Abstract
DAMTC (7,8-diacetoxy-4-methylcoumarin) is a thioderivative of 4-methyl coumarin, and previously we have shown that DAMTC is a potent inhibitor of cell growth and an inducer of apoptosis in non-small cell lung cancer (A549) cells. It induces apoptosis through mitochondrial pathway by modulating NF-κB, mitogen-activated protein kinase (MAPK) and p53 pathways. Herein, we explored the genome-wide effects of DAMTC in A549 cells using the concerted approach of transcriptomics and proteomics. In addition to apoptotic pathways, which have been validated earlier, the bioinformatic analysis of microarray data identified small GTPase-mediated signal transduction among the significantly altered biological processes. Interestingly, we observed significant downregulation of some members of the Rho family GTPases in the proteomics data too. Downregulation of Rho GTPases (RhoGDIα (Rho GDP dissociation inhibitor-α, also known as ARHGDIA), Ras homolog family member A, Ras-related C3 botulinum toxin substrate 1 and cell division cycle 42) was validated by western blotting. The Rho protein family is implicated in maintaining the actin filament assembly and cell motility, and we also observed that DAMTC treatment causes actin cytoskeletal reorganization, promotes filopodia formation and inhibits cell motility in A549 cells. The effect of DAMTC treatment on cytoskeleton was reversed after the overexpression of RhoGDIα. In addition, DAMTC augmented the apoptotic effect of etoposide, a proapoptotic chemotherapeutic drug. This elucidation of the mechanism behind DAMTC-induced apoptosis and inhibition of cell motility in A549 cells may make it a potential therapeutic for lung cancer.
Keywords: DAMTC, apoptosis, RhoGDIα, Rho family GTPase, cytoskeletal reorganization
Lung cancer is the second leading cause of cancer deaths worldwide. Among lung cancers, non-small cell lung cancer accounts for 80% of total lung cancer cases.1, 2, 3 Although, there has been a significant improvement in the early diagnosis and treatment of non-small cell lung cancer (NSCLC), its 5-year survival rate remains a paltry 10–20%.4, 5, 6 At present, there is increasing interest in finding natural products that can target multiple cellular pathways and generally be non-toxic to the normal cells.
Coumarins are polyphenolic compounds, which have been shown to possess various biological activities. Studies from our own laboratory have shown that DAMTC (7,8-diacetoxy-4-methylcoumarin) is a potent inhibitor of cell growth and an inducer of apoptotic cell death. Our previous results also suggested that the inhibition of MAPK signaling pathway and the activation of NF-κB and p53 pathway may participate in the anti-proliferative activity of DAMTC in NSCLC (A549) cells.7 However, the detailed molecular mechanism(s) by which DAMTC inhibits cell growth and induces apoptosis is still unknown. Hence, the present study was undertaken to identify the comprehensive effects of DAMTC treatment in NSCLC (A549) cells.
Although transcriptional profiling of control and treated samples using oligonucleotide microarray allows the simultaneous analysis of the expression of thousands of genes in a high-throughput setting,8, 9 proteomic profiling using two-dimensional gel electrophoresis allows identification of differentially expressed proteins in large sets of complex protein mixtures.10 Using both transcriptome- and proteome-based approaches, we have shown that in addition to inducing apoptosis, DAMTC causes actin cytoskeletal reorganization, promotes filopodia formation and inhibits cell motility in A549 cells. The Rho family GTPases (RhoGDIα (Rho GDP dissociation inhibitor-α, also known as ARHGDIA), RhoA (Ras homolog family member A), Rac1 (Ras-related C3 botulinum toxin substrate 1) and Cdc42 (cell division cycle 42)) has been implicated as mediators of actin rearrangements. The molecular effectors of signaling pathways leading to filopodia formation are yet to be completely defined.
Results
Genome-wide expression profiling of A549 cells after DAMTC treatment
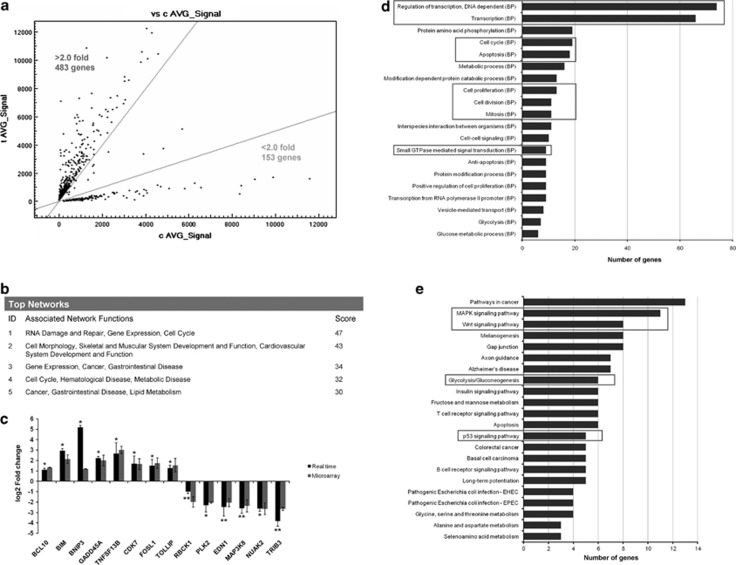
The changes in gene expression profile after DAMTC treatment in NSCLC (A549) cells were determined by a microarray experiment using Illumina human WG-6 v.3.0 beadchip (Illumina Inc., San Diego, CA, USA). The analysis of microarray data using Bead Studio software identified that DAMTC treatment in NSCLC (A549) cells differentially regulate 650 genes (P≤0.05). Among the differentially expressed genes, 497 were found to be upregulated and 153 genes were found to be downregulated. A scatter plot between the average signal of differentially expressed genes in vehicle-treated control samples and DAMTC-treated samples shows that most of the differentially expressed genes are more than two-fold upregulated or downregulated (Figure 1a). The list of differentially expressed genes along with their respective fold changes is given in Supplementary Table 1.
Figure 1.
(a) Scatter plot of average signal of differentially expressed genes in microarray experiment. The y axis represents average signal of differentially expressed genes in NSCLC (A549) cells treated with 160 μg/ml of DAMTC; the x axis represents average signal of these differentially expressed genes in vehicle-treated NSCLC (A549) control cells. (b) The list of top five networks along with their respective score obtained from the IPA analysis. (c) Real-time PCR analysis of selected genes. The y axis represents log2-fold change in relative expression level of genes in DAMTC-treated NSCLC (A549) cells compared with vehicle-treated NSCLC (A549) cells. Cells were treated with 160 μg/ml DAMTC and the expression levels of genes were measured using gene-specific primers given in Supplementary Table 3 by real-time PCR. Expression level of 18s rRNA was taken as internal control for normalization of real-time PCR data. Experiment was performed in triplicate for each gene, and the error bar represents s.d. *P<0.05 compared with vehicle-treated control cells, and **P<0.001 compared with vehicle-treated control cells. (d) Bar diagram showing the distribution of differentially expressed genes in DAMTC-treated NSCLC (A549) cells into different GO classes. The list of differentially expressed genes was uploaded into the GeneCodis software, and the list of significant biological processes enriched was generated. The y axis represents the significantly enriched biological processes. The x axis represents number of genes in each biological process. (e) Bar diagram showing the distribution of differentially expressed genes in DAMTC-treated NSCLC (A549) cells into KEGG pathways obtained after the analysis of differentially expressed genes by GeneCodis software. The x axis represents the number of genes in each enriched KEGG pathway and y axis represents significantly enriched pathways
Functional analysis of the transcriptomics data
We next carried out the functional analysis of the differentially expressed genes using Ingenuity Pathway Analysis (IPA) tool (IPA version 7.5; IPA Tool; Ingenuity Systems, Redwood City, CA, USA; http://www.ingenuity.com). Out of 650 differentially expressed genes, 553 genes mapped to the IPA database. Forty networks were found to be highly significant, out which 29 had a score >10 in NSCLC (A549) cells (Supplementary Table 2). The score is the probability that a collection of genes equal to or greater than the number in a network could be achieved by chance alone. A score of 3 indicates a 1/1000 chance that the focus genes are in a network not due to random chance. Therefore, a score of 3 or higher has a 99.9% confidence level of not being generated by random chance alone. This score is the cut-off value for identifying gene networks.
