Abstract
A 3,3′-diindolylmethane (DIM) is a major in vivo condensation product of indole-3-carbinol (I3C), which are present in cruciferous vegetables. Although these compounds have been widely implicated in anti-tumorigenic and pro-apoptotic properties in animal as well as in vitro models of cancer, the underlying cellular mechanisms regulated by DIM are only partially understood. Activating transcription factor 3 (ATF3) is a member of the ATF/CREB subfamily of the basic-region leucine zipper (bZIP) family and has been known to induce apoptosis in human colorectal cancer cells. The present study was performed to elucidate the molecular mechanism of ATF3 induction by DIM in human CRC cells. The DIM treatment induced apoptosis and induced ATF3 gene expression at protein and mRNA levels. DIM increased ATF3 promoter activity and the region of −84 to +34 within ATF3 promoter was responsible for promoter activation by DIM. This region contained an ATF binding site. Deletion and point mutation of the ATF binding site (−23 to −16) abolished ATF3 promoter activation by DIM and overexpression of ATF4 enhanced ATF3 transactivation. Chromatin immunoprecipitation (ChIP) assay confirmed the binding of ATF4 in the ATF3 promoter. Inhibition of ATF4 expression by siRNA results in repression of DIM-induced ATF3 expression. The current study demonstrates that DIM stimulates ATF3 expression through ATF4-mediated pathway and subsequently induces apoptosis in human colorectal cancer cells.
Keywords: DIM, ATF3, ATF4, colon cancer
1. Introduction
Epidemiological studies have shown that the level of consumption of cruciferous vegetables is inversely associated with incidences of colon cancers , and in experimental animals, cruciferous vegetables have been shown to inhibit chemically-induced colon cancer .
3,3′-diindolylmethane (DIM) is a major acid condensation product of indole-3-carbinol (I3C), a naturally occurring component of Brassica species or cruciferous vegetables (cabbage, broccoli, cauliflower, and Brussels sprouts) . DIM is readily detected in the livers and feces of rodents fed I3C, whereas the parent I3C compound has not been detected in tissues of these rodents . Thus, the biological effects of I3C are attributable to DIM, which exhibits anti-tumorigenic activities in vivo and in vitro by inhibiting the growth of colon, prostate, and breast cancer cells . DIM and its derivatives also suppress cell proliferation and induce apoptosis in colon cancer cells , as well as in other types of cancer cells including prostate , breast , bladder , pancreas , and hepatoma . Further, DIM induces apoptosis by inactivating the AKT pathway in breast cancer cells and activating TRAIL expression and Nur77 in colon cancer cells . At the transcriptional level, DIM induces anti-tumorigenic genes IFNγ and caveolin via a PPARγ-dependent pathway, and p21WAF1/CIP1 expression by Sp1-mediated activation . DIM also inhibits VEGF-induced neovascularization by inhibiting Ras signaling in endothelial cells and suppresses MMP2- and MMP9-mediated metastasis . Like other phytochemicals, DIM affects many different pathways and thus knowledge of additional mechanisms by which DIM controls the apoptosis pathway could provide a better understanding of DIM’s effect on anti-tumorigenesis.
Activating transcription factor 3 (ATF3) is a member of the ATF/CREB subfamily of the basic-region leucine zipper (bZIP) family, and its expression is dramatically induced in response to a variety of stress conditions in many different tissues . For the incidence and development of cancer, ATF3 exhibits dual functions (tumor suppressor or tumor promoter), depending on cell context. For example, the expression of ATF3 was repressed in human colorectal tumors compared to normal adjacent tissue , and overexpression of ATF3 protein induced apoptosis in colon cancer cells . Furthermore, ATF3 enhances p53 activation , inhibits Ras-mediated tumorigenesis, and down-regulates cyclin D1 and matrix metalloproteinase-2 expression . ATF3 also mediates therapeutic activity of Hsp90 inhibition and anti-metastatic activity of NDRG1 and KAI1 . In the previous studies, we and others reported that ATF3 was induced by treatment of cells with the anti-tumorigenic compounds indole-3-carbinol , conjugated linoleic acid , epicatechin gallate , and resveratrol , as well as tolfenamic acid and PI3 kinase inhibitor . Moreover, ATF3 is a target of some anti-cancer drugs such as cisplatin and bortezomib . On the other hand, ATF3 is rapidly induced in cells treated with growth stimulators such as serum and growth factor . ATF3 induces DNA synthesis and expression of cyclin D1 in hepatocytes and is involved in serum-induced cell proliferation as a target gene of c-myc . In breast cancer, ATF3 enhances cancer-cell initiating features and is associated with activation of the canonical Wnt/β-catenin pathway.
In a previous study, we reported that DIM induces expression of ATF3 and subsequently activates the pro-apoptotic protein NSAID-activated gene-1 (NAG-1) and apoptosis in HCT-116 human colorectal adenocarcinoma cells . In this report, we further investigated the molecular mechanism of DIM-induced ATF3 expression and found that ATF4 activates in DIM-induced ATF3 transcription.
2. Materials and Methods
2.1. Materials
Human colorectal adenocarcinoma cells HCT-116, SW480, HT-29, LoVo, and Caco-2, were purchased from American Type Culture Collection (Manassas, VA). Culture media were purchase from the followings: McCoy’s 5A (Bio Whittaker, Rockland ME), RPMI1640 (Mediatech, Herndon, VA), Ham’s F-12 (HyClone, Logan, UT), and DMEM (Invitrogen, Carlsbad, CA). DIM and cycloheximide were purchased from Sigma (St. Louis, MO). Antibodies for ATF3, ATF4, and actin and siRNAs for ATF3 and ATF4 were purchased from Santa Cruz (Santa Cruz, CA). All chemicals were purchased from Fisher Scientific, unless otherwise specified.
2.2. Plasmid
Human ATF3 promoter constructs (−1850 to +34 and −84 to +34) were previously reported . The internal-deleted or point-mutated constructs of the ATF3 promoter were constructed from wild-type ATF3-Luc (pATF3−84/+34) using the QuikChange II mutagenesis kit (Stratagene, La Jolla, CA). Expression vectors for C/EBPα, β, δ, CHOP, CREB, ATF3, and ATF2 were described previously . ATF4 expression vector was purchased from OriGene (Rockville, MD).
2.3. Cell culture
HCT-116 and HT-29 cells were maintained in McCoy’s 5A medium. SW480 cells and LoVo cells were maintained in RPMI and Ham’s F-12 medium, respectively. Caco-2 cells were maintained in DMEM. All culture medium was supplemented with 10% fetal bovine serum (FBS), and 100 units/mL penicillin, 100 μg/mL streptomycin.
2.4. Analysis of colorectal cancer cell proliferation and apoptosis
Cell proliferation was measured according to the manufacturer’s instruction for the Cell Proliferation Assay system (Promega, Madison, WI). Briefly, the cells were seeded onto 96-well culture plates and then treated with 0, 12.5, 25, or 50 μM of DIM in culture media containing 1% FBS for 0, 1, 2, or 4 d. The cells were treated with 20 μL of CellTiter96 Aqueous One solution for 1 h at 37°C and absorbance (A490) was compared in an ELISA plate reader (Bio-Tek Instruments Inc, Winooski, VT). Apoptosis of DIM-treated cells was measured as previously described .
2.5. Transient transfections
Transient transfections were performed using the Lipofectamine (Invitrogen) or PolyJet DNA transfection reagent (SignaGen Laboratories, Ijamsville, MD) according to the manufacturers’ instruction. HCT-116 cells were plated in 12-well plates at the concentration of 2×105 cells/well. After growth overnight, plasmid mixtures containing 0.5 μg of ATF3 promoter linked to luciferase and 0.05 μg of pRL-null vector were transfected for 5 h (for lipofectamine) and 16 h (for Polyjet). The transfected cells were cultured in the absence or presence of DIM for 24 h. The cells were then harvested in 1X luciferase lysis buffer, and luciferase activity was normalized to the pRL-null luciferase activity using a dual luciferase assay kit (Promega). For the co-transfection experiment, 0.25 μg of ATF3 promoter and 0.25 μg of expression vectors were co-transfected with 0.05 μg of pRL-null vector.
2.6. Isolation and analysis of RNA and RTPCR
Total RNA was prepared using an RNA isolation kit (Eppendorf, Hamburg, Germany) and treated with DNase I (Invitrogen) prior to cDNA synthesis. Total RNA (1 μg) was reverse-transcribed with iScript cDNA kit (BioRad, Hercules, CA) according to the manufacturer’s instruction. PCR was carried out using ReadyMix Taq polymerase (Sigma) with primers for human ATF3 and GAPDH as follows: ATF3: forward 5′-gtttgaggattttgctaacctgac-3′, and reverse 5′-agctgcaatcttatttctttctcgt-3′; GAPDH: forward 5′- gggctgcttttaactctggt-3′, and reverse 5′- tggcaggtttttctagacgg-3′.
2.7. Western Blot analysis
Cells were washed with PBS, and cell lysates were isolated in RIPA buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS) supplemented with protease inhibitors (1 mM PMSF, 5 μg/ml aprotinin, 5 μg/ml leupeptin) and phosphatase inhibitors (1 mM Na3VO4, 1 mM NaF) and centrifuged at 10,000×g for 5 min at 4°C. Protein concentration was determined by the BCA protein assay (Pierce, Rockford, IL) using BSA as the standard. The proteins were separated on SDS-PAGE and transferred to nitrocellulose membranes (Osmonics, Minnetonka, MN). The membranes were incubated with a specific primary antiserum in TBS containing 0.05% Tween 20 (TSB-T) and 5% nonfat dry milk at 4°C overnight. After three washes with TBS-T, the blots were incubated with peroxidase-conjugated IgG for 1 h at room temperature, visualized using ECL (Amersham Biosciences, Piscataway, NJ) and quantified by Scion Image Software (Scion Corp., Frederick, MD).
2.8. RNA Interference
HCT-116 cells were transfected with control small interference RNA (siRNA) or ATF4 siRNA at a concentration of 10 nM, using PepMute siRNA transfection reagent (SignaGen) for 24 h according to the manufacturer’s protocol. After serum starvation for overnight, the cells were treated with DMSO or 25 μM DIM for 6 h.
2.9. Chip Assay
The ChIP assay was performed as we described previously . Briefly, HCT-116 cells were treated with 25 μM DIM for 6 h. After cross-linkage with 1% formaldehyde for 10 min at 37°C, cells were rinsed twice with ice-cold PBS, resuspended in 200 μL of SDS lysis buffer, and incubated on ice for 10 min. Cell lysates were sonicated four times for 10 s and centrifuged for 10 min. Immunoprecipitation was performed with 5 μg of specific antibodies for ATF4 at 4°C for overnight. Immune complexes were then mixed with Protein A/G Agarose/Salmon Sperm DNA at 4°C. The chromatin-associated DNA was eluted and reverse cross-linked by heating at 65°C for 4 h. DNA was purified by adding proteinase and phenol/choloroform extraction. Precipitated DNA was resuspended in 50 μl of TE and analyzed by PCR with 30 cycles using the following primer pairs: forward, 5′- ccgaacttgcatcaccagt-3′ and reverse, 5′-cgttgcatcaccccttttat-3′. PCR products were electrophresed through a 2% agroase gel and visualized by ethidium bromide staining.
2.10. Statistical analysis
SAS for windows (v9.2; SAS institute, Inc.) statistical analysis software was used. For multiple group comparisons, analysis of variance with Tukey’s multiple comparison test was used to compare mean values. The Student t test was used to analyze differences between samples. Results were considered statistically significant at * P < 0.05, ** P < 0.01, and *** P < 0.001.
3. Results
3.1. Effect of DIM on cell proliferation, apoptosis, and ATF3 expression
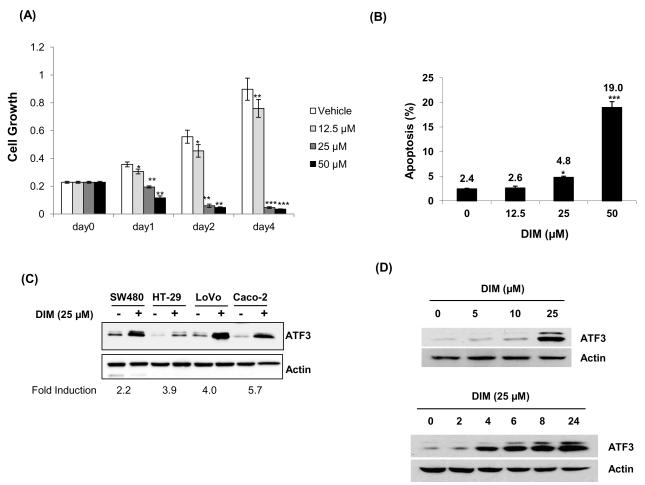
To investigate whether DIM affects cell growth in colorectal cancer cells, HCT-116 cells were incubated with 0, 12.5, 25, and 50 μM of DIM for 0, 1, 2, and 4 d, and cell proliferation was measured. As shown in Fig. 1A, cell growth significantly decreased in dose-dependent manner after treatment with DIM. Subsequently, HCT-116 cells were incubated with 0, 12.5, 25, and 50 μM DIM for 24 h and apoptosis measured. As shown in Fig. 1B, percentage of apoptotic cells were 2.4, 2.6, 4.8 and 19.0% in HCT-116 cells treated with 0, 12.5, 25 and 50 μM DIM, respectively. There is growing evidence that ATF3 is linked to cell growth arrest and apoptosis in colorectal tumorigenesis. To investigate whether DIM induces ATF3 in human colorectal cancer cells, SW480, HT-29, LoVo, and Caco-2 cells were treated with 25 μM DIM for 24 h. ATF3 induction by DIM was observed in human colorectal cancer cells, indicating that ATF3 induction by DIM is a common phenomenon in colorectal cancer cells tested here (Fig. 1C). Finally, HCT-116 cells were treated with DIM and ATF3 expression was measured. In agreement with the previous report , DIM induces ATF3 expression in a dose and time dependent manner (Fig. 1D).
Fig. 1. Induction of ATF3 expression and apoptosis in DIM-treated human CRC cells.
(A) HCT-116 cells were treated with 0, 12.5, 25, and 50 μM of DIM for 0, 1, 2, and 4 d. Cell growth was measured using CellTiter96 Aqueous One Solution Cell Proliferation Assay. Values are expressed as mean ± SD of four replicates. *P < 0.05, **P < 0.01, ***P < 0.001 versus Vehicle (DMSO)-treated cells. (B) HCT-116 cells were treated with 0, 12.5, 25, and 50 μM of DIM for 24 h. Apoptosis was analyzed using PI/Annexin V staining. Values are expressed as mean ± SD of three replicates. *P < 0.05, ***P < 0.001 versus 0 μM cells. (C) Human colorectal cancer cells (SW480, HT-29, LoVo, and Caco-2) were treated with 25 μM DIM for 24 h in serum-free media and Western blot analysis was performed using ATF3 and Actin antibodies. A representative blot was shown in the top and average fold induction over the vehicle-treated samples from three independent experiments is shown in the bottom. (D) HCT-116 cells were exposed to DIM in different concentrations and times as indicated. Cell lysates were isolated and subjected to Western analysis.
3.2. Effect of DIM on ATF3 promoter activity
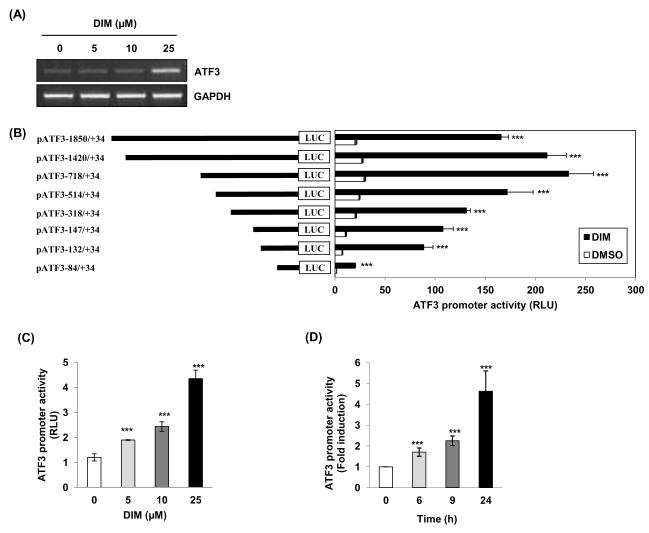
To elucidate a molecular mechanism by which DIM increases ATF3 expression in human colorectal cancer cells, first we measured ATF3 mRNA levels to see whether increasing ATF3 protein is associated with transcriptional regulation. ATF3 mRNA in cells increased at 25 μM DIM treatment (Fig. 2A), consistent with protein expression (Fig. 1D). To investigate whether DIM affects transcriptional regulation of the ATF3 gene, promoter activity was measured using different sizes of ATF3 promoter luciferase constructs (pATF3−1850/+34, pATF3−1420/+34, pATF3−718/+34, pATF3−514/+34, pATF3−318/+34, pATF3−147/+34, pATF3−132/+34 and pATF3−84/+34) . These constructs were transfected into HCT-116 cells and treated with 25 μM DIM for 24 h. As shown in Fig. 2B, DIM treatment resulted in an increase of promoter activity. The fold induction was 7.9, 7.7, 7.8, 7.0, 6.3, 9.9, 11.9, and 11.6 in pATF3−1850/+34, pATF3−1420/+34, pATF3−718/+34, pATF3−514/+34, pATF3−318/+34, pATF3−147/+34, pATF3−132/+34 and pATF3−84/+34, respectively. Because fold inductions of luciferase activities by DIM were highest in cells transfected with pATF3−132/+34 and pATF3−84/+34 constructs, we then continued to study using pATF3−84/+34 construct. Induction of ATF3 promoter activity by DIM showed dose- and time-dependence in cells transfected with the pATF3−84/+34 construct (Fig. 2C and 2D).
Fig. 2. The effect of DIM on ATF3 gene promoter activity.
(A) The cells were treated with indicated concentrations of DIM for 24 h, and RT-PCR was performed using specific primers as described under “materials and methods”. (B) The ATF3 promoter constructs (0.5 g) with different lengths were co-transfected with pRL-null vector (0.05 μg) into HCT-116 cells as described in “materials and methods.” The cells were treated with DMSO or DIM (25 μM each) for 24 h and luciferase activity was measured. RLU indicates relative luciferase unit. *** P < 0.001 versus 0 μM-treated cells. (C) The pATF3−84/+34 construct was co-transfected with pRL-null vector (0.05 μg) into HCT-116 cells. The cells were treated with different concentrations of DIM for 24 h and luciferase activity was measured. The results show mean ± SD of 3 separate transfections. *** P < 0.001 versus DMSO-treated cells. (D) The pATF3−84/+34 construct was co-transfected with pRL-null vector (0.05 μg) into HCT-116 cells. The cells were treated with 25 μM of DIM for 0, 6, 9, and 24 h and luciferase activity was measured. Fold induction refers to ratio of luciferase activity of DIM-treated cells at each time point compared to 0 time point. The results show mean ± SD of three separate transfections. *** P < 0.001 versus DMSO-treated cells.
