Abstract
BACKGROUND AND PURPOSE
Telomerase is the enzyme responsible for extending G-strand telomeric DNA and represents a promising target for treatment of neoplasia. Inhibition of telomerase can be achieved by stabilization of G-quadruplex DNA structures. Here, we characterize the cellular effects of a novel G-quadruplex stabilizing compound, 3,6-bis(4-methyl-2-vinylpyrazinium iodine) carbazole (BMVC4).
EXPERIMENTAL APPROACH
The cellular effects of BMVC4 were characterized in both telomerase-positive and alternative lengthening of telomeres (ALT) cancer cells. The molecular mechanism of how BMVC4 induced senescence is also addressed.
KEY RESULTS
BMVC4-treated cancer cells showed typical senescence phenotypes. BMVC4 induced senescence in both ALT and telomerase-overexpressing cells, suggesting that telomere shortening through telomerase inhibition might not be the cause for senescence. A large fraction of DNA damage foci was not localized to telomeres in BMVC4-treated cells and BMVC4 suppressed c-myc expression through stabilizing the G-quadruplex structure located at its promoter. These results indicated that the cellular targets of BMVC4 were not limited to telomeres. Further analyses showed that BMVC4 induced DNA breaks and activation of ataxia telangiectasia-mutated mediated DNA damage response pathway.
CONCLUSIONS AND IMPLICATIONS
BMVC4, a G-quadruplex stabilizer, induced senescence by activation of pathways of response to DNA damage that was independent of its telomerase inhibitory activity. Thus, BMVC4 has the potential to be developed as a chemotherapeutic agent against both telomerase positive and ALT cancer cells.
Keywords: G-quadruplex, telomere, telomerase, senescence, c-myc, carbazole
Introduction
Telomeres are the physical ends of eukaryotic chromosomes. They protect chromosomes from degradation and end-to-end fusions (O'Sullivan and Karlseder, 2010). Telomeric DNA consists of short, simple and tandem repeats of DNA sequences with one strand rich in guanine. For example, telomeric DNA in vertebrates is comprised of the 6 bp sequence TTAGGG/CCCTAA. In addition to double-stranded duplex sequences, telomeres also have single-stranded 3′ protruding overhangs (Makarov et al., 1997; Wright et al., 1997). These overhangs can adopt a variety of different non-B DNA conformations in vitro, such as intramolecular fold-backs and tetra-stranded DNA structures through guanine rich sequences (Sen and Gilbert, 1991). Telomerase is an RNA-dependent DNA polymerase that extends the telomeric TTAGGG repeats (Collins and Mitchell, 2002). In human cells, the enzyme comprises a high-molecular-weight complex with a RNA template subunit hTR and protein components including the catalytic subunit human telomerase reverse transcriptase, hTERT.
Telomerase activity has been identified in immortalized cell lines and in 80–90% of human cancer specimens representing a range of cancer types (Shay and Bacchetti, 1997) and has been directly implicated in cellular immortalization and tumorigenesis (Bodnar et al., 1998; Hahn et al., 1999a). In most normal human cells, telomerase activity is low or not detectable, and telomeric DNA is progressively lost at a rate of 30–120 bp with each replication cycle (Hastie et al., 1990; Counter et al., 1992). Eventually, telomeres shorten to a critical length that leads to a growth arrest state termed replicative senescence (Counter et al., 1992; Blasco et al., 1997). Indeed, it has been proposed that telomere length could serve as a counting mechanism for cell divisions in normal cells. In cancer cells, however, the reactivation of telomerase is thought to stabilize telomere length, thereby compensating for the cell division-related telomere erosion and providing unlimited proliferative capacity to malignant cells (Counter et al., 1992). Inhibition of telomerase can result in telomere shortening followed by arrest of proliferation and cell death by apoptosis (Zhang et al., 1999; Hahn et al., 1999b). Thus, the inhibition of telomerase in tumour cells could disrupt telomere maintenance and return malignant cells to proliferative crisis followed by senescence or cell death (Harley et al., 1990; Counter et al., 1992).
Several strategies to inhibit telomerase activity have been reported (Rezler et al., 2002). These include peptide nucleic acids and 2′-O-MeRNA oligonucleotides directed towards the telomerase RNA template, low MW compounds that target telomeric DNA such as cationic porphyrins, anthraquinones and some planar aromatic chromophores, and nucleosidic reverse transcriptase inhibitors (Hsu and Lin, 2005). A non-nucleoside compound BIBR1532 was also reported to inhibit telomerase non-competitively with an IC50 of ∼100 nM (Damm et al., 2001; Pascole et al., 2002). It is one of the most potent telomerase inhibitors reported up to this time. Another approach made the use of dominant-negative alleles of hTERT, expression of which resulted in cell death of telomerase-positive cancer cell lines (Zhang et al., 1999; Hahn et al., 1999b).
The tetra-stranded DNA structures formed by guanine rich sequences, G-quadruplexes, have been shown to inhibit telomerase activity (Raghuraman and Cech, 1990; Zahler et al., 1991). Thus, the G-quadruplex stabilizers have been proposed as promising telomerase inhibitory agents (Mergny and Helene, 1998). Indeed, a number of small molecules that bind to and stabilize the folded quadruplex were shown to inhibit telomerase activity in vitro (Hsu and Lin, 2005). These compounds were reported to induce senescence in cancer cells through inhibiting telomerase (Tauchi et al., 2003; 2006; Incles et al., 2004; Burger et al., 2005; Gomez et al., 2006; Zhou et al., 2006; Salvati et al., 2007; Huang et al., 2008; Rizzo et al., 2009), making them potentially valuable candidates for anticancer therapy. However, several compounds interacting with G-quadruplex structures have also been shown to inhibit proliferation in alternative lengthening of telomeres (ALT) cell lines (Gowan et al., 2001; Riou et al., 2002; Kim et al., 2003; Pennarun et al., 2005), suggesting that the cellular effects of G-quadruplex stabilizers might not be limited to telomeres.
We have identified a carbazole derivative, 3,6-bis(1-methyl-4-vinylpyridium iodide) carbazole (BMVC), that increases the melting temperature (Tm) of G-quadruplexes formed by human telomeric DNA sequences by over 10°C (Chang et al., 2003). BMVC inhibits telomerase activity, accelerates telomere shortening and shows inhibitory effects on tumorigenesis (Huang et al., 2008). Several of the BMVC-related compounds also inhibited telomerase activity at the sub-µM level, suggesting a high potential for these compounds in anti-telomerase inhibitor development. Here, we report the characterization of another carbazole derivative, 3,6-bis(4-methyl-2-vinylpyrazinium iodide) carbazole (BMVC4), and its effects on cancer cells. We found that BMVC4 selectively inhibited telomerase activity in vitro, induced progressive telomere shortening and caused senescence in telomerase-positive cancer cells. Remarkably, telomerase inhibition might not be the primary cause of BMVC4-induced senescence. We showed that BMVC4 also targeted a G-quadruplex forming sequence (QFS) located at the promoter region of c-myc and reduced its expression. Moreover, breaks in DNA and the response to DNA damage, mediated by the ataxia telangiectasia-mutated (ATM) kinase pathway were induced in BMVC4-treated cells. Thus, BMVC4 induced senescence in both telomerase-positive and telomerase-negative ALT cancer cells.
Methods
Senescence-associated β-galactosidase staining
Detection of senescence-associated (SA) β-galactosidase (SA-β-Gal) followed the standard protocol (Dimri et al., 1995). Briefly, cells were washed with PBS, fixed in 2% formadehyde/0.2% glutaldehyde (or 3% formaldehyde) and incubated at 37°C with 1 mg·mL−1 of freshly prepared SA-β-Gal stain solution. Staining was evident in 2–4 h and maximal in 24–48 h.
PCR stop assays
The PCR primers were designed to amplify the region covering the −148 bp and −914 bp of the c-myc promoter. The sequences of the primers were 5′-AGGGGATTTGTCTCTTCTGA-3′ and 5′-ATCCTCTCTCGCTAATCTCC-3′. Plasmid pc-MycPro-Luc and its mutants were used as the templates for the reactions. Assays were performed in 20 mM Tris pH 8.8 buffer with 10 mM KCl, 1.5 mM MgCl2, 10 mM (NH4)2SO4, 0.1% Triton X-100, 100 nmol of plasmid DNA, 7.5 pmol of each primer, 0.5 mM dNTPs, 2.5 U of Taq polymerase and the indicated amount of BMVC4. Reaction mixtures were incubated in a thermocycler with the following cycling conditions: 94°C for 5 min, followed by 30 cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 2 min. Amplified products were resolved on a 1% agarose gel and stained with ethidium bromide.
Alkaline comet assay
Cells were treated with 10 µM carbazole or BMVC4 for 6 and 12 days and subjected to alkaline comet assays to detect DNA breaks. Briefly, the BMVC4-treated cells were suspended and mixed with low-melting-point agarose to cast the cells on a microscope slide. The embedded cells were lysed with alkaline lysis buffer (2.5 M NaCl, 120 mM EDTA, 10 mM Tris pH 10, 10 % DMSO and 1 % Triton X-100) at 4°C overnight. Electrophoresis was performed in denaturing buffer (1 mM EDTA and 0.3 N NaOH) at 25V and 300 mA for 30 min and then neutralized in buffer containing 400 mM Tris–HCl, pH 7.5. Visualization of the fragmented chromosomal DNA was achieved by staining the cells with SYBR Green. The images were captured under an Olympus fluorescent microscope (Hamburg, Germany) and processed using Metavue Software. Quantification of the relative length and intensity of SYBR Green-stained DNA was measured and presented as the Olive tail moment using CASP software (Comet Assay Software Project).