The top five networks identified in the IPA analysis were associated with RNA damage and repair, gene expression, cell cycle, cell morphology, and cancer and metabolic disease having a score of 47, 43, 34, 32 and 30, respectively (Figure 1b). These top networks (Supplementary Figure 1) converged on NF-κB, p38MAPK, ERK and AKT (V-akt murine thymoma viral oncogene homolog 1, AKT1), which strengthened our previous findings where we had reported the effect of DAMTC on these genes.7 We validated some genes from these networks and some proapoptotic genes identified in the microarray by real-time PCR. The expression of BCL10 (B-cell CLL/lymphoma 10), BCL2L11 (BCL-2 like 11, also known as BIM), BNIP3 (BCL2/adenovirus E1B 19 kDa interacting protein 3), GADD45A (growth arrest and DNA-damage-inducible-α), TNFSF13B (tumor necrosis factor (ligand) superfamily, member 13b), CDK7 (cyclin-dependent kinase 7), FOSL1 (FOS-like antigen 1) and TOLLIP (Toll-interacting protein) increased by 1.1, 2.9, 5.1, 2.2, 2.65, 1.69, 1.5 and 1.24 log2-fold, respectively, in NSCLC (A549) cells after DAMTC treatment as compared with vehicle-treated cells (P≤0.05). Expression of RBCK1 (RanBP-type and C3HC4-type zinc finger containing 1), PLK2 (polo-like kinase 2), EDN1 (endothelin 1), MAP3K8 (MAPK kinase kinase 8), NUAK2 (SNF1-like kinase, 2) and TRIB3 (tribbles homolog 3) were reduced by 0.99, 2.3, 2.47, 2.61, 2.64 and 3.84 log2-fold, respectively (P≤0.05), in DAMTC-treated NSCLC (A549) compared with vehicle-treated cells (Figure 1c). A good correlation was observed between the microarray and the real-time data.
Simultaneously, we also conducted functional classification of differentially expressed genes on the basis of Gene Ontology (GO) terms using a web-based program GeneCodis. This program looks for the enrichment of genes from a particular class in the GO database of NCBI in comparison with the NCBI genes as the reference list. The data was analyzed for biological processes and for KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways. We found that the biological processes showing significant enrichment after the DAMTC treatment in NSCLC (A549) cells were transcription (66 genes), regulation of transcription (74 genes), cell cycle (19 genes) and apoptosis (18 genes; Figure 1d). Apart from these, another biological process that showed a significant enrichment was small GTPase-mediated signal transduction. We next checked whether the differentially expressed genes fall into some predefined pathways. Figure 1e depicts the bar diagram representing the enriched KEGG pathways in NSCLC (A549) cells after DAMTC treatment with a P≤0.05. The pathways that were highly enriched in KEGG database were as follows: MAPK signaling pathway (11 genes), glycolysis pathway (6 genes), Wnt signaling pathway (8 genes) and p53 signaling pathway (5 genes).
It is well known that the transcriptome profiling were performed only for catalog transcripts at the cells' disposal for mounting a response to changing growth conditions. Most intracellular and intercellular biochemical processes are affected by protein–protein or other protein–substrate interactions;11 hence, we next performed proteome profiling following DAMTC treatment in NSCLC (A549) cells.
Identification of differentially expressed proteins by two-dimensional DIGE
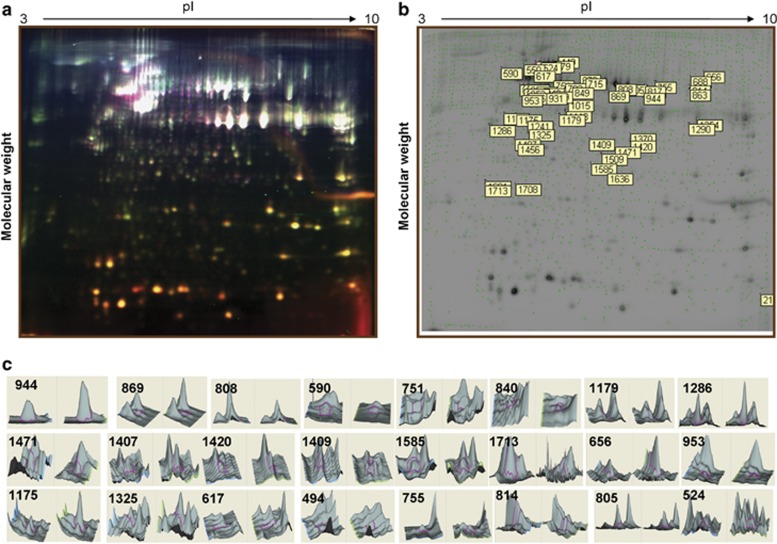
Decyder analysis of the analytical gels showed that the maximum number of detected spots in the master gel were around 2000. Statistical analysis was done using Student's t-test, and proteins of interests were manually confirmed to be significantly regulated using the 3D simulation of protein spot intensity. Fifty-six proteins were considered to be differentially expressed in DAMTC-treated NSCLC (A549) cells in comparison with vehicle-treated NSCLC (A549) cells. A representative difference gel electrophoresis (DIGE) image showing a large number of spots labeled with Cy2, Cy3 and Cy5 is presented in Figure 2a. The gel showing differentially expressed spots in A549 cells is given in Figure 2b. A preparatory gel containing an unlabeled protein was run and the spots were matched with the master gel. Among the 56 differentially expressed spots, 32 matched with the preparatory gel. The matched spots were excised from the gel and they were taken for mass spectrometric analysis. We were able to identify 24 spots corresponding to 20 unique proteins. The molecular weight and isoelectric point of the identified proteins were also matched with their corresponding positions in the gel and were found to be well correlated. The 3D view of all the identified proteins is presented in Figure 2c.
Figure 2.
Two-dimensional (2D) DIGE analysis of vehicle-treated control cells and DAMTC-treated NSCLC (A549) cells. (a) Representative 2D-DIGE image containing the Cy2, Cy3 and Cy5-labeled vehicle-treated control cells and DAMTC-treated protein lysates. (b) Master gel showing the differentially expressed spots between vehicle-treated control cells and DAMTC-treated NSCLC (A549) cells. The number in boxes represents the master spot number. (c) 3D view of all the differentially expressed proteins identified by 2D-DIGE analysis of NSCLC (A549) cells treated with DAMTC
Functional analysis of the proteomics data
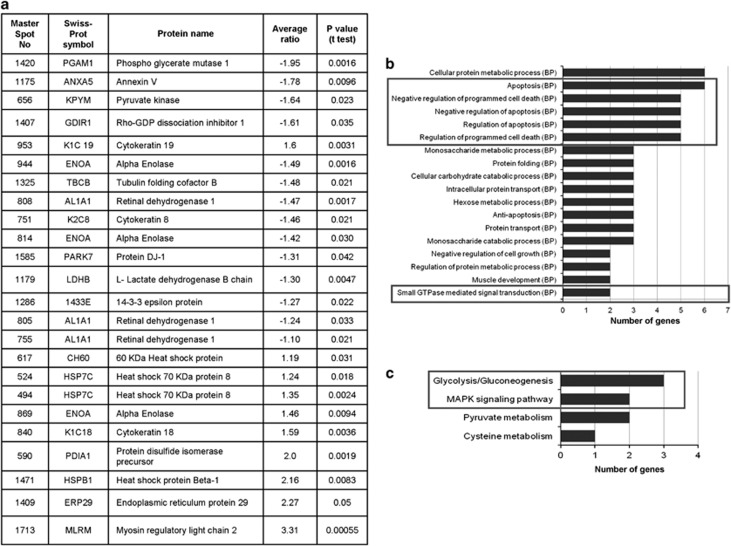
Among the altered proteins, we found several metabolic enzymes, heat-shock proteins, cytokeratins, RhoGDIα, DJ-1 (also known as PARK7, Parkinson protein 7) and 14-3-3 epsilon (also known as YWHAE, tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon). The list of all the identified proteins along with their P-values and the average ratio is given in Figure 3a. The differentially expressed proteins were analyzed using the GeneCodis software, which grouped these proteins into predefined biological processes and the KEGG pathways. As shown in Figure 3b, the enriched biological processes were apoptosis, regulation of apoptosis and small GTPase-mediated signal transduction. The enriched KEGG pathways included MAPK signaling pathway and glycolysis pathway (Figure 3c). Interestingly, the same GO categories and pathways were found to be enriched in the transcriptome analysis, suggesting their importance in the DAMTC-induced changes in NSCLC (A549) cells.
Figure 3.
Differentially expressed proteins in DAMTC-treated NSCLC (A549) cells and their distribution into different functional categories. NSCLC (A549) cells were treated with DAMTC (160 μg/ml) for 24 h, and the total cellular proteins were separated by two-dimensional difference in gel electrophoresis. Differentially expressed proteins were identified by MS/MS analysis in DAMTC-treated NSCLC (A549) cells with a P-value<0.05 (a). These differentially expressed proteins were distributed into different GO classes of biological processes (b) and KEGG pathways (c) using GeneCodis software. The x axis represents the number of proteins. The y axis shows the significantly enriched biological process or KEGG pathway
There is enough literature, which shows that members of the Rho family of small GTPases proteins are involved in cell cytoskeleton organization, migration, transcription and proliferation through their interaction with multiple target proteins.12 IPA analysis of the transcriptomics data found several genes, which are structural constituents of the cytoskeleton, and some of them are also regulated by Rho-GTPases (e.g., TUBB, TUBB2A, TUBB3, TUBB2C, KLHL21, KLH18, LNX2, DCTN5, CSRP2, FNBP1, SH3GLB1, KATNA1, MAPRE2, ABLIM3, SDC4 and HK2; Supplementary Table 2). As both the bioinformatic analysis (which predicts important altered pathways) and the proteomics data (the final outcome) showed involvement of small GTPases, we selected them for further validation.
Validation of the proteomics data using real-time PCR and western blot analysis
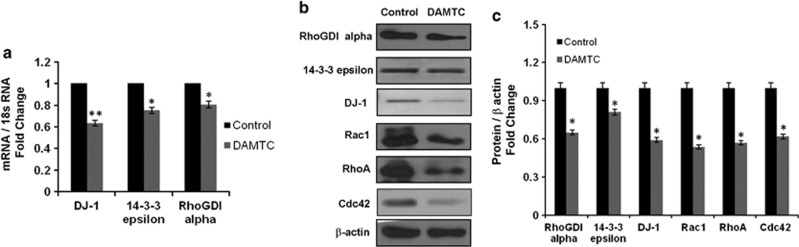
As shown in Figure 4a, transcript levels of DJ-1, 14-3-3 epsilon and RhoGDIα decreased by 1.57- ,1.33- and 1.25-fold, respectively, in DAMTC-treated cells as compared with vehicle-treated NSCLC (A549) cells (P≤0.05). In accordance with our proteomics data, the protein levels of 14-3-3 epsilon, RhoGDIα and DJ-1 decreased by 1.23- ,1.53- and 1.68-fold, respectively, in DAMTC-treated NSCLC (A549) cells as compared with vehicle-treated control cells (Figures 4b and c). We also performed western blot analysis for Rac1, RhoA and Cdc42 (the substrates for RhoGDIα), and observed 1.87-, 1.75- and 1.62-fold decrease in Rac1, RhoA and Cdc42 as compared with vehicle-treated NSCLC (A549) cells, respectively (P≤0.05; Figure 4b and c).
Figure 4.
(a) Real-time PCR for 14-3-3 epsilon, RhoGDIα and DJ-1. Expression levels of 14-3-3 epsilon, RhoGDIα and DJ-1 was measured using quantitative real-time PCR with gene-specific primers given in Supplementary Table 3. Expression level of 18s was taken as an internal control for normalization of real-time PCR data. Experiments were performed in triplicate for each gene and the error bar represents s.d. *P<0.05 compared with vehicle-treated control cells and **P<0.001 compared with vehicle-treated control cells. (b) Western blot analysis of 14-3-3 epsilon, RhoGDIα, DJ-1, RhoA, Rac1 and Cdc42. NSCLC (A549) cells were treated with DAMTC (160 μg/ml) for 24 h. Total cell lysates were prepared and protein (40–60 μg) was subjected to SDS-PAGE, followed by immunoblot analysis using specific antibodies and secondary horseradish peroxidase-conjugated or alkaline phosphatase-conjugated antibodies. β-actin was used as internal control to ensure that equal amount of proteins were loaded in each lane. Blots are representative of three independent experiments with similar results. (c) Densitometric analysis of western blots was done as described in ‘Materials and Methods'. Data shown is representative of three independent experiments. The error bar represents s.d. *P<0.05 compared with vehicle-treated control cells
DAMTC induces changes in the cytoskeleton and migration ability of (NSCLC) A549 cells
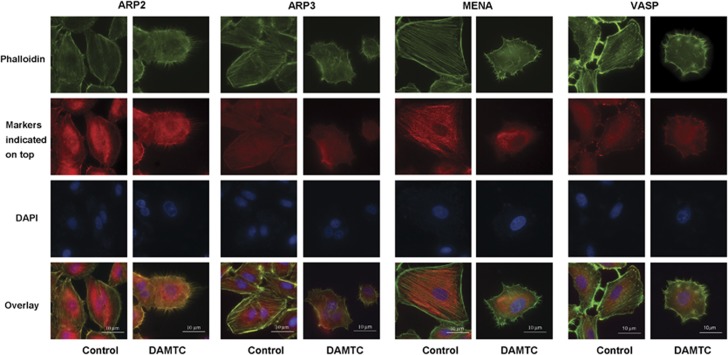
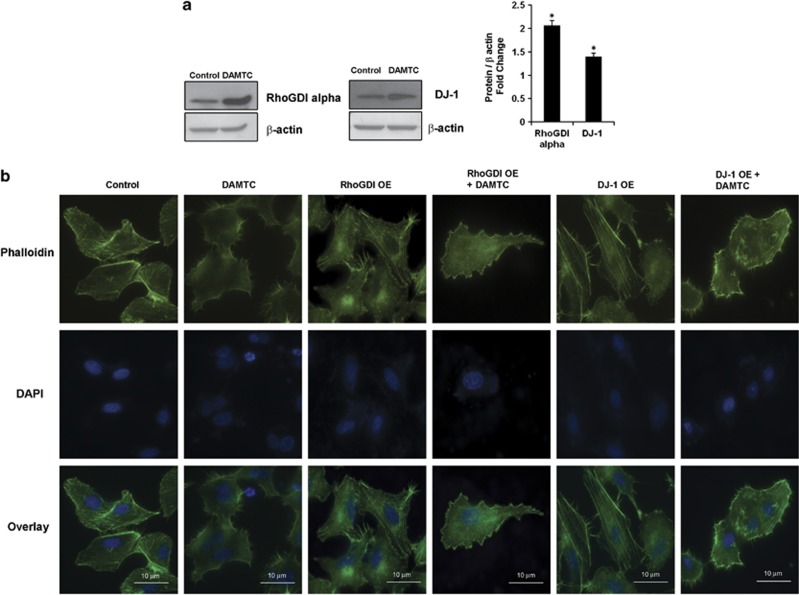
Small GTPases of the Rho-GTPase family (RhoA, Rac1, Cdc42) are known to act directly on the cytoskeleton and are responsible for the development of membrane ruffles, stress fibers, lamellipodia and filopodia. As we observed downregulation of RhoGDIα, Cdc42, Rac1 and RhoA, we sought to see if there were any changes in the cytoskeleton after DAMTC treatment. For this, the cells were stained for Arp2 (actin-related protein 2)/Arp3 (actin-related protein 3)/Vasp (vasodilator-stimulated phosphoprotein)/Mena (ENAH, enabled homolog; markers for filopodia and lamellipodia) and phalloidin. Vehicle-treated cells had characteristic actin stress fibers throughout the cytoplasm, whereas in DAMTC-treated cells there seemed to be clear qualitative differences in the intracellular distribution of these actin filaments. The stress fibers had changed into an unorganized pattern inside the cytosol, and cells possessed filopodia-like structures extending from the membrane, which were absent in the vehicle-treated NSCLC (A549) cells (Figure 5). Similar filopodia-like structures were also observed in the DAMTC-treated NCI-H460 cells (Supplementary Figure 2). We also observed that Arp2, Arp3 and Vasp localize along the length of the filopodia-like structures after DAMTC treatment in NSCLC (A549) cells; however, no significant changes were observed in the localization of Mena before and after DAMTC treatment (Figure 5). Furthermore, to check whether these effects of DAMTC were due to reduced expression of RhoGDIα, we overexpressed RhoGDIα using a cDNA clone (Figure 6a) and observed the reversal of the effects of DAMTC treatment on cytoskeleton in NSCLC (A549) cells (Figure 6b). This reversal was not observed after overexpression of DJ-1 (another differentially expressed protein).
Figure 5.
Double staining with phalloidin (green) and anti-Arp2/anti-Arp3/anti-Mena/anti-Vasp antibodies (red) in vehicle-treated NSCLC (A549) cells and DAMTC-treated NSCLC (A549) cells. The Arp2, Arp3 and Vasp can be clearly observed along with phalloidin (merging of green and red) in the membrane protrusions, whereas the Mena protein is concentrated in the cytosol and absent from the membrane protrusions in DAMTC-treated cells. The blue dye represents the DAPI-stained nucleus. The corresponding localization of these proteins is also shown for the vehicle-treated NSCLC (A549) cells
Figure 6.
(a) NSCLC (A549) cells were transiently transfected with RhoGDIα or DJ-1 cDNA clone. There was 2.1-fold increase in RhoGDIα expression and 1.4-fold increase in DJ-1 expression. Data shown is representative of three independent experiments. The error bar represents s.d. *P<0.05 compared with vehicle-treated control cells. (b) Microscopic images of phalloidin (green)- and DAPI (blue)-stained vehicle-treated and DAMTC-treated NSCLC (A549) cells clearly indicate the changes in cytoskeleton induced by DAMTC treatment. The effect of DAMTC treatment on the cytoskeleton of NSCLC (A549) was reversed by overexpression of RhoGDIα as can be seen by shortening of the membrane protrusions. This reversal was not observed when DJ-1 was overexpressed. OE in the figure implies overexpression
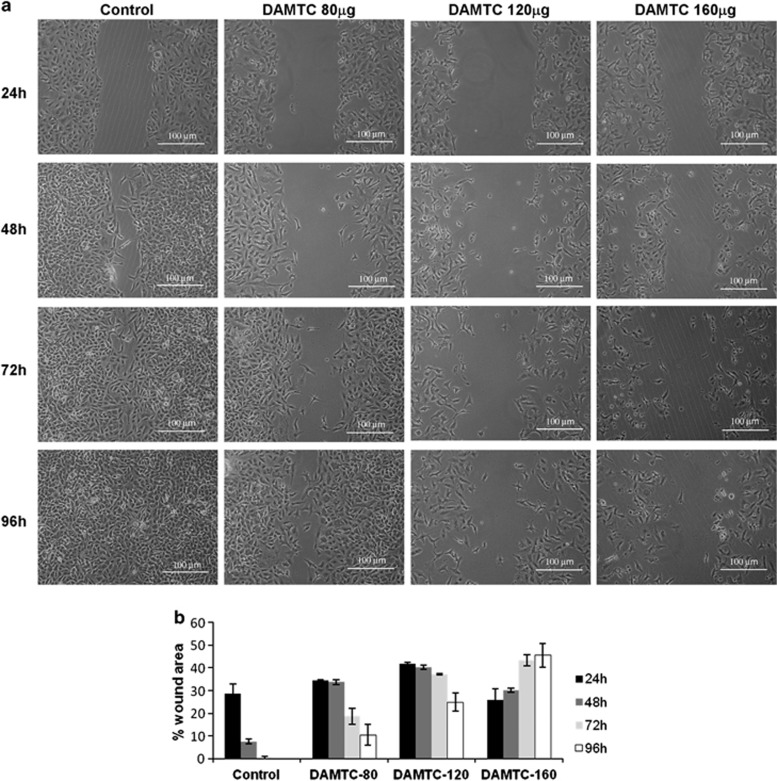
RhoGTPases not only function as cytoskeletal regulators but also regulates cellular motility;12, 13 hence, we next performed wound-healing assay in a dose-dependent manner. As expected, the migration of NSCLC (A549) cells was considerably reduced after DAMTC treatment (Figure 7a). In the vehicle-treated cells, the wound area was fully healed after 96 h, whereas in the DAMTC-treated cells the rate of cell migration decreased considerably and the filling of the wound area was dose dependent, thereby indicating that DAMTC treatment severely alters the migration ability of the cells. The quantitative values of the wound size as determined by the Wimasis online software are graphically depicted in Figure 7b. This experiment was also performed in NCI-H460 cells with similar results (Supplementary Figure 3).
Figure 7.
(a) Wound-healing assay of DAMTC-treated NSCLC (A549) cells. The number of cells migrating in the wound increased in vehicle-treated NSCLC (A549) cells, whereas fewer cells migrated in the wound area in DAMTC-treated cells and this migration was also dose-dependent. (b) Graphical representation of the quantitative values of the wound size at different doses of DAMTC and at different time points post-DAMTC treatment as determined by the WimScratch module of Wimasis online software. Data shown are representatives of three independent experiments. The error bar represents s.d.
DAMTC augments the apoptotic effect of etoposide, a proapoptotic chemotherapeutic drug in (NSCLC) A549 cells
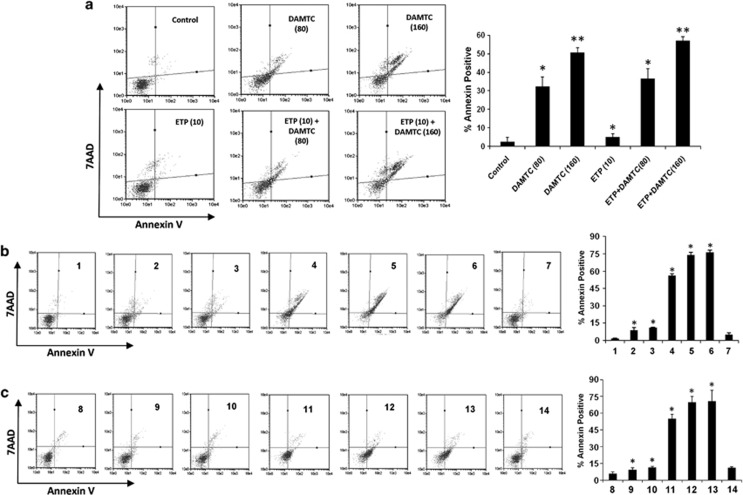
The literature suggests that downregulation of RhoGDIα and DJ-1 enhances the sensitivity to other chemotherapeutic drugs.14, 15 As DAMTC treatment in NSCLC (A549) cells led to downregulation of both DJ-1 and RhoGDIα, we next checked whether DAMTC treatment enhances the effect of etoposide on NSCLC (A549) cells. The cells were treated with DAMTC (80 or 160 μg/ml) or DAMTC in combination with etoposide (10 μM), and percentage apoptosis was measured using annexin assay. We found that DAMTC (80 μg/ml) alone resulted in 32.37% apoptotic cells (P=0.024), DAMTC (160 μg/ml) alone resulted in 50.57% apoptotic cells (P=0.002) and etoposide (10 μM) alone resulted in 5% apoptotic cells (P=0.013). However, DAMTC (80 μg/ml) in combination with etoposide resulted in 36.57% apoptotic cells (P=0.016), whereas DAMTC (160 μg/ml) in combination with etoposide resulted in 57.1% apoptotic cells (P=0.001) as compared with vehicle-treated cells (Figure 8a). These results indicated that loss of RhoGDIα and DJ-1 expression augmented the etoposide-induced apoptosis in NSCLC (A549) cells.
Figure 8.
(a) DAMTC enhances the apoptotic effect of etoposide. NSCLC (A549) cells were treated with DAMTC (80/160 μg/ml) alone or in combination with etoposide for 24 h, and percentage apoptosis was measured using annexin assay. Data shown is a representative of experiments repeated four times. The x axis represents annexin-V PE fluorescence and y axis represents 7-AAD fluorescence. Bar graph shows the percentage of annexin-positive cells. Data is mean±s.e.m. of four independent experiments. *P<0.05 compared with vehicle-treated control cells, and **P<0.001 compared with vehicle-treated control cells. ETP in the figure implies etoposide. (b) Effect of RhoGDIα and DJ-1 silencing on DAMTC-induced apoptosis. NSCLC (A549) cells were transiently transfected with RhoGDIα or DJ-1 siRNA alone or with DAMTC, and percentage apoptosis was measured using annexin assay. Data shown is representative of experiments repeated at least three times. The x axis represents annexin-V PE fluorescence and y axis represents 7-AAD fluorescence. Bar graph shows the percentage of annexin-positive cells. Data is mean±s.d. of three independent experiments. *P<0.05 compared with vehicle-treated control cells (1: vehicle-treated control cells; 2: DJ-1 siRNA; 3: RhoGDIα siRNA; 4: DAMTC; 5: DAMTC+DJ-1 siRNA; 6: DAMTC+RhoGDIα siRNA; 7: negative control siRNA). (c) Effect of RhoGDIα and DJ-1 overexpression on DAMTC-induced apoptosis. NSCLC (A549) cells were transiently transfected with RhoGDIα or DJ-1 cDNA clone alone or with DAMTC, and percentage apoptosis was measured using annexin assay. Data shown is representative of experiments repeated at least three times. The x axis represents annexin-V PE fluorescence and y axis represents 7-AAD fluorescence. Bar graph shows the percentage of annexin-positive cells. Data is mean±s.d. of three independent experiments. *P<0.05 compared with vehicle-treated control cells (8: vehicle-treated control cells; 9: DJ-1 clone; 10: RhoGDIα clone; 11: DAMTC; 12: DAMTC+DJ-1 cDNA; 13: DAMTC+RhoGDIα cDNA; 14: negative control plasmid)
DAMTC-induced apoptosis: role of small GTPases
To elucidate the role of RhoGDIα and DJ-1 in DAMTC-induced apoptosis, we transiently transfected NSCLC (A549) cells with siRNA against RhoGDIα and DJ-1, and examined the effect of RhoGDIα and DJ-1 depletion on apoptosis. The silencing of RhoGDIα and DJ-1 after siRNA transfection was confirmed by western blotting analysis (data not shown). The annexin assay revealed that suppression of RhoGDIα exhibited an increase in apoptosis to 11.1% as compared with 1.65% apoptosis in vehicle-treated cells (Figure 8b). Similarly, suppression of DJ-1 also exhibited an increase in apoptosis to 8.65% as compared with vehicle-treated cells. DAMTC treatment alone resulted in 56% apoptotic cells, whereas DAMTC-treated cells transfected with either DJ-1 or RhoGDIα siRNA resulted in 74 and 76% apoptotic cells, respectively, as compared with vehicle-treated cells (P<0.05). Simultaneously, we performed annexin assays in the presence or absence of a plasmid construct harboring RhoGDIα cDNA or DJ-1 cDNA in NSCLC (A549) cells. Overexpression of RhoGDIα and DJ-1 after transfection of their respective cDNA clones was checked by western blotting (Figure 6a). Surprisingly, at 24-h post transfection, the overexpression of RhoGDIα cDNA or DJ-1 cDNA actually leads to a slight increase in apoptotic cell death in A549 cells (Figure 8c). This suggests that RhoGDIα and DJ-1 may not be the only effector molecules that have a role in DAMTC-induced apoptosis. Further studies are necessary to discover the key target molecule of DAMTC-induced apoptosis.
Discussion
The transcriptome and proteome profiling led to the identification of several differentially expressed genes and proteins following DAMTC treatment in NSCLC (A549) cells. The IPA analysis of the transcriptomics data suggested the importance of NF-κB, p38, MAPK, ERK and AKT in DAMTC-induced apoptosis in NSCLC (A549) cells, which strengthened our previously published findings7 (Supplementary Figure 1).
The GeneCodis analysis of transcriptomics data showed that in addition to other pathways, there was a significant enrichment of small GTPase-mediated signal transduction. Small GTP-binding proteins (also known as small GTPases) are one of the largest families of signaling proteins, with over 100 family members. These signaling proteins are essential for multiple cellular processes, such as cell proliferation (Ras), cytoskeleton dynamics (Rho and Rop), membrane trafficking (Arf and Rab) and nucleo-cytoplasmic transport (Ran). They have gained a special emphasis in cancer research within the last decade as a large number of small GTPases have been associated with the development of all types of tumors. Rho GTPases have been reported to contribute to cancer initiation and progression, including the acquisition of unlimited proliferation potential, survival and evasion from apoptosis, angiogenesis, tissue invasion and the establishment of metastases.12 Interestingly, in our proteomics data, we observed significant downregulation of RhoGDIα after DAMTC treatment in A549 cells.
RhoGDIα is frequently overexpressed in human tumors and has been characterized as a cell-death mediator and as a potential anti-cancer target.14, 16 Zhang et al,15 have also shown that overexpression of RhoGDIα increases resistance of cancer cells to chemotherapeutic drugs, whereas silencing of RhoGDIα using siRNA increases sensitivity to drug-induced apoptosis. As DAMTC downregulates the expression of RhoGDIα, it emphasizes its potential as an anti-cancer drug. Literature suggests that signals transmitted through Rho family GTPase are coupled to MAPK pathway,17, 18, 19 and our previous study, as well as IPA analysis, shows that MAPK has a role in DAMTC-induced apoptosis, thus suggesting that Rho family GTPases could be the central target molecule of DAMTC responsible for its action.
RhoGDIα is a member of the RhoGDI family of proteins, which regulate Rho subfamily GTPases including RhoA, Rac1 and Cdc42.20, 21 RhoA, Rac1 and Cdc42 proteins are known to regulate the formation of stress fibers, membrane ruffles/lamellipodia and filopodia, respectively,22 which affect cellular morphology, movement, attachment, phagocytosis and cytokinesis.13, 14 In the current study, we observed significant downregulation of these proteins after DAMTC treatment. We demonstrate here for the first time that DAMTC treatment not only leads to alterations in A549 cell morphology, including membrane protrusions, but also induces filopodia formation. Also, this effect of DAMTC on cytoskeleton was shown to be reversed after overexpression of RhoGDIα. We further demonstrated the presence of Arp2/3 complex and Vasp expression in the membrane protrusions, but we did not find any change in the expression of Mena before and after DAMTC treatment in A549 cells. Consistent with our findings, there is enough evidence in the literature where fliopodia formation has been observed in the absence of Vasp and/or Mena.23, 24 Traditionally, Arp2/3 complex has been described as lamellipodial markers and Vasp/ Mena as filopodial markers, but over the last few years the role of Arp2/3 complex has also been implicated in filopodial formation.13, 25
We also demonstrated that DAMTC inhibits cell motility in A549 cells. DAMTC can thus be considered as a promising anti-cancer drug, as it not only induces apoptosis but can also prevent the migration and angiogenesis of cancer cells.
In addition to the Rho family GTPases, we found other interesting leads from the study, which need further investigation. There was downregulation of 14-3-3 epsilon protein (anti-apoptotic protein) after DAMTC treatment. This protein is downregulated during apoptosis of JURL-MK1 and K562 cells.26 We also observed increased expression of cytokeratin 18 (a marker of cell death) and cytokeratin 19, and decreased expression of cytokeratin 8 after DAMTC treatment. Cytokeratins are important constituent of intermediate filaments and several investigators have shown an important role of these cytokeratins in signal transduction and apoptosis pathways.27, 28, 29 The glycolytic enzymes, such as pyruvate kinase isoenzyme M2, α-enolase and phosphoglycerate mutase 1, were also downregulated after DAMTC treatment. All these enzymes are associated with lung cancer; for instance, the α-enolase expression increases progressively with the clinical stages of lung cancer,30, 31 whereas pyruvate kinase isoenzyme M232 and phosphoglycerate mutase 133, 34 are overexpressed in lung cancer. Moreover, there was downregulation of ℒ-lactate dehydrogenase B chain after DAMTC treatment. Chen et al.35 has shown that elevation of the ℒ-lactate dehydrogenase B is correlated with the clinical stage of lung cancer, thereby suggesting the effectiveness of DAMTC. Furthermore, our study showed that DAMTC downregulates DJ-1 in DAMTC-treated A549 cells. DJ-1 is an oncogene that cooperates with H-Ras and transforms cells by increasing cell proliferation and resistance to cell cycle arrest.36, 37 Increased DJ-1 levels had also been found in primary lung tumors, and its expression was correlated negatively with clinical outcomes in NSCLC patients.14, 37, 38 Gu et al.39 have also shown that DJ-1 interacts with the ERK pathway and change the susceptibility of cells to oxidative stress. As we have shown previously the involvement of ERK in DAMTC-induced apoptosis in A549, we propose that DJ-1 may be the upstream target of DAMTC-mediating apoptosis. The literature also suggests that DJ-1 promotes metastasis of pancreatic cancer cells by modulating the ERK pathway.40
Conclusion
The high-throughput probing of DAMTC-induced effects in A549 has revealed important insights. DAMTC not only induces apoptosis in A549 cells but also downregulates RhoA, Rac1 and Cdc42, which have an important role in increased motility, invasion and metastasis. These revelations further emphasize the potential of this molecule as an anti-cancer therapeutic. Future studies in this direction may provide a more conclusive answer.
Materials and Methods
Cell culture
Human lung adenocarcinoma cells (NSCLC, A549) were obtained from the National Center for Cell Science, Pune, India, and maintained in DMEM medium containing 10% (v/v) fetal calf serum, 100 units/ml penicillin, 100 μg/ml streptomycin and 0.25 μg/ml amphotericin in a humidified 5% CO2 atmosphere. DAMTC was synthesized as described earlier.41 DAMTC was dissolved at a concentration of 10 mg/ml in DMSO and stored at −20 °C. A549 cells were treated with DAMTC at a concentration of 160 μg/ml for 24 h. For transient transfection experiments, A549 cells were grown in tissue culture plates. For knockdown experiments, 100 nM of RhoGDIα or DJ-1 siRNA (Santa Cruz Biotechnology, Santa Cruz, CA, USA) were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. For overexpression experiments, 3 μg of RhoGDIα (catalog number SC117424) or DJ-1 cDNA clone (catalog number SC115623; OriGene Technologies, Rockville, MD, USA) were transfected using Lipofectamine LTX and Plus Reagent (Invitrogen) according to the manufacturer's protocol. The media was replaced with complete DMEM media 6-h post transfection, and DAMTC (160 μg/ml) was added for another 24 h wherever indicated. DMSO-treated cells (0.1%) were taken as vehicle-treated controls.
RNA isolation and cRNA preparation
Total RNA from vehicle-treated cells and DAMTC-treated A549 cells was isolated using Trizol reagent according to the manufacturer's protocol, with minor modification. Isolated RNA was further purified using the RNA easy MinElute cleanup kit (Qiagen, Valencia, CA, USA) by essentially following the manufacturer's instructions. cRNA preparation was done using Illumina TotalPrep RNA Amplification Kit (Ambion Inc., Austin, TX, USA) following manufacturer's instructions. Briefly, 500 ng of total RNA was reverse transcribed with an oligo dT primer using ArrayScript enzyme (Ambion Inc.). The cDNA synthesized was column-purified, and used for amplification and cRNA synthesis using in vitro transcription. The cRNA was labeled during in vitro transcription reaction with biotinylated UTP and further column-purified.
Microarray experiment using Illumina Beadarray Chips
The genome-wide gene expression profiling of DAMTC-treated and -untreated A549 cells was done using Bead Arrays (Illumina Inc.) according to manufacturer's protocol. In brief, 1.5 μg of labeled cRNA from three biological replicates was hybridized to Illumina Genome-Wide Expression BeadChips (Human Ref-6v.3.0; Illumina Inc.) representing ∼43 000 human transcripts at 58 °C overnight. The array was washed and blocked, and hybridized beads were stained with Streptavidin Cy3 (Sigma, St. Louis, MO, USA). The chips were scanned using Illumina scanner iScan.
Microarray data analysis
The data were analyzed using Analysis module of Illumina Bead Studio-3 Software. The data was average-normalized, and the Illumina Custom error model was used for computation of false-discovery rate. The genes having detection P≤0.01 and differential P≤0.05 were considered to be differentially expressed genes and selected for further investigation. The data obtained in this experiment have been deposited in NCBI's Gene Expression Omnibus42 and is accessible through the Gene Expression Omnibus Series accession number GSE29698 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE29698).
IPA and GeneCodis analysis
IPA analysis was used for annotating genes into biological networks and for evaluation of functional significance as described in earlier studies.43, 44 Data sets representing genes with altered expression profile derived from microarray analyses were imported into the IPA Tool (Ingenuity Systems; http://www.ingenuity.com). The basis of the IPA program consists of the Ingenuity Pathway Knowledge Base, which is derived from known functions and interactions of genes published in literature. Thus, the IPA Tool allows the identification of biological networks, global functions and functional pathways of a particular data set. The program also gives the significance value of the genes, the other genes with which it interacts and how the products of the genes directly or indirectly act on each other, including those not involved in the microarray analysis. The networks created are ranked depending on the number of significantly expressed genes they contain and also list diseases that were most significant.
The list of differentially expressed genes/proteins obtained after the microarray/proteomic analysis of DAMTC-treated A549 cells were also imported into the GeneCodis software. In our analysis, we used the default settings of GeneCodis, which employs hypergeometric test for calculating P-values and false-discovery rate for P-values correction.45, 46
Real-time PCR
Total RNA was isolated using Trizol reagent, and reverse transcription was carried out with M-MuLV reverse transcriptase (Fermentas, Glen Burnie, MA, USA) according to the manufacturer's protocol. Real-time PCR was done using SYBR Green PCR master mix (Applied Biosystems, Foster City, CA, USA) in ABI 7500 real-time PCR instrument. Sequence for primers used for real-time PCR is given in Supplementary Table 3. 18s rRNA was used as endogenous control for normalization. Relative quantitation of gene expression was carried out using the Pfaffl method.47
Two-dimensional DIGE
Sample preparation
Four biological replicates of vehicle-treated A549 control cells and DAMTC (160 μg/ml for 24 h)-treated A549 cells were harvested and subsequently solublized in DIGE lysis buffer (7 M urea, 2 M thiourea, 4% CHAPS, 30 mM Tris and protease inhibitor cocktail, pH 8.5), and kept on ice for 10 min. Samples were sonicated and centrifuged at 13 000 r.p.m. for 45 min to remove any cellular debris. The supernatant was collected and stored at −80 °C until use. Protein estimation was done by Bradford assay (Sigma) using BSA as a standard.
Labeling with Cy dye DIGE flours
A549 cell lysates were labeled with Cy2, Cy3 and Cy5 dye following the protocol given in the Ettan Dige User Manual (GE Healthcare, Little Chalfont, UK) with minor modifications. Briefly, 50 μg of protein either from vehicle-treated control A549 lysate or DAMTC-treated lysate was labeled with 400 pmol of Cy3 and Cy5 dye. The samples were vortexed, spinned and incubated at 4 °C in dark for 45 min. Labeling reaction was quenched by an addition of 10 mℳ lysine for 10 min. The pooled standard containing equal amount of protein from all the samples was labeled with 400 pmol of Cy2 to serve as an internal standard for 2D-DIGE. To remove any dye biasness, two of the vehicle-treated control cells were labeled with Cy3 and two of them were labeled with Cy5; similarly for the treated cells, two of them were labeled with Cy3 and remaining two were labeled with Cy5. Gel multiplexing was obtained by mixing 50 μg each of one Cy3- and one Cy5-stained samples supplemented with 50 μg of Cy2-stained internal standard. Thus, each gel contained 150 μg of total protein. In total, four gels were run for four biological replicates simultaneously. The experimental design for vehicle-treated control cells and DAMTC-treated biological replicate is shown in Supplementary Table 4. For preparatory gel, a total of 800 μg protein containing the pooled sample from all the samples was used.
Two-dimensional electrophoresis
The volume of each mixed sample was adjusted to 350 μl with rehydration buffer (8 M urea, 2% CHAPS, 20 mM DTT, 0.5% IPG buffer and 0.002% Bromo-Phenol Blue). IPG strips (18 cm, pH 3–10, linear) were rehydrated for 16 h with rehydration buffer containing sample. Isoelectric focusing was successively carried out at 200 V for 30 min, 500 V for 3 h, 4 kV for 2 h, 8 kV for 2 h, 10 kV for 2 h and 3 kV for 10 h at 20 °C, and a maximum current setting of 50 μA per strip in an IPGphor 3 isoelectric focusing unit (GE Healthcare). Once the isoelectric focusing is over, the strips were stored at −80 °C until use. Before the second dimensional separation, IPG strips were equilibrated first with equilibration buffer (75 mM Tris, 6 M urea, 30% glycerol and 2% SDS) containing DTT (1%), and then with iodoacetamide (2.5%) for 20 min each to reduce and alkylate the proteins. The second dimensional separation was conducted on 1.5-mM thick 15% polyacrylamide gels using the Protean-II electrophoresis cell (BioRad, Hercules, CA, USA). The strips were carefully placed over the top of the gel and then sealed with 0.5% agarose gel containing bromophenol blue. The gel was run at 30 mA per gel for the first 30 min, and followed by 40 mA per gel in the dark for the remaining part of the run. Once the dye front reached the bottom of the gel, the run was stopped and gels were immediately removed for scanning. For preparatory gel, the same running conditions were adopted, and after the run was over the gel was fixed with 40% ethanol and 10% acetic acid for 30 min and then stained with colloidal Coomassie solution overnight. Gel was washed several times with Milli-Q water to remove the background stain. The colloidal Coomassie-stained preparatory gel was then scanned in a typhoon scanner using the Cy5 wavelength.
Image acquisition and analysis of protein spots
All analytical gels were scanned using the Typhoon Trio variable mode imager (GE Healthcare) using optimal excitation/emission wavelength for each DIGE fluor: Cy2 (488/520 nm), Cy3 (532/580 nm) and Cy5 (633/670 nm). The analysis of the gels was done using DeCyder differential analysis software version 6.5 (GE Healthcare). For individual gel analysis, spots were detected, quantified and normalized according to the volume ratio of corresponding spots detected in the Cy2 image of the pooled-sample internal standard using the differential in-gel analysis module. All normalized differential in-gel analysis data sets from four gels were collectively analyzed employing the biological variation analysis module, which enables matching of multiple images from different gels to provide statistical data on average abundance for each protein spot among the DIGE gels in the study. Student's t-test was used to assess the statistical significance of differential spots. On the basis of a standardized average spot volume ratio, spots between vehicle-treated control cells and treatment at 95% confidence level (P<0.05) were considered to be significant. Preparatory gel image was also loaded into the Decyder software and then spots were detected and matched to the master gel. The matched differential spots were manually cut from the colloidal Coomassie-stained preparatory gel and in-gel trypsin digestion was done. In brief, spot was destained, dehydrated, reduced and alkylated with 50 mM iodoacetamide. To each sample, sequencing grade-modified trypsin was added and tryptic peptides from gel pieces were used for mass spectrometric analysis.
Mass spectrometry analysis and database searching
Peptide digests from each gel spot were spotted onto a 384-well MALDI sample plate and mixed with equal volume of CHCA matrix solution (10 mg/ml α-cyano-4-hydroxy-cinnamic acid in 0.1% TFA, 50% ACN) in triplicates. Peptide mass spectra was obtained using a MALDI-TOF/TOF (ABSCIEX TOF/TOF 5800, Applied Biosystems) mass spectrometer. The MS data was acquired in a window of a mass to charge ratio (m/z) 700–4000. The mass spectrometric automatic data acquisition was performed in positive reflector mode. For calibration and tuning purposes, 4700 Cal Mix (Applied Biosystems/MDS Sciex, Concord, ON, Canada) containing 6 calibrant peptides in the m/z range of 904–3658 were spotted on 13 dedicated calibration spot locations distributed over the MALDI target plate. Automatic jobs were created for MALDI spots. For MS analysis, 1000 shots were acquired and averaged over an m/z window of 700–4000. From each MS spectrum, up to 50 of the strongest precursor peaks with a minimal signal-to-noise ratio of 10 were selected for MS/MS acquisition. For each precursor, 3000 shots were acquired and averaged. All MS/MS fragmentation was performed using post-source decay.
The mass spectrometer spectra were analyzed using Protein Pilot software ABSCI EX's (version 3.0; Applied Biosystems) with Mascot search engine (MASCOT V2.2) in Human SWISS-PROT databases (release version 51.6 containing 257 964 sequences, 93 947 433 residues). Maximum number of missed cleavages was set to 1. The search parameters allowed oxidation of methionine as a variable modification and carboxyamidomethylation of cysteine as a fixed modification. Precursor mass tolerance was set at 100 p.p.m. and MS/MS fragment mass tolerance was set at 0.4 Da. Mowse score that signifies P<0.05 was considered significant for successful protein identification (see Supplementary Table 5).
Western blotting
A549 cells were treated with DAMTC (160 μg/ml) for 24 h, trypsinized and the cell pellet was lysed with modified RIPA buffer (50 mM Tris-HCl, pH 7.4 150 mM NaCl, 1 mM EDTA, 1% NP40, 0.25% Na deoxycholate, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin, 1 mM PMSF, 1 mM sodium orthovandate and 1 mM sodium fluoride) and kept in ice for 30 min. After centrifugation at 15 000 × g for 5 min, the cell lysates were collected. Western blotting was performed as previously described.48 β-actin was used as normalizing control.
Annexin assay
A549 cells were plated on a 24-well tissue culture plate and were treated with DAMTC (80/160 μg/ml), etoposide (10 μM) alone or in combination for 24 h. Percentage apoptosis was measured with annexin assay according to manufacturer's protocol using Guava Nexin Assay kit (Guava Technologies, Hayward, CA, USA). Annexin V–PE fluorescence was analyzed by cytosoft software (Guava Technologies).
Statistical analysis
Statistical analysis was performed with Student's two-tailed t-test; P<0.05 was considered statistically significant. Results are presented as mean of three independent experiments±s.d./s.e.m. Measurement of signal intensity on PVDF membranes after western blotting with various antibodies was performed using AlphaImager 3400 (Alpha InnoTech Corporation, San Leandro, CA, USA). The integrated density values were calculated as the density values of the specific protein bands/β-actin density values. The fold change was then calculated by dividing this value for DAMTC-treated cells by the value for vehicle-treated control cell samples.
Migration assay
A549 cells were plated on a six-well tissue culture plate. The next day, cells were treated with 80/120/160 μg/ml of DAMTC. After 24 h, the wound was artificially created by scraping with a sterilized pipette tip and the fresh media was added to the cells. The wound area was monitored for the next 3 days at an interval of 24 h. The microscopy images of the cells migrating in the wound area were taken at × 10 magnification by an inverted microscope (Nikon Eclipse Ti, Nikon, Tokyo, Japan). The quantitative values of the wound size were determined by web-based WimScratch module of Wimasis online software.
Immunocytochemistry and phalloidin staining
A549 cells were seeded and grown on 25 × 25-mM cover slips in 6-well tissue culture plates. The next day, cells were treated with 160 μg/ml of DAMTC. After 24 h, the cells were fixed in 4% formaldehyde solution in PBS for 15 min. Cells were permeabilized for 5 min in 0.1% Triton X-100 followed by 3 washes with 1 × PBS. Cells were incubated in blocking buffer (1% BSA and 0.1% Triton X-100 in 1 × PBS) for 1 h, primary antibody (anti-Arp2, Arp3, Mena, Vasp (Cell Signaling Technology, Danvers, MA, USA) for 2 h followed by TRITC-conjugated secondary antibody (Invitrogen) for 1 h and phalloidin (Alexa Fluor 488; Invitrogen) staining for 20 min at room temperature. The cells were then washed three times with 1 × PBS. In some experiments, the cells were fixed, permeabilized and stained with phalloidin alone. The nuclei were stained with mounting media containing DAPI solution (Invitrogen). The fluorescence images were captured using Nikon microscope (Ti Eclipse) at × 60 magnification under corresponding excitation and emission wavelengths.
Acknowledgments
This work was supported by the grants from Council of Scientific and Industrial Research (CSIR; EXP0011).
Glossary
- 14-3-3 epsilon
YWHAE, tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon
- AKT
V-akt murine thymoma viral oncogene homolog 1, AKT1
- Arp2
actin-related protein 2
- Arp3
actin-related protein 3
- Cdc42
cell division cycle 42
- DJ-1
PARK7, Parkinson protein 7
- m/z
mass to charge ratio
- Mena
ENAH, enabled homolog
- Rac1
Ras-related C3 botulinum toxin substrate 1
- RhoA
Ras homolog family member A
- RhoGDIα
Rho GDP dissociation inhibitor-α, also known as ARHGDIA
- Vasp
vasodilator-stimulated phosphoprotein
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Disease website (http://www.nature.com/cddis)
Supplementary Material
References
- Spiro SG, Silvestri GA. One hundred years of lung cancer. Am J Respir Crit Care Med. 2005;172:523–529. doi: 10.1164/rccm.200504-531OE. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnero A. Targeting the cell cycle for cancer therapy. Br J Cancer. 2002;87:129–133. doi: 10.1038/sj.bjc.6600458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang GC, Chen KC, Yang TY, Yin MC, Lin CP, Kuo BI, et al. Activity of gefitinib in advanced non-small-cell lung cancer with very poor performance status. Invest New Drugs. 2005;23:73–77. doi: 10.1023/B:DRUG.0000047108.39129.7c. [DOI] [PubMed] [Google Scholar]
- Wright G, Manser RL, Byrnes G, Hart D, Campbell DA. Surgery for non-small cell lung cancer: systematic review and meta-analysis of randomised controlled trials. Thorax. 2006;61:597–603. doi: 10.1136/thx.2005.051995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel A, Prasad AK, Parmar VS, Ghosh B, Saini N. Apoptogenic effect of 7,8-diacetoxy-4-methylcoumarin and 7,8-diacetoxy-4-methylthiocoumarin in human lung adenocarcinoma cell line: role of NF-kappaB, Akt, ROS and MAP kinase pathway. Chem Biol Interact. 2009;179:363–374. doi: 10.1016/j.cbi.2008.10.060. [DOI] [PubMed] [Google Scholar]
- Wang Y, Barbacioru C, Hyland F, Xiao W, Hunkapiller KL, Blake J, et al. Large scale real-time PCR validation on gene expression measurements from two commercial long-oligonucleotide microarrays. BMC Genomics. 2006;7:59. doi: 10.1186/1471-2164-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CS, Sohn SH, Jