations, screening revealed the occurrence of six mutations (p.E542K, p.E545A, p.E545G, and p.E545K in exon 9; p.M1043I and p.H1047R in exon 20), which have been widely reported in mutation databases (HGMD and COSMIC; see above) as commonly associated with CRC, with a recognized functional role of the corresponding mutated proteins. The variant p.E545A was the mutation with the highest frequency in our series (detected in 54/384 [14%] cases) (Table 3). Mutations of PIK3CA and KRAS were found to coexist in 15/384 (3.9%) of cases.
Table 3.
Prevalence of somatic mutations inPIK3CAgene
| Exon | Mutated cases(%)* | Protein | DNA |
|---|---|---|---|
| 9 |
1 (1.5%) |
E542K |
c.1624 G > A |
| |
54 (80.6%) |
E545A |
c.1634A > C |
| |
4 (6.0%) |
E545G |
c.1634A > G |
| |
1 (1.5%) |
E545K |
c.1633 G > A |
| 20 |
1 (1.5%) |
M1043I |
c.3129 G > T |
| 6 (8.9%) | H1047R | c.3140A > G |
*percentages are referred to the series of 67 positive cases.
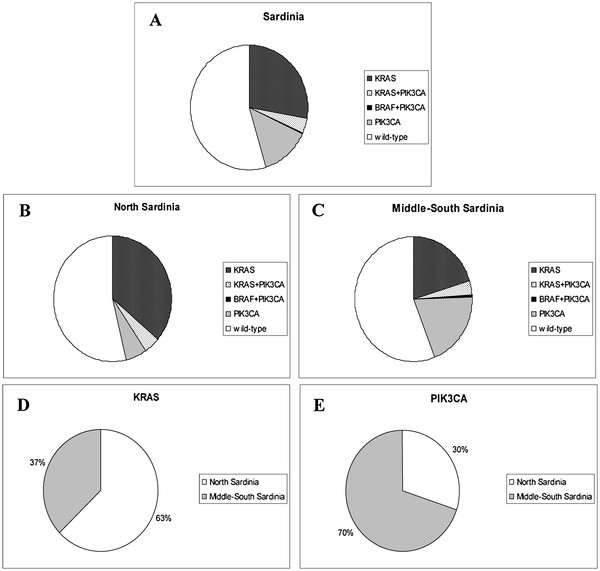
Table 4 summarizes the distribution and relationship of the somatic mutations identified in the series of 384 CRC tumours for all three candidate genes. Altogether, a mutation of at least one gene was discovered in about half (174/384; 45.3%) of CRC cases; in other words, 54.7% (N = 210) primary tumours displayed a wild-type genetic status in these three genes (Table 4).
Table 4.
Frequencies of somatic mutations in the series of 384 patients screened for all three genes, according to the geographical origin
|
Patients’ origin |
KRAS |
|
|
PIK3CA |
wild-type |
|---|---|---|---|---|---|
| |
|
KRAS + PIK3CA |
BRAF + PIK3CA |
|
|
| % | % | % | % | % | |
|
North Sardinia (N = 183) |
66 |
8 |
0 |
11 |
98 |
|
36.1 |
4.4 |
0 |
6.0 |
53.5 |
|
|
Middle-South Sardinia (N = 201) |
41 |
7 |
1 |
40 |
112 |
|
20.4 |
3.5 |
0.5 |
19.9 |
55.7 |
|
|
Total (N = 384) |
107 |
15 |
1 |
51 |
210 |
| 27.8 | 3.9 | 0.3 | 13.3 | 54.7 |
Considering the patients’ origin, PIK3CA mutations were found to be inversely distributed as compared to the KRAS mutations: 19 (10%) out of 183 patients from North Sardinia versus 48 (24%) out of 201 patients from Middle-South Sardinia were found to carry mutations in exons 9 and 20 of the PIK3CA gene (Table 4; Figure 1). As for KRAS, such a heterogeneous distribution of PIK3CA mutations was found highly significant [p<0.001 (OR, 2.45; 95% CI, 2.36-2.55); p normalized scientific notation = 5.9 x 10-4]. To avoid any putative artefact, such discrepancies were again confirmed in independent duplicate sequencing experiments. As shown in Figure 2, majority (63%) of KRAS mutations were found in patients from North Sardinia whereas more than two thirds (70%) of PIK3CA mutations were detected in patients from Middle-South Sardinia. No difference in distribution of KRAS and PIK3CA mutations between rural and urban areas, both globally and within the two (North vs. Middle-South) geographical regions was observed.