Tm measurement
The Tm was measured by monitoring the circular dichroism (CD) maximum at 295 nm on a Jasco (Great Dunmow, Essex, UK) J-715 spectropolarimeter by ramping the temperature from 5 to 90°C at a rate of 0.8°C·min−1. Oligonucleotide d(TTAGGG)4 was purchased from (Life Technologies-Applied Biosystems, Carlsbad, CA, USA). Solutions of 10 mM Tris–HCl (pH 7.5) and 150 mM NaCl were mixed with DNA and heated to 90°C for 2 min, cooled slowly to room temperature and then stored for 42 days at 4°C before use. The molar concentration of DNA was determined by monitoring the 260 nm absorbance. The d(T2AG3)4 DNA forms a G-quadruplex structure at room temperature as indicated by the 295 nm positive CD band detected at 25°C.
Telomerase activity assay
The ability of agents to inhibit telomerase in a cell-free assay was assessed with a modified TRAP-G4 for G-quadruplex-induced telomerase activity assay (Gomez et al., 2002). Telomerase-extended products were resolved by 10% polyacrylamide gel electrophoresis and visualized by SYBER Green I staining of the gel. Total cell lysates derived from H1299 lung cancer cell line were used as a source of telomerase. Protein concentrations of the lysates were determined by a Bio-Rad (Hercules, CA, USA) assay kit using BSA as standards.
Cell viability assay
Cells were grown in 96-well plates (∼2000 cells per well) in the presence of 5% CO2 at 37°C. To examine the short-term cytotoxic effect, cells were then incubated with different concentrations of BMVC4 for 72 h. The cytotoxicity was determined by MTT assays and was analysed spectrophotometrically at the absorbance of 570 nm.
Population doubling study
Cells were treated with BMVC4 or DMSO (drug vehicle control) and were grown in T25 tissue culture flasks at 3 × 105 cells per flask for 3 or 4 days. They were then trypsinized and counted. Each time, 3 × 105 cells were replaced in the new culture flask with fresh BMVC4 or an equivalent volume of DMSO. The experiments were continued until there were fewer than 3 × 105 cells available for reseeding. The results were obtained from at least three independent experiments.
Telomere length determination
Cells were treated with 10 µM BMVC4 or DMSO (drug vehicle control) for 3, 6 or 9 days and total genomic DNA was prepared using the extraction kit purchased from BD Biosciences Clontech (San Jose, CA, USA). To determine the telomere length, genomic DNA was digested with HinfI and RsaI and separated by 1% agarose gel electrophoresis. After electrophoresis, the DNA was transferred onto a Hybond N+ (GE Healthcare-Amersham, Little Chalfont, Buckinghamshire, UK) membrane and hybridized with 32P-labeled ∼800 bp (TTAGGG)n fragments. Telomeric smears were visualized and quantified with a PhosphorImager (Molecular Dynamics, Sevenoaks, Kent, UK).
Immunofluorescence analysis
H1299 cells were treated with BMVC4, fixed and incubated with anti-γ-H2AX antibody (Upstate, JBW301, Merk Millipore, Billerica, MA, USA) or anti-trimethyl-Histone H3-Lys9 antibody (H3K9Me3, Upstate). Visualization of γ-H2AX or H3K9Me3 was achieved by addition of Rhodamine-conjugated anti-mouse IgG (Jackson, Rhodamine Red™-X-conjugated, West Grove, PA, USA) and observed under fluorescence microscopy (Olympus BX50). Confocal analysis was conducted using anti-γ-H2AX antibody (Abcam, ab2893, Cambridge, UK) and anti-TRF2 antibody (Imgenex, 4A794-15, San Diego, CA, USA). Visualization of γ-H2AX and TRF2 were achieved by addition of FITC-conjugated anti-rabbit IgG (Jackson, FITC-conjugated) and Rhodamine-conjugated anti-mouse IgG (Jackson, Rhodamine Red™-X-conjugated) and then observed under laser confocal microscopy (Olympus FV1000).
Quantification of c-myc mRNA
For RT-PCR analysis, total RNA was isolated using Trizol reagent (Sigma) and reversely transcribed using random hexamers with a cDNA reverse transcription kit (Applied Biosystems). The resulting cDNA was mixed with the PCR reaction mixture and amplified using c-myc (forward primer 5′-ATTCTCTGCTCTCCTCGACG-3′ and reverse primer 5′-AGGATAGTCCTTCCGAGTGGA-3′) specific primers. The PCR cycle number was set up to permit the distinction between mRNA expression profiles among the samples. PCR products were visualized on a 1% agarose gel. The mRNA level of GAPDH was used as an internal control (forward primer 5′- GACCACAGTCCATGCCATCAC-3′ and reverse primer 5′- TCCACCACCCTGTTGCTGTAG-3′). The real-time quantitative RT-PCR was also used to determine the mRNAs of c-myc and hTERT. The parameter Ct is defined as the fractional cycle number at which the fluorescence is generated by the Cyber green I system (FastStart Universal SYBR Green Master, Roche, Indianapolis, IN, USA). Relative levels of c-myc were determined by normalizing the Ct values of c-myc with the Ct of GAPDH. The values were obtained from three independent experiments.
Immunoblotting analysis
The BMVC4-treated cells were washed twice with PBS and suspended in lysis buffer [10 mM Tris–HCl (pH 7.4), 150 mM NaCl, 5 mM EDTA, 1% Triton X100, 100 µM phenylmethylsulphonyl fluoride, leupeptin 1 mg·mL−1, aprotinin 1 mg·mL−1, and 25 mM dithiothreitol]. After 30 min on ice, the lysates were cleared by centrifugation. Samples corresponding to 50–100 µg of protein (Bio-Rad protein assay) were separated on 10% SDS-PAGE gels and transferred to nitrocellulose membranes. Immunoblotting analysis was carried out according to standard procedures using ECL detection (Perkin Elmer, Waltham, MA, USA). The membranes were hybridized with the following antibodies: anti-phospho-Rb (rabbit, pSer780, Sigma-Aldrich, St Louis, MO, USA), anti-c-Myc (mAb, 9E10, Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-Sp1 (rabbit, Sc-59, Santa Cruz Biotechnology), anti-hTERT (rabbit, H-231, Santa Cruz Biotechnology), anti-ATR phosphor-S428 (rabbit, Cell Signaling Technology, Danvers, MA, USA) and anti-ATM-phosphor-S1981 (rabbit, EP1890Y, GeneTex, Irvine, CA, USA). The equivalent loading of lanes was verified by hybridization with anti-GAPDH (mAb, ab9482-200, Abcam). Horseradish peroxidase-conjugated donkey anti-rabbit or sheep anti-mouse antibodies (Amersham) were used as the secondary antibodies.
Site-directed mutagenesis
Plasmid pc-MycPro-Luc (KN) carrying the 1260 bp c-myc promoter upstream from a luciferase reporter gene was used as the template for mutagenesis (a gift from Yan-Hwa Wu Lee, National Ying-Ming University, Taipei). PCR mutagenesis was used to generate mutations on QFS1 (KNQ1) and/or QFS2 (KNQ2) using Pfu DNA polymerase (Agilent Technologies, Santa Clara, CA, USA). The resulting mutations were verified by DNA sequencing of the plasmids.
Luciferase assay
H1299 cells were co-transfected with pc-MycPro-Luc plasmids or the mutated plasmids and pCMVβ plasmids using Lipofectamin™ 2000 (Life Technologies-Invitrogen, Grand Island, NY, USA). One day after transfection, the transfected cells were treated with 10 µM of BMVC4 for a further 3 days. The firefly luciferase activities were measured and normalized with β-Gal activity.
Small interference RNAs
The siRNA used to knock down ATM was synthesized by Sigma-Proligo with sequences: 5′-AAGCGCCTGATTCGAGATCCT-3′ (Chou et al., 2008). Oligofectamine (Invitrogen) was used to transfect siRNA into H1299 cells.
Statistical analysis
Student's t-test was applied to assess whether the means of two groups are statistically different from each other. Here, we consider P < 0.05 as significant.
Materials
Carbazole and 3,6-dibromocarbazole were purchased from (Sigma-Aldrich) and used without further purification. The synthesis of BMVC4 has been described previously (Chang et al., 2003). General chemicals used in this study were purchased from (Sigma-Aldrich). Cell lines (human lung cancer H1299; breast cancer MCF7; cervical cancer HeLa; VA13; SaoS2; U2OS) were obtained from Bioresource Collection and Research Center, Taiwan
Results
BMVC4 is a potent G-quadruplex stabilizer and telomerase inhibitor
Carbazole derivatives that stabilized G-quadruplex DNA structure formed by human telomeric sequence have been previously designed and synthesized (Huang et al., 2008). Here, we examined another carbazole derivative, BMVC4, that has cationic charges on the two pendant groups of pyridinium rings of carbazole (Figure 1A). The effect of BMVC4 on the G-quadruplex DNA of human telomeric sequences d(T2AG3)4 (Hum) was first evaluated using circular dichroism (CD) to monitor the Tm of Hum quadruplexes. The 295 nm CD band was measured as a function of temperature to determine the Tm of Hum quadruplexes in the presence of carbazole and BMVC4. We found that the Tm of Hum quadruplexes could be increased more than 10°C upon interaction with BMVC4, suggesting that it could thermally stabilize the Hum quadruplexes (Figure 1B). The effects of G-quadruplex stabilizer on telomerase were next analysed. The direct inhibitory effect on telomerase was also evaluated by conventional TRAP assays (Kim et al., 1994). Our results indicated that BMVC4 has an IC50 value against telomerase at ∼0.2 µM (Figure 1C, left panel). In the standard TRAP assay, telomerase extension of single-stranded DNA substrate is followed by PCR amplification of the extended products. As BMVC4 was added early in the reaction, it may affect both telomerase and Taq polymerase (De Cian et al., 2007). To rule out the possible inhibitory effects on Taq polymerase, BMVC4 was added after the telomerase extension step of the TRAP assays (Figure 1C, right panel). A complete inhibition of the assay was observed at concentrations of BMVC4 >3 µM. Thus, although BMVC4 also affected the PCR reactions, a higher concentration of BMVC4 was required. Our results indicated that BMVC4 was a potent G-quadruplex stabilizer and selectively inhibited telomerase in vitro.
