Summary
Chemotaxis by Bacillus subtilis requires the CheD protein for proper function. In a cheD mutant when McpB was the sole chemoreceptor in B. subtilis, chemotaxis to asparagine was quite good. When McpC was the sole chemoreceptor in a cheD mutant, chemotaxis to proline was very poor. The reason for the difference between the chemoreceptors is because CheD deamidates Q609 in McpC and does not deamidate McpB. When mcpC-Q609E is expressed as the sole chemoreceptor in a cheD background, chemotaxis is almost fully restored. Concomitantly, in vitro McpC activates the CheA kinase poorly, whereas McpC-Q609E activates it much more. Moreover, CheD, which activates chemoreceptors, binds better to McpC-Q609E compared with unmodified McpC. Using hydroxyl radical susceptibility in the presence or absence of CheD, the most likely sites of CheD binding were the modification sites where CheD, CheB, and CheR carry out their catalytic activities. Thus, CheD appears to have two separate roles in B. subtilis chemotaxis - to bind to chemoreceptors to activate them as part of the CheC/CheD/CheYp adaptation system and to deamidate selected residues to activate the chemoreceptors and enable them to mediate amino acid chemotaxis.
Introduction
The Bacillus subtilis chemotaxis pathway employs a modified two-component system involving a CheA histidine kinase and a CheY response regulator. CheA forms ternary complexes with the chemoreceptors and CheW adaptor proteins (Hanlon et al., 1992, Fuhrer & Ordal, 1991, Hanlon & Ordal, 1994). These complexes are membrane-associated and found to cluster predominantly at the poles of the cell (Wu et al., 2011). The binding of attractants such as amino acids, sugars, and oxygen to these chemoreceptors increases the rate of CheA autophosphorylation (Hanlon & Ordal, 1994, Muller et al., 1997, Garrity et al., 1998, Kristich et al., 2003, Hou et al., 2000). The phosphoryl group on CheA is then transferred to CheY, a cytoplasmic protein (Garrity & Ordal, 1997, Bischoff & Ordal, 1991). Phosphorylated CheY (CheYp) can then bind to the flagella and cause them to rotate counter clockwise (Garrity & Ordal, 1997). This process is opposite to what occurs in the analogous two-component system in the Escherichia coli chemotaxis pathway, where attractants decrease the rate of CheA autophosphorylation (Rao et al., 2008, Borkovich et al., 1989). However, CheYp causes clockwise rotation of the flagella in E. coli (Wolfe et al., 1987). The effect, therefore, is the same in the two organisms – attractants cause the flagella to rotate counter clockwise.
A defining feature of bacterial chemotaxis is adaptation, in which bacteria return to their prestimulus distribution of clockwise and counterclockwise flagellar rotation following stimulation with attractants (Szurmant & Ordal, 2004). Adaptation enables cells to sense chemical gradients. The B. subtilis chemotaxis pathway involves three distinct systems for adaptation as opposed to the one in E. coli (Rao et al., 2008). Interestingly, the sole adaptation system in E. coli is also found in B. subtilis. This system utilizes methylation to tune the activity and sensitivity of the chemoreceptors to attractants and repellents. Two enzymes – the CheR methyltransferase and the CheB methylesterase – add or remove, respectively, methyl groups to or from conserved glutamate residues on the chemoreceptors (Burgess-Cassler et al., 1982, Goldman & Ordal, 1984, Goldman et al., 1984). In E. coli, methylation increases the chemoreceptor’s ability to activate CheA and reduces its sensitivity to attractants (Hazelbauer et al., 2008). In B. subtilis, the effect of methylation is site specific, where methylation of some glutamates increases the activity and/or sensitivity of chemoreceptors while methylation of others decreases them (Glekas et al., 2011, Zimmer et al., 2000). The second adaptation system present in B. subtilis but not E. coli involves CheV (Karatan et al., 2001). This hybrid protein consists of a CheW-like adaptor domain and a CheY-like response-regulator domain. In coupling the chemoreceptors to CheA, it is functionally redundant to CheW and is phosphorylated by CheA (Karatan et al., 2001, Rosario et al., 1994). CheV phosphorylation not only affects the ability of the chemoreceptors to activate CheA but also influences where CheV localizes within the cell (Wu et al., 2011). The third system, also not present in E. coli, involves two interacting proteins, CheC and CheD (Rao et al., 2008). CheC is a weak CheYp phosphatase and CheD is a chemoreceptor deamidase that targets specific glutamines on the chemoreceptors (Szurmant et al., 2004, Kristich & Ordal, 2002). CheD also binds to CheC and increases its phosphatase activity, and CheYp increases the strength of this interaction (Chao et al., 2006, Muff & Ordal, 2007). In addition, CheD is thought to increase CheA activity, presumably by binding and activating the chemoreceptors (Rosario et al., 1995).
CheC and CheD are believed to function in a feedback loop with CheYp. According to this model, CheC is believed to compete with the chemoreceptors for CheD (Rao et al., 2008). When CheYp levels are high following the addition of attractant, CheC is a better binding partner for CheD (Muff & Ordal, 2007). As a consequence, less CheD is bound to the chemoreceptors and more is bound to CheC. This reduces CheYp levels, because the CheA kinase is less active due to reduced binding of CheD to the chemoreceptors. This feedback loop, in which CheYp controls the binding of CheD to the chemoreceptors through their mutual interaction with CheC, forms the third adaptation system in B. subtilis, hereafter referred to as the CheC/CheD/CheYp adaptation system.
The participation of CheC in this adaptation system appears to be distinct from its enzymatic activity. Indeed, CheC’s phosphatase activity was previously found to play a minor role in chemotaxis whereas its ability to bind CheD was found to play a major one (Muff & Ordal, 2007). The main CheYp phosphatase in B. subtilis chemotaxis is FliY, a bifunctional protein that is also an integral component of the flagellar C-ring (Szurmant et al., 2003). FliY is a much stronger CheYp phosphatase than CheC, suggesting that the main role of the active site in CheC is to bind CheYp rather than to catalyze the hydrolysis of the aspartyl-phosphate bond (Szurmant et al., 2004). In the case of CheD, there is no redundant deamidase. While the CheB methylesterase also deamidates conserved glutamines on the chemoreceptors in E. coli and potentially in B. subtilis as well, these glutamines are distinct from those targeted by CheD (Glekas et al., 2011). What role CheD-mediated deamidation plays in B. subtilis chemotaxis, however, is not known.
The present study further investigates the role of CheD in B. subtilis chemotaxis by addressing the following questions: 1) which glutamines does CheD deamidate; 2) what is the effect of these covalent modifications on chemotaxis; 3) how does deamidation affect CheA kinase activity; 4) how is this effect distinct from CheD’s role as an allosteric regulator; and 5) where does CheD bind the chemoreceptors? Answers to these questions support a model where CheD plays two separate roles in B. subtilis chemotaxis. The first is to bind to the chemoreceptors and increase their activity as part of the CheC/CheD/CheYp adaptation system; the second is to activate some chemoreceptors and make them functional for amino-acid chemotaxis by deamidating specific glutamine residues.
Results
The McpC chemoreceptor requires CheD
B. subtilis possesses ten chemoreceptors. Of these, McpB, the sole chemoreceptor for asparagine, and McpC, the sole chemoreceptor for proline, have been the most extensively studied (Hanlon & Ordal, 1994, Muller et al., 1997). Previous work found that CheD is required for taxis towards proline but not for taxis towards asparagine, as determined using a capillary assay (Kirby et al., 2001). These experiments were performed in the presence of all ten chemoreceptors, and signaling was stimulated with an attractant that is known to use only McpB (asparagine) or McpC (proline) as the sole chemoreceptor. While these results showed that signaling through McpC had a greater requirement for CheD, the presence of other chemoreceptors that also require CheD may have had an effect on the overall chemotactic ability. The chemoreceptors in B. subtilis are known to interact with one another and affect chemotactic ability, and we wished to eliminate confounding effects (Zimmer et al., 2002). The first goal of this work was to employ strains where only a single chemoreceptor was expressed to eliminate potential crosstalk with heterologous chemoreceptors.
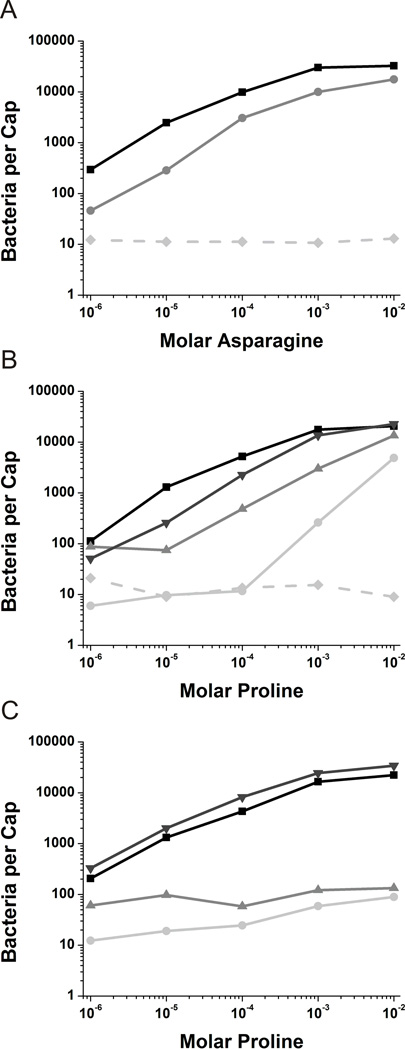
When McpB was the sole chemoreceptor expressed (Δ10mcp amyE::mcpB), the number of cells that accumulated in the capillary increased with the concentration of asparagine, as expected and in a similar fashion to the wild-type strain (Figures 1A and 1C). The same general trend was observed when cheD was also deleted (Δ10mcp cheD amyE::mcpB) except that accumulation was reduced roughly two-fold to three-fold (Figure 1A). Cells expressing only the McpC chemoreceptor (Δ10mcp amyE:mcpC) also showed normal accumulation over a range of proline concentrations of 1 µM to 0.1 M (Figure 1B). However, when cheD was deleted (Δ10mcp cheD amyE::mcpC), no significant accumulation was observed at concentrations less than 0.1 mM proline (Figure 1B). Only at concentrations greater than 0.1 mM proline did the cells accumulate within capillary, and even at these higher concentrations accumulation was roughly twenty-fold lower. These results show that McpC has a much greater requirement for CheD than does McpB, much more so than previously shown, though McpB also requires CheD for optimal taxis.