Figure 2.
Geographical distribution of mutation carriers in the series of 384 patients screened for all three genes. (A) Entire island. (B) Northern and (C) Central-Southern regions. Prevalence of (D) KRAS- and (E) PIK3CA-mutated patients according to the geographical origin.
Both KRAS and PIK3CA mutations were evaluated for association with several pathological parameters: sex, age at diagnosis, anatomical location of primary CRC, tumour grading, AJCC stage of the disease. In our series, no significant correlation was found between the occurrence of KRAS or PIK3CA mutations and all analyzed parameters. However, a non-significant trend for PIK3CA mutations to be associated with a older age of onset and a higher tumour grade was observed (see Table 1). Absence of any association between the different variables was also confirmed by a multivariate analysis.
Finally, we preliminarily evaluated the association between PIK3CA mutations and response to the treatment with anti-EGFR monoclonal antibodies in patients with wild-type KRAS. Among the 118 KRAS mutation negative patients for whom results were available (105 [89%] treated with cetuximab in association with systemic chemotherapy and 13 [11%] with panitumumab alone), the objective response rate at first evaluation was 30% (N = 35; no complete clinical response was registered) (Table 5). In this subset of patients, a slight but not significant difference in rates of partial responses was observed between PIK3CA-mutated (7/29; 24%) and PIK3CA-wild-type (28/89; 31%) cases (Table 5).
Table 5.
Clinical response to anti-EGFR therapy inKRASwild-type patients
| |
|
KRASwt |
|
|---|---|---|---|
| KRASwt (N = 118) | PI3Kwt (N = 89) | PI3Kmut (N = 29) | |
|
PR |
35 (30%) |
28 (31%) |
7 (24%) |
|
SD |
58 (49%) |
45 (51%) |
13 (45%) |
| PD | 25 (21%) | 16 (18%) | 9 (31%) |
PR, partial response; SD, stable disease; PD, progression disease. wt, wild-type.
Discussion
In this study, we have reported the prevalence of somatic mutations in KRASPIK3CA, and BRAF genes among patients with advanced colorectal carcinoma from Sardinia, whose population shows genetic peculiarity due to geographical isolation and strong genetic drift [25]. Prevalence of candidate gene mutations may vary among distinct populations due to concurrence of different environmental factors and genetic backgrounds. Furthermore, patients’ geographical origin within the same population may also account for different mutation rates in pathogenetic cancer genes, as already demonstrated for different types of cancer in Sardinian population by our group [15-17]. In summary, we observed a moderate rate of KRAS mutations (145/478; 30.3%) or PIK3CA mutations (67/384; 17.4%) and a very low rate of BRAF mutations (1/384; 0.3%) in a cohort of primary colorectal carcinomas.
Prevalence of KRAS mutations in our series is consistent with data from literature, indicating that such an alteration can be found in 30-40% of colorectal carcinomas [6]. Controversial data have been instead published about the PIK3CA mutation rates, ranging from 7% [26] to 30% [27] of CRC cases presenting a mutated PIK3CA. In the present study, all detected PIK3CA variants have been previously demonstrated to be oncogenic in CRC cellular models [28] and commonly associated with colorectal carcinoma (17%; see Table 3); the mutation rate was therefore comparable with that described in majority of previous reports.