3.3. Identification of cis-acting element responsible for DIM-induced ATF3 expression
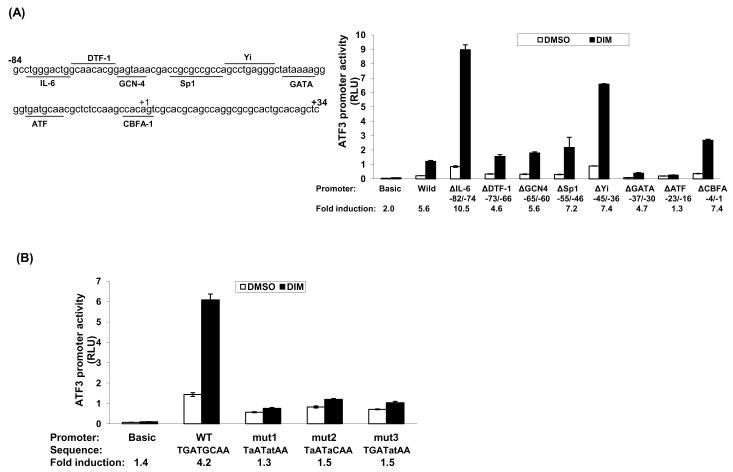
The ATF3 gene promoter within the −84 and +34 region (pATF3−84/+34) contains multiple transcription factor-binding sites including IL-6, DTF-1, GCN-4, Sp1, Yi, GATA, ATF, and CBFA-1 (Fig. 3A, left panel) . To confirm the responsible site for the transactivation of the ATF3 gene by DIM, we constructed deletion clones lacking each binding site. HCT-116 cells were transfected with deletion constructs and then exposed to DMSO or 25 μM DIM for 24 h. As shown in Fig. 3A (right panel), transfection of the wild-type pATF3−84/+34 promoter increased luciferase activity 5.6 fold, whereas transfection of the promoter lacking ATF binding site increased luciferase activity 1.3 fold, which is similar to basal induction (2 fold) by DIM found in cells transfected with empty vector (pGL3-Basic). It is interesting that deletion of IL-6, Sp-1, Yi and CBFA binding sites increased promoter activities, whereas lack of DTF-1 and GATA slightly decreased promoter activity by DIM treatment, compare to wild-type transfected cells in fold induction. Next, to obtain further evidence that the ATF binding site is responsible for activation of ATF3 transcription by DIM, we constructed three point mutation clones replacing two or three nucleotides within the ATF binding site as described in Fig. 3B. Wild-type pATF3−84/+34 resulted in 4.2-fold induction of luciferase activity. However, all clones having point mutations in the ATF binding site completely blocked induction of luciferase activity by DIM, which is comparable with empty vector-transfected cells. These results strongly indicate that the region spanning −23 and −16 in the promoter of ATF3 plays an essential role in mediating the effect of DIM.
Fig. 3. Identification of regulatory element for DIM-induced ATF3 promoter activation.
(A) The putative transcription factor binding sites within the −84 to +34 region of the ATF3 promoter (left panel). The internal deletion clones of the ATF3 promoter (0.5 μg) lacking binding site of indicated transcription factor were co-transfected with 0.05 μg of pRL-null vector using Lipofectamine. After growth overnight with fresh media, the cells were treated with 25 μM of DIM for 24 h. Luciferase activity was measured as a ratio of firefly luciferase signal/renilla luciferase signal and was shown as mean ± S.D. of three independent transfections. (B) Luciferase assay with point mutation clones. Point mutations are indicated by small letters. The cells were co-transfected with 0.5 μg of point mutated clones of ATF3 promoter and 0.05 μg of pRL-null vector using Lipofectamine and treated with 25 μM of DIM for 24 h. The results are presented as mean ± S.D. of three independent transfections.
3.4. Identification of ATF4 as positive regulator of ATF3 transcription
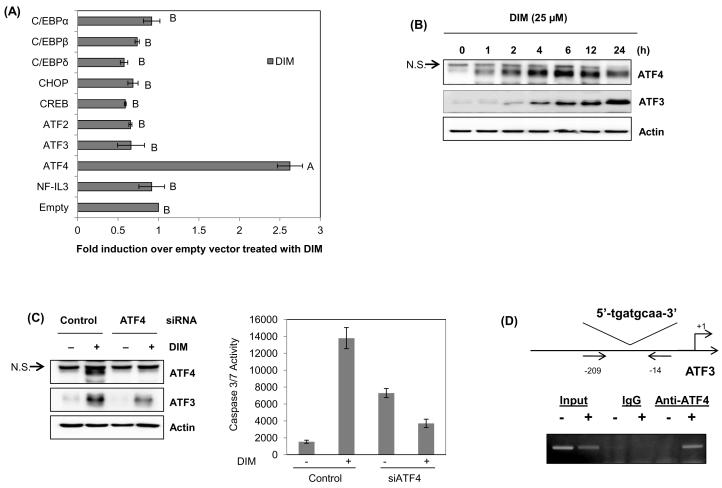
The core sequence (tgatgcaa) of the ATF binding site is potentially associated with binding to ATF, CREB, C/EBP, and NF-IL3. Thus, to obtain evidence of what transcription factor regulates ATF3 transactivation induced by DIM, ATF3 promoter clones were co-transfected with expression vectors indicated in Fig. 4A and luciferase activity was determined. Expression of transfected vectors was confirmed by Western blot as described previously , and overexpression of ATF4 was confirmed (data not shown). As a result, overexpression of C/EBPβ, C/EBPδ, CHOP, CREB, ATF2 and ATF3 led to decreased DIM-induced ATF3 transactivation, compared with empty vector-transfected cells, suggesting that these transcription factors play a significant role in repressing ATF3 expression. However, C/EBPα and NF-IL3 expression did not affect DIM-mediated ATF3 promoter activity. Interestingly, ATF4 overexpression induced transcriptional activity of ATF3 gene in basal and DIM-treated groups (Fig. 4B). Because ATF4 expression is associated with ATF3 transactivation, we examined whether DIM affects ATF4 expression. As shown in Fig. 4B, DIM began to increase the ATF4 protein level after 1 h, which was followed by ATF3 induction. Subsequently, we tested if ATF4 is responsible for DIM-induced ATF3 expression. Knockdown of ATF4 suppressed DIM-induced ATF3 overexpression and an increased caspase 3/7 activity was not observed in cells transfected with ATF4 siRNA (Fig. 4C). These data indicate that ATF4 mediates DIM-induced ATF3 expression and apoptosis. Finally, to see whether ATF4 directly binds to the ATF3 promoter including the ATF binding site, we performed chromatin immunoprecipitation (ChIP) assay using PCR primers amplifying the ATF3 promoter. Chromatin-bound proteins were immunoprecipitated with specific antibodies for IgG and ATF4 using nuclear proteins of HCT-116 cells treated with DIM for 6 h. Then, DNA fragments associated with immunoprecipitated proteins were amplified by PCR using primers containing ATF binding site on ATF3 promoter. As shown in Fig. 4D, immunoprecipitation of chromatin-protein complex with anti-ATF4 amplified 196 bp size of PCR products whereas no bands were found in the presence of IgG. An aliquot (2%) of the total chromatin DNA was used for input. This results support that endogenous ATF4 binds to ATF binding site on ATF3 promoter. Taken together, DIM-induced ATF3 expression is mediated by ATF4 expression.
Fig. 4. Effect of ATF4 overexpression on DIM-induced ATF3 transactivation.
(A) Cells were co-transfected with wild-type ATF3 promoter in the presence of each expression vector using Lipofectamine. After growth overnight with fresh media, the cells were treated with 25 μM of DIM for 24 h. The results are presented as the mean ± S.D. of three independent transfections. Data were analyzed using Tukey’s multiple comparison test; mean with same letters indicate no significance (P < 0.05). (B) Cells were treated with 25 μM of DIM for 0, 1, 2, 4, 6, 12, and 24 h and Western blot was performed with ATF4, ATF3 and actin antibodies. N.S., no-specific. (C) Left panel. Cells were transfected with ATF4 siRNA using PepMute siRNA transfection reagent for 24 h, and then treated with 25 μM of DIM for 6 h. N.S., no-specific. Right panel. The same cell lysates were examined caspase 3/7 activity (Caspase-Glo 3/7 Assay kit, Promega). The results are presented as mean ± S.D. of three independent transfections. (D) Binding of ATF4. The ChIP assay was performed using HCT-116 cells treated with DIM (25 μM) for 6 h as described in “Material and methods.” The chromatin-associated DNA was incubated in the absence or in the presence of specific antibodies for IgG and ATF4. An aliquot (2%) of the total chromatin DNA was used for input. Immnoprecipitates were subjected to PCR with a primer-pair specific to the ATF3 promoter that amplified a 196 bp fragment. After 30 cycles of amplification, PCR products were electrophresed through a 2% agarose gel and visualized by ethidium bromide staining.
4. Discussion
The anti-tumorigenic effects of I3C and its condensation product, DIM, in a variety of experimental cancer models have raised an important question regarding the underlying molecular mechanisms. Here, we demonstrate direct evidence that DIM activates ATF3 gene expression via an ATF4 dependent pathway in human colorectal cancer cells. ATF3 also mediates endoplasmic reticulum (ER) stress-induced cell death of human colorectal cancer cells in response to DIM . These results demonstrate that understanding regulation of cell death pathways in response to ER stress-inducing drugs such as DIM provide the potential to elucidate novel therapeutic targets for chemoprevention and chemotherapeutics.
Luciferase analysis using comprehensive internal deletion clones, and point mutation of the ATF binding site (-23/-16) of the ATF3 promoter indicates that potential protein binding to this sequence (TGATGCAA) may modulate DIM-induced ATF3 transcriptional activity. ChIP assay demonstrated that ATF4 directly binds to this site, and co-transfection with ATF4 enhanced DIM-induced ATF3 transactivation. ATF4 (also known as CREB-2) belongs to the ATF/CREB family of bZIP protein and is induced by several factors and stressors, such as hypoxia ER stress and amino acid deprivation . ATF4 expression is regulated transcriptionally, translationally, or posttranslationally. In particular, ATF4 is degraded by SCFβ-TrCP in phosphorylation-dependent interaction although the specific phosphorylation site is not known. Recent data suggest that ATF4 plays an important role in drug resistance and oncogenic process ; however, our data indicate that ATF4 may play a role in anti-tumorigenesis of human colorectal cancer since it is up-regulated by the anti-cancer compound DIM and controls tumor-suppressor ATF3 expression. In addition, knockdown of ATF4 using siRNA ameliorated DIM-induced ATF3 expression. Thus, the exact biological activity of ATF4 in carcinogenesis remains to be elucidated. On the other hand, we also observed that overexpression of ATF3, C/EBPs, and CREB suppressed DIM-induced ATF3 transactivation (Fig. 4A). Because ATF3, CREB and C/EBP are able to potentially bind to this site, we speculated that it could be due to sequestration of ATF4 against binding of other inhibitory bZIP proteins. Indeed, it has been reported that many transcription factors bind to the ATF binding site with comparative or synergistic manner .
Our group recently reported that tolfenamic acid induced ATF3 expression via MAPK-mediated ATF2 phosphorylation . However, in the present study, we observed that ATF2 significantly suppressed DIM-induced ATF3 transactivation as with other C/EBPs, CREB and ATF3. Although it is unclear why two compounds activate ATF3 expression differently, it is likely that DIM modulates a different signaling pathway to activate the same ATF3 gene. Unlike tolfenamic acid, DIM acts as a PPAR gamma ligand and suppresses growth of colon cancer cells in a PPARγ-dependent manner . However, our results indicated that ATF3 induction by DIM is PPARγ–independent (data not shown). Therefore, there are distinct mechanisms between DIM and tolfenamic acid with respect to ATF3 expression, and thus elucidating molecular mechanisms by which ATF3 is induced by different anti-cancer compounds will likely provide a better understanding of anti-tumorigenic mechanisms in colorectal cancer cells.
It has been reported that several DIM derivatives are synthesized and characterized their anti-cancer activity . Two different groups of DIM derivatives, ring-substituted DIM (methyl-substituted DIM) and C-substituted DIM (methylene-substituted DIM), exhibit different binding affinity to PPARγ and AhR nuclear receptors . In our study, these DIM derivatives also showed ATF3 induction at lower concentrations compared to parent DIM (data not shown), suggesting potential use of these derivatives as a chemopreventive and chemotherapeutic agent. These results also indicate that a specific structure plays an important role in a specific pathway and furthermore would validate the importance in human health as well as sanction its use as a template for further structural development of more effective and safe chemopreventive compounds.
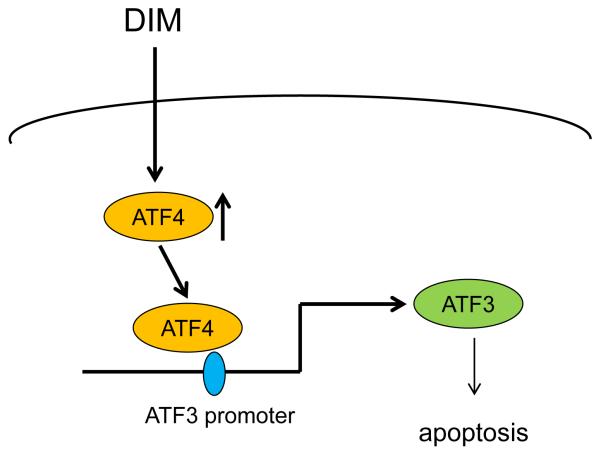
In conclusion, the current study provides information on molecular events of pro-apoptotic activity by DIM. DIM-induced ATF4 expression and the resulting ATF3 activation inhibits proliferation and induces apoptosis in human colon cancer cells (Fig. 5).
Fig. 5. Schematic diagram of DIM’s action in colorectal cancer cells.
DIM increases an ATF4 transcriptional factor that selectively binds to the ATF site in the ATF3 promoter. This in turn results in the increased expression of ATF3. Increased ATF3 expression results in an increase of apoptosis in colorectal cancer cells.
Acknowledgements
We thank Misty Bailey (University of Tennessee) for her critical reading of this manuscript. Financial support for XZ was provided by Program in Organizational or Personal Cooperation with Foreign Counterparts (2010630161), China Scholarship Council, China. This work was supported by NIH grant RO1CA108975, and The University of Tennessee Center of Excellence in Livestock Diseases and Human Health to SJB.
Abbreviations
- DIM
3,3′-diindolylmethane
- I3C
indole-3-carbinol
- ATF3
activating transcription factor3
- ATF4
activating transcription factor4
- CRC
colorectal cancer
Footnotes
Conflict of Interest No conflict of interest exists in the submission of this manuscript
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Witte JS, Longnecker MP, Bird CL, Lee ER, Frankl HD, Haile RW. Relation of vegetable, fruit, and grain consumption to colorectal adenomatous polyps. Am J Epidemiol. 1996;144:1015–25. doi: 10.1093/oxfordjournals.aje.a008872. [DOI] [PubMed] [Google Scholar]
- [2].Kassie F, Uhl M, Rabot S, Grasl-Kraupp B, Verkerk R, Kundi M, et al. Chemoprevention of 2-amino-3-methylimidazo[4,5-f]quinoline (IQ)-induced colonic and hepatic preneoplastic lesions in the F344 rat by cruciferous vegetables administered simultaneously with the carcinogen. Carcinogenesis. 2003;24:255–61. doi: 10.1093/carcin/24.2.255. [DOI] [PubMed] [Google Scholar]
- [3].Chen I, Safe S, Bjeldanes L. Indole-3-carbinol and diindolylmethane as aryl hydrocarbon (Ah) receptor agonists and antagonists in T47D human breast cancer cells. Biochem Pharmacol. 1996;51:1069–76. doi: 10.1016/0006-2952(96)00060-3. [DOI] [PubMed] [Google Scholar]
- [4].Anderton MJ, Manson MM, Verschoyle RD, Gescher A, Lamb JH, Farmer PB, et al. Pharmacokinetics and tissue disposition of indole-3-carbinol and its acid condensation products after oral administration to mice. Clin Cancer Res. 2004;10:5233–41. doi: 10.1158/1078-0432.CCR-04-0163. [DOI] [PubMed] [Google Scholar]
- [5].Chang YC, Riby J, Chang GH, Peng BC, Firestone G, Bjeldanes LF. Cytostatic and antiestrogenic effects of 2-(indol-3-ylmethyl)-3,3′-diindolylmethane, a major in vivo product of dietary indole-3-carbinol. Biochem Pharmacol. 1999;58:825–34. doi: 10.1016/s0006-2952(99)00165-3. [DOI] [PubMed] [Google Scholar]
- [6].Oganesian A, Hendricks JD, Williams DE. Long term dietary indole-3-carbinol inhibits diethylnitrosamine-initiated hepatocarcinogenesis in the infant mouse model. Cancer Lett. 1997;118:87–94. doi: 10.1016/s0304-3835(97)00235-8. [DOI] [PubMed] [Google Scholar]
- [7].Srivastava B, Shukla Y. Antitumour promoting activity of indole-3-carbinol in mouse skin carcinogenesis. Cancer Lett. 1998;134:91–5. doi: 10.1016/s0304-3835(98)00247-x. [DOI] [PubMed] [Google Scholar]
- [8].Yoshida M, Katashima S, Ando J, Tanaka T, Uematsu F, Nakae D, et al. Dietary indole-3-carbinol promotes endometrial adenocarcinoma development in rats initiated with N-ethyl-N’-nitro-N-nitrosoguanidine, with induction of cytochrome P450s in the liver and consequent modulation of estrogen metabolism. Carcinogenesis. 2004 doi: 10.1093/carcin/bgh225. [DOI] [PubMed] [Google Scholar]
- [9].Kim DJ, Shin DH, Ahn B, Kang JS, Nam KT, Park CB, et al. Chemoprevention of colon cancer by Korean food plant components. Mutat Res. 2003;523-524:99–107. doi: 10.1016/s0027-5107(02)00325-1. [DOI] [PubMed] [Google Scholar]
- [10].Frydoonfar HR, McGrath DR, Spigelman AD. Inhibition of proliferation of a colon cancer cell line by indole-3-carbinol. Colorectal Dis. 2002;4:205–7. doi: 10.1046/j.1463-1318.2002.00346.x. [DOI] [PubMed] [Google Scholar]
- [11].Zhang J, Hsu BAJ, Kinseth BAM, Bjeldanes LF, Firestone GL. Indole-3-carbinol induces a G1 cell cycle arrest and inhibits prostate-specific antigen production in human LNCaP prostate carcinoma cells. Cancer. 2003;98:2511–20. doi: 10.1002/cncr.11844. [DOI] [PubMed] [Google Scholar]
- [12].Rahman KM, Li Y, Sarkar FH. Inactivation of akt and NF-kappaB play important roles during indole-3-carbinol-induced apoptosis in breast cancer cells. Nutrition and cancer. 2004;48:84–94. doi: 10.1207/s15327914nc4801_12. [DOI] [PubMed] [Google Scholar]
- [13].Chintharlapalli S, Papineni S, Baek SJ, Liu S, Safe S. 1,1-Bis(3′-indolyl)-1-(p-substitutedphenyl)methanes are peroxisome proliferator-activated receptor gamma agonists but decrease HCT-116 colon cancer cell survival through receptor-independent activation of early growth response-1 and nonsteroidal anti-inflammatory drug-activated gene-1. Mol Pharmacol. 2005;68:1782–92. doi: 10.1124/mol.105.017046. [DOI] [PubMed] [Google Scholar]
- [14].Chintharlapalli S, Smith R, 3rd, Samudio I, Zhang W, Safe S. 1,1-Bis(3′-indolyl)-1-(p-substitutedphenyl)methanes induce peroxisome proliferator-activated receptor gamma-mediated growth inhibition, transactivation, and differentiation markers in colon cancer cells. Cancer research. 2004;64:5994–6001. doi: 10.1158/0008-5472.CAN-04-0399. [DOI] [PubMed] [Google Scholar]
- [15].Nachshon-Kedmi M, Yannai S, Haj A, Fares FA. Indole-3-carbinol and 3,3′-diindolylmethane induce apoptosis in human prostate cancer cells. Food Chem Toxicol. 2003;41:745–52. doi: 10.1016/s0278-6915(03)00004-8. [DOI] [PubMed] [Google Scholar]
- [16].Rahman KW, Sarkar FH. Inhibition of nuclear translocation of nuclear factor-{kappa}B contributes to 3,3′-diindolylmethane-induced apoptosis in breast cancer cells. Cancer research. 2005;65:364–71. [PubMed] [Google Scholar]