Fig. 1.
Capillary assays in cheD genetic backgrounds.
A. Capillary assays towards asparagine with mcpB expressed in the Δ10mcp background (■) and the Δ10mcp cheD background ( ). The Δ10mcp strain (
). The Δ10mcp strain ( ) is included as a negative control.
) is included as a negative control.
B. Capillary assays towards proline with mcpC expressed in the Δ10mcp background (■), mcpC expressed in the Δ10mcp cheD background ( ), mcpC-Q609E expressed in the Δ10mcp background (▼) and mcpC-Q609E expressed in the Δ10mcp cheD background (
), mcpC-Q609E expressed in the Δ10mcp background (▼) and mcpC-Q609E expressed in the Δ10mcp cheD background ( ). The Δ10mcp strain (
). The Δ10mcp strain ( ) is included as a negative control.
) is included as a negative control.
C. Capillary assays towards proline with all other nine chemoreceptors present. Wild-type strain OI1085 (■), mcpC expressed in the cheD mcpC knockout ( ), mcpC-Q609E expressed in the mcpC knockout (▼) and mcpC-Q609E expressed in the cheD mcpC knockout (
), mcpC-Q609E expressed in the mcpC knockout (▼) and mcpC-Q609E expressed in the cheD mcpC knockout ( ).
).
Identification of CheD-mediated deamidation sites
CheD was shown to enzymatically deamidate glutamines at residues 593, 594, and a third unidentified residue on the McpA chemoreceptor (Kristich & Ordal, 2002). CheD can also deamidate glutamine residues on T. maritima chemoreceptor cytoplasmic fragments (Chao et al., 2006). With the exception of the unidentified residue on McpA, these glutamine residues adhere to a well-conserved consensus sequence (Table 1). We sought to identify whether CheD deamidates any glutamine residues on McpB and McpC. Identifying these sites would potentially explain the disparity of CheD’s effect on MpcB and MpcC and also allow us to decouple the effect of CheD binding on chemoreceptor activation from its deamidase function.
Table 1.
CheD modification substrate sites.
| Chemoreceptor | Site | Sequence |
|---|---|---|
| McpA | 586 | |
| 593/4 | ||
| McpC | 609 |
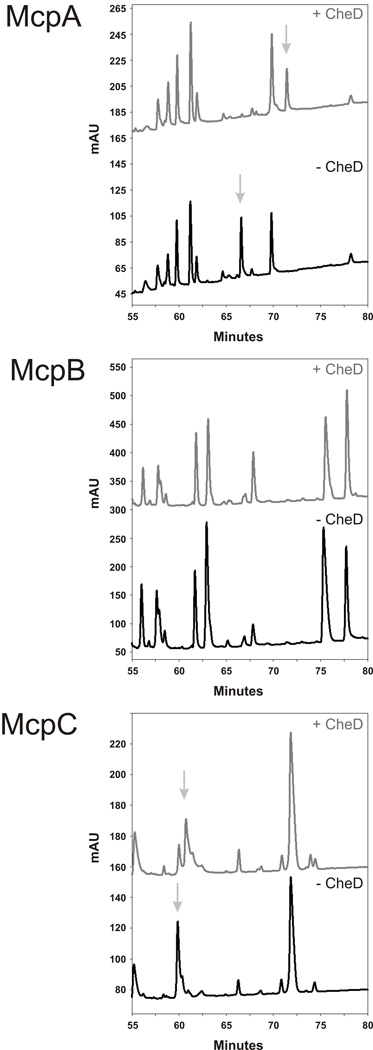
Previous work on E. coli chemoreceptors demonstrated that deamidated peptides exhibit different elution times on a reverse-phase HPLC column and that this difference can be used as a criterion for isolating deamidated chemoreceptor fragments (Kehry et al., 1983). A similar approach was employed here to identify the sites of deamidation. Briefly, purified cytoplasmic fragments of the chemoreceptors were incubated in the presence or absence of CheD and then digested with trypsin. These tryptic peptides were then separated and collected using C18 reverse-phase HPLC chromatography. Any tryptic peptides that showed a difference in elution time in the presence of CheD were submitted for both Edman degradation and tandem MS sequencing in order to identify the deamidated residues. The full tryptic digestion was also submitted for tandem MS, and showed identical results (data not shown).
The HPLC trace from McpA and McpC both showed a change in elution time for one of the peptides in the presence of CheD (Figure 2). The McpB fragments, however, did not show any changes in elution times, suggesting that this chemoreceptor is not deamidated by CheD in vitro. By sequencing the deamidated fragments from McpA and McpC, we found that the third deamidated residue on McpA is residue 586 (Table 2) and, as expected, it does not adhere to the consensus modification sequence. The other two previously identifiedsites of deamidation on McpA were also verified. In addition, we found that the deamidation site on McpC is residue 609 (Table 2), and this site adheres to the consensus sequence. These results show that CheD has the ability to enzymatically deamidate McpA and McpC in B. subtilis but not McpB.
Fig. 2.
Reverse-phase HPLC separation of tryptically digested McpA, McpB and McpC cytoplasmic chemoreceptor fragments. The presence (grey trace) or absence (black trace) of CheD is indicated, and any elution peaks that change profile due to deamidation are noted with arrows.
Table 2.
Strains and plasmids used in this study
| Strain or Plasmid | Relevant genotype or description | Reference |
|---|---|---|
| OI1085 | Che+, trpF7 hisH2 metC133 | (Ullah & Ordal, 1981) |
| OI2934 | cheD1::cat | (Rosario et al., 1995) |
| OI3280 | mcpC4::erm | (Muller et al., 1997) |
| OI3545 | Δ(mcpA mcpB tlpA tlpB)101::cat mcpC4::erm tlpC::cat yvaQ::erm yfmS::erm yoaH::erm yhfV::erm | (Hou et al., 2000) |
| OI3547 | OI3545 amyE5720::mcpB | (Zimmer et al., 2000) |
| OI3628 | OI3545 cheD1::cat | (Kristich & Ordal, 2002) |
| OI3629 | OI3545 cheD1::cat amyE5720::mcpB | (Zimmer et al., 2002) |
| OI3669 | OI3545 cheD1::cat amyE5720::mcpC | (Kristich & Ordal, 2002) |
| OI3736 | cheD1::cat mcpC4::erm | This work |
| OI4007 | OI3545 amyE5720::mcpC | (Kristich & Ordal, 2002) |
| OI4418 | OI3628 amyE5720::mcpC-Q609E | This work |
| OI4419 | OI3736 amyE5720::mcpC-Q609E | This work |
| OI4489 | OI3545 amyE5720::mcpC-Q609E | This work |
| OI4490 | OI3280 amyE5720::mcpC-Q609E | This work |
| BL-21 | E. coli protease deficient expression host | Amersham |
| BL-21 (DE3) | E. coli protease deficient expression host | Novagen |
| TG-1 | E. coli cloning host | Amersham |
| RP3098 | E. coli Δ(flhD-flhB)4, che | J.S.Parkinson |
| pGEX-6P-2 | GST-Tag Expression plasmid (AmpR) | Amersham |
| pUSH1 | B. subtilis-E. coli shuttle vector for His-tag fusions | (Schon & Schumann, 1994) |
| pAIN750 | B. subtilis amyE integration vector | (Kristich & Ordal, 2002) |
| pAIN620 | pUSH1 expressing 6xHis-McpA cytoplasmic fragment | (Kristich & Ordal, 2002) |
| pAIN153 | pEB112 co-expressing 6xHis-McpA and CheD | (Kristich & Ordal, 2002) |
| pGEX-6P-2::CheD | GST-CheD overexpression plasmid | (Muff & Ordal, 2007) |
| pGEX-6P-2::CheA | GST-CheA overexpression plasmid | This work |
| pGEX-6P-2::CheW | GST-CheW overexpression plasmid | This work |
| pGG100 | pUSH1 expressing 6xHis-McpB cytoplasmic fragment | This work |
| pGG102 | pEB112 co-expressing 6xHis-McpB and CheD | This work |
| pGG105 | pUSH1 expressing 6xHis-McpC cytoplasmic fragment | This work |
| pGG107 | pEB112 co-expressing 6xHis-McpC and CheD | This work |
| pGG147 | pAIN750 containing mcpC Q609E | This work |
A deamidated McpC chemoreceptor partially restores chemotaxis in a cheD mutant
Having identified the deamidation sites on McpA and McpC, we next sought to determine how they affect chemotaxis. In particular, does mutating these residues to glutamates remove the CheD requirement? There is no known ligand for McpA, so only McpC could be tested. We mutated gln609 to a glutamate, effectively mimicking enzymatic deamidation, and then integrated the mutated chemoreceptor into strains lacking the cheD gene, both in the Δ10mcp and mcpC backgrounds. These strains were then examined for chemotaxis towards proline using the capillary assay.