The most surprising finding was the nearly lack of BRAF mutations in our series. In recent meta-analyses, the BRAF V600E mutation - which represents the most common mutation in BRAF gene (more than 90% of cases) - was detected in about 9% of primary colorectal carcinomas [29,30]. One could speculate that the very low frequency of BRAF mutation detected in our series may be somehow due to patients’ origin or, in other words, to the peculiarity of the genetically-isolated Sardinian population. On this regard, it cannot be excluded that different pathogenetic mechanisms of transformation could occur in different populations. Microsatellite instability (MSI), a recognized marker of a tendency for replication errors in human cancers, has been widely indicated as a factor associated with higher frequency of mutations in BRAF gene among colorectal carcinomas [29,31,32]. Although such an analysis was not conducted in the present study, Sardinian CRC population has been previously demonstrated to present an incidence of MSI similar to that observed in other CRC populations from Western countries [33,34]. Therefore, this factor could not explain the striking discrepancy on the BRAF mutation prevalence in our series.
Considering the two prevalent alterations, KRAS and PIK3CA mutations were more or less equally distributed among the different patients’ subsets, and no statistically significant correlation with sex, onset age (though patients with older age at diagnosis and a higher tumour grade are more likely to present with a PIK3CA mutation), disease stage, primary CRC location, or tumour grading was observed (see Table 1).
As schematically represented in Figure 2, we made comparisons between prevalence of KRAS and PIK3CA mutations within different geographical areas of the island. In a population sharing a quite similar lifestyle and diet habit across the island (moreover, smokers were homogeneously distributed among patients of different origin - though such an information from medical records was available in only about 70% of cases from our series), the observation that a higher frequency of KRAS mutations was found in CRC cases from North Sardinia (43% vs. 21%) whereas a higher prevalence of PIK3CA mutations was found in patients from South Sardinia (24% vs. 10%) strongly suggest that different “genetic background” may also induce discrepant penetrance and distribution of somatic mutations in candidate cancer genes. As for similar data reported by our group in breast cancer and melanoma, the geographical distribution of the genetic variants in the island seems to be related to the specific large areas of Sardinia, which reflect its ancient history: the North area, delimited by the mountain chain crossing Sardinia and linguistically different from the rest of the island; and the Middle-South area, land of the ancient Sardinian population and domain of pastoral culture. Nevertheless, our findings clearly indicate that mutation frequency for any candidate cancer gene needs to be accurately evaluated in each geographical area within every single population. Due to this unexpected heterogeneity in distribution of somatic mutations in such few main genes, we already started the collection of tumour DNA samples from Sardinian colorectal cancer patients in order to perform a whole-exome sequencing and define a more comprehensive pattern of mutations in this population.
Finally, although performed on a limited number of CRC cases, our preliminary data seemed to indicate no correlation between PIK3CA mutations and response to the anti-EGFR treatment in patients with wild-type KRAS (objective responses were considered at first evaluation only) (see Table 5). A second study focused on evaluating such clinical aspects in a larger subset of patients is ongoing.
KRAS mutations are considered as an early event in the sequential accumulation of molecular alterations underlying the progression from colorectal adenoma to malignant carcinoma, resulting in an important tumour growth advantage. During the recent past years, a targeted therapy with monoclonal antibodies (cetuximab and panitumumab), blocking the EGFR-driven cell proliferation signals, has been introduced into the therapy of metastatic colorectal cancer [9,35]. No significant response to therapy with anti-EGFR antibodies have been observed in colorectal cancer patients exhibiting KRAS mutations [36]. Moreover, majority of colorectal carcinomas exhibiting wild-type KRAS do not respond to such therapies either [9]. These phenomena are expected from the molecular point of view, since EGFR tyrosine kinase transmits proliferation signals via RAS-GTPase on the cell membrane inner surface, which in turn can bind effector proteins such as RAF or PIK3CA kinases [37]. Therefore, the occurrence of functional integrity of the RAS-driven pathways - BRAF-MEK-ERK and PIK3CA-AKT - is necessary in order to really interfere with tumour cell growth through inhibition of EGFR target. In other words, the assessment of mutational status of BRAF and PIK3CA genes into the KRAS wild-type population may indeed improve the selection of patients presenting such a functional integrity of the RAS-driven pathways (though we are aware that additional alterations in downstream effectors may intervene). In contrast to KRAS, the heterogeneity of BRAF and PIK3CA mutations has not been adequately investigated in colorectal cancer thus far.