- [17].Kassouf W, Chintharlapalli S, Abdelrahim M, Nelkin G, Safe S, Kamat AM. Inhibition of bladder tumor growth by 1,1-bis(3′-indolyl)-1-(p-substitutedphenyl)methanes: a new class of peroxisome proliferator-activated receptor gamma agonists. Cancer research. 2006;66:412–8. doi: 10.1158/0008-5472.CAN-05-2755. [DOI] [PubMed] [Google Scholar]
- [18].Abdelrahim M, Newman K, Vanderlaag K, Samudio I, Safe S. 3,3′-Diindolylmethane (DIM) and derivatives induce apoptosis in pancreatic cancer cells through endoplasmic reticulum stress-dependent upregulation of DR5. Carcinogenesis. 2005 doi: 10.1093/carcin/bgi270. [DOI] [PubMed] [Google Scholar]
- [19].Gong Y, Firestone GL, Bjeldanes LF. 3,3′-Diindolylmethane (DIM) is a novel topoisomerase II{alpha} catalytic inhibitor that induces S phase retardation and mitotic delay in human hepatoma HepG2 cells. Mol Pharmacol. 2005 doi: 10.1124/mol.105.018978. [DOI] [PubMed] [Google Scholar]
- [20].Cho SD, Yoon K, Chintharlapalli S, Abdelrahim M, Lei P, Hamilton S, et al. Nur77 agonists induce proapoptotic genes and responses in colon cancer cells through nuclear receptor-dependent and nuclear receptor-independent pathways. Cancer research. 2007;67:674–83. doi: 10.1158/0008-5472.CAN-06-2907. [DOI] [PubMed] [Google Scholar]
- [21].Xue L, Firestone GL, Bjeldanes LF. DIM stimulates IFNgamma gene expression in human breast cancer cells via the specific activation of JNK and p38 pathways. Oncogene. 2005;24:2343–53. doi: 10.1038/sj.onc.1208434. [DOI] [PubMed] [Google Scholar]
- [22].Hong C, Kim HA, Firestone GL, Bjeldanes LF. 3,3′-Diindolylmethane (DIM) induces a G(1) cell cycle arrest in human breast cancer cells that is accompanied by Sp1-mediated activation of p21(WAF1/CIP1) expression. Carcinogenesis. 2002;23:1297–305. doi: 10.1093/carcin/23.8.1297. [DOI] [PubMed] [Google Scholar]
- [23].Chang X, Tou JC, Hong C, Kim HA, Riby JE, Firestone GL, et al. 3,3′-Diindolylmethane inhibits angiogenesis and the growth of transplantable human breast carcinoma in athymic mice. Carcinogenesis. 2005;26:771–8. doi: 10.1093/carcin/bgi018. [DOI] [PubMed] [Google Scholar]
- [24].Rajoria S, Suriano R, George A, Shanmugam A, Schantz SP, Geliebter J, et al. Estrogen induced metastatic modulators MMP-2 and MMP-9 are targets of 3,3′-diindolylmethane in thyroid cancer. PLoS One. 6:e15879. doi: 10.1371/journal.pone.0015879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hai T, Hartman MG. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene. 2001;273:1–11. doi: 10.1016/s0378-1119(01)00551-0. [DOI] [PubMed] [Google Scholar]
- [26].Bottone FG, Jr., Martinez JM, Alston-Mills B, Eling TE. Gene modulation by Cox-1 and Cox-2 specific inhibitors in human colorectal carcinoma cancer cells. Carcinogenesis. 2004;25:349–57. doi: 10.1093/carcin/bgh016. [DOI] [PubMed] [Google Scholar]
- [27].Yamaguchi K, Lee SH, Kim JS, Wimalasena J, Kitajima S, Baek SJ. Activating transcription factor 3 and early growth response 1 are the novel targets of LY294002 in a phosphatidylinositol 3-kinase-independent pathway. Cancer research. 2006;66:2376–84. doi: 10.1158/0008-5472.CAN-05-1987. [DOI] [PubMed] [Google Scholar]
- [28].Yan C, Lu D, Hai T, Boyd DD. Activating transcription factor 3, a stress sensor, activates p53 by blocking its ubiquitination. Embo J. 2005;24:2425–35. doi: 10.1038/sj.emboj.7600712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wang H, Mo P, Ren S, Yan C. Activating transcription factor 3 activates p53 by preventing E6-associated protein from binding to E6. The Journal of biological chemistry. 285:13201–10. doi: 10.1074/jbc.M109.058669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lu D, Wolfgang CD, Hai T. Activating transcription factor 3, a stress-inducible gene, suppresses Ras-stimulated tumorigenesis. The Journal of biological chemistry. 2006;281:10473–81. doi: 10.1074/jbc.M509278200. [DOI] [PubMed] [Google Scholar]
- [31].Chen HH, Wang DL. Nitric oxide inhibits matrix metalloproteinase-2 expression via the induction of activating transcription factor 3 in endothelial cells. Mol Pharmacol. 2004;65:1130–40. doi: 10.1124/mol.65.5.1130. [DOI] [PubMed] [Google Scholar]
- [32].Hackl C, Lang SA, Moser C, Mori A, Fichtner-Feigl S, Hellerbrand C, et al. Activating transcription factor-3 (ATF3) functions as a tumor suppressor in colon cancer and is up-regulated upon heat-shock protein 90 (Hsp90) inhibition. BMC cancer. 10:668. doi: 10.1186/1471-2407-10-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Liu W, Iiizumi-Gairani M, Okuda H, Kobayashi A, Watabe M, Pai SK, et al. KAI1 Gene Is Engaged in NDRG1 Gene-mediated Metastasis Suppression through the ATF3-NF{kappa}B Complex in Human Prostate Cancer. The Journal of biological chemistry. 286:18949–59. doi: 10.1074/jbc.M111.232637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lee SH, Kim JS, Yamaguchi K, Eling TE, Baek SJ. Indole-3-carbinol and 3,3′-diindolylmethane induce expression of NAG-1 in a p53-independent manner. Biochem Biophys Res Commun. 2005;328:63–9. doi: 10.1016/j.bbrc.2004.12.138. [DOI] [PubMed] [Google Scholar]
- [35].Lee SH, Yamaguchi K, Kim JS, Eling TE, Safe S, Park Y, et al. Conjugated linoleic acid stimulates an anti-tumorigenic protein NAG-1 in an isomer specific manner. Carcinogenesis. 2006;27:972–81. doi: 10.1093/carcin/bgi268. [DOI] [PubMed] [Google Scholar]
- [36].Baek SJ, Kim JS, Jackson FR, Eling TE, McEntee MF, Lee SH. Epicatechin gallate-induced expression of NAG-1 is associated with growth inhibition and apoptosis in colon cancer cells. Carcinogenesis. 2004;25:2425–32. doi: 10.1093/carcin/bgh255. [DOI] [PubMed] [Google Scholar]