The deamidated chemoreceptor (Q609E) partially restored chemotaxis to proline in a Δ10mcp cheD background when compared to the wild-type receptor in the same genetic background (Figure 1B). When the deamidated chemoreceptor was expressed in a Δ10mcp (cheD+) background, chemotaxis to proline was nearly fully restored. These results suggest that deamidation of residue 609 places the chemoreceptor in a more functional state, though CheD is still required for full chemotactic function.
When the deamidated chemoreceptor (mcpCQ609E) was expressed in an mcpC cheD background with the other nine chemoreceptors present, chemotaxis to proline was still very poor (Figure 1C). This defect is presumably due to the presence of other chemoreceptors that still require CheD to deamidate them and/or simply to be present.
Chemoreceptor deamidation enhances CheA activation
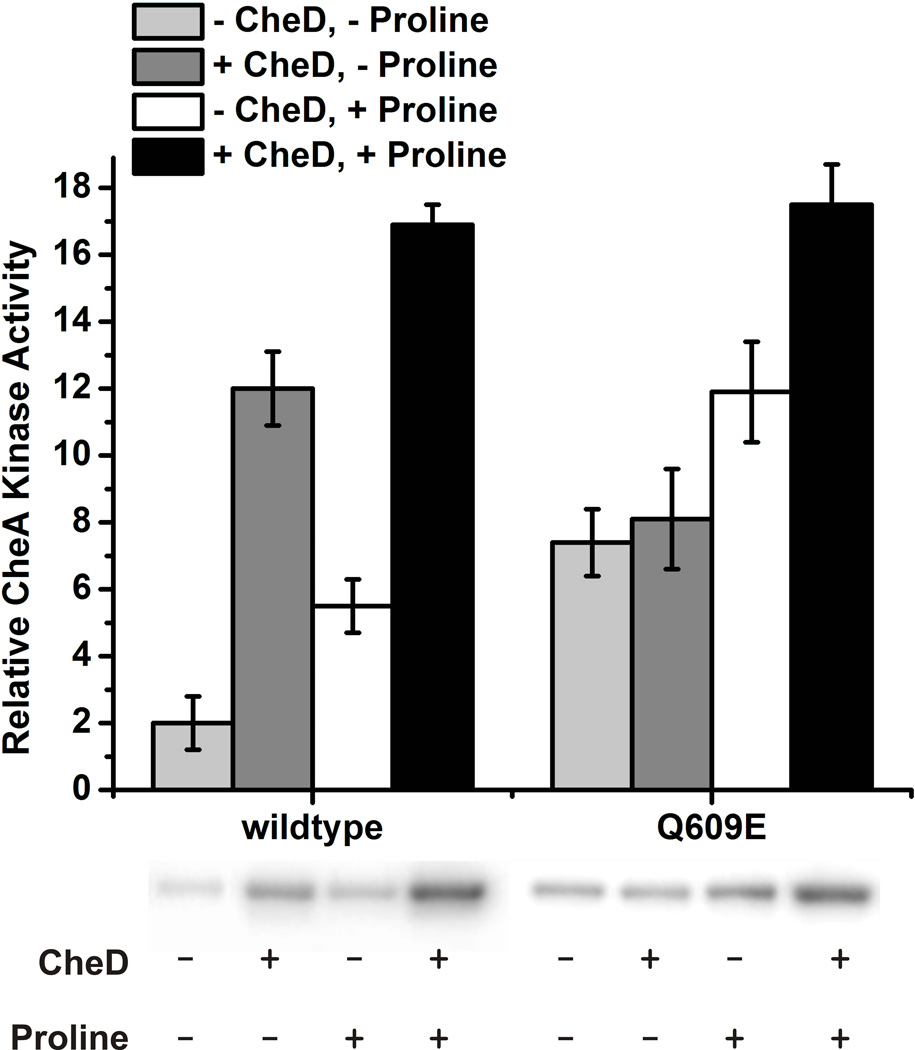
The capillary assay results suggest that CheD-mediated deamidation of residue Q609 may activate McpC. This activation, in turn, would lead to higher levels of phosphorylated CheA kinase (CheAp). To test this hypothesis, we measured CheAp levels in purified membranes containing wild-type or deamidated (Q609E) McpC chemoreceptor in the presence of CheW and CheA using an in vitro kinase assay. This assay allowed us to test how chemoreceptor deamidation affects the ability of proline to activate the CheA kinase.
Briefly, purified membranes from cells expressing either wild-type or deamidated (Q609E) McpC chemoreceptor were homogenized using sonication, and 6 µM of chemoreceptor was mixed with 2 µM CheA, 2 µM CheW, 2 µM CheD (when applicable), and 1 mM proline (when applicable). Western blots ensured that wild-type and deamidated McpC (Q609E) were added in equal amounts (Figure S1). The reaction was initiated by adding 0.1 mM [γ32P]ATP. After 15 seconds, the reaction was quenched using 2×Laemmli sample buffer containing 25 mM EDTA. The relative levels of [γ32P]ATP-labeled CheAp were measured using SDS-PAGE gel electrophoresis and phosphorimaging.
We note that this assay differs significantly from the analogous ones in E. coli. First, it was not necessary to over-express McpC. Wild-type level expression of the McpC chemoreceptor was sufficient to activate CheA kinase, although this is not the case with some other B. subtilis chemoreceptors (data not shown). The second major difference was that we did not need to add CheY. The rate of CheA autophosphorylation is relatively slow in B. subtilis, taking at least 20 seconds before CheAp levels reach their steady-state levels (Figure S2). In E. coli, the process is much more rapid (Borkovich & Simon, 1990). Moreover, CheAp levels do not saturate in B. subtilis – we can measure the rate-enhancement of CheA autophosphorylation by proline or CheD simply by measuring [γ32P]ATP-labeled CheAp. These differences persisted regardless of how long we allowed the reaction to proceed (Figure S2). Based on these results, we chose to run the reaction for 15 seconds as this time provided the maximal resolution. However, any choice of time would have sufficed for the purposes of this experiment, where the goal was to see how CheD and deamidation affect chemoreceptor-mediated CheA activation by proline.
Using this assay, we observed that proline increases the levels of CheAp roughly three-fold with membranes expressing wild-type McpC (Figure 3). The addition of CheD increases CheA phosphorylation roughly six-fold in the absence of proline and roughly three-fold in the presence of proline. When the analogous experiments were performed with deamidated McpC (Q609E), we found that CheD does not increase the levels of CheAp in the absence of proline (Figure 3). However, in the presence of proline, CheD activates the deamidated chemoreceptor roughly 1.4-fold, slightly less than the wild-type chemoreceptor. In addition, we also tested the effect of CheC and found that CheC inhibited the ability of CheD to activate the chemoreceptors, consistent with our model for the CheC/CheD/CheYp adaptation system (Figure S3).
Fig. 3.
In vitro kinase assays show that the deamidated McpC leads to higher CheA-P levels than the wild-type fragment. Densitometry measurements of the phosphorimage of SDS-PAGE gel bands corresponding to CheA-P are plotted above the gel. The amount of phosphorylated CheA as visualized in the phosphorimage (by relative density) corresponds to CheA activity. The experiments were performed in triplicate and error bars represent standard deviation.
Three main conclusions can be derived from these data. First, in the absence of CheD, the wild-type chemoreceptor activates CheA less than the deamidated chemoreceptor, as was hypothesized based on the capillary assay results. Second, in the absence of proline, the wild-type chemoreceptor is greatly affected by the presence of CheD, while the CheD effect on CheA activation of the deamidated chemoreceptor is less pronounced. Third, both the wild-type and deamidated chemoreceptor need both CheD and proline in order to fully activate the CheA kinase. Taken together, these observations show that both deamidation and CheD binding enhance CheA activation and are separate yet additive effects.
