Abstract
This work illustrates potential adverse effects linked with the expression of proteinase inhibitor (PI) in plants used as a strategy to enhance pest resistance. Tobacco (Nicotiana tabacum L. cv Xanthi) and Arabidopsis [Heynh.] ecotype Wassilewskija) transgenic plants expressing the mustard trypsin PI 2 (MTI-2) at different levels were obtained. First-instar larvae of the Egyptian cotton worm (Spodoptera littoralis Boisd.) were fed on detached leaves of these plants. The high level of MTI-2 expression in leaves had deleterious effects on larvae, causing mortality and decreasing mean larval weight, and was correlated with a decrease in the leaf surface eaten. However, larvae fed leaves from plants expressing MTI-2 at the low expression level did not show increased mortality, but a net gain in weight and a faster development compared with control larvae. The low MTI-2 expression level also resulted in increased leaf damage. These observations are correlated with the differential expression of digestive proteinases in the larval gut; overexpression of existing proteinases on low-MTI-2-expression level plants and induction of new proteinases on high-MTI-2-expression level plants. These results emphasize the critical need for the development of a PI-based defense strategy for plants obtaining the appropriate PI-expression level relative to the pest's sensitivity threshold to that PI.
PIs are widely spread throughout the plant kingdom. They are known to be involved in several physiological processes, such as reserve control and defense against pathogens and pests (Koiwa et al., 1997). In the latter case, PIs have been shown to be developmentally expressed in seeds and reserve organs (Birk, 1994; Koiwa et al., 1997) or induced by wounding in leaves (Schaller and Ryan, 1995). The natural protective role of PIs against phytophageous insects and the availability of PI-encoding sequences encouraged the development of pest-resistance programs based on PI expression in transgenic plants (Ryan, 1990).
Despite several reports of successful protection of plants and trees against phytophageous insects from several taxonomic orders, mainly Lepidoptera (Jongsma and Bolter, 1997; Gatehouse, 1998; Jouanin et al., 1998; Schuler et al., 1998), defense strategies based on PI expression in plants have not resulted in any commercial application so far. This relates to two distinct problems: (a) the pests' capacity to react to PI consumption, and (b) the PI-expression levels in transgenic plants.
The first point is exemplified by the fact that PIs shown to be potent inhibitors of insect gut proteinases in in vitro assays failed to produce any deleterious effect when fed to larvae (Baker et al., 1984; Purcell et al., 1992). Recent data provide some insights into the insect pests' ability to overcome the potential deleterious effects caused by PI consumption. Several mechanisms were reported: the inactivation of PI by insensitive proteinases (Jongsma and Bolter, 1997; Michaud, 1997) and the synthesis of novel proteinases insensitive to the PI ingested (Jongsma et al., 1996; Jongsma and Bolter, 1997). To counter such resistance mechanisms, several strategies have been proposed for the design of more efficient pest-resistant transgenic plants: the expression in the same plant of several genes encoding entomopathogenic molecules (PI and other types of molecules such as Bacillus thuringiensis toxins) and/or the use of engineered inhibitors with increased specificity or a larger spectrum (Jongsma et al., 1996; Jongsma and Bolter, 1997; Michaud 1997; Reeck et al., 1997).
In comparison with the pests' adaptation capacities, little attention has been given to potential problems linked to the level of expression of PI genes in transgenic plants. Reports have shown that to cause deleterious effects on pests, PI-expression levels have to be relatively high, on the order of 1% of total soluble protein, as previously shown in bioassays of artificial diets (Hilder et al., 1987; Leplé et al., 1995). Nevertheless, to our knowledge, no attempt has been made to estimate the possible occurrence of adverse effects linked to inadequate PI-expression levels in transgenic plants. In view of a possible field release of transgenic plants expressing PI, this point appears to be of crucial importance.
To test the possibility of the occurrence of negative effects linked to PI-expression levels, tobacco (Nicotiana tabacum L. cv Xanthi) and Arabidopsis L. (Heynh.) ecotype Wassilewskija transgenic plants constitutively expressing MTI-2 were obtained. MTI-2 was isolated from seeds of white mustard (Menegatti et al., 1992) and is related (70% amino acid homology) to the rapeseed trypsin inhibitor (Ceciliani et al., 1994). Both PIs have primary structure characteristics that are uncommon to PI families described so far (Reeck et al., 1997). In mustard, MTI-2 exhibits expression patterns (Ceci et al., 1995) similar to that of PIs known to be part of plant-defense mechanisms in potato (Suh et al., 1991) and tomato (Wingate and Ryan, 1991). Therefore, MTI-2 is thought to be a good candidate for enhancing pest resistance in plants. In this paper we report the effect of consumption by larvae of a lepidopteran pest, the Egyptian cotton worm (Spodoptera littoralis Boisd.), of leaves of tobacco and Arabidopsis plants expressing both low and high levels of MTI-2.
MATERIALS AND METHODS
Construction of Plasmid Vector pKY-MTI-2
The mti-2 cDNA (accession no. Y16190) was obtained by PCR amplification of a full-length cDNA inserted in plasmid pUC19 (L.R. Ceci, unpublished data) by using the M13 forward oligonucleotide as an upstream primer and the specific oligonucleotide 5′-GGC AGC CTC TAG AAA CTC AAA TGC CAC CTC TTA G-3′ as a downstream primer. This primer contains the UGA stop codon and an XbaI restriction site. The fragment obtained by SacI-XbaI restriction of the amplification product was inserted in the pKYLX71–35S2 binary plasmid (Maiti et al., 1993), downstream of the 35S2 promoter. The resulting construct was named pKY-MTI-2 (Fig. 1). It also carries a kanamycin-resistance gene under the control of the nopaline synthase promoter for selection of transformed plants.
Figure 1.
Structure of the pKY-MTI-2 T-region. Pnos, Tnos, Nopaline synthase promoter and terminator, respectively. nptII, Neomycin phosphotransferase-coding sequence; mti-2, MTI-2-coding sequence; P35S2, 35S RNA promoter with a doubled enhancer; trbcS, pea Rubisco terminator; LB, RB, left and right border sequences, respectively. The arrows indicate the direction of transcription of the mti-2 and nptII genes.
Plant Transformation and Regeneration
pKY-MTI-2 and pKYLX71–35S2 were transferred to Agrobacterium tumefaciens strain GV301(pMP90) by triparental mating (Van Haute et al., 1983) and used for tobacco (Nicotiana tabacum L. cv Xanthi) and Arabidopsis L. (Heynh.) ecotype Wassilewskija transformation. Transformation of tobacco plants was carried out according to the leaf-disc method (Mathis and Hinchee, 1994), and transgenic Arabidopsis plants were obtained by infiltration (Bechtold et al., 1993). The progeny of tobacco and Arabidopsis transformed lines was obtained by self-pollination of plants. The segregation of the introduced gene was observed by selection on kanamycin-containing medium (100 mg/L).
Plant Preparation
Plants used for biochemical tests and bioassays were grown simultaneously in the greenhouse with the following conditions: 13-h day/7-h night, 8000 lux of natural light supplemented by sodium vapor lamps, maximum (day)/minimum (night) temperature at 23°C/14°C. Ten and 20 plants per line were prepared, respectively, for tobacco and Arabidopsis so that each plant was used only once for leaf sampling, either for MTI-2-expression analysis or for insect feeding, to avoid bias due to the systemic induction of endogenous PIs.
MTI-2 Expression in Leaves of Transgenic Plants
The MTI-2-expression level was measured, in parallel with the bioassay, in leaves of the same developmental stage as those used for larvae feeding. Tests were performed on extracts prepared as described by Leplé et al. (1995), with the following changes: after the initial centrifugation step the supernatant was not heated but was centrifuged again (144,000g, 1 h, 4°C). The resulting supernatant was collected for tests. The MTI-2 expression in plant leaves was monitored using two different protocols: gelatin/PAGE for rapid detection/plant comparison and azocasein tests for quantification. Gelatin/PAGE was conducted essentially as described by Michaud et al. (1993) on 15% (w/v) acrylamide/0.6% (w/v) bis-acrylamide gels containing 0.1% gelatin (porcine, type A). After the proteins were renatured, the gel was incubated with trypsin (0.5 μg/mL in 0.1 m Tris, pH 8.0) for 15 min at room temperature and then for 4 h at 37°C. Staining was with Coomassie brilliant blue. Azocasein tests used to quantify the level of inhibition of β-bovine trypsin by leaf-soluble protein extracts are based on the quantification of the proteolysis products of azocasein. Protein extracts were incubated for 2 h at 37°C in 0.02 m CaCl2, 0,01 m Tris-HCl, pH 7.5, 2.5 mg/mL trypsin, and 0.5% azocasein. Reactions were stopped and readings made as described by Leplé et al. (1995). The remaining trypsin activity was expressed as a percentage of the control activity without extract.
Feeding Bioassays
Thirty late, first-instar (just before the change to second instar) larvae of the Egyptian cotton worm (Spodoptera littoralis) were placed in a box on detached tobacco leaves. Boxes were kept at 22°C. Damp absorbing paper provided sufficient humidity in the boxes. Leaves were replaced by fresh ones every 2 d. The development of larvae was monitored throughout the bioassay by noting the larval stage of each insect every 2 d. At the end of the test, after 10 d, insects were individually weighed and kept at −80°C until needed or were immediately dissected.
Gut pH Determination
pH was measured using narrow-range (6.5–10.0) pH indicator paper. Two measurements were obtained from each of three independent homogenates. Each homogenate was prepared from guts isolated from two larvae.
Proteinase Activity in S. littoralis
Gut extracts were prepared from larvae surviving the 10-d feeding bioassay. At least six extracts were made for each condition tested. Each extract was prepared using the guts of two medium-sized larvae. All insects used for gut preparation were third-instar larvae. Larval gut extracts and subsequent proteinase activity tests were performed using azoalbumin as a substrate, as described by Leplé et al. (1995). When used, PIs (0.1 mg/mL BBI, 0.1 mm E64) were preincubated with proteinases for 5 min before the addition of azoalbumin.
Detection of Digestive Proteinase in S. littoralis by Two-Step Gelatin/PAGE
Proteins extracted from S. littoralis larval guts were resolved on SDS-PAGE and then transferred to a gelatin/polyacrylamide gel, as described by Michaud (1998). Transfer was carried out at 4°C for 40 min at 45 V. Renaturation was achieved by incubating the gelatin/polyacrylamide gel at 4°C for 30 min in a 2.5% Triton X-100 solution. Subsequent gelatin digestion was carried out at 37°C for 2 h by incubating the gelatin/polyacrylamide gel in a 10 mm ethanolamine, 10 mm phosphate buffer (pH 11.0) containing 8% Triton X-100. Proteinases were visualized as clear bands on a blue background after the gel was stained with Coomassie blue. For inhibition studies, BBI (0.5 mg/mL) or leaf-protein extracts (0.3 mg/mL) was added to the renaturing and reaction solutions.
Leaf-Surface Consumption Analysis
Leaves fed to the larvae during d 9 and 10 were collected. The remaining leaf surface was measured by a computer-aided image analyzer (Morphostar software, Imstar Co., Paris) and compared with a reconstruction of the intact leaf.
Statistics
Mean weights of larvae were compared using a Student's t test modified for small samples. Mortality was compared using a χ2 test. For each test, the statistical significance is given in parentheses.
RESULTS
Plant Transformation and in Vitro Analysis of MTI-2 Expression
Transgenic tobacco and Arabidopsis plants bearing the T-DNA from either pKY-MTI-2 (MTI-2-expressing plants) or pKYLX71–35S2 (control plants) were obtained by A. tumefaciens-mediated transformation. T-DNA integration and copy number were determined by PCR and Southern-blot hybridization (results not shown). Zygotic status and T-DNA transmission to the progeny were checked by in vitro segregation analysis on kanamycin-containing medium (results not shown). Of the transformed lines obtained, one homozygous line of each tobacco (F1) and Arabidopsis (F3) control plant bearing one copy of the T-DNA was chosen and named CT and CA, respectively. MTI-2 expression in plants bearing the pKY-MTI-2 T-DNA was measured by in vitro inhibition of β-bovine trypsin by leaf-soluble protein extracts, using azocasein as a substrate. Two single-copy homozygous lines of transgenic tobacco were chosen that exhibited either a high (line 2T) or low (line 4T) MTI-2 level of expression in leaves (Table I).
Table I.
Trypsin inhibition by leaf extracts from selected transgenic lines
| Line | Trypsin Inhibition |
|---|---|
| mg/g soluble proteins | |
| Tobacco (F1) | |
| Control | Not detected |
| 2T | 111.6 |
| 4T | 37.2 |
| Arabidopsis (F3) | |
| Control | 17 |
| 7A | 122.5 |
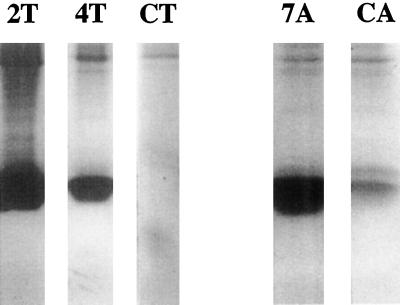
By comparing on activity gels the trypsin inhibition by leaf extracts of MTI-2-expressing plants with the trypsin inhibition by soybean trypsin inhibitor, Kunitz type, we estimated that the high-expression line (2T) expressed MTI-2 at about 1.6% of leaf-soluble proteins, whereas in the low-expression line (4T) MTI-2 accounted for 0.5% of the soluble proteins. All Arabidopsis lines expressed MTI-2 at a comparable level, similar to that observed in tobacco line 2T (Table I). Therefore, only one single-copy homozygous line, noted 7A, was selected. In both tobacco and Arabidopsis, northern-blot analysis gave results concurrent with azocasein tests (results not shown). Figure 2 shows a comparison of β-bovine trypsin inhibitory activity of the selected transgenic lines. In Arabidopsis, a weak trypsin inhibition activity was observed in line CA, at a molecular size similar to that of MTI-2. This may be related to the presence in the Arabidopsis genome of a PI gene (accession no. AC002355) with high sequence homology to MTI-2. All selected lines were subsequently used in S. littoralis larvae-feeding bioassays.
Figure 2.
Visualization of MTI-2 in transgenic plants by gelatin/PAGE. Plant extracts were separated in acrylamide gels containing gelatin. Gelatin was subsequently degraded by incubation of the gel in a trypsin-containing solution. Trypsin inhibition by 10 μg (tobacco) and 5 μg (Arabidopsis) of soluble proteins from leaf extract was visualized by the undegraded blue-stained gelatin located where the PI initially migrated.
Bioassays on S. littoralis Larvae
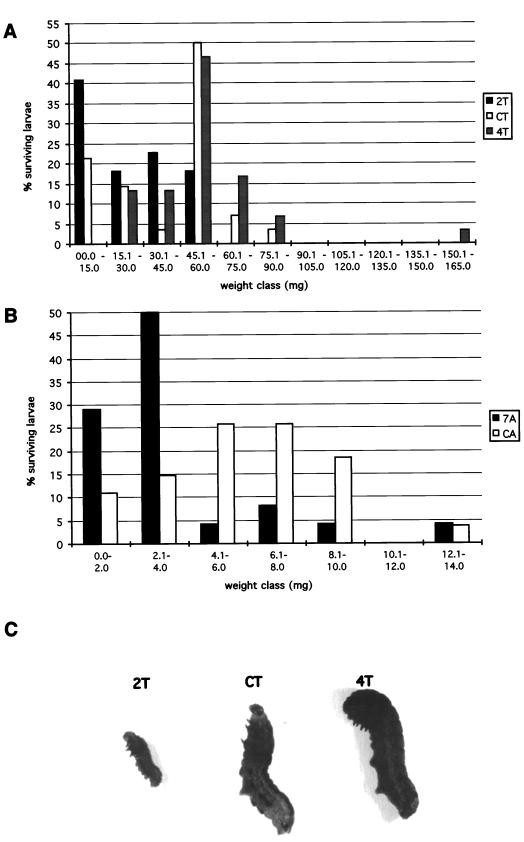
The effect of feeding on leaves from MTI-2-expressing plants was tested on late first-instar larvae of S. littoralis. Larvae were grown on detached leaves of tobacco and Arabidopsis transgenic lines for 10 and 7 d, respectively. The results obtained with tobacco are summarized in Table II. Larvae fed on tobacco line 2T showed a significant increase in mortality (27% versus 7% in control, P < 5%), and surviving larvae were smaller after 10 d (24.5 mg versus 40.6 mg, P < 1%) when compared with larvae fed on control plants (Fig. 3C). Similar results were obtained with Arabidopsis, with 23% mortality on line 7A (10% on control) and a mean weight reduction of 37% (4 mg instead of 6.4 mg), comparable to the 39% weight reduction observed on tobacco line 2T. The damage observed on tobacco leaves was reduced by 30% on the line expressing high levels of MTI-2 (Table II). In contrast, larvae fed on the tobacco line expressing low levels of MTI-2 showed no significant difference in mortality (0% versus 7%) and had an increased mean weight (54.0 mg versus 40.6 mg, P < 5%) by the end of the bioassay when compared with larvae fed on control plants (Fig. 3C). Damage to the leaves was also increased by 26% compared with the control (Table II). Differences observed in mean weight between larvae fed on lines 2T and 4T (54.0 mg versus 24.5 mg, P < 0.1%; Table II; Fig. 3C) proceeded both from a tendency to have, respectively, a slower or faster development than the control (Table II) and a decreased or increased weight, respectively, compared with the control (Fig. 3A).
Table II.
Bioassays using S. littoralis larvae fed on transgenic tobacco plants
| Line | Mortality at 10 d | Larvae
Instar at 10 d
|
Mean Wt at 10 d | Leaf Surface Eaten | ||
|---|---|---|---|---|---|---|
| L2 | L3 | L4 | ||||
| % of initial | % of surviving larvae | mg | cm2 per larvae | |||
| Control | 7 | 4 | 85 | 11 | 40.6 | 4.18 |
| 2T | 27 | 9 | 91 | 0 | 24.5 | 2.92 |
| 4T | 0 | 0 | 80 | 20 | 54.0 | 5.26 |
Assays were performed on groups of 30 pre-L2 instar larvae fed for 10 d on transgenic tobacco leaves. Mortality at the end of the assay is reported as the percentage of initial larvae number. Larval instar at the end of the assay is given as a percentage of surviving larvae. L2, L3, and L4 refer to instar 2, 3, and 4, respectively. Leaf-surface damage was measured after the last 2 d of the assay (d 9 and 10).
Figure 3.
Comparison of weight of S. littoralis larvae fed on transgenic plant leaves for 10 d. A and B, Weight distribution in larvae fed on tobacco (A) or Arabidopsis (B); C, larvae grown on tobacco lines 2T, 4T, and CT. Each larvae shown here had a weight nearly equal to the mean weight observed in the insect pool to which it belonged.
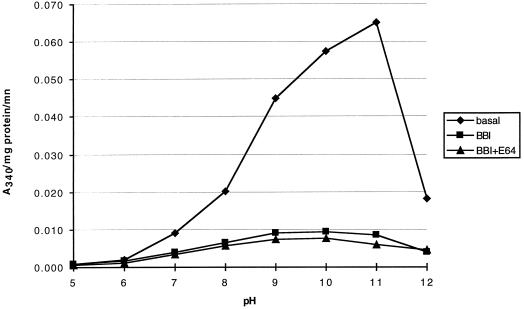
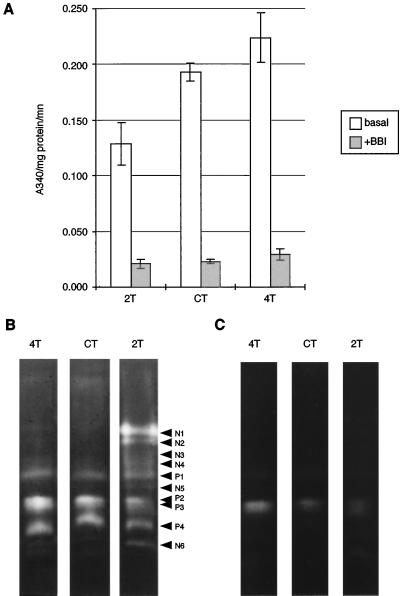
To assess potential effects of MTI-2 consumption on the gut proteinase pool in S. littoralis larvae, proteinase activity was first analyzed in fourth-instar larvae grown on control plants. Activity profiles showed a peak of activity at pH 11.0 (Fig. 4), which is in agreement with previously published data for this insect (Ishaaya et al., 1971). This result is in accordance with the mean pH value observed for gut homogenate, i.e. pH 9.3. The overall activity showed a high sensitivity to the trypsin/chymotrypsin inhibitor BBI (Fig. 4), with 90% of activity inhibited at the optimum pH (Fig. 5A). Proteinase activity insensitive to BBI is likely to include elastase-like proteinase(s), since degradation of N-succinyl-Ala-Ala-Pro-Leu-p-nitroanilide, shown to be a substrate for gut chymotrypsin and elastase-like proteinase in other lepidopteran pests (Johnston et al., 1995), was only partly inhibited by the specific chymotrypsin inhibitor N-tosyl-l-Phe chloromethyl ketone (result not shown). E64, a potent inhibitor of Cys proteinases, had no significant effect on BBI-insensitive proteolytic activity (Fig. 4). The proteolytic complex was found to include at least four proteinases active at pH 11.0 (Fig. 5B). These proteinases were completely inhibited by BBI (result not shown), whereas protein extract from MTI-2-expressing plants inhibited all but P2 proteinase, which showed reduced activity (Fig. 5C). Overall, the digestive proteolytic system we observed in S. littoralis is consistent with previously published data for Noctuidae (Christeller et al., 1992), which had a proteinase activity that relied mainly on Ser proteinases, with a major component being trypsin-like proteinases (although chymotrypsin- and elastase-like proteinases are also present). The proteinase P2, which is partly inhibited by MTI-2, may account for part of this non-trypsin-like activity.
Figure 4.
Determination of digestive proteinase activity in S. littoralis gut. Gut proteinase activity was monitored using azoalbumin, a general proteinase substrate, at different pHs. The activity was measured by the amount of liberated oligo- and polypeptides by reading A340. The activity was monitored without (basal) or with PIs: 0.1 mg/mL of the trypsin/chymotrypsin BBI or 0.1 mg/mL of BBI and 0.1 mm of the Cys PI E64 (BBI + E64).
Figure 5.
Comparison of digestive proteinase activity (pH 11.0) of S. littoralis larvae fed on transgenic tobacco plants for 10 d. Gut extracts were labeled according to the transgenic plants they fed on. A, Activity monitored without (basal) or with BBI at 0.1 mg/mL (+BBI). Each point is the mean of two repetitions for at least six extracts. The confidence interval was computed at 10%. B and C, Gelatin/PAGE assays of gut protein extracts (5 μg of protein) incubated with protein-leaf extract (1 μg/μL) from the control (B) or the MTI-2-expressing plant line 2T (C). Proteinases present in control larvae are denoted P1 to P4, whereas proteinases induced in larvae fed line 2T tobacco plants are denoted N1 to N6.
Consumption of leaves from tobacco plants expressing low levels of MTI-2 resulted in an increase in total proteinase activity (Fig. 5A). This higher activity stemmed from an increased expression of proteinases already present in control insects (Fig. 5, B and C). The sensitivity of these newly induced proteinases to BBI (result not shown) and MTI-2 (Fig. 5C) appeared to be unchanged when compared with proteinases extracted from control larvae. On the contrary, the consumption of tobacco expressing high levels of MTI-2 resulted in a 33% decrease in the proteolytic activity at pH 11.0 (Fig. 5A), although six new proteinases appeared on the proteolytic profile (Fig. 5B). These new proteinases were completely inhibited by BBI (result not shown) and MTI-2 (Fig. 5C). On activity gels, there was no evidence for an increase in proteinase P3 (which is incompletely inhibited by MTI-2) activity compared with control larvae. It is interesting that, whatever the level of expression of MTI-2 in leaves, the amount of proteinase activity insensitive to BBI did not change significantly (Fig. 5A).
DISCUSSION
This work illustrates the risks associated with a strategy for pest control based on PI expression in transgenic plants. It concentrates on the occurrence of unexpected adverse effects as the result of inadequate PI expression. A variation of PI-expression levels within the range previously shown to provide insect resistance to transgenic plants (Hilder et al., 1987; McManus et al., 1994; Leplé et al., 1995; Duan et al., 1996; Xu et al., 1996; Gatehouse et al., 1997; Yeh et al., 1997) resulted in dramatic changes in the reaction of S. littoralis larvae to transgenic leaf consumption. When the trypsin PI MTI-2 was expressed at the highest level in tobacco and Arabidopsis transgenic plants, deleterious effects were observed on insect larvae, together with a reduction of leaf damage, thus providing effective pest resistance to the plant. These results are in agreement with those obtained by Yeh et al. (1997) on the closely related pest, Spodoptera litura F, when expressing a sweet potato trypsin inhibitor in tobacco. On the contrary, when MTI-2 was expressed at lower levels in tobacco plants, larvae developed faster, were bigger than on control plants, and caused more damage to the leaves. These results were related to physiological changes in the proteolytic profile of the digestive tract of S. littoralis larvae.
The growth-enhancing effect of PI-containing leaves has been previously reported in a few different studies. The Noctuidae Thysanoplusia orichalcea F., closely related to S. littoralis, showed an increased growth when fed tobacco expressing a chymotrypsin inhibitor inconsistently (McManus et al., 1994). The cabbage seed weevil (Coleoptera Psylliodes chrysocephala L.), raised on oilseed rape expressing the Cys PI OCI, exhibited an increased weight compared with controls, together with a doubling of both OCI-sensitive and -insensitive preexisting proteinases (Girard et al., 1998c). Similarly, our results show that the consumption by S. littoralis of leaves expressing MTI-2 at the lowest level induced an increase in preexisting proteinase expression in gut, although not as large as found with P. chrysocephala. The mechanisms underlying this increase in proteinase production and linking it to an increase in leaf consumption are still unknown. Proteinase overproduction associated with a reduced growth has often been reported when insects were fed diets containing PI (Jongsma and Bolter, 1997). Broadway and Duffey (1986) proposed that PI-induced deleterious effects are caused by stimulating proteinase overproduction aimed at compensating the inhibition of a proteolytic activity, thus inducing a shortage of some amino acids. Such a compensation mechanism may explain results obtained from tobacco plants expressing low levels of MTI-2, even though such a level of PI expression failed to induce deleterious effects on S. littoralis. The increase in leaf-surface consumption observed on low MTI-2-expressing plants may be a consequence of the decrease in the diet's quality due to the presence of MTI-2 and/or to the increase in gut proteolytic capacity of the larvae. Global proteinase activity enhancement could provide sufficient MTI-2-insensitive proteinases to counter, at least partly, the effect of the amount of MTI-2 present in the larvae's diet, e.g. by degrading this PI, as recently reported in other insects (Girard et al., 1998b; Giri et al., 1998).
The proteolytic activity observed in larvae fed leaves expressing high levels of MTI-2 differed greatly from that observed in larvae fed leaves with low MTI-2-expression levels. The 30% decrease in overall proteinase activity, compared with the control, was shown to coincide with the appearance of new proteinases. The induction of new forms of proteinase after feeding on a PI-containing diet has been previously described in other insects (Broadway, 1996; Jongsma and Bolter, 1997). Newly induced proteinases have been shown to exhibit a lower sensitivity toward the PI consumed by the insect. In S. littoralis we observed that all of the newly induced proteinases were still sensitive to MTI-2, suggesting that the deleterious effects observed on growth and mortality could be linked to a shortage of available amino acids because of extensive proteinase synthesis. Leaf damage was reduced the same way as overall proteinase activity, suggesting that the consumed leaf area is at least partly determined by the proteolytic activity in the gut.
The dramatic differences observed in larvae fed leaves expressing high or low levels of MTI-2 suggest the occurrence of a sensitivity threshold in S. littoralis toward MTI-2. This threshold represents the minimum concentration of MTI-2 in the larvae's diet necessary to induce deleterious effects. Below this threshold value, larvae are able to overcome the inhibition caused by PI ingestion by overexpressing digestive proteinases. Above this threshold, PI ingestion induces strong adaptive reactions, such as the production of new proteinases, mobilizing enough of the larvae resources to impair its growth. If the threshold is not reached, adverse effects such as damage enhancement and/or increased pest growth can occur.
This work addresses questions that must be considered on a case-by-case basis with regard to two different parameters: the target pest and the in planta expression characteristics. On the one hand, the sensitivity threshold for a same PI is likely to differ from insect to insect, since different insects have been shown to exhibit various sensitivities to a given PI and single insects have been shown to exhibit different sensitivities to various PIs (Larocque and Houseman, 1990; Christeller et al., 1992; McManus et al., 1994). The design of a pest-defense strategy for a crop using a particular PI is therefore dependent on the assessment of the sensitivity threshold of the target pest toward the chosen PI. In addition, care must be taken for the deployment of PI transgenic plants in the field, since different populations of the insect pests can exhibit different sensitivity levels to a same PI, as reported recently (Girard et al., 1998a). However, the stability of expression is paramount to the successful use of PI to protect plants against attack by insect pests. Aside from the problem of the choice of an adequate promoter, the development of a successful defense strategy against a given pest, based on PI expression in planta, must take into account potential variations in PI-expression levels in a plant (as a result of age, physiological status, etc.) and between plants of the same line (Elkind et al., 1995) to ensure that the sensitivity threshold of this pest for the PI is always reached. With regard to this requirement, the use of PI presenting a high affinity to an insect digestive proteinase is highly desirable, as lower PI-expression levels could be sufficient to impair development. Strategies aimed at improving the affinity of a PI for a target pest's major proteinase before its expression in plants (Jongsma et al., 1996) should therefore receive special attention.
ACKNOWLEDGMENTS
We thank Dr. J. Chaufaux (Station de lutte biologique, Institut National de la Recherche Agronomique, La Minière, 78285 Guyancourt cedex, France) for providing S. littoralis larvae and Dr. M. LeMétayer (Laboratoire de Neurobiologie Comparée des Invertébrés, Institut National de la Recherche Agronomique, BP23, 91440 Bures sur Yvette, France) for technical assistance during image-analysis sessions. The authors are also grateful to Dr. M. Giband for critical reading of the manuscript.
Abbreviations:
- 2T
MTI-2-expressing tobacco line 2
- 4T
MTI-2-expressing tobacco line 4
- 7A
Arabidopsis MTI-2-expressing line 7
- BBI
soybean Bowman Birk Inhibitor
- CA
Arabidopsis control line
- CT
tobacco control line
- E64
trans-epoxysuccinyl-l-leucylamido(4-guanidino)-butane
- MTI-2
mustard trypsin inhibitor 2
- PI
proteinase inhibitor
Footnotes
This work was partially funded by Ministero per le Polotiche Agrarie. F.D.L. is a PhD student partially sponsored by Consorzio Interuniveritario per le Biotechnologie. The French-Italian collaboration was funded by a Galileo project.
LITERATURE CITED
- Baker J, Woo S, Mullen M. Distribution of proteinases and carbohydrases in the midgut of larvae of the sweet potato weevil Cylas elegantulus and response of proteinases to inhibitors from sweet potato. Entomol Exp Appl. 1984;36:97–105. [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. CR Acad Sci Ser III Sci Vie. 1993;316:1194–1199. [Google Scholar]
- Birk Y. Protein proteinase inhibitors in legume seeds–overview. Arch Latinoam Nutr. 1994;44:26S–30S. [PubMed] [Google Scholar]
- Broadway RM. Dietary proteinase inhibitors alter complement of midgut proteases. Arch Insect Biochem Physiol. 1996;32:39–53. [Google Scholar]
- Broadway R, Duffey S. Plant proteinase inhibitors: mechanism of action and effect on the growth and digestive physiology of larval Heliothis virescens and Spodoptera exigua. J Insect Physiol. 1986;32:827–833. [Google Scholar]
- Ceci LR, Spoto N, de Virgilio M, Gallerani R. The gene coding for the mustard trypsin inhibitor-2 is discontinuous and wound-inducible. FEBS Lett. 1995;364:179–181. doi: 10.1016/0014-5793(95)00386-n. [DOI] [PubMed] [Google Scholar]
- Ceciliani F, Bortolotti F, Menegatti E, Ronchi S, Ascenzi P, Palmieri S. Purification, inhibitory properties, amino acid sequence and identification of the reactive site of a new serine proteinase inhibitor from oil-rape (Brassica napus) seeds. FEBS Lett. 1994;342:221–224. doi: 10.1016/0014-5793(94)80505-9. [DOI] [PubMed] [Google Scholar]
- Christeller JT, Laing WA, Markwick NP, Burgess EPJ. Midgut protease activities in 12 phytophageous lepidopteran larvae: dietary and protease inhibitor interactions. Insect Biochem Mol Biol. 1992;22:735–746. [Google Scholar]
- Duan X, Li X, Xue Q, Abo-El-Saad M, Xu D, Wu R. Transgenic rice plants harboring an introduced potato proteinase inhibitor II gene are insect resistant. Nature Biotechnol. 1996;14:494–498. doi: 10.1038/nbt0496-494. [DOI] [PubMed] [Google Scholar]
- Elkind Y, Nir B, Nadler-Hassar T. Quantitative analysis of the transgene variability among primary tobacco transformants. Transgenic Res. 1995;4:30–38. [Google Scholar]
- Gatehouse AMR (1998) Biotechnological application of plant genes in the production of insect resistant crops. In SL Clement, SS Quinsenberry, eds, Global Plant Genetic Resources for Insect Resistant Crops, Chap 15. CRC Press, Boca Raton, FL (in press)
- Gatehouse AMR, Davison GM, Newell CA, Merryweather A, Hamilton WDO, Burgess EPJ, Gilbert RJC, Gatehouse JA. Transgenic potato plants with enhanced resistance to the tomato moth, Lacanobia oleracea: growth room trials. Mol Breeding. 1997;3:49–63. [Google Scholar]
- Girard C, Bonadé-Bottino M, Pham-Delègue MH, Jouanin L. Two strains of cabbage seed weevil (Coleoptera: Curculionidae) exhibit differential susceptibility to a transgenic oilseed rape expressing oryzacystatin I. J Insect Physiol. 1998a;44:569–577. doi: 10.1016/s0022-1910(98)00050-x. [DOI] [PubMed] [Google Scholar]
- Girard C, Le Métayer M, Bonadé-Bottino M, Pham-Delègue MH, Jouanin L. High level of resistance to proteinase inhibitors may be conferred by proteolytic cleavage in beetle larvae. Insect Biochem Mol Biol. 1998b;28:229–237. doi: 10.1016/s0965-1748(98)00027-7. [DOI] [PubMed] [Google Scholar]
- Girard C, Le Métayer M, Zaccomer B, Bartlet E, William I, Bonadé-Bottino M, Pham-Delègue MH, Jouanin L. Growth stimulation of beetle larvae reared on a transgenic oilseed rape expressing a cysteine proteinase inhibitor. J Insect Physiol. 1998c;44:263–270. doi: 10.1016/s0022-1910(97)00142-x. [DOI] [PubMed] [Google Scholar]
- Giri AP, Harsulkar AM, Deshpande VV, Sainani MN, Gupta VS, Ranjekar PK. Chickpea defensive proteinase inhibitors can be inactivated by podborer gut proteinases. Plant Physiol. 1998;116:393–401. [Google Scholar]
- Hilder VA, Gatehouse AMR, Sheerman SE, Barker RF, Boulter D. A novel mechanism of insect resistance engineered into tobacco. Nature. 1987;300:160–163. [Google Scholar]
- Ishaaya I, Moore I, Joseph D. Protease and amylase activity in larvae of the Egyptian cotton worm, Spodoptera littoralis. J Insect Physiol. 1971;17:945–953. [Google Scholar]
- Johnston KA, Lee MJ, Brough C, Hilder VA, Gatehouse AMR, Gatehouse J. Protease activities in the larval midgut of Heliothis virescens: evidence for trypsin and chymotrypsin-like enzymes. Insect Biochem Mol Biol. 1995;25:375–383. [Google Scholar]
- Jongsma MA, Bolter C. The adaptation of insect to plant protease inhibitors. J Insect Physiol. 1997;43:885–896. doi: 10.1016/s0022-1910(97)00040-1. [DOI] [PubMed] [Google Scholar]
- Jongsma MA, Stiekema WJ, Bosch D. Combatting inhibitor-insensitive proteases of insect pests. Trends Biotechnol. 1996;14:331–333. [Google Scholar]
- Jouanin L, Bonadé-Bottino M, Girard C, Morrot G, Giband M. Transgenic plants for insect resistance. Plant Sci. 1998;131:1–11. [Google Scholar]
- Koiwa H, Bressan RA, Hasegawa PM. Regulation of protease inhibitors and plant defense. Trends Plant Sci. 1997;2:379–384. [Google Scholar]
- Larocque AM, Houseman JG. Effect of ingested soybean, ovomucoid and corn protease inhibitors on digestive processes of the European corn borer, Ostrinia nubilalis (Lepidoptera: pyralydae) J Insect Physiol. 1990;36:691–697. [Google Scholar]
- Leplé JC, Bonadé-Bottino M, Augustin S, Pilate G, Dumanois G, Le Tan VD, Delplanque A, Cornu D, Jouanin L. Toxicity to Chrysomela tremulae (Coleoptera: Chrysomelidae) of transgenic poplars expressing a cysteine proteinase inhibitor. Mol Breeding. 1995;1:319–328. [Google Scholar]
- Maiti IB, Murphy JF, Shaw AG. Plants that express a potyvirus VPg-proteinase gene are resistant to virus infection. Proc Natl Acad Sci USA. 1993;90:6110–6114. doi: 10.1073/pnas.90.13.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis NL, Hinchee MAW. Agrobacterium inoculation techniques for plant tissues. In: Gelvin SB, Schilperoort RA, editors. Plant Molecular Biology Manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 1–9. [Google Scholar]
- McManus MT, White DWR, McGregor PG. Accumulation of a chymotrypsin inhibitor in transgenic tobacco can affect the growth of insect pests. Transgenic Res. 1994;3:50–58. [Google Scholar]
- Menegatti E, Tedeschi G, Ronchi S, Bortolotti F, Ascenzi P, Thomas RM, Bolognesi M, Palmieri S. Purification, inhibitory properties and amino acid sequence of a new serine proteinase inhibitor from white mustard (Sinapis alba L.) seeds. FEBS Lett. 1992;301:10–14. doi: 10.1016/0014-5793(92)80199-q. [DOI] [PubMed] [Google Scholar]
- Michaud D. Avoiding protease-mediated resistance in herbivorous pests. Trends Biotechnol. 1997;15:4–6. [Google Scholar]
- Michaud D (1998) Gel electrophoresis of proteolytic enzymes. Anal Chim Acta (in press)
- Michaud D, Faye L, Yelle S. Electrophoretic analysis of plant cysteine and serine proteinases using gelatin-containing polyacrilamide gels and class-specific proteinase inhibitors. Electrophoresis. 1993;14:94–98. doi: 10.1002/elps.1150140117. [DOI] [PubMed] [Google Scholar]
- Purcell JP, Greenplate JT, Sammons RD. Examination of midgut luminal proteinase activities in six economically important insects. Insect Biochem Mol Biol. 1992;22:41–47. [Google Scholar]
- Reeck GR, Kramer KJ, Baker JE, Kanost MR, Fabrick JA, Behnke CA (1997) Proteinase inhibitors and resistance of transgenic plants to insects. In N Carozzi, M Koziel, eds, Advances in Insect Control. The Role of Transgenic Plants. Taylor and Francis, London, pp 157–183.
- Ryan CA. genes for improving defences against insects and pathogens Annu Rev Phytopathol. 1990;28:425–449. [Google Scholar]
- Schaller A, Ryan CA. Systemin—a polypeptide defense signal in plants. Bioessays. 1995;18:27–33. doi: 10.1002/bies.950180108. [DOI] [PubMed] [Google Scholar]
- Schuler TH, Poppy GM, Kerry BR, Denholm I. Insect-resistant transgenic plants. Trends Biochem Sci. 1998;16:168–175. doi: 10.1016/S0167-7799(98)01298-0. [DOI] [PubMed] [Google Scholar]
- Suh SG, Stiekema WJ, Hannapel DJ. Proteinase-inhibitor activity and wound-inducible gene expression of the 22-kDa potato-tuber proteins. Planta. 1991;184:423–430. doi: 10.1007/BF00197888. [DOI] [PubMed] [Google Scholar]
- Van Haute E, Joos H, Maes M, Warren G, Van Montagu M, Schell J. EMBO J. 1983;2:411–417. doi: 10.1002/j.1460-2075.1983.tb01438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingate VPM, Ryan CA. Uniquely regulated proteinase inhibitor I gene in a wild tomato species. Inhibitor I family gene is wound-inducible in leaves and developmentally regulated in fruit. Plant Physiol. 1991;97:496–501. doi: 10.1104/pp.97.2.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Xue Q, McElroy D, Mawal Y, Hilder VA, Wu R. Constitutive expression of a cowpea trypsin inhibitor gene CpTI in transgenic rice plants confers resistance to two major insect pests. Mol Breeding. 1996;2:167–173. [Google Scholar]
- Yeh KW, Lin MI, Tuan SJ, Chen YM, Lin CY, Kao SS. Sweet potato (Ipomoea batatas) trypsin inhibitors expressed in transgenic tobacco plants confer resistance against Spodoptera litura. Plant Cell Rep. 1997;16:696–699. doi: 10.1007/s002990050304. [DOI] [PubMed] [Google Scholar]