Figure 1.
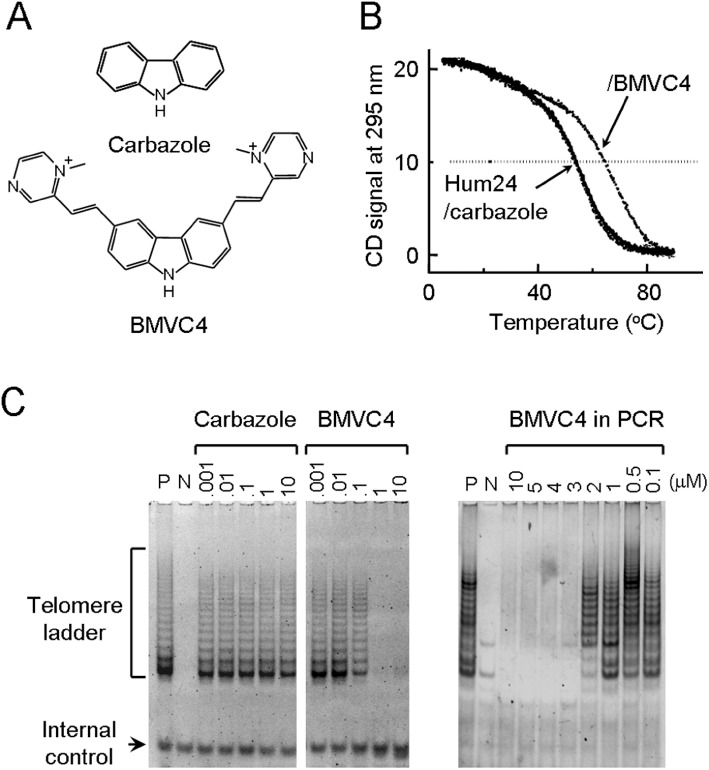
Structure of BMVC4 and its activity as an inhibitor of telomerase. (A) Structure of carbazole and BMVC4. (B) BMVC4 enhances the Tm of Hum quadruplex. Temperature-dependent CD signals at 295 nm of quadruplex d(T2AG3)4 (Hum24) and upon interaction with carbazole and BMVC4 were measured, respectively. (C) BMVC4 inhibits telomerase activity. The effects of BMVC4 on telomerase were measured using TRAP-G4 (left panel) assay (see Methods). In the assays, BMVC4 at various concentrations were incubated with telomerase-active cell extracts for 5 min at room temperature before the telomerase extension reactions. Telomerase-active cell extracts were prepared from H1299 cells (P). RNase A-treated extracts were used as negative controls (N). The positions of telomerase ladders and internal controls are indicated. The inhibitory effects of BMVC4 on Taq polymerase were also determined (right panel). Indicated amounts of BMVC4 were added to the TRAP reactions after the telomerase extension steps. PCR reactions were then conducted as in standard assays.
BMVC4 induced senescence in telomerase-positive cancer cells
We next investigated the effect of BMVC4 on telomerase-positive cancer cells. Short-term (48 h) cell viability assays were performed on a human lung cancer cell line, H1299. Survival of cells after incubation with different concentrations of BMVC4 was measured. We found that BMVC4 caused almost negligible cytotoxicity to H1299 cells at concentrations up to 40 µM (Figure 2A). The senescence-related phenotypes were then analysed after long-term treatment of H1299 cells with BMVC4 at non-toxic concentrations, 0.5, 1 or 10 µM. We found that the H1299 cells stopped proliferating after a lag period (Figure 2B). The length of lag period appeared to be dependent on the dose of BMVC4. The delayed growth inhibitory effect was not limited to non-small cell lung carcinoma H1299 cells as two other telomerase-positive cancer cells, breast cancer MCF7 and cervical cancer HeLa cells, also showed similar growth effects of BMVC4 (Supporting Information Figure S1). The morphology of the BMVC4-treated cells was also observed. At the late passage, the cells became enlarged with irregular cell shapes and more distinct nuclei (data not shown). These cells also showed significant elevation of SA-β-Gal activity (Figure 2C). Almost 30% of the 10 µM BMVC4-treated cells stained positive for SA-β-gal activities. In contrast, the percentage of SA-β-gal-positive cells was similar for the DMSO-treated control (∼2%) and BMVC4-treated early passage cells (∼3–4%). The results clearly indicated that BMVC4 induced the senescence program in telomerase-positive cancer cells. We further analysed the presence of senescence-associated heterochromatin foci (SAHF) using antibody against trimethylated H3 at Lys9 (H3K9me3). SAHF is a distinct DNA-dense heterochromatic structure that accumulates in senescent human fibroblasts. Typically, it bears the hallmarks of heterochromatin, such as the H3K9me3 (Narita et al., 2003; Zhang et al., 2005). As shown in Figure 2D, the BMVC4-treated H1299 cells displayed increasing immunostaining for H3K9me3. Approximately 45% of H3K9me3-positive cells were visible after 9 days of the treatments (Figure 2D, right panel). The results demonstrated that the senescence program was initiated in response to BMVC4.
Figure 2.
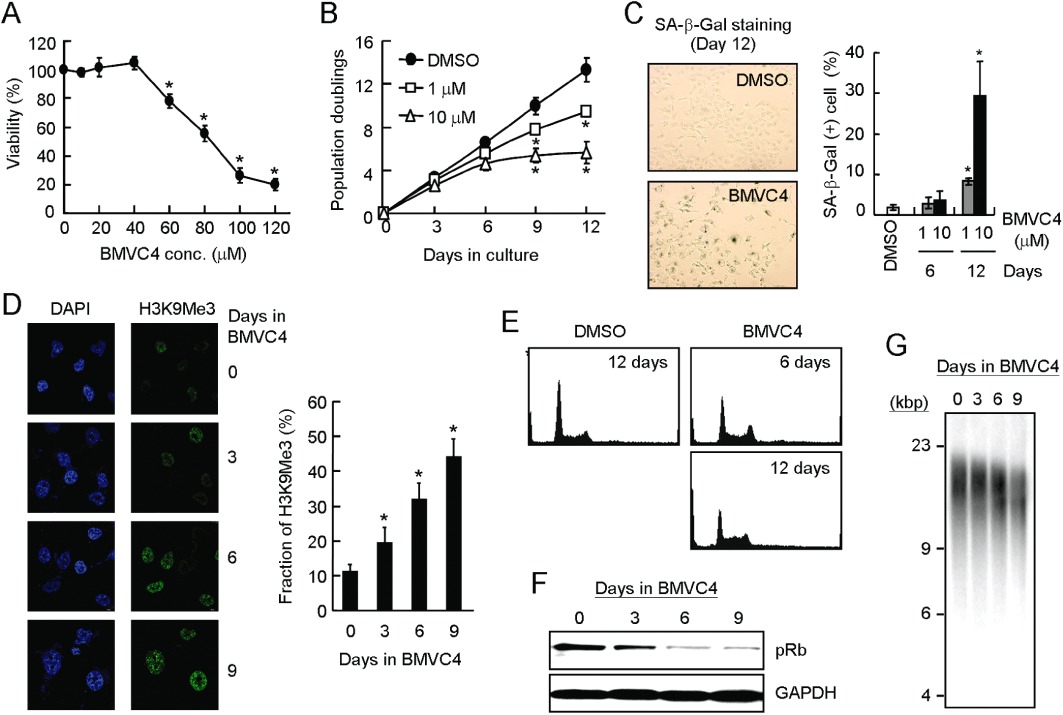
Senescent phenotype and telomeres shortening in BMVC4-treated cancer cells. (A) About 2 × 103 H1299 cells were seeded in 96-well plates and incubated with BMVC4 at various concentrations for another 48 h. Cell growth was then determined using the MTT assay. The values are obtained from three experiments using the values for untreated cells as 100%. *P < 0.05 (P= 1 × 10−5, 5 × 10−12, 5 × 10−16 and 4 × 10−17 for BMVC4 at 60, 80, 100 and 120 µM, respectively), significant effects of BMVC4. (B) Delayed anti-proliferation activity of BMVC4. H1299 cells were treated with 1 or 10 µM of BMVC4. The cells were counted during the passages and the population doubling was determined. Results were obtained from the average of three independent experiments. *P < 0.05 (P= 0.0009 and 9 × 10−6 for 1 µM BMVC4 at days 9 and 12, respectively; P= 7 × 10−6 and 3 × 10−6 for 10 µM BMVC4 at days 9 and 12, respectively), significant effects of BMVC4. (C) Detection of SA-β-galactosidase in BMVC4-treated H1299 cells. H1299 cells were treated with 1 or 10 µM of BMVC4 and subjected to staining for SA-β-galactosidase. Photographs of the 10 µM BMVC4-treated and X-gal stained cells are shown (left). The levels of SA-β-galactosidase positive cells were also measured (right). *P < 0.05 (P= 0.027 and 0.015 for 1 and 10 µM BMVC4 at day 12, respectively), significant effects of BMVC4. (D) Accumulation of H3K9Me3 in BMVC4-treated cells. Confocal images of indirect immunofluorescence of histone H3 tri-methylation on Lys9 (H3K9Me3) in BMVC4-treated H1299 cells (left). The DNA was also stained by DAPI. Quantification of the H3K9Me3 foci positive cells was conducted (right). *P < 0.05 (P= 0.03, 0.003 and 0.006 for days 3, 6 and 9, respectively), significant effects of BMVC4. (E) The H1299 cells were treated with 10 µM BMVC4 for 6 or 12 days and then analysed by flow cytometry. Histograms of the PI-stained cells are presented. (F) Reduction of pRB by BMVC4. Cell extracts were prepared after treating BMVC4 for indicated days and then analysed by immunoblotting assays using antibodies against phospho-Rb (pRb) or GAPDH. Bound antibodies were visualized by chemiluminescence using an ECL kit (Amersham-Pharmacia). (G) Telomere shortening in BMVC4 treated H1299 cells. Effects of BMVC4 on telomere lengths. H1299 cells were treated with 10 µM BMVC4 and total genomic DNA were prepared and analysed for telomere length by Southern blotting using telomeric DNA as the probe.