CheD binds deamidated chemoreceptors more tightly than unmodified, wild-type chemoreceptors
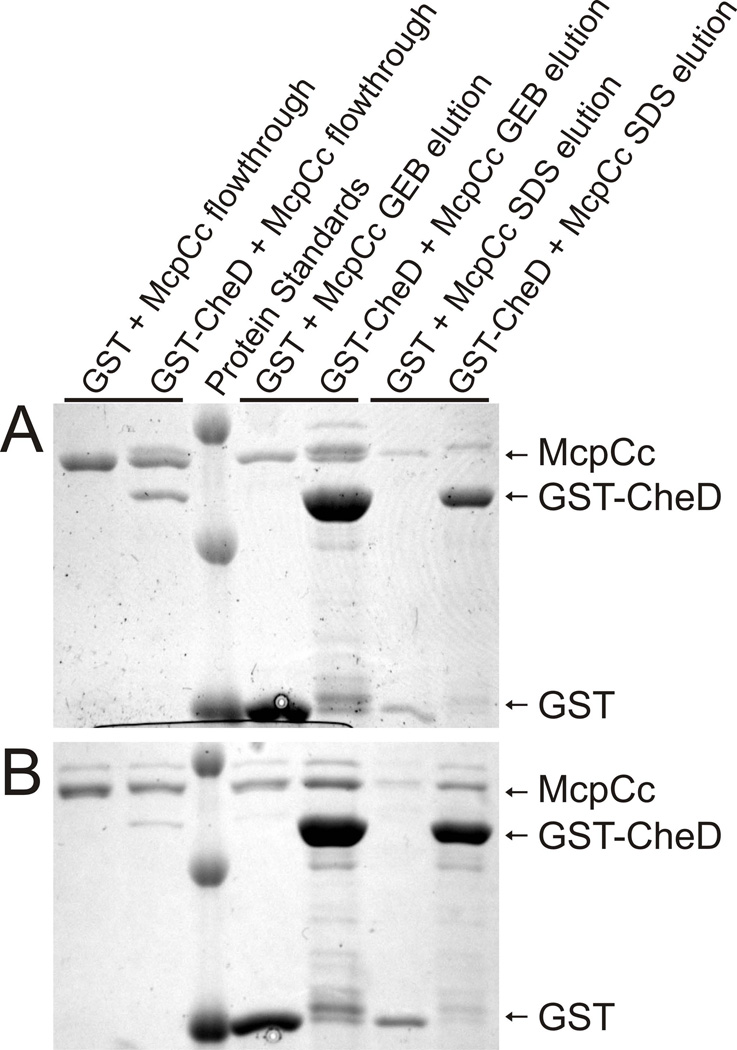
The preceding results established that CheD activates McpC, both by deamidating the chemoreceptor and also by binding it. However, these experiments did not address whether CheD preferentially binds the deamidated chemoreceptor. To test whether CheD binds more tightly to the deamidated chemoreceptors than the unmodified chemoreceptors, we employed a quantitative GST pulldown assay. In this assay, CheD was fused to GST and incubated for 15 minutes with equimolar amounts of the cytoplasmic fragment of McpC. This mixture was then incubated for an additional 15 minutes with pre-washed glutathione beads. The unbound fraction was then removed by centrifugation, and two elution steps using glutathione elution buffer (GEB) and 1% SDS ensured that all protein was removed. Note that there were no intervening washing steps. All three fractions were then analyzed using SDS-PAGE and visualized by Coomassie staining (Figure 4). The resulting bands were then quantified. Standard curves were then generated to determine the amount of protein in each band. From these quantities, the amount of chemoreceptor that bound GST alone in each step was subtracted. Using these results, we calculated the fraction of chemoreceptors bound to CheD.
Fig. 4.
Quantitative GST-pulldown assays. A. Wild-type McpC cytoplasmic chemoreceptor fragment. B. Deamidated (Q609E) cytoplasmic chemoreceptor fragment. Upper bands of the chemoreceptor fragment correspond to deamidation. GEB buffer contains glutathione for elution from the GST beads, and the SDS elution is a second elution that removes any proteins still on the beads.
The results from this assay were that CheD bound 8.4% of the unmodified, wild-type chemoreceptors and 13.2% of the deamidated chemoreceptors. These results demonstrate that CheD binds more tightly to the deamidated chemoreceptors than to the unmodified chemoreceptors, and this difference is statistically significant as determined using a two-sided t-test (p=0.018). If we assume there is a single CheD binding site on the chemoreceptors, then these results would imply a KD of 202 µM for the unmodified chemoreceptor and 117 µM for the deamidated (Q609E) chemoreceptor. Similar experiments were performed with the cytoplasmic fragment of McpB, and no quantifiable binding was observed (data not shown).
CheD binds McpC at the modification site
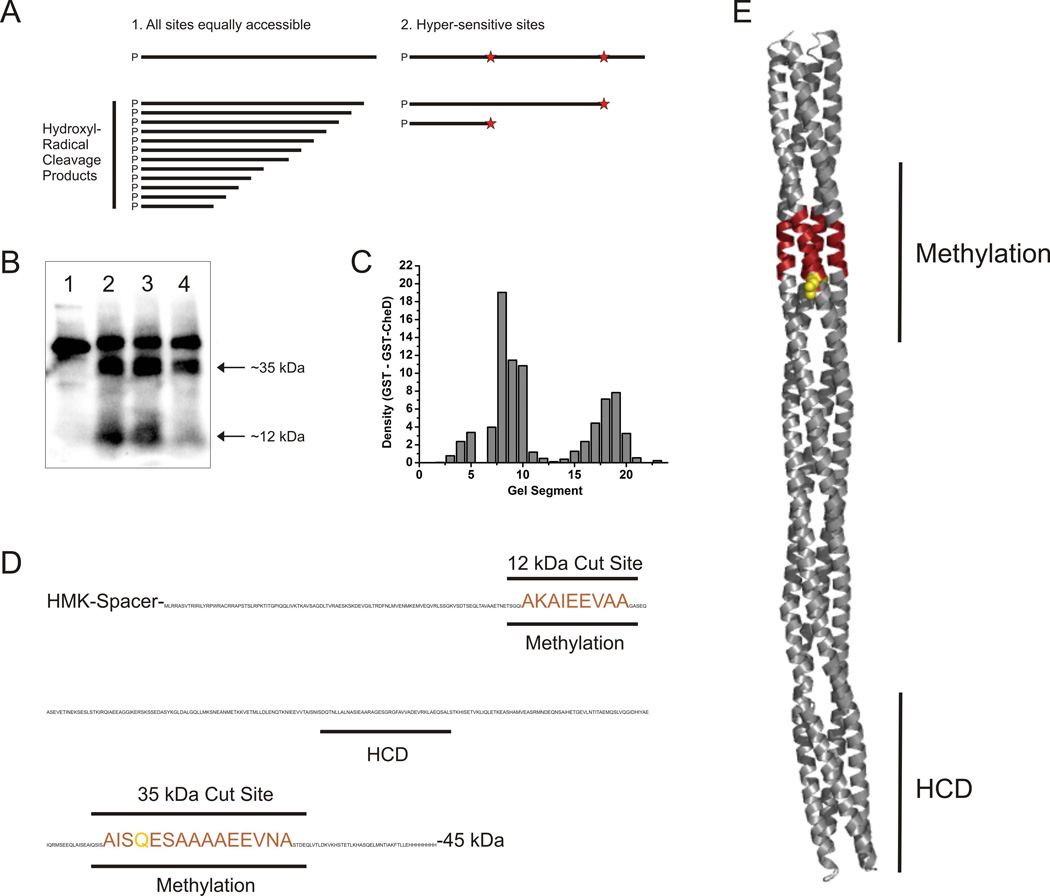
A previous study suggested CheD may bind the HAMP domain of McpC (Kristich & Ordal, 2004). However, work in T. maritima showed that CheD can bind a chemoreceptor fragment that does not contain the HAMP domain (Chao et al., 2006). To identify where CheD binds the chemoreceptor, we employed hydroxyl radical protein footprinting (Loizos, 2004). Briefly, the C-terminal fragment of McpC, between the second transmembrane-spanning region and the C-terminus of the chemoreceptor (McpCc), was fused to the site of phosphorylation of heart muscle kinase at its N-terminus and 8xHis at its C-terminus. The protein was then end-labeled with [γ32P]ATP and purified using nickel agarose beads. The labeled protein was subjected to hydroxyl radical cleavage in the presence or absence of GST-CheD or the GST control. The resulting peptide fragments were then separated by SDS-PAGE and analyzed using phosphorimaging.
In its ideal form, this method would generate a uniform distribution of fragments from very small to approximately the molecular weight of the uncut McpCc (Figure 5A). What was observed instead were two bands of approximately 35 kDa and 12 kDa, with some intensity at other sizes (Figure 5B). We assume that these bands represent hypersensitive cut sites or clusters of peptide bonds unusually susceptible to hydroxyl radical cleavage. GST-CheD protected these sites from hydroxyl-radical cleavage, especially the site at 12 kDa (Figure 5B). To quantitate these results, the region just below where the uncut chemoreceptor runs to the dye front was visualized and divided into 23 equal fragments using Labworks software. The density of each of the regions was quantitated. The quantitation confirmed what was concluded from visual inspection of the gel (Figure 5C). The changes caused by the presence of GST-CheD resulted from protection of hypersensitive cut sites in the region of residues 375–393 and 608–620, the second of which corresponds to the CheD-mediated site of deamidation. When mapped onto both the sequence and a structural model of the McpC cytoplasmic domain the cut sites overlap at the site of deamidation/methylation (Figure 5D and E). We conclude that within the resolution of this method, CheD interacts with McpC at the sites of deamidation/methylation and not at the HAMP domain. Furthermore, the methylation region appears to be much more susceptible to hydroxyl-radical cleavage, which suggests that the structure in this region is much more open than the rest of the chemoreceptor.
Fig. 5.
Hydroxyl radical protein footprinting experiments reveal the segments of the McpC cytoplasmic chemoreceptor fragment that interact with CheD.
A. A diagram of the potential cutting patterns arising from hydroxyl radical peptide cleavage.
B. The phosphor-imaging scan of an SDS-PAGE gel containing amino-terminally γ32P-labeled McpC cytoplasmic fragments. The four lanes in the gel correspond to: (1) Un-cut McpCc. (2) McpCc subjected to hydroxyl-radical cleavage. (3) McpCc + GST subjected to hydroxyl-radical cleavage. (4) McpCc + GST-CheD subjected to hydroxyl-radical cleavage.
C. The phosphor-imaging scan was segmented and the difference between GST and GST-CheD lanes plotted as a function of band density.
D. The hydroxyl-radical cleavage sites protected in the presence of GST-CheD compared to the GST control are shown on the McpCc protein sequence and highlighted in red. The previously uncovered deamination site is shown in green.
E. These cleavage sites are mapped onto the homology model McpCc structure. The protected regions are highlighted in red and the deamidation site is in green. HCD denotes the highly conserved domain of the chemoreceptor that interacts with CheA, CheW, and CheV.
Discussion
CheD plays a key role in B. subtilis chemotaxis. It activates the chemoreceptors using two separate mechanisms. The experiments presented here seek to further understand these two mechanisms. First, the McpC chemoreceptor was shown to require CheD for normal chemotactic function, while the McpB chemoreceptor was found to require it much less so. By identifying the CheD-mediated sites of deamidation, we were able to mimic enzymatic deamidation using the point mutant McpCQ609E. When this deamidated chemoreceptor was expressed in a strain lacking cheD and the other chemoreceptors, the mutant phenotype was partially rescued, although not to wild-type levels. Furthermore, in vitro kinase assays showed that both deamidation and CheD binding lead to increases in CheA kinase activation. Finally, CheD’s interaction with the McpC chemoreceptor was characterized using quantitative pulldowns and hydroxyl-radical footprinting. Taken together, these results clearly show that CheD can activate chemoreceptors in bacterial chemotaxis and does so by binding to and enzymatically modifying the chemoreceptor at the same location.
The role of deamidation
CheD enzymatically deamidated a glutamine residue to a glutamate on two of the three chemoreceptors tested here. Other than site 586 on McpA, all the deamidation sites found adhered well to the previously established consensus modification sequence motif (A/S-X-X-Q/E-Q/E-X-X-A/S). The fact that CheD did not deamidate McpB is not totally unexpected based on the capillary assay results. However, previous evidence showed that CheD was required for deamidation of McpB (Kristich & Ordal, 2002). We note that these experiments were performed in a strain that also contained CheB. One possibility is that McpB is deamidated by CheB and that somehow this reaction requires CheD, perhaps by altering the conformation of the chemoreceptor. This conclusion is plausible because the chemoreceptors in cheD mutants are poorly methylated. The second possibility, which we cannot entirely discount at this time, is that McpB is not deamidated by CheD in vitro but is in vivo. What is clear is that the methylation sites on McpB are not deamidated by CheD. The only consensus deamidation site on McpB is residue 371, which is subject to methylation following deamidation (presumably by CheB), and this site is not deamidated by CheD (Glekas et al., 2011).
The direct effect of CheD-mediated deamidation of McpC was seen in both the capillary assays and the in vitro kinase assays. These results showed that deamidation of McpC results in a more active chemoreceptor, even in the absence of CheD and proline. In the capillary assay, deamidation was not sufficient to fully rescue the cheD phenotype. The kinase assays also showed higher CheA activity with the deamidated chemoreceptor in the absence of CheD, but full CheA activation was only achieved when both proline and CheD were present. These results suggest that, although deamidation does increase chemoreceptor activation, CheD is required for more than just its enzymatic function.
While a deamidated residue can enhance chemoreceptor activity, we do not know whether these residues are subsequently methylated. The only chemoreceptor known to contain a consensus glutamine residue subject to methylation following deamidation is residue 371 on McpB (Zimmer et al., 2000). As argued above, this site is most likely deamidated by CheB. CheD-mediated deamidation at residue Q609 was shown to activate the McpC chemoreceptor. However, if this site is subsequently methylated, it would suggest that CheD also plays an indirect role in the methylation system by providing more sites for methylation. For a long time the prevailing dogma was that only the second residue in the Q/E-Q/E pair in the consensus modification site was methylated (Le Moual & Koshland, 1996). However, recent in vitro work in T. maritima shows that the first Q/E in this pair can get methylated (Perez et al., 2006). So, it is possible that residue Q609 on McpC might get methylated. At the very least, the deamidation site is very close to the three putative sites of methylation (Figure 5), suggesting that the deamidation might have an indirect effect on methylation either way. Future work that includes a detailed analysis of the methylation sites on McpA and McpC will lead to a comprehensive understanding of the CheD-mediated deamidation, an undertaking beyond the scope of this work.
The CheC/CheD/CheYp adaptation system
Our results show that CheD is required for more than just its enzymatic function. Its role in the CheC/CheD/CheYp adaptation system is required for full chemotactic function, as shown by the capillary assay results. The addition of CheD in the kinase assay shows that it enhances CheA activation. It had been assumed that CheD was necessary for activation of the CheA kinase since a cheD mutant exhibits such a low bias in a tethered cell assay (Kirby et al., 2001). The data presented here can now explain that phenotype. They also explain the role of CheD in the CheC/CheD/CheYp adaptation system. Once the CheC/CheYp complex reduces CheD binding to the chemoreceptors, CheAp levels will decrease because the chemoreceptor is less active. This decrease is one aspect of CheD’s role in adaptation. The CheC:CheD complex can better de-phosphorylate CheYp, so, once CheD is bound to CheC, CheY-P will get de-phosphorylated more rapidly. The decrease in CheYp levels is the second aspect of CheD’s role in adaptation.
The pulldowns suggest a modest CheD:McpC interaction with an estimated KD of only 202 µM for the wild-type chemoreceptor. Isothermal titration calorimetry studies in T. maritima showed that CheD interacts with CheC at a lower KD of roughly 1 µM, indicating much stronger binding (Park et al., 2004). In the presence of CheY-P, the interaction is even stronger. So, how is CheD even available to interact with the chemoreceptors and perform its various functions if it binds CheC much more tightly? We previously found that there are more copies of CheD (roughly 1200) in the cell than CheC (roughly 770 copies), and that could be one reason that CheD would still be available to bind at the chemoreceptor complex (Cannistraro et al., 2011). Moreover, there are many more chemoreceptors in the cell than CheC, roughly 59,000 per cell. Although the CheD:Mcp interaction is weaker, there are many more binding targets compared with CheC, which should compensate for the lower affinity.
The CheD binding site
While CheD’s separate roles both lead to CheA kinase activation, we did not previously know whether CheD interacts with the chemoreceptor in different regions to accomplish its separate functions. Previous work with chimeric chemoreceptors demonstrated that the CheD requirement for McpC could be assigned to the HAMP domain and suggested that this is the site where CheD binds (Kristich & Ordal, 2004). However, work in T. maritima showed that CheD could fully modify and strongly bind cytoplasmic chemoreceptor fragments lacking a HAMP domain (Chao et al., 2006). The final two experiments of this study sought to resolve this discrepancy and locate the CheD binding site on McpC. All the cytoplasmic fragments of McpC used in this study contain the entire HAMP domain, meaning that all possible CheD binding sites would be accessible. Two conclusions became evident. First, CheD binds the deamidated chemoreceptor more tightly. Second, CheD binds only at the region involved in chemoreceptor modification, and not at the HAMP domain. This region on McpC also encompasses the three putative methylation sites (Figure 5E). While we definitely cannot rule out the possibility that CheD binds the HAMP domain as well in full-length receptors, the HAMP domain is physically connected to the modification region and has been shown to affect other regions of the chemoreceptors (Swain et al., 2009, Zhou et al., 2009, Zhou et al., 2011), so the HAMP requirement is not totally unexpected in light of our findings.
The effect of other chemoreceptors
Our results demonstrate that heterologous chemoreceptors requiring CheD can negatively affect the signaling through other chemoreceptors, as seen when the deamidated McpC chemoreceptor was tested in the presence of other chemoreceptors. According to these results, the deamidation effect, which leads to a more active chemoreceptor, is reduced by the other chemoreceptors. While only one type of chemoreceptor is needed to bind an attractant, the entire chemoreceptor ensemble participates in signaling (Zimmer et al., 2002). If some of the chemoreceptors in the ensemble are much less active, they can lead to a decrease in the ability of the entire complex to excite the CheA kinase. The importance of heterologous chemoreceptors in B. subtilis chemotaxis has been shown before with respect to the methylation system where other chemoreceptors can “rescue” a non-methylatable McpB chemoreceptor (Zimmer et al., 2002). While the work presented here shows that less active chemoreceptors can exert a negative effect, it is clear that a more detailed understanding of the role of heterologous chemoreceptors is needed for a fuller understanding of chemotactic signaling in B. subtilis.
Experimental procedures
Bacterial stains and plasmids
All B. subtilis strains used in this study are derived from the chemotactic strain (Che+) OI1085 (Ullah & Ordal, 1981). All cloning and plasmid propagation was performed in E. coli strains TG1 (GE Healthcare Life Sciences). All strains and plasmids used in this study are summarized in Table 2.