In the present study, because some somatic mutations (in BRAF and PIK3CA genes or in KRAS and PIK3CA genes) occurred concomitantly in a given patient, which is in line with literature, a total of about 45% of all patients showed at least one mutation in any of these three genes (see Table 4). Therefore, our data suggest that including mutation analyses for BRAF and PIK3CA in addition to KRAS into a standard diagnostic setting of colorectal cancer would allow the identification of an additional fraction (in our case, about 15%) of patients who cannot be considered as “true wild-type” for such main proliferation-controlling genes. However, whether or not these additional patients might benefit from EGFR-specific antibody therapy has to be verified in prospective clinical studies.
Conclusions
Although Sardinian population is considered genetically homogeneous, the results obtained in the present study may represent a clear indication that: a) differences into the genetic background - related to distinct patients’ origin within the island - may account for different mutation rates in candidate cancer genes (in our series, KRAS and PIK3CA), even at somatic level; and b) mutation frequency for any candidate cancer gene needs to be accurately evaluated in each geographical area.
Abbreviations
COSMIC: Catalogue of somatic mutations in cancer; CRC: Colorectal carcinoma; EGFR: Epidermal growth factor receptor; FFPE: Formalin-fixed paraffin-embedded; HGMD: Human gene mutation database; MSI: Microsatellite instability; PCR: Polymerase chain reaction.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
GPa, performed mutation analysis, data analysis and interpretation, helped to draft the manuscript; MC, carried out mutation analysis; ACon, BM, GB, AP, MI, FC, VT, TS, and GS participated in patients' collection and data acquisition; FT, performed quality control of pathological data; MB, performed statistical analysis; GPi, performed data analysis and interpretation, participated in the design of the study, drafted the manuscript; Acos, performed pathological review and data interpretation, conceived of the study. All authors read and approved the final manuscript.
Contributor Information
Grazia Palomba, Email: graziap68@yahoo.it.
Maria Colombino, Email: colombinom@yahoo.it.
Antonio Contu, Email: acontu@aslsassari.it.
Bruno Massidda, Email: masbru@medicina.unica.it.
Giovanni Baldino, Email: giovannibaldino@tiscali.it.
Antonio Pazzola, Email: apazzola@aslsassari.it.
MariaTeresa Ionta, Email: mtionta@yahoo.it.
Francesca Capelli, Email: francapelli@tiscali.it.
Vittorio Trova, Email: oncalghero@libero.it.
Tito Sedda, Email: tito.sedda@alice.it.
Giovanni Sanna, Email: gisanna@uniss.it.
Francesco Tanda, Email: tandaf@uniss.it.
Mario Budroni, Email: mariobudroni@tiscalinet.it.
Giuseppe Palmieri, Email: gpalmieri@yahoo.com.
Antonio Cossu, Email: cossu@uniss.it.
Acknowledgements
Other investigators of the Sardinian Translational Oncology Group involved in this study and considered as co-authors: Marta Contu, Angelo Cuccu, Antonio Farris, Antonio Macciò, Giuseppe Mameli, Nina Olmeo, Salvatore Ortu, Elisabetta Petretto, Valeria Pusceddu, Luciano Virdis.
Authors are grateful to patients for their important contribution to this study. Work was supported by the Italian Ministry of Health “Progetto Ricerca Finalizzata” and Sardinian Regional Government (Regione Autonoma della Sardegna).