- [37].Whitlock NC, Bahn JH, Lee SH, Eling TE, Baek SJ. Resveratrol-induced apoptosis is mediated by early growth response-1, Kruppel-like factor 4, and activating transcription factor 3. Cancer Prev Res (Phila) 4:116–27. doi: 10.1158/1940-6207.CAPR-10-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lee SH, Bahn JH, Whitlock NC, Baek SJ. Activating transcription factor 2 (ATF2) controls tolfenamic acid-induced ATF3 expression via MAP kinase pathways. Oncogene. 2010;29:5182–92. doi: 10.1038/onc.2010.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].St Germain C, Niknejad N, Ma L, Garbuio K, Hai T, Dimitroulakos J. Cisplatin induces cytotoxicity through the mitogen-activated protein kinase pathways and activating transcription factor 3. Neoplasia. 12:527–38. doi: 10.1593/neo.92048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bruning A, Burger P, Vogel M, Rahmeh M, Friese K, Lenhard M, et al. Bortezomib treatment of ovarian cancer cells mediates endoplasmic reticulum stress, cell cycle arrest, and apoptosis. Invest New Drugs. 2008 doi: 10.1007/s10637-008-9206-4. [DOI] [PubMed] [Google Scholar]
- [41].Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JC, et al. The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283:83–7. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- [42].Allan AL, Albanese C, Pestell RG, LaMarre J. Activating transcription factor 3 induces DNA synthesis and expression of cyclin D1 in hepatocytes. The Journal of biological chemistry. 2001;276:27272–80. doi: 10.1074/jbc.M103196200. [DOI] [PubMed] [Google Scholar]
- [43].Tamura K, Hua B, Adachi S, Guney I, Kawauchi J, Morioka M, et al. Stress response gene ATF3 is a target of c-myc in serum-induced cell proliferation. Embo J. 2005;24:2590–601. doi: 10.1038/sj.emboj.7600742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Yin X, Wolford CC, Chang YS, McConoughey SJ, Ramsey SA, Aderem A, et al. ATF3, an adaptive-response gene, enhances TGF{beta} signaling and cancer-initiating cell features in breast cancer cells. J Cell Sci. 123:3558–65. doi: 10.1242/jcs.064915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Yan L, Della Coletta L, Powell KL, Shen J, Thames H, Aldaz CM, et al. Activation of the canonical Wnt/beta-catenin pathway in ATF3-induced mammary tumors. PLoS One. 6:e16515. doi: 10.1371/journal.pone.0016515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Lee SH, Bahn JH, Choi CK, Whitlock NC, English AE, Safe S, et al. ESE-1/EGR-1 pathway plays a role in tolfenamic acid-induced apoptosis in colorectal cancer cells. Molecular cancer therapeutics. 2008;7:3739–50. doi: 10.1158/1535-7163.MCT-08-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Cho KN, Sukhthankar M, Lee SH, Yoon JH, Baek SJ. Green tea catechin (−)-epicatechin gallate induces tumour suppressor protein ATF3 via EGR-1 activation. Eur J Cancer. 2007;43:2404–12. doi: 10.1016/j.ejca.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lee SH, Krisanapun C, Baek SJ. NSAID-activated gene-1 as a molecular target for capsaicin-induced apoptosis through a novel molecular mechanism involving GSK3{beta}, C/EBP{beta}, and ATF3. Carcinogenesis. 2010;31:719–28. doi: 10.1093/carcin/bgq016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Yang H, Park SH, Choi HJ, Moon Y. The integrated stress response-associated signals modulates intestinal tumor cell growth by NSAID-activated gene 1 (NAG-1/MIC-1/PTGF-beta) Carcinogenesis. 2010;31:703–11. doi: 10.1093/carcin/bgq008. [DOI] [PubMed] [Google Scholar]
- [50].Lei P, Abdelrahim M, Cho SD, Liu S, Chintharlapalli S, Safe S. 1,1-Bis(3′-indolyl)-1-(p-substituted phenyl)methanes inhibit colon cancer cell and tumor growth through activation of c-jun N-terminal kinase. Carcinogenesis. 2008;29:1139–47. doi: 10.1093/carcin/bgn103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ye J, Kumanova M, Hart LS, Sloane K, Zhang H, De Panis DN, et al. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J. 2010;29:2082–96. doi: 10.1038/emboj.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Roybal CN, Yang S, Sun C-W, Hurtado D, Vander Jagt DL, Townes TM, et al. Homocysteine Increases the Expression of Vascular Endothelial Growth Factor by a Mechanism Involving Endoplasmic Reticulum Stress and Transcription Factor ATF4. Journal of Biological Chemistry. 2004;279:14844–52. doi: 10.1074/jbc.M312948200. [DOI] [PubMed] [Google Scholar]
- [53].Lassot I, Segeral E, Berlioz-Torrent C, Durand H, Groussin L, Hai T, et al. ATF4 degradation relies on a phosphorylation-dependent interaction with the SCF(betaTrCP) ubiquitin ligase. Mol Cell Biol. 2001;21:2192–202. doi: 10.1128/MCB.21.6.2192-2202.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Igarashi T, Izumi H, Uchiumi T, Nishio K, Arao T, Tanabe M, et al. Clock and ATF4 transcription system regulates drug resistance in human cancer cell lines. Oncogene. 2007;26:4749–60. doi: 10.1038/sj.onc.1210289. [DOI] [PubMed] [Google Scholar]
- [55].Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. Journal of Cell Science. 2004;117:5965–73. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- [56].Chintharlapalli S, Papineni S, Safe SH. 1,1-Bis(3′-indolyl)-1-(p-substitutedphenyl)methanes Inhibit Growth, Induce Apoptosis, and Decrease the Androgen Receptor in LNCaP Prostate Cancer Cells through PPAR{gamma}-independent Pathways. Mol Pharmacol. 2006 doi: 10.1124/mol.106.028696. [DOI] [PubMed] [Google Scholar]
- [57].Lei P, Abdelrahim M, Cho SD, Liu X, Safe S. Structure-dependent activation of endoplasmic reticulum stress-mediated apoptosis in pancreatic cancer by 1,1-bis(3′-indoly)-1-(p-substituted phenyl)methanes. Molecular cancer therapeutics. 2008;7:3363–72. doi: 10.1158/1535-7163.MCT-08-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]