The 6xHis fusion pUSH plasmids that contained the cytoplasmic chemoreceptor fragments were made by amplifying the carboxy-terminal region of the mcpB or mcpC genes from OI1085 genomic DNA with engineered BamHI restriction sites. This resulting fragment was then ligated into the pUSH1 plasmid. The pEB112 derivatives for co-expression of the chemoreceptor fragment and cheD were made as described (Kristich & Ordal, 2002). Briefly, plasmids containing the chemoreceptor and CheD in pEB112 were digested with NotI and BamHI, and the 1100 bp fragment that contained the chemoreceptor fragment gene was ligated into the bigger pEB112 derivative. The 6xHis-fusion expression plasmids (pUSH1 and pEB112 variants) were expressed in the E. coli RP3098 strain, which is devoid of all chemotaxis proteins.
The GST-fusion pGEX-6P-2 plasmids were made by amplifying cheW or cheA from OI1085 genomic DNA by PCR with engineered 5’ EcoRI and 3’ NotI sites. This fragment was then ligated into pGEX-6P-2. The plasmids were propagated in the E. coli strain BL-21 (Amersham) for protein over-expression.
The pMP4463 plasmid was constructed using a multi-step process. A double stranded oligonucleotide containing the recognition sequence (LRRASV) for a heart muscle kinase HMK sequence (Loizos, 2004) and 5’-NdeI and 3’-SpeI overhands was ligated into pET42b(+) (Novagen), which contains a 6xHis tag. The cytoplasmic domain of McpC (McpCc) was then amplified from OI1085 genomic DNA with engineered 5’ ScaI and 3’ NcoI restriction sites and ligated into the pET-HMK plasmid created above. Unfortunately, placing both the HMK site and the 6xHis-tag on the amino-terminal end of McpCc did not create a protein that was usable in the protein footprinting experiments. The McpCc fragment was excised from the plasmid using the SpeI and BamHI restriction enzymes and a new cytoplasmic fragment of McpC was amplified with 5’ EagF and 3’ XhoI restriction sites and then ligated into the resulting plasmid. This process yielded an expressed protein with an amino-terminal HMK site, a 23 amino acid spacer, then McpCc and finally a carboxy-terminal 8xHis-tag (HMK-McpCc-His8). The 23 amino acid spacer between the HMK site and the McpCc allows for cleavage products of less than 20 amino acids to be visualized using the hydroxyl-radical footprinting technique (see below).
A cheD mcpC double knockout strain was created by crossing the cheD knockout strain OI2934 (Rosario et al., 1995) with the mcpC knockout strain OI3280 (Muller et al., 1997) and selecting for erythromycin resistance. The deamidated McpC-Q609E chemoreceptor strains were created by using Quickchange (Stratagene) mutagenesis on the pAIN750mcpC plasmid, which contains the full-length mcpC chemoreceptor gene under control of its native promoter (Kristich & Ordal, 2002). This mutated chemoreceptor was then integrated back into the amyE locus of the cheD mcpC strain, the Δ10mcp strain OI3545 (Hou et al., 2000), which is missing all ten B. subtilis chemoreceptors, and the Δ10mcp cheD strain 3628 (Kristich & Ordal, 2002). Expression was confirmed using Western blots, and expression levels were found to be similar to the wild-type McpC (data not shown).
Capillary assay
The capillary assay was used to quantitatively measure the chemotactic ability of various strains (Adler, 1973, Zimmer et al., 2002). Briefly, strains were grown overnight at 30°C on TBAB (1% tryptone, 0.3% beef extract, 0.5% NaCl, 1.5% agar) plates. Cells were then scraped from the plate and resuspended to an A600nm of 0.03 in capillary assay minimal media (50 mM K3PO4, pH 7.0, 1.2 mM MgCl2, 0.14 mM CaCl2, 1 mM (NH4)2SO4, 0.01 mM MnCl2, 20 mM sorbitol and 0.02% tryptone, supplemented with 50 µg/ml histidine, methionine, and tryptophan). The cultures were grown to an A600nm of 0.4 at 37°C and 250 rpm shaking, after which 50 µl of a 5% glycerol-0.5 M sodium lactate solution was added, and then the cells were incubated a further 15 min. The cells were then washed three times with chemotaxis buffer (10 mM K3PO4, pH 7.0, 0.14 mM CaCl2, 0.3 mM (NH4)2SO4, 0.1 mM EDTA, 5 mM sodium lactate, 0.05% (v/v) glycerol (Ordal & Goldman, 1975)), diluted to an A600nm of 0.001 and aliquoted into 0.3 ml ponds on a temperature-controlled plate at 37°C. Closed-end capillary tubes filled with the appropriate chemoattractant (asparagine or proline) were inserted into the pond. After 30 minutes, the cells in the capillaries were harvested and transferred to 0.5 ml top agar (1% tryptone, 0.8% NaCl, 0.8% agar, 0.5 mM EDTA) and plated onto TBr (1% tryptone, 0.5% NaCl, 1.5% agar) plates. These plates were incubated at 37°C for 16 h at which point colonies were counted to derive the data. Experiments were performed in triplicate and on two different days to assure reproducibility.
Protein purification
The chemoreceptor carboxy-terminal fragments were purified as 6xHis-fusions under denaturing conditions as previously described (Kristich & Ordal, 2002). For the pEB112 derivatives, cells were grown at 37° C in LB (1% tryptone, 0.5% yeast extract, 0.5% NaCl) plus 100 µg/ml ampicillin to mid-log phase, expression was induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG), and incubation was continued for 3 hours at 37° C. For the pUSH1 derivatives, cells were grown at 37°C in LB plus 10 µg/ml chloramphenicol for 9 hours. Cells were then harvested by centrifugation at 5,000 × g for 5 min, resuspended in Buffer B (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris, pH 8) and incubated at room temperature for 1 h with mixing. The supernatants were clarified by two serial centrifugations (7,000 × g, 5 min; 40,000 × g, 1 hour) and loaded onto a 5 mL Hi-Trap Chelating column charged with 0.1 M NiSO4 paired to an AKTA Prime FPLC system (GE Healthcare). The column was washed with 10 volumes Buffer B, followed by 10 washes with Buffer C (Buffer B at pH 6.3). Elution was performed with 25 mL Buffer E (Buffer B at pH 4.5). These samples were dialyzed against three changes of 25 mM NH4HCO3 at 4 °C, and aliquots were frozen at −80°C.
The GST-fusion proteins (CheA, CheD and CheW) were purified per the manufacturer’s protocol (GE Healthcare). To purify the GST fusion proteins, pGEX-6P-2 with the assorted chemotaxis proteins cloned into the multiple-cloning site were grown in 2 liters of LB with 100 µg/ml ampicillin at 37°C and shaking at 250 rpm until the A600nm was equal to 0.8. Expression was then induced by the addition of 1 mM IPTG, and the culture was grown at 25°C with 250 rpm shaking for 16 h. For CheA, the cultures were induced at 37°C for 4 h. Cells were collected by centrifugation at 7,000 × g for 10 min. The cell pellet was resuspended in 3 ml TBS (50 mM Tris, pH 7.5, 150 mM NaCl) + 1% Triton X100 + 1 mM dithiothreitol (DTT) for every 1 g cell pellet. The cells were then disrupted by sonication, and cell debris was removed by centrifugation at 7,000 × g for 10 min, and further clarified with centrifugation for 1 h at 40,000 × g. The cell lysate was then passed through a 5 ml GSTrap column (GE Healthcare) and washed with at least 5 bed volumes of TBS. The fusion proteins were eluted from the column with 20 ml GEB (50 mM Tris, pH 8.0, 10 mM glutathione). If necessary, the GST tag was removed by digestion with 100 units of PreScission protease for 12 h at 4°C. This solution was again passed over the GSTrap column to remove the GST and protease. The pure Che protein flow-through was collected and stored in TKMD (50 mM Tris, pH 8.0, 50 mM KCl, 5 mM MgCl2, 0.1 mM DTT, 10% (v/v) glycerol) buffer at −80°C.
Chemoreceptor modification reactions
Deamidation reactions were performed in a 1:1 molar ratio of chemoreceptor to enzyme (CheD) in TKMD buffer, at a final concentration of 50 µM. Reactions were initially visualized with SDS-PAGE gels to ensure chemoreceptor modification. All reactions were performed at room temperature, although other conditions were tried and found to be less efficient, and for at least 1 h to ensure complete modification. Both in vitro reactions with GST-CheD and co-expression of CheD and the chemoreceptor fragments produced a fully deamidated chemoreceptor fragment (data not shown).
Chemoreceptor digestion and HPLC peptide separation
Deamidation reactions, or chemoreceptor fragments that were co-expressed in the presence of CheD, were digested in a 25:1 molar ratio of protein to trypsin for 24 hours at 37°C. Samples were then injected into a Shimadzu VP Series HPLC, incorporating a Waters Symmetry 300 Reversed Phase C18 column, and running Shimadzu VP-EZStart software, which was subsequently used for all HPLC analysis. A 0–55% gradient of water:acetonitrile, both with 0.1% trifluoroacetic acid, run over 75 min, at 1 ml/min and 35° C, was used to elute tryptic peptides. Peptides were visualized with a UV detector set at 220 nm. Peaks were collected and sent for MALDI MS for initial mass observation. Any peptides that were found to show a different elution profile were sent for both Edman degradation and MS/MS for further identification of modification sites. MALDI and MS/MS analysis was performed by the staff at the UIUC Mass Spectrometry Laboratory and Edman degradation was performed at the UIUC Protein Sciences Facility.
Preparation of bacterial membranes
The bacterial strains were grown on TBAB plates and incubated overnight at 30°C. Colonies from these plates were then used to start 50-ml day cultures in capillary assay minimal medium with an initial A600nm of 0.02. The cells were incubated at 37°C with aeration until they reached mid-exponential phase (approximately 6 h.). The cells were then diluted 1:10 (vol/vol) into 50 ml of capillary assay minimal medium (with only 0.02% tryptone) and further incubated at 37°C until they reached mid-exponential phase (approximately 6 h.). The cells were then diluted an additional time to an A600nm of 0.01 and were incubated until they reached mid-exponential phase (approximately 10 h) in cap assay minimal (without tryptone). The culture was then diluted 1:10 (vol/vol) into multiple flasks (to a total volume of 50 ml) and returned to the incubator until an A600nm of 0.6 was reached in cap assay minimal (without tryptone).
The cells were then harvested by pelleting at 9,900 × g for 15 minutes and washed three times with 1 M KCl to remove extracellular proteases. Cells were resuspended in sonication buffer+ (10 mM potassium phosphate buffer pH 7, 10 mM MgCl2, 1 mM EDTA, 0.3 mM DTT, 20 mM KCl, 1 mM glutamate, 2 mM phenylmethanesulphonyl fluoride and 20% glycerol). The EDTA and phenylmethanesulphonyl fluoride were added as protease inhibitors. Cells were then sonicated and debris removed by centrifugation at 17,600 × g for 15 minutes.
Bacterial membranes were removed by centrifugation at 120,000 × g for 2 hr in a Beckman 70 Ti rotor. Pelleted membranes were resuspended in MT buffer (10 mM potassium phosphate buffer pH 7, 1 mM MgCl2, 0.1 mM EDTA, and 1 mM 2-mercaptoethanol) and homogenized in a glass / Teflon homogenizer and re-centrifuged. This step was repeated once more. Finally, the membranes were homogenized in MT buffer at a concentration of 32 mg/ml and membranes were stored in small aliquots at −80°C.
In vitro assay for chemoreceptor-coupled kinase activity
Reactions consisted of isolated chemoreceptor-containing B. subtilis membranes (see above) to which purified proteins were added to yield the following monomeric concentrations: ~6 µM chemoreceptor, 2 µM CheW, up to 2 µM CheD (when indicated) and 2 µM CheA kinase. The reaction buffer contained 50 mM Tris base, pH 7.5, 50 mM KCl, and 5 mM MgCl2, with or without 1 mM proline (as indicated). Twenty microliter reactions were pre-incubated at 23°C for 40–60 min to permit formation of the chemoreceptor–kinase complex. CheA autophosphorylation was initiated by the addition of [γ32P]ATP (4000–8000 cpm/pmol) to a final concentration of 0.1 mM. Five microliter aliquots were quenched at 15 s by mixing with 15 µL of 2 × Laemmli sample buffer containing 25 mM EDTA at room temperature, essentially fixing the level of phospho-CheA. Initial phospho-CheA formation rates were analyzed by 12% SDS–PAGE. Gels were dried down immediately after electrophoresis and phospho-CheA was quantitated by phosphor-imaging (Molecular Dynamics).
Quantitative GST pulldown assay
These experiments were all performed at 4° C to try and minimize chemoreceptor deamidation by CheD. Initially, 5 nmol GST (control) or GST-CheD plus 5 nmol of chemoreceptor cytoplasmic fragment (McpCc) were mixed to a final volume of 250 µl in PBS-TX buffer (150 mM NaCl, 20 mM Na2HPO4 pH 7.4, 1.0% Triton X-100) and incubated for 15 minutes. This mixture was then added to 150 µl of pre-washed glutathione beads (GE Healthcare) and incubated for another 15 minutes. The samples were then added to a Handee spin cup column (Pierce) and centrifuged at 500 × g. Initial elution was performed by adding 250 µl of GEB buffer, and then a second elution was performed with 1% sodium dodecyl sulfate (SDS) to ensure that all the protein was captured.
Aliquots from each step of the procedure were run on 10 % SDS-PAGE gels, as were standard amounts of GST-CheD (100 to 3.2 pmol) and McpCc (50 to 1.6 pmol). The gels were visualized with Coomassie stain and imaged with a UVP EpiChem3 Darkroom and LabWorks Image Acquisition and Analysis Software. Using the Labworks software, the density of each standard band was calculated and the standards were used to prepare standard curves for the amounts of the GST-CheD and McpCc. These standard curves were then used to calculate the amount of protein in the sample lanes, and from these calculations the total amount of McpCc bound to GST was subtracted from the amount bound to GST-CheD to determine the total amount of fragment bound to GST-CheD. Since all the protein was recovered and the starting concentrations of all the proteins were known, the total amount of unbound GST-CheD and McpCc as well as the amount of each bound to one another could be determined. The dissociation constant of this protein interaction, KD, is defined as [GST-CheDFREE][McpCcFREE]/[GST-CheD:McpCc], and is presented here in µM units.
Hydroxyl radical footprinting experiments
These experiments were based on the previously published protocol with one simple alteration (Loizos, 2004). Briefly, 40 µg of HMK-McpCc-His8 was end-labeled with [γ32P]ATP, purified using Ni2+-NTA agarose beads (Qiagen) and concentrated with a 10K MWCO Amicon Ultra centrifugal filter. The only deviation from the published protocol was that 8 M urea was added to the binding and elution buffers, and then removed with dialysis. Once the labeled protein was purified, it was subjected to hydroxyl radical cleavage by adding 2 µl each of iron-EDTA (10 mM ammonium iron (II) sulfate, 20 mM EDTA), 0.2 M sodium ascorbate and 10 mM hydrogen peroxide in footprinting reaction buffer (10 mM MOPS, pH 7.2, 200 mM NaCl, 10 mM MgCl2). Three separate reactions were run: HMK-McpCc-His8 alone, HMK-McpCc-His8 + GST and HMK-McpCc-His8 + GST-CheD. Each reaction contained 13 µl chemoreceptor (65 pmol) and 1 µl GST or GST-CheD (65 pmol). The reactions were terminated by adding 5 µl 4X SDS-PAGE loading buffer, and the samples were run on on a BioRad TGX 4–12% SDS-PAGE gel. The gels were visualized as described above using a phosphor-imager. Each lane on the gel was segmented into 23 equal pieces using the Labworks software (see above), and the density of each segment was analyzed. The difference in density of the GST control and the GST-CheD reactions for each segment was plotted on a difference plot, and regions that were protected in the presence of GST-CheD were identified.
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health Grant GM054365 to G.W.O and C.V.R. C.V.R. is an investigator with the Energy Biosciences Institute.
References
- Adler J. A method for measuring chemotaxis and use of the method to determine optimum conditions for chemotaxis by Escherichia coli. J Gen Microbiol. 1973;74:77–91. doi: 10.1099/00221287-74-1-77. [DOI] [PubMed] [Google Scholar]
- Bischoff DS, Ordal GW. Sequence and characterization of Bacillus subtilis CheB, a homolog of Escherichia coli CheY, and its role in a different mechanism of chemotaxis. J Biol Chem. 1991;266:12301–12305. [PubMed] [Google Scholar]
- Borkovich KA, Kaplan N, Hess JF, Simon MI. Transmembrane signal transduction in bacterial chemotaxis involves ligand-dependent activation of phosphate group transfer. Proc Natl Acad Sci U S A. 1989;86:1208–1212. doi: 10.1073/pnas.86.4.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich KA, Simon MI. The dynamics of protein phosphorylation in bacterial chemotaxis. Cell. 1990;63:1339–1348. doi: 10.1016/0092-8674(90)90429-i. [DOI] [PubMed] [Google Scholar]
- Burgess-Cassler A, Ullah AH, Ordal GW. Purification and characterization of Bacillus subtilis methyl-accepting chemotaxis protein methyltransferase II. J Biol Chem. 1982;257:8412–8417. [PubMed] [Google Scholar]
- Cannistraro VJ, Glekas GD, Rao CV, Ordal GW. Cellular stoichiometry of the chemotaxis proteins in Bacillus subtilis. J Bacteriol. 2011;193:3220–3227. doi: 10.1128/JB.01255-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao X, Muff TJ, Park SY, Zhang S, Pollard AM, Ordal GW, Bilwes AM, Crane BR. A receptor-modifying deamidase in complex with a signaling phosphatase reveals reciprocal regulation. Cell. 2006;124:561–571. doi: 10.1016/j.cell.2005.11.046. [DOI] [PubMed] [Google Scholar]
- Fuhrer DK, Ordal GW. Bacillus subtilis CheN, a homolog of CheA, the central regulator of chemotaxis in Escherichia coli. J Bacteriol. 1991;173:7443–7448. doi: 10.1128/jb.173.23.7443-7448.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity LF, Ordal GW. Activation of the CheA kinase by asparagine in Bacillus subtilis chemotaxis. Microbiology. 1997;143:2945–2951. doi: 10.1099/00221287-143-9-2945. [DOI] [PubMed] [Google Scholar]
- Garrity LF, Schiel SL, Merrill R, Reizer J, Saier MH, Jr, Ordal GW. Unique regulation of carbohydrate chemotaxis in Bacillus subtilis by the phosphoenolpyruvate-dependent phosphotransferase system and the methyl-accepting chemotaxis protein McpC. J Bacteriol. 1998;180:4475–4480. doi: 10.1128/jb.180.17.4475-4480.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glekas GD, Cates JR, Cohen TM, Rao CV, Ordal GW. Site-specific methylation in Bacillus subtilis chemotaxis: effect of covalent modifications to the chemotaxis receptor McpB. Microbiology. 2011;157:56–65. doi: 10.1099/mic.0.044685-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman DJ, Nettleton DO, Ordal GW. Purification and characterization of chemotactic methylesterase from Bacillus subtilis. Biochemistry. 1984;23:675–680. doi: 10.1021/bi00299a014. [DOI] [PubMed] [Google Scholar]
- Goldman DJ, Ordal GW. In vitro methylation and demethylation of methyl-accepting chemotaxis proteins in Bacillus subtilis. Biochemistry. 1984;23:2600–2606. doi: 10.1021/bi00307a010. [DOI] [PubMed] [Google Scholar]
- Hanlon DW, Marquez-Magana LM, Carpenter PB, Chamberlin MJ, Ordal GW. Sequence and characterization of Bacillus subtilis CheW. J Biol Chem. 1992;267:12055–12060. [PubMed] [Google Scholar]
- Hanlon DW, Ordal GW. Cloning and characterization of genes encoding methyl-accepting chemotaxis proteins in Bacillus subtilis. J Biol Chem. 1994;269:14038–14046. [PubMed] [Google Scholar]
- Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou S, Larsen RW, Boudko D, Riley CW, Karatan E, Zimmer M, Ordal GW, Alam M. Myoglobin-like aerotaxis transducers in Archaea and Bacteria. Nature. 2000;403:540–544. doi: 10.1038/35000570. [DOI] [PubMed] [Google Scholar]
- Karatan E, Saulmon MM, Bunn MW, Ordal GW. Phosphorylation of the response regulator CheV is required for adaptation to attractants during Bacillus subtilis chemotaxis. J Biol Chem. 2001;276:43618–43626. doi: 10.1074/jbc.M104955200. [DOI] [PubMed] [Google Scholar]
- Kehry MR, Engstrom P, Dahlquist FW, Hazelbauer GL. Multiple covalent modifications of Trg, a sensory transducer of Escherichia coli. J Biol Chem. 1983;258:5050–5055. [PubMed] [Google Scholar]
- Kirby JR, Kristich CJ, Saulmon MM, Zimmer MA, Garrity LF, Zhulin IB, Ordal GW. CheC is related to the family of flagellar switch proteins and acts independently from CheD to control chemotaxis in Bacillus subtilis. Mol Microbiol. 2001;42:573–585. doi: 10.1046/j.1365-2958.2001.02581.x. [DOI] [PubMed] [Google Scholar]
- Kristich CJ, Glekas GD, Ordal GW. The conserved cytoplasmic module of the transmembrane chemoreceptor McpC mediates carbohydrate chemotaxis in Bacillus subtilis. Mol Microbiol. 2003;47:1353–1366. doi: 10.1046/j.1365-2958.2003.03375.x. [DOI] [PubMed] [Google Scholar]
- Kristich CJ, Ordal GW. Bacillus subtilis CheD Is a Chemoreceptor Modification Enzyme Required for Chemotaxis. J Biol Chem. 2002;277:25356–25362. doi: 10.1074/jbc.M201334200. [DOI] [PubMed] [Google Scholar]
- Kristich CJ, Ordal GW. Analysis of chimeric chemoreceptors in Bacillus subtilis reveals a role for CheD in the function of the McpC HAMP domain. J Bacteriol. 2004;2002;186:5950–5955. doi: 10.1128/JB.186.17.5950-5955.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Moual H, Koshland DE., Jr Molecular evolution of the C-terminal cytoplasmic domain of a superfamily of bacterial receptors involved in taxis. J Mol Biol. 1996;261:568–585. doi: 10.1006/jmbi.1996.0483. [DOI] [PubMed] [Google Scholar]
- Loizos N. Identifying Protein Interactions by Hydroxyl-Radical Protein Footprinting. Current Protocols in Protein Science. 2004:9.1–9.11. doi: 10.1002/0471140864.ps1909s38. [DOI] [PubMed] [Google Scholar]
- Muff TJ, Ordal GW. The CheC phosphatase regulates chemotactic adaptation through CheD. J Biol Chem. 2007;282:34120–34128. doi: 10.1074/jbc.M706432200. [DOI] [PubMed] [Google Scholar]
- Muller J, Schiel S, Ordal GW, Saxild HH. Functional and genetic characterization of mcpC, which encodes a third methyl-accepting chemotaxis protein in Bacillus subtilis. Microbiology. 1997;143(Pt 10):3231–3240. doi: 10.1099/00221287-143-10-3231. [DOI] [PubMed] [Google Scholar]
- Ordal GW, Goldman DJ. Chemotaxis away from uncouplers of oxidative phosphorylation in Bacillus subtilis. Science. 1975;189:802–805. doi: 10.1126/science.808854. [DOI] [PubMed] [Google Scholar]
- Park SY, Chao X, Gonzalez-Bonet G, Beel BD, Bilwes AM, Crane BR. Structure and function of an unusual family of protein phosphatases: the bacterial chemotaxis proteins CheC and CheX. Mol Cell. 2004;16:563–574. doi: 10.1016/j.molcel.2004.10.018. [DOI] [PubMed] [Google Scholar]
- Perez E, Zheng H, Stock AM. Identification of methylation sites in Thermotoga maritima chemotaxis receptors. J Bacteriol. 2006;188:4093–4100. doi: 10.1128/JB.00181-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao CV, Glekas GD, Ordal GW. The three adaptation systems of Bacillus subtilis chemotaxis. Trends Microbiol. 2008;16:480–487. doi: 10.1016/j.tim.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario MM, Fredrick KL, Ordal GW, Helmann JD. Chemotaxis in Bacillus subtilis requires either of two functionally redundant CheW homologs. J Bacteriol. 1994;176:2736–2739. doi: 10.1128/jb.176.9.2736-2739.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario MM, Kirby JR, Bochar DA, Ordal GW. Chemotactic methylation and behavior in Bacillus subtilis: role of two unique proteins, CheC and CheD. Biochemistry. 1995;34:3823–3831. doi: 10.1021/bi00011a040. [DOI] [PubMed] [Google Scholar]
- Schon U, Schumann W. Construction of His6-tagging vectors allowing single-step purification of GroES and other polypeptides produced in Bacillus subtilis. Gene. 1994;147:91–94. doi: 10.1016/0378-1119(94)90044-2. [DOI] [PubMed] [Google Scholar]
- Swain KE, Gonzalez MA, Falke JJ. Engineered socket study of signaling through a four-helix bundle: evidence for a yin-yang mechanism in the kinase control module of the aspartate receptor. Biochemistry. 2009;48:9266–9277. doi: 10.1021/bi901020d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szurmant H, Bunn MW, Cannistraro VJ, Ordal GW. Bacillus subtilis hydrolyzes CheY-P at the location of its action, the flagellar switch. J Biol Chem. 2003 doi: 10.1074/jbc.M306180200. [DOI] [PubMed] [Google Scholar]
- Szurmant H, Muff TJ, Ordal GW. Bacillus subtilis CheC and FliY are members of a novel class of CheY-P hydrolyzing proteins in the chemotactic signal transduction cascade. J Biol Chem. 2004 doi: 10.1074/jbc.M311497200. [DOI] [PubMed] [Google Scholar]
- Szurmant H, Ordal GW. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol Mol Biol Rev. 2004;68:301–319. doi: 10.1128/MMBR.68.2.301-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah AH, Ordal GW. In vivo and in vitro chemotactic methylation in Bacillus subtilis. J Bacteriol. 1981;145:958–965. doi: 10.1128/jb.145.2.958-965.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AJ, Conley MP, Kramer TJ, Berg HC. Reconstitution of signaling in bacterial chemotaxis. J Bacteriol. 1987;169:1878–1885. doi: 10.1128/jb.169.5.1878-1885.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Walukiewicz HE, Glekas GD, Ordal GW, Rao CV. Attractant binding induces distinct structural changes to the polar and lateral signaling clusters in Bacillus subtilis chemotaxis. J Biol Chem. 2011;286:2587–2595. doi: 10.1074/jbc.M110.188664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Ames P, Parkinson JS. Mutational analyses of HAMP helices suggest a dynamic bundle model of input-output signalling in chemoreceptors. Mol Microbiol. 2009;73:801–814. doi: 10.1111/j.1365-2958.2009.06819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Ames P, Parkinson JS. Biphasic control logic of HAMP domain signalling in the Escherichia coli serine chemoreceptor. Mol Microbiol. 2011;80:596–611. doi: 10.1111/j.1365-2958.2011.07577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer MA, Szurmant H, Saulmon MM, Collins MA, Bant JS, Ordal GW. The role of heterologous receptors in McpB-mediated signalling in Bacillus subtilis chemotaxis. Mol Microbiol. 2002;45:555–568. doi: 10.1046/j.1365-2958.2002.03035.x. [DOI] [PubMed] [Google Scholar]
- Zimmer MA, Tiu J, Collins MA, Ordal GW. Selective methylation changes on the Bacillus subtilis chemotaxis receptor McpB promote adaptation. J Biol Chem. 2000;275:24264–24272. doi: 10.1074/jbc.M004001200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.