References
- Tanaka H, Deng G, Matsuzaki K, Kakar S, Kim GE, Miura S, Sleisenger MH, Kim YS. BRAF mutation, CpG island methylator phenotype and microsatellite instability occur more frequently and concordantly in mucinous than non-mucinous colorectal cancer. Int J Cancer. 2006;118:2765–2771. doi: 10.1002/ijc.21701. [DOI] [PubMed] [Google Scholar]
- Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- Tol J, Nategaal ID, Punt CJA. BRAF mutation in metastatic colorectal cancer. N Engl J Med. 2009;361:98–99. doi: 10.1056/NEJMc0904160. [DOI] [PubMed] [Google Scholar]
- Peltomäki P. Mutations and epimutations in the origin of cancer. Exp Cell Res. 2012;318:299–310. doi: 10.1016/j.yexcr.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Heinemann V, Stintzing S, Kirchner T. et al. Clinical relevance of EGFR- and KRAS-status in colorectal cancer patients treated with monoclonal antibodies directed against the EGFR. Cancer Treat Rev. 2009;35:262–271. doi: 10.1016/j.ctrv.2008.11.005. [DOI] [PubMed] [Google Scholar]
- Qiu LX, Mao C, Zhang J, Zhu XD, Liao RY, Xue K, Li J, Chen Q. Predictive and prognostic value of KRAS mutations in metastatic colorectal cancer patients treated with cetuximab: a meta-analysis of 22 studies. Eur J Cancer. 2010;46:2781–2787. doi: 10.1016/j.ejca.2010.05.022. [DOI] [PubMed] [Google Scholar]
- Roth AD, Tejpar S, Delorenzi M, Yan P, Fiocca R, Klingbiel D, Dietrich D, Biesmans B, Bodoky G, Barone C, Aranda E, Nordlinger B, Cisar L, Labianca R, Cunningham D, Van Cutsem E, Bosman F. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60–00 trial. J Clin Oncol. 2010;28:466–474. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- Samowitz WS, Sweeney C, Herrick J, Albertsen H, Levin TR, Murtaugh MA, Wolff RK, Slattery ML. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res. 2005;65:6063–6069. doi: 10.1158/0008-5472.CAN-05-0404. [DOI] [PubMed] [Google Scholar]
- Bardelli A, Siena S. Molecular mechanisms of resistance to cetuximab and panitumumab in colorectal cancer. J Clin Oncol. 2010;28:1254–1261. doi: 10.1200/JCO.2009.24.6116. [DOI] [PubMed] [Google Scholar]
- Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- Baldus SE, Schaefer KL, Engers R, Hartleb D, Stoecklein NH, Gabbert HE. Prevalence and heterogeneity of KRAS, BRAF, and PIK3CA mutations in primary colorectal adenocarcinomas and their corresponding metastases. Clin Cancer Res. 2010;16:790–799. doi: 10.1158/1078-0432.CCR-09-2446. [DOI] [PubMed] [Google Scholar]
- Janku F, Lee JJ, Tsimberidou AM, Hong DS, Naing A, Falchook GS, Fu S, Luthra R, Garrido-Laguna I, Kurzrock R. PIK3CA mutations frequently coexist with RAS and BRAF mutations in patients with advanced cancers. PLoS One. 2011;6:e22769. doi: 10.1371/journal.pone.0022769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benvenuti S, Sartore-Bianchi A, Di Nicolantonio F, Zanon C, Moroni M, Veronese S, Siena S, Bardelli A. Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti–epidermal growth factor receptor antibody therapies. Cancer Res. 2007;67:2643–2648. doi: 10.1158/0008-5472.CAN-06-4158. [DOI] [PubMed] [Google Scholar]
- Budroni M, Cesaraccio R, Pirino D, Sechi O, Oggiano M, Piras D, Sechi A, Cossu A, Palmieri G, Tanda F. In: Cancer Incidence in Five Continents, Vol. IX, International Agency for Research on Cancer (IARC) Scientific Publications, No. 160. Curado MP, Edwards B, Shin HR, Storm H, Ferlay J, Heanue M, Boyle P, editor. Lyon, IARC; 2007. Cancer incidence in Sassari Province (1998–2002) [Google Scholar]
- Palomba G, Pisano M, Cossu A, Budroni M, Dedola MF, Farris A, Contu A, Baldinu P, Tanda F, Palmieri G. Spectrum and prevalence of BRCA1 and BRCA2 germline mutations in Sardinian breast cancer patients through a hospital-based screening. Cancer. 2005;104:1172–1179. doi: 10.1002/cncr.21298. [DOI] [PubMed] [Google Scholar]
- Palomba G, Loi A, Uras A, Fancello P, Piras G, Gabbas A, Cossu A, Budroni M, Contu A, Tanda F, Farris A, Orrù S, Floris C, Pisano M, Lovicu M, Santona MC, Landriscina G, Crisponi L, Palmieri G, Monne M. A role of BRCA1 and BRCA2 germline mutations in breast cancer susceptibility within Sardinian population. BMC Cancer. 2009;9:245. doi: 10.1186/1471-2407-9-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casula C, Muggiano A, Cossu A, Budroni M, Caracò C, Ascierto PA, Pagani E, Stanganelli I, Canzanella S, Sini MC, Palomba G, Palmieri G. The Italian Melanoma Intergroup (IMI) Role of key-regulator genes in melanoma susceptibility and pathogenesis among patients from South Italy. BMC Cancer. 2009;9:352. doi: 10.1186/1471-2407-9-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene FL, Page DL, Fleming ID, editor. American Joint Committee on Cancer Staging Manual. 6. Springer, Philadelphia; 2002. [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J. et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Casula M, Colombino M, Satta MP, Cossu A, Ascierto PA, Bianchi-Scarrà G, Castiglia D, Budroni M, Rozzo C, Manca A, Lissia A, Carboni A, Petretto E, Satriano SMR, Botti G, Mantelli M, Ghiorzo P, Stratton MR, Tanda F, Palmieri G. BRAF gene is somatically mutated but does not make a major contribution to malignant melanoma susceptibility. J Clin Oncol. 2004;22:286–292. doi: 10.1200/JCO.2004.07.112. [DOI] [PubMed] [Google Scholar]
- Nosho K, Kawasaki T, Ohnishi M, Suemoto Y, Kirkner GJ, Zepf D, Yan L, Longtine JA, Fuchs CS, Ogino S. PIK3CA mutation in colorectal cancer: relationship with genetic and epigenetic alterations. Neoplasia. 2008;10:534–541. doi: 10.1593/neo.08336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Institute of Medical Genetics in Cardiff. The Human Gene Mutation Database. http://archive.uwcm.ac.uk.
- Wellcome Trust Sanger Institute. Catalogue of Somatic Mutations in Cancer (COSMIC) http://www.sanger.ac.uk/genetics/CGP/cosmic/
- Sharma SG, Gulley ML. BRAF mutation testing in colorectal cancer. Arch Pathol Lab Med. 2010;134:1225–1228. doi: 10.5858/2009-0232-RS.1. [DOI] [PubMed] [Google Scholar]
- Caramelli D, Vernesi C, Sanna S, Sampietro L, Lari M, Castrì L, Vona G, Floris R, Francalacci P, Tykot R, Casoli A, Bertranpetit J, Lalueza-Fox C, Bertorelle G, Barbujani G. Magnitude and distribution of linkage disequilibrium in population isolates and implications for genome-wide association studies. Nat Genet. 2006;38:556–560. doi: 10.1038/ng1770. [DOI] [PubMed] [Google Scholar]
- Naguib A, Cooke JC, Happerfield L, Kerr L, Gay LJ, Luben RN, Ball RY, Mitrou PN, McTaggart A, Arends MJ. Alterations in PTEN and PIK3CA in colorectal cancers in the EPIC Norfolk study: associations with clinicopathological and dietary factors. BMC Cancer. 2011;11:123. doi: 10.1186/1471-2407-11-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C, Yang ZY, Hu XF, Chen Q, Tang JL. PIK3CA exon 20 mutations as a potential biomarker for resistance to anti-EGFR monoclonal antibodies in KRAS wild-type metastatic colorectal cancer: a systematic review and meta-analysis. Ann Oncol. 2012;23:1518–1525. doi: 10.1093/annonc/mdr464. [DOI] [PubMed] [Google Scholar]
- Samuels Y, Diaz LA, Schmidt-Kittler O, Cummins JM, Delong L, Cheong I, Rago C, Huso DL, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Mao C, Liao RY, Qiu LX, Wang XW, Ding H, Chen Q. BRAF V600E mutation and resistance to anti-EGFR monoclonal antibodies in patients with metastatic colorectal cancer: a meta-analysis. Mol Biol Rep. 2011;38:2219–2223. doi: 10.1007/s11033-010-0351-4. [DOI] [PubMed] [Google Scholar]
- De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P, Penault-Llorca F, Rougier P, Vincenzi B, Santini D, Tonini G, Cappuzzo F, Frattini M, Molinari F, Saletti P, De Dosso S, Martini M, Bardelli A, Siena S, Sartore-Bianchi A, Tabernero J, Macarulla T, Di Fiore F, Gangloff AO, Ciardiello F, Pfeiffer P. et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- Lubomierski N, Plotz G, Wormek M, Engels K, Kriener S, Trojan J, Jungling B, Zeuzem S, Raedle J. BRAF mutations in colorectal carcinoma suggest two entities of microsatellite-unstable tumors. Cancer. 2005;104:952–961. doi: 10.1002/cncr.21266. [DOI] [PubMed] [Google Scholar]
- Kumar K, Brim H, Giardiello F, Smoot DT, Nouraie M, Lee EL, Ashktorab H. Distinct BRAF (V600E) and KRAS mutations in high microsatellite instability sporadic colorectal cancer in African Americans. Clin Cancer Res. 2009;15:1155–1161. doi: 10.1158/1078-0432.CCR-08-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombino M, Cossu A, Manca A, Dedola MF, Giordano M, Scintu F, Curci A, Avallone A, Comella G, Amoruso M, Margari A, Bonomo GM, Castriota M, Tanda F, Palmieri G. Prevalence and prognostic role of microsatellite instability in patients with rectal carcinoma. Ann Oncol. 2002;13:1447–1453. doi: 10.1093/annonc/mdf240. [DOI] [PubMed] [Google Scholar]
- Colombino M, Cossu A, Arba A, Manca A, Curci A, Avallone A, Comella G, Botti G, Scintu F, Amoruso M, D’Abbicco D, d’Agnessa MR, Spanu A, Tanda F, Palmieri G. Microsatellite instability and mutation analysis among Southern Italian patients with colorectal carcinoma: detection of different alterations accounting for MLH1 and MSH2 inactivation in familial cases. Ann Oncol. 2003;14:1530–1536. doi: 10.1093/annonc/mdg402. [DOI] [PubMed] [Google Scholar]
- Rodríguez J, Viúdez A, Ponz-Sarvisé M, Gil-Aldea I, Chopitea A, García-Foncillas J, Gil-Bazo I. Improving disease control in advanced colorectal cancer: Panitumumab and cetuximab. Crit Rev Oncol Hematol. 2010;74:193–202. doi: 10.1016/j.critrevonc.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Linardou H, Dahabreh IJ, Kanaloupiti D, Siannis F, Bafaloukos D, Kosmidis P, Papadimitriou CA, Murray S. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: A systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol. 2008;9:962–972. doi: 10.1016/S1470-2045(08)70206-7. [DOI] [PubMed] [Google Scholar]
- Giehl K. Oncogenic Ras in tumor progression and metastasis. Biol Chem. 2005;386:193–205. doi: 10.1515/BC.2005.025. [DOI] [PubMed] [Google Scholar]