The progression of cell cycle in BMVC4-treated cells was analysed using flow cytometry. BMVC4 induced a decrease in the percentage of G0/G1-phase cells and a slight increase in the percentage of S-phase cells. The percentage of sub-G1-phase cells was not markedly increased after 12-day treatment (Figure 2E). The cell cycle profile was similar to that exhibited by senescence induced by overexpressing Raf-1 oncogene or DNA-damaging agents (Chang et al., 1999; Olsen et al., 2002). We have also analysed the expression pattern of pRb using immunoblotting analysis. In H1299 cells, treatment with BMVC4 consistently inhibited Rb protein phosphorylation (Figure 2F). These results suggested that the reduction of Rb protein phosphorylation was involved in BMVC4-induced senescence.
The effect of BMVC4 on telomere length was also determined. Genomic DNAs were prepared from H1299 cells treated with 10 µM BMVC4 at different time intervals and then analysed by Southern hybridization using radioactive telomeric DNA as the probe. In BMVC4-treated cells, we observed telomere shortening at a rate of ∼230 bp per population doubling (Figure 2G), a rate that is higher than the rate of around 30–120 bp per population doubling, in human normal cells (Hastie et al., 1990; Counter et al., 1992).
BMVC4-induced senescence in ALT and hTERT-overexpressing cells
We have shown that BMVC4 inhibited telomerase and affected telomere lengths. To determine if telomere shortening through telomerase inhibition is the cause of senescence, ALT cells with telomere maintenance independent of telomerase were used for further analysis. If telomere shortening caused by telomerase inhibition is the cause of senescence, the ALT cells should not be induced into senescence by BMVC4 treatment. The ALT cells were treated with BMVC4 and analysed for the growth inhibitory effects. All three ALT lines showed growth inhibitory effects, similar to that observed in telomerase-positive cancer cells (Figure 3, left panels). Decreased cell viability was also observed in VA13 and SaoS2 cells after long-term BMVC4 treatments. Moreover, all these three ALT cells showed SA-β-Gal activities upon BMVC4 treatments, although the levels of senescence varied in different cells (Figure 3, right panels). Why SA-β-Gal activity did not increase in SaoS2 cells is not clear to us; nevertheless, our results showed that BMVC4 also induced senescence in telomerase-negative cancer cells.
Figure 3.
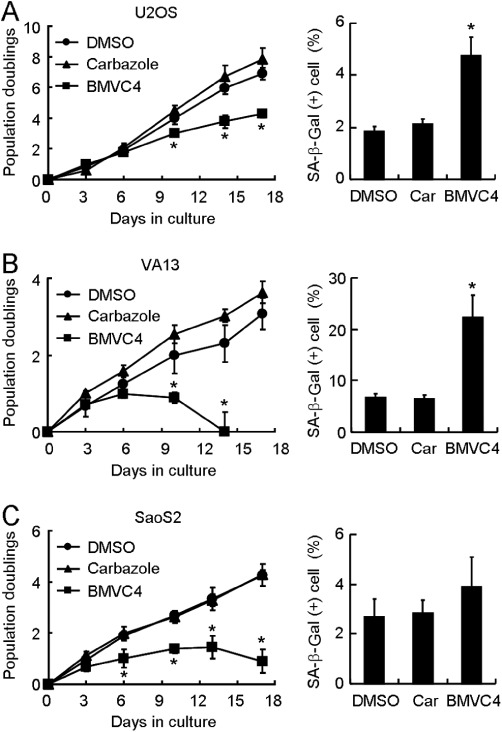
Senescent phenotype in BMVC4-treated ALT cells. (A) U2OS, (B) VA13 or (C) SaoS2 cells were treated with 10 µM of BMVC4. The cells were counted during the passages and the population doubling was determined (left). Results were obtained from the average of three independent experiments. *P < 0.05 (P= 0.03, 0.006 and 0.003 for U2OS at days 10, 14 and 17, respectively; P= 0.03 and 0.006 for VA13 at days 6 and 10, respectively; P= 0.03, 0.001, 0.01 and 0.001 for SaoS2 at days 6, 10, 13 and 17, respectively), significant effects of BMVC4. The levels of SA-β-galactosidase positive cells were also measured at day 9 (right). *P < 0.05 (P= 0.002 and 0.003 for U2OS and VA13, respectively), significant effects of BMVC4.
To further determine the involvement of telomerase in BMVC4-induced senescence, we increased the expression level of hTERT through transfecting plasmid carrying hTERT under the control of CMV promoter into H1299 cells. We assumed that if telomerase is involved in BMVC4-induced senescence, forced hTERT expression to increase telomerase level might rescue the senescence phenotype in BMVC4-treated cells. As shown in Figure 4A, the hTERT protein level was significantly elevated in cells harbouring the plasmid pCMV-hTERT. However, the hTERT overexpression did not appear to rescue the growth of H1299 cells after BMVC4 treatments. In addition, the senescent phenotypes were similar in both BMVC4-treated control and hTERT-overexpressing cells (Figure 4B). We have also tested the effects of BMVC4 in ALT cell overexpressing hTERT (Supporting Information Figure S2). Our results showed that BMVC4 inhibited cell growth regardless of hTERT overexpression in U2OS cells. Taken together, these results indicate that senescence induced by BMVC4 is not likely to be caused by pathways related to telomerase inhibition.
Figure 4.
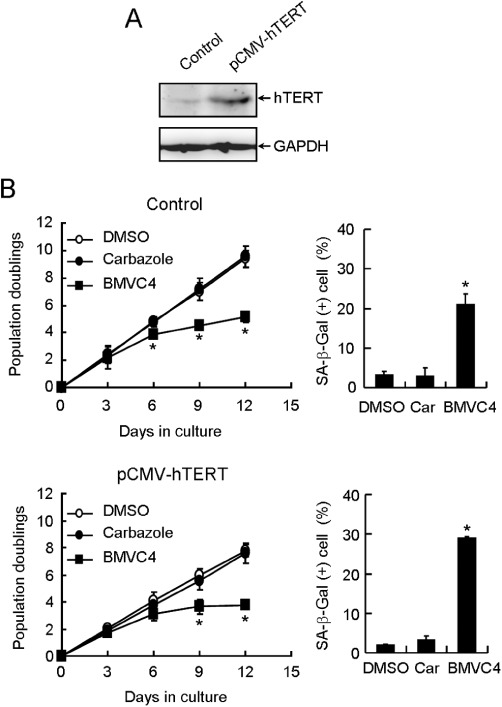
BMVC4 induced senescence in hTERT-overexpressing H1299 cells. (A) Total cell extracts prepared from H1299 cells harbouring vector (Control) or hTERT gene under the control of CMV (pCMV-hTERT) promoter were analysed for the hTERT and GAPDH levels using immunoblots. (B) Control (top panels) or hTERT-overexpressing (bottom panels) H1299 cells were treated with 10 µM of BMVC4. The cells were counted during the passages and the population doubling was determined (left). Results were obtained from the average of three independent experiments. *P < 0.05 (P= 0.02, 0.002 and 0.0004 for vector control at days 6, 9 and 12, respectively; P= 0.004 and 0.0001 for pCMV-hTERT at days 9 and 12, respectively), significant effects of BMVC4. The levels of SA-β-galactosidase positive cells were also measured 12 days after treatments (right). *P < 0.05 (P= 0.012 and 0.00006 for pCMV control and pCMV-hTERT, respectively), significant effects of BMVC4.
Only a small fraction of BMVC4-induced DNA damage foci was localized to telomeres
Telomere shortening eventually leads to telomere uncapping that induces responses to DNA damage that signal for cell cycle arrest (Takai et al., 2003). The effect of BMVC4 on the induction of such signals from DNA damage was further analysed using the nuclear accumulation of γ-H2AX. As shown in Figure 5A, a significant amount of γ-H2AX was accumulated after 6 days of treatment with BMVC4 (Figure 5A). The increased levels of γ-H2AX became more apparent after treating the cells for 12 days. At day 12, up to 38% of the BMVC4-treated cells showed γ-H2AX staining (Figure 5B, left panel). This result indicated that reponses to DNA damage were induced in cancer cells treated with BMVC4 for a longer time. To test if the foci of DNA damage were caused by telomere uncapping, the localization of the DNA damage foci and telomeres were determined by co-staining of γ-H2AX and telomere binding protein TRF2 using confocal microscopy. Surprisingly, a large fraction of the γ-H2AX DNA damage foci was not localized to telomeres (Figure 5A). Quantification of these images showed that only ∼20% of the γ-H2AX foci were localized to telomeres after MVC4 treatment (Figure 5B, right panel). These results indicated that although BMVC4 induced DNA damage foci on telomeres, probably through perturbation of telomere replication, significantly higher levels of DNA damage were induced in other loci in the chromosomes. Thus, the cellular targets of BMVC4 were not limited to telomeres.
Figure 5.
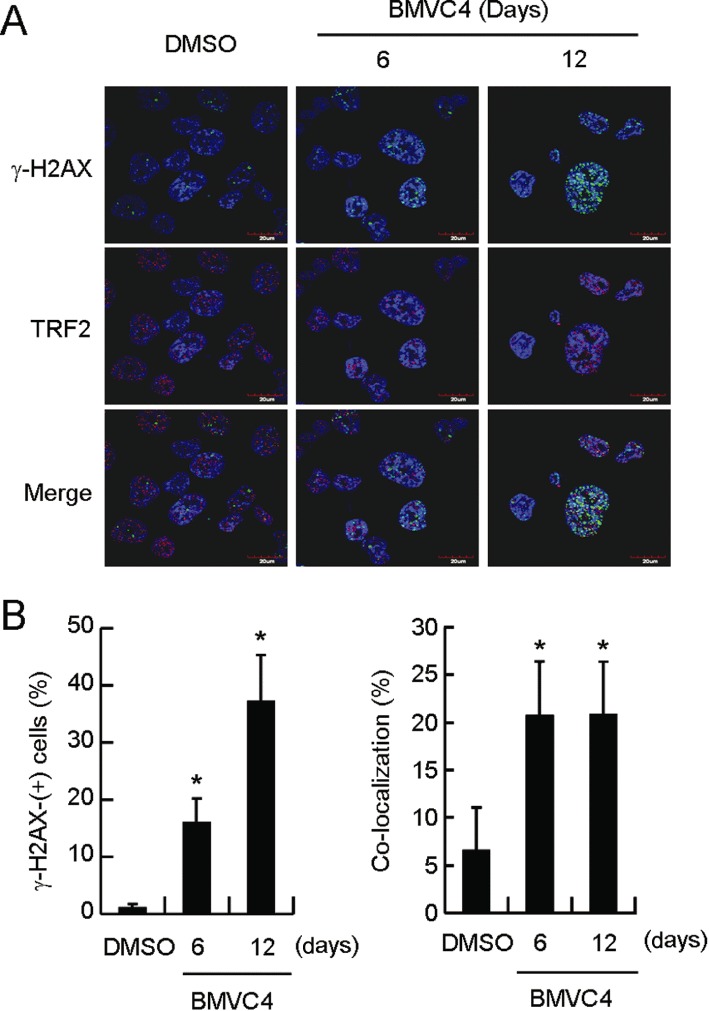
DNA-damage foci formation in BMVC4-treated H1299 cells. (A) Induction of responses to DNA damage after treatment with BMVC4. H1299 cells were treated with 10 µM of BMVC-4 for 6 or 12 days and stained with antibodies against γ-H2AX and TRF2. Staining of DNA by Hoechst 33258 was employed to locate the positions of nuclei. (B) Quantification of γ-H2AX-positive cells. The left panel shows the percentage of γ-H2AX-positive cells. *P < 0.05 (P= 0.014 and 0.013 for day 6 and 12, respectively), significant effects of BMVC4. The right panel shows the percentage of co-localized γ-H2AX and TRF2 foci. *P < 0.05 (P= 0.0007 and 0.027 for day 6 and 12, respectively), significant effects of BMVC4.
BMVC4 suppressed c-myc expression through induction of G-quadruplex structure formation at its promoter
In addition to telomeres, QFS are present in human genome. Bioinformatic analysis indicated that about 375 000–377 000 potential intramolecular QFS were identified within the human genome (Huppert and Balasubramanian, 2005; Todd et al., 2005). Significantly, localization of several QFS at the promoter regions was shown to affect the expression of their downstream genes. For example, the expression of c-myc (Siddiqui-Jain et al., 2002), c-kit (Phan et al., 2007; Hsu et al., 2009; Kuryavyi et al., 2010), KRAS (Cogoi and Xodo, 2006), HIF1α (De Armond et al., 2005), Rb1 (Xu and Sugiyama, 2006), VEGF (Sun et al., 2005; 2011), hTERT (Palumbo et al., 2009), c-myb (Palumbo et al., 2008), PDGFR-β (Qin et al., 2010) and PDGF-A (Qin et al., 2007) was affected by G-quadruplex stabilizing ligands. These QFS are potential targets for BMVC4. Here, expression analyses were conducted to test if c-myc expression was affected by BMVC4. Real-time PCR analysis showed that levels of mRNA for c-myc were significantly decreased by BMVC4 treatment (Figure 6A). Consequently, the c-myc protein level was also decreased by BMVC4 treatment (Figure 6B). In contrast, the expression of Sp1, whose promoter does not include QFS, was slightly activated by both carbazole and BMVC4. The level of a downstream target of c-myc, the telomerase catalytic subunit hTERT (Wang et al., 1998; Wu et al., 1999), was also decreased, further supporting the repression of c-myc by BMVC4 (Figure 6B).
Figure 6.
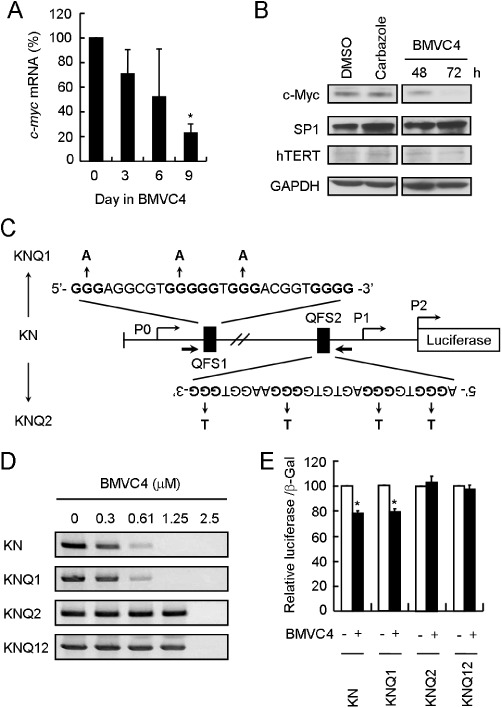
BMVC4 represses c-myc expression through the formation of quadruplex-forming sequence within the QFS2 of the c-myc promoter. (A) Expression of c-myc on RNA level was determined by real-time RT-PCR in H1299 cells incubated with 10 µM of BMVC4 for 3, 6 or 9 days. *P < 0.05 (P= 0.005), significant effects of BMVC4. (B) The protein levels of c-myc and hTERT were decreased in H1299 cells treated with 10 µM of BMVC4 for 48 or 72 h and then analysed for the protein levels using immunoblotting. (C) Schematic diagrams showed the mutation sites of MycPro-luciferase. The G-quadruplex-forming sequences in wild-type (KN), QFS1 mutations (KNQ1) and QFS2 mutations (KNQ2) were indicated. (D) BMVC4 stabilized G-quadruplex structure on c-myc promoter. The PCR stop assays were performed in the presence of BMVC4 at various concentrations (0, 0.3, 0.61, 1.25, 2.5 µM). (E) QFS2 is required for BMVC4 to suppress c-myc expression. c-myc reporter plasmid constructs carrying the indicated mutations were transfected into H1299 cells. The cells were then incubated with 10 µM of BMVC4 for 3 days and the luciferase activity was determined. The value of luciferase activity in DMSO-treated cells was defined as 100%. *P < 0.05 (P= 0.001 for both KN and KNQ1), significant effects of BMVC4.
We next determined if c-myc repression by BMVC4 is achieved by stabilizing the QFS located at its promoter region. Two QFS were identified within the c-myc promoter, QFS1 and QFS2 (Figure 6C). These sequences are located at 148 bp (non-coding strand, QFS2) and 914 bp (coding strand, QFS1) upstream of the transcription start site. Mutations in these two QFS that disrupt their G-quadruplex forming abilities were generated and analysed by the PCR stop assays (Gomez et al., 2004). In the assay, the QFS are stabilized into G-quadruplex structures that block hybridization with a complementary strand overlapping with the last G-repeat. Thus, formation of the PCR product by Taq polymerase is inhibited. Here, the c-myc wild-type (KN) and mutant (KNQ1, KNQ2 and KNQ12) promoter constructs were incubated with increasing concentrations of BMVC4 in the presence of their complementary strand for 30 PCR cycles. As shown in Figure 6D, BMVC4 added to wild-type promoter sequences inhibited formation of the DNA product in a dose-dependent manner, with IC50 values ∼0.2 µM. Although the QFS1 mutation (KNQ1) required a similar concentration of BMVC4 to inhibit the PCR reaction, compared with wild-type sequences, the QFS2 mutation (KNQ2) required ∼10-fold more BMVC4 to achieve the same effect. Mutations on both motifs did not further increase the amount of BMVC4 required to inhibit the PCR reaction (KNQ12). The results indicated that BMVC4 selectively stabilized the G-quadruplex structure formed by the QFS2 sequence.
To confirm this conclusion, a reporter construct with the luciferase gene fused downstream to the c-myc promoter was used to assess the expression efficiency of wild-type and mutant promoters. Plasmid constructs containing wild-type or mutant promoters were transfected into H1299 cells and the luciferase activities were determined. The luciferase activity expressed by the wild-type c-myc promoter was reduced by ∼20% after 24 h of BMVC4 treatments (Figure 6E). Mutation in the QFS1 region (KNQ1) also showed a similar BMVC4-induced repression. In contrast, promoters containing the QFS2 mutations (KNQ1 and KNQ12) failed to respond to BMVC4 treatments. Thus, selective binding and stabilizing of G-quadruplex structure formed by QFS2 sequences suppresses the expression efficiency of c-myc promoters. These results clearly demonstrated that, in addition to telomeres, BMVC4 also targeted the QFS located at the c-myc promoter.
To determine if the reduced c-myc expression accounts for the effects on growth of BMVC4-treated cells, we used c-myc overexpressing cells. As shown in Supporting Information Figure S3, BMVC4 still induced growth inhibitory effects in c-myc overexpressing cells suggesting that these effects were not due to repression of c-myc levels.
Induction of chromosome breaks and ATM-mediated DNA damage response by BMVC4
The results in Figure 5 show that responses to DNA damage response were activated by BMVC4. To determine the source of the DNA damage induced by BMVC4, alkaline comet assays were conducted to evaluate the levels of DNA strand breaks on chromosomes (Singh et al., 1988). The assay enables the detection of DNA breaks at the single cell level. It involves encapsulation of cells in a low-melting-point agarose suspension, lysis of the cells in alkaline conditions and electrophoresis of the suspended lysed cells. The undamaged DNA strands are too large and do not leave the cell cavity, whereas the DNA fragments that were induced by DNA breaks are free to move in a given period of time. Therefore, the amount of DNA that leaves the cell cavity is a measure of the amount of DNA damage in the cell. As shown in Figure 7A, while solvent DMSO and carbazole did not cause detectable strand breaks, greater numbers of strand breaks were observed in cells treated with BMVC4 for 12 days.
Figure 7.
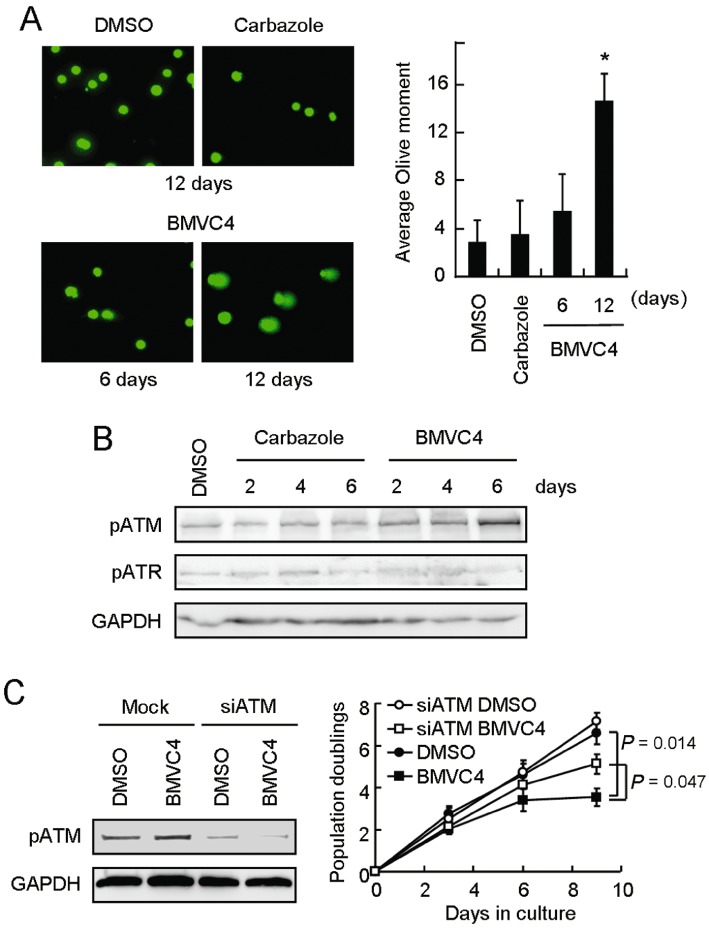
DNA strand breaks and ATM-mediated DNA damage response in BMVC4-treated cells. (A) DNA strand breaks in BMVC4-treated cells. H1299 cells were treated with 10 µM carbazole or BMVC4 for 6 days or 12 days and then analysed by comet assays. Single-cell electrophoresis was conducted and stained with SYBR Green. The images were taken (left panels) and the relative comet tail moment was measured (right panel). The histograms represent the average of five independent experiments (n > 100). *P < 0.05 (P= 0.001), significant effects of BMVC4. (B) Activation of ATM in BMVC4-treated cells. H1299 cells were treated with 10 µM carbazole or BMVC4 for 2, 4 or 6 days and then analysed by immunoblotting assays. (C) H1299 cells were transfected with siRNA targeting ATM. Total cell extracts were prepared and immunoblotting analysis was conducted using antibodies against ATM or GAPDH (left). The siATM treated-H1299 cells were also treated with 10 µM of BMVC4. The cells were counted during the passages and the population doubling was determined. Results were obtained from the average of three independent experiments (right).
We next analysed the molecular pathway of responses to DNA damage activated by BMVC4 treatment. The detection of DNA damage in cells induces a series of cellular responses that result in DNA repair, cell cycle arrest, senescence or cell death (Branzei and Foiani, 2008). In mammals, the ATM and ATR (ATM and Rad3-related) kinases are critical regulators of the cellular response to DNA damage. ATM and ATR are both protein kinases with overlapping substrate specificities that are activated in response to distinct, as well as partially overlapping, damage signals (Matsuoka et al., 2007). Using immunoblotting analyses, we found that ATM was phosphorylated on Ser1981, while no detectable enhancement of ATR phosphorylation on Ser428 was observed, in BMVC4-treated H1299 cells (Figure 7B). These results suggest that ATM is activated in the response to DNA damage, triggered by BMVC4.
To evaluate the role of ATM in the response to BMVC4-induced DNA damage, we used siRNA to knock down the expression of ATM. As shown in Figure 7C, immunoblotting showed that the ATM levels werer decreased by treatment of H1299 cells with siRNA targeting ATM (left panel). Significantly, the BMVC4-induced delayed growth proliferation was partly suppressed upon knocking down the expression of ATM (right panel). The results strongly suggest that the observed cellular effects of BMVC4 were mediated through a ATM-dependent pathway in response to DNA damage. The involvement of ATR in BMVC4-induced DNA damage response is uncertain, although we did not detect apparent enhancement of ATR phosphorylation by BMVC4. More studies are required to determine whether the response to DNA damage is exclusively mediated through the ATM pathway.
Discussion and conclusion
In the present manuscript, we have characterized the cellular effects of a carbazole derivative, BMVC4. BMVC4 stabilized G-quadruplex DNA structure formed by human telomeric DNA sequences and effectively inhibited telomerase with an IC50 at 0.2 µM. Treatment of telomerase-positive cancer cells with BMVC4 induced senescence that was accompanied by progressive telomere shortening. Interestingly, BMVC4 also induced senescence in telomerase-overexpressing cells and ALT cells in which telomere maintenance is not mediated by telomerase. Thus, senescence induced by BMVC4 did not appear to be caused by telomerase inhibition. Consistent with this observation, most of the DNA damage foci induced by BMVC4 are not localized to telomeres. Taking into account the G-quadruplex DNA binding properties of BMVC4, we also showed that BMVC4 suppressed expression of c-myc through stabilizing the G-quadruplex structure, QFS2, located at the c-myc promoter region. The results indicated that BMVC4 could target other QFS located within chromosomes. Thus, BMVC4 had an additional cellular effect that was independent of its telomerase-inhibitory effect. We further showed that BMVC4 could achieve this effect through inducing DNA breaks on chromosomes and the consequent response to DNA damage might be mediated through the ATM-dependent pathway. Thus, although BMVC4 inhibited telomerase in vitro and reduced telomere length in vivo, it induced s senescence through a DNA damage response pathway that was independent of its telomerase-inhibition activity. Of note, the present analysis was conducted at BMVC4 concentrations that were sufficient to cause senescence after a short incubation time. Because BMVC4 inhibited telomerase with IC50 of ∼200 nM, the possibility that extended times of incubation with lower concentrations of BMVC4 may result in senescence mediated by telomere shortening, cannot be excluded.
Several lines of evidence support a hypothesis that non-telomere pathways were involved in BMVC4-induced senescence. First, immunostaining analysis showed that ∼80% of the γ-H2AX DNA damage foci did not localize to telomeres. Second, alkaline comet assays showed that DNA breaks were induced after long-term BMVC4 treatment. Third, BMVC4 also induced senescence in ALT cells, similar to that in telomerase-positive cells. Fourth, forced telomerase expression did not rescue the senescence phenotype induced by BMVC4. These results cannot be simply explained by telomerase inhibition. Based on its ability to bind G-quadruplex structures, BMVC4 is likely to target QFS located within the chromosomes and to affect cancer cells in a telomere-independent manner. Our finding of DNA breaks accumulated after BMVC4 treatment might explain how this compound induced senescence in cancer cells.
Another G-quadruplex stabilizing agent, RHPS4, has been shown to induce an ATR-dependent replication stress pathway in drug-treated cells (Salvati et al., 2007). Further, phosphorylation of ATR kinase was induced and co-localized with damaged telomeres (Salvati et al., 2007; Rizzo et al., 2009). As the unwinding activities of several RecQ helicases, including WRN, BLM and FANCJ, can be inhibited by G-quadruplex structure stabilizers such as the porphyrin compounds NMM and T4 (Li et al., 2001; Huber et al., 2002; Wu et al., 2008), it was postulated that a G-quadruplex stabilizing agent might bind and stabilize the G-quadruplex DNA formed at telomeres and inhibit RecQ helicases during telomere replication. The DNA replication stress then induces DNA breaks on chromosomes to activate the DNA damage response pathways (d'Adda di Fagagna, 2008). Although the detailed mechanism of how BMVC4 induced responses to DNA damage is unclear, we consider that the perturbation of telomere replication might not be the major source for DNA damage. It is generally accepted that ATM plays the primary role in responding to double-strand breaks (DSBs), whereas ATR plays a secondary role but is the primary mediator of responses to UV-induced damage and stalled replication forks (Branzei and Foiani, 2008). Here, we found that only a small fraction of DNA damage foci was localized to telomeres. Moreover, DNA breaks were induced and the ATM-mediated response to DNA damage was activated following BMVC4 treatment. Thus, while the involvement of telomeres cannot be completely ruled out, it is likely that a DSB-activated, DNA damage-induced, senescence pathway is responsible for the senescence induced by BMVC4.
Our results showed that the cellular effects of BMVC4 were not limited to telomeres. Here, we showed that BMVC4 selectively inhibited telomerase and targeted the G-quadruplex-forming sequences (QFS) located at the c-myc promoter. These selective effects are likely to be related to the G-quadruplex binding activity of BMVC4. Here, we consider that the binding of BMVC4 to non-telomeric G-quadruplex-forming sequences might cause the telomere-independent cellular effects of BMVC4. Thus, although BMVC4 was also shown to bind duplex DNA, the cellular effects cannot be attributed to the duplex DNA binding activity of BMVC4. It is also interesting to note that although two QFS were identified at the c-myc promoter, only one, QFS2, was affected by BMVC4. Thus, BMVC4 might have preferences towards certain sequences, as the QFS1 and QFS2 are not identical. Additionally, local and/or global chromosomal contexts might contribute to the preferential binding of BMVC4 to these G-quadruplex structures. It is also interesting to note that BMVC, another carbazole derivative that differs from BMVC4 at the pyridinium ring, did not affect c-myc expression (data not shown). Thus, G-quadruplex stabilizers might also show selectivity towards G-quadruplex structures formed by different sequences.
Acknowledgments
We thank Dr Kou-Juey Wu for providing plasmids pMT2T and pMT2T-cMyc and Dr Yan-Hwa Wu Lee for providing plasmid pc-MycPro-Luc. This work is supported by Academia Sinica (AS-98-TP-A04), the National Science Council under grants 99–3112-B-010-001 and 97–2311-B-010-005-MY3, and the National Health Research Institute grant NHRI-EX100-10050SI.
Glossary
- ALT
alternative lengthening of telomeres
- ATM
ataxia telangiectasia-mutated
- ATR
ATM and Rad3-related
- BMVC
3,6-bis(1-methyl-4-vinylpyridium iodide) carbazole
- BMVC4
3,6-bis(4-methyl-2-vinylpyrazinium iodide) carbazole
- hTERT
human telomerase reverse transcriptase
- QFS
G-quadruplex forming sequences
- SA-β-Gal
senescence-associated (SA) β-galactosidase
Statement of conflicts of interest
None.
Supporting information
Additional Supporting Information may be found in the online version of this article:
Figure S1 Senescent phenotype of BMVC4-treated cancer cells. MCF7 and HeLa cells were treated with 1 or 10 μM of BMVC4. The cells were counted during the passages and the population doubling was determined. Results were obtained from the average of three independent experiments.
Figure S2 BMVC4 induced senescence in hTERT-overexpressing U2OS cells. (A) Total cell extracts prepared from U2OS cells harbouring vector (Control) or hTERT gene under the control of CMV (pCMV-hTERT) promoter were analysed for the hTERT and GAPDH levels using immunoblots. (B) Control or hTERT-overexpressing U2OS cells were treated with 10 μM of BMVC4. The cells were counted during the passages and the population doubling was determined.
Figure S3 BMVC4 induced senescence in c-myc-overexpressing VA13 cells. (A) VA13 cells harbouring vector (MT2T) or c-myc gene under the control of CMV (MT2T-Myc) were analysed for the c-myc and GAPDH levels using immunoblots. (B) The cells were treated with 10 μM of BMVC4, counted during the passages, and the population doubling was determined.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Blasco MA, Lee H-W, Hande MP, Samper E, Lansdorp PM, DePinho RA, et al. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu C-P, Morin GB, et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- Burger AM, Dai F, Schultes CM, Reszka AP, Moore MJ, Double JA, et al. The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res. 2005;65:1489–1496. doi: 10.1158/0008-5472.CAN-04-2910. [DOI] [PubMed] [Google Scholar]
- Chang B-D, Xuan Y, Broude EV, Zhu H, Schott B, Fang J, et al. Role of p53 and p21waf1/cip1 in senescence-like terminal proliferation arrest induced in human tumor cells by chemotherapeutic drugs. Oncogene. 1999;18:4808–4818. doi: 10.1038/sj.onc.1203078. [DOI] [PubMed] [Google Scholar]
- Chang C-C, Wu J-Y, Chang T-C. A carbazole derivative synthesis for stabilizing the quadruplex structure. J Chin Chem Soc. 2003;50:185–188. [Google Scholar]
- Chou W-C, Wang H-C, Wong F-H, Ding S, Wu P-E, Shieh S-Y, et al. Chk2-dependent phosphorylation of XRCC1 in the DNA damage response promotes base excision repair. EMBO J. 2008;27:3140–3150. doi: 10.1038/emboj.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoi S, Xodo LE. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006;34:2536–2549. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K, Mitchell JR. Telomerase in the human organism. Oncogene. 2002;21:564–579. doi: 10.1038/sj.onc.1205083. [DOI] [PubMed] [Google Scholar]
- Counter CM, Avilion AA, LeFeuvre CE, Stewart NG, Greider CW, Harley CB, et al. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Adda di Fagagna F. Living on a break: cellular senescence as a DNA-damage response. Nat Rev Cancer. 2008;8:512–522. doi: 10.1038/nrc2440. [DOI] [PubMed] [Google Scholar]
- Damm K, Hemmann U, Garin-Chesa P, Hauel N, Kauffmann I, Priepke H, et al. A highly selective telomerase inhibitor limiting human cancer cell proliferation. EMBO J. 2001;20:6958–6968. doi: 10.1093/emboj/20.24.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Armond R, Wood S, Sun D, Hurley LH, Ebbinghaus SW. Evidence for the presence of a guanine quadruplex forming region within a polypurine tract of the hypoxia inducible factor 1α promoter. Biochemistry. 2005;44:16341–16350. doi: 10.1021/bi051618u. [DOI] [PubMed] [Google Scholar]
- De Cian A, Cristofari G, Reichenbach P, De Lemos E, Monchaud D, Teulade-Fichou M-P, et al. Reevaluation of telomerase inhibition by quadruplex ligands and their mechanisms of action. Proc Natl Acad Sci U S A. 2007;104:17347–17352. doi: 10.1073/pnas.0707365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez D, Mergny J-L, Riou JF. Detection of telomerase inhibitors based on G-quadruplex ligands by a modified telomeric repeat amplification protocol assay. Cancer Res. 2002;62:3365–3368. [PubMed] [Google Scholar]
- Gomez D, Lemarteleur T, Lacroix L, Mailliet P, Mergny J-L, Riou J-F. Telomerase downregulation induced by the G-quadruplex ligand 12459 in A549 cells is mediated by hTERT RNA alternative splicing. Nucleic Acids Res. 2004;32:371–379. doi: 10.1093/nar/gkh181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez D, O'Donohue M-F, Wenner T, Douarre C, Macadre J, Koebel P, et al. The G-quadruplex ligand telomestatin inhibits POT1 binding to telomeric sequences in vitro and induces GFP-POT1 dissociation from telomeres in human cells. Cancer Res. 2006;66:6908–6912. doi: 10.1158/0008-5472.CAN-06-1581. [DOI] [PubMed] [Google Scholar]
- Gowan SH, Heald R, Stevens MFG, Kelland LR. Potent inhibition of telomerase by small-molecule pentacyclic acridines capable of interacting with G-quadruplexes. Mol Pharmacol. 2001;60:981–988. doi: 10.1124/mol.60.5.981. [DOI] [PubMed] [Google Scholar]
- Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. Creation of human tumour cells with defined genetic elements. Nature. 1999a;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- Hahn WC, Stewart SA, Brooks MW, York SG, Eaton E, Kurachi A, et al. Inhibition of telomerase limits the growth of human cancer cells. Nat Med. 1999b;5:1164–1170. doi: 10.1038/13495. [DOI] [PubMed] [Google Scholar]
- Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- Hastie ND, Dempster M, Dunlop MG, Thompson AM, Green DK, Allshire RC. Telomere reduction in human colorectal carcinoma and with ageing. Nature. 1990;346:866–868. doi: 10.1038/346866a0. [DOI] [PubMed] [Google Scholar]
- Hsu S-TD, Varnai P, Bugaut A, Reszka AP, Neidle S, Balasubramanian S. A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-tetrad dynamics. J Am Chem Soc. 2009;131:13399–13409. doi: 10.1021/ja904007p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y-H, Lin J-J. Telomere and telomerase as targets for anti-cancer and regeneration therapies. Acta Pharmacol Sin. 2005;26:513–518. doi: 10.1111/j.1745-7254.2005.00098.x. [DOI] [PubMed] [Google Scholar]
- Huang F-C, Chang C-C, Lou P-J, Kuo I-C, Chien C-W, Chen C-T, et al. G-quadruplex stabilizer 3,6-bis(1-methyl-4-vinylpyridinium)carbazole diiodide induces accelerated senescence and inhibits tumorigenic properties in cancer cells. Mol Cancer Res. 2008;6:955–964. doi: 10.1158/1541-7786.MCR-07-0260. [DOI] [PubMed] [Google Scholar]
- Huber MD, Lee DC, Maizels N. G4 DNA unwinding by BLM and Sgs1p: substrate specificity and substrate-specific inhibition. Nucleic Acids Res. 2002;30:3954–3961. doi: 10.1093/nar/gkf530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert J, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incles CM, Schultes CM, Kempski H, Koehler H, Kelland LR, Neidle S. A G-quadruplex telomere targeting agent produces p16-associated senescence and chromosomal fusions in human prostate cancer cells. Mol Cancer Ther. 2004;3:1201–1206. [PubMed] [Google Scholar]
- Kim M-Y, Duan W, Gleason-Guzman M, Hurley LH. Design, synthesis, and biological evaluation of a series of fluoroquinoanthroxazines with contrasting dual mechanisms of action against topoisomerase II and G-quadruplexes. J Med Chem. 2003;46:571–583. doi: 10.1021/jm0203377. [DOI] [PubMed] [Google Scholar]
- Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PLC, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- Kuryavyi V, Phan AT, Patel DJ. Solution structures of all parallel-stranded monomeric and dimeric G-quadruplex scaffolds of the human c-kit2 promoter. Nucl Acids Res. 2010;38:6757–6773. doi: 10.1093/nar/gkq558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J-L, Harrison RJ, Reszka AP, Brosh RM, Jr, Bohr VA, Neidle S, et al. Inhibition of the Bloom's and Werner's syndrome helicases by G-quadruplex interacting ligands. Biochemistry. 2001;40:15194–15202. doi: 10.1021/bi011067h. [DOI] [PubMed] [Google Scholar]
- Makarov VL, Hirose Y, Langmore JP. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell. 1997;88:657–666. doi: 10.1016/s0092-8674(00)81908-x. [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, III, Hurov KE, Luo J, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Mergny J-L, Helene C. G-quadruplex DNA: a target for drug design. Nature Med. 1998;4:1366–1367. doi: 10.1038/3949. [DOI] [PubMed] [Google Scholar]
- Narita M, Nunez S, Heard E, Narita M, Lin AW, Hearn SA, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- O'Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol. 2010;11:171–181. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen CL, Gardie B, Yaswen P, Stampfer MR. Raf-1-induced growth arrest in human mammary epithelial cells is p16-independent and is overcome in immortal cells during conversion. Oncogene. 2002;21:6328–6339. doi: 10.1038/sj.onc.1205780. [DOI] [PubMed] [Google Scholar]
- Palumbo SL, Memmott RM, Uribe DJ, Krotova-Khan Y, Hurley LH, Ebbinghaus SW. A novel G-quadruplex-forming GGA repeat region in the c-myb promoter is a critical regulator of promoter activity. Nucleic Acids Res. 2008;36:1755–1769. doi: 10.1093/nar/gkm1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbo SL, Ebbinghaus SW, Hurley LH. Formation of a unique end-to-end stacked pair of G-quadruplexes in the hTERT core promoter with implications for inhibition of telomerase by G-quadruplex-interactive ligands. J Am Chem Soc. 2009;131:10878–10891. doi: 10.1021/ja902281d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascole E, Wenz C, Lingner J, Hauel N, Priepke H, Kauffmann I, et al. Mechanism of human telomerase inhibition by BIBR1532, a synthetic, non-nucleosidic drug candidate. J Biol Chem. 2002;277:15566–15572. doi: 10.1074/jbc.M201266200. [DOI] [PubMed] [Google Scholar]
- Pennarun G, Granotier C, Gauthier LR, Gomez D, Hoffschir F, Mandine E, et al. Apoptosis related to telomere instability and cell cycle alterations in human glioma cells treated by new highly selective G-quadruplex ligands. Oncogene. 2005;24:2917–2928. doi: 10.1038/sj.onc.1208468. [DOI] [PubMed] [Google Scholar]
- Phan AT, Kuryavyi V, Burge S, Neidle S, Patel DJ. Structure of an unprecedented Gquadruplex scaffold in the human c-kit promoter. J Am Chem Soc. 2007;129:4386–4392. doi: 10.1021/ja068739h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Rezler EM, Gokhale V, Sun D, Hurley LH. Characterization of the G-quadruplexes in the duplex nuclease hypersensitive element of the PDGF-A promoter and modulation of PDGF-A promoter activity by TMPyP4. Nucleic Acids Res. 2007;35:7698–7713. doi: 10.1093/nar/gkm538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Fortin JS, Tye D, Gleason-Guzman M, Brooks TA, Hurley LH. Molecular cloning of the human platelet-derived growth factor receptor β (PDGFR-β) promoter and drug targeting of the G-quadruplex-forming region to repress PDGFR-β expression. Biochemistry. 2010;49:4208–4219. doi: 10.1021/bi100330w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman MK, Cech TR. Effect of monovalent cation-induced telomeric DNA structure on the binding of Oxytricha telomeric protein. Nucleic Acids Res. 1990;18:4543–4552. doi: 10.1093/nar/18.15.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezler EM, Bearss DJ, Hurley LH. Telomeres and telomerases as drug targets. Curr Opin Pharmacol. 2002;2:415–423. doi: 10.1016/s1471-4892(02)00182-0. [DOI] [PubMed] [Google Scholar]
- Riou JF, Guittat L, Mailliet P, Laoui A, Renou E, Petitgenet O, et al. Cell senescence and telomere shortening induced by a new series of specific G-quadruplex DNA ligands. Proc Natl Acad Sci U S A. 2002;99:2672–2677. doi: 10.1073/pnas.052698099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo A, Salvati E, Porru M, D'Angelo C, Stevens MF, D'Incalci M, et al. Stabilization of quadruplex DNA perturbs telomere replication leading to the activation of an ATR-dependent ATM signaling pathway. Nucleic Acids Res. 2009;37:5353–5364. doi: 10.1093/nar/gkp582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvati E, Leonetti C, Rizzo A, Scasella M, Mottolese M, Galati R, et al. Telomere damage induced by the G-quadruplex ligand RHPS4 has an antitumor effect. J Clin Invest. 2007;117:3236–3247. doi: 10.1172/JCI32461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen D, Gilbert W. The structure of telomeric DNA: DNA quadruplex formation. Curr Opin Struct Biol. 1991;1:435–438. [Google Scholar]
- Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer. 1997;33:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci U S A. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- Sun D, Guo K, Rusche JJ, Hurley LH. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005;33:6070–6080. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Guo K, Shin Y-J. Evidence of the formation of G-quadruplex structures in the promoter region of the human vascular endothelial growth factor gene. Nucleic Acids Res. 2011;39:1256–1265. doi: 10.1093/nar/gkq926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- Tauchi T, Shin-ya K, Sashida G, Sumi M, Nakajima A, Shimamoto T, et al. Activity of a novel G-quadruplex-interactive telomerase inhibitor, telomestatin (SOT-095), against human leukemia cells: involvement of ATM-dependent DNA damage response pathways. Oncogene. 2003;22:5338–5347. doi: 10.1038/sj.onc.1206833. [DOI] [PubMed] [Google Scholar]
- Tauchi T, Shin-ya K, Sashida G, Sumi M, Okabe S, Ohyashiki JH, et al. Telomerase inhibition with a novel G-quadruplex-interactive agent, telomestatin: in vitro and in vivo studies in acute leukemia. Oncogene. 2006;25:5719–5725. doi: 10.1038/sj.onc.1209577. [DOI] [PubMed] [Google Scholar]
- Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Xie LY, Allan S, Beach D, Hannon GJ. Myc activates telomerase. Genes Dev. 1998;12:1769–1774. doi: 10.1101/gad.12.12.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright WE, Tesmer VM, Huffman KE, Levene SD, Shay JW. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 1997;11:2801–2809. doi: 10.1101/gad.11.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K-J, Grandori C, Amacker M, Simon-Vermot N, Polack A, Lingner J, et al. Direct activation of TERT transcription by c-MYC. Nature Genet. 1999;21:220–224. doi: 10.1038/6010. [DOI] [PubMed] [Google Scholar]
- Wu Y, Shin-ya K, Brosh RM., Jr FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol Cell Biol. 2008;28:4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Sugiyama H. Formation of the G-quadruplex and i-motif structures in retinoblastoma susceptibility genes (Rb) Nucleic Acids Res. 2006;34:949–954. doi: 10.1093/nar/gkj485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structure. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- Zhang R, Poustovoitov MV, Ye X, Santos HA, Chen W, Daganzo SM, et al. Formation of macroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev Cell. 2005;8:19–30. doi: 10.1016/j.devcel.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Zhang X, Mar V, Zhou W, Harrington L, Robinson MO. Telomere shortening and apoptosis in telomerase-inhibited human tumor cells. Genes Dev. 1999;13:2388–2399. doi: 10.1101/gad.13.18.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JM, Zhu X-F, Lu Y-J, Deng R, Huang Z-S, Mei Y-P, et al. Senescence and telomere shortening induced by novel potent G-quadruplex interactive agents, quindoline derivatives, in human cancer cell lines. Oncogene. 2006;25:503–511. doi: 10.1038/sj.onc.1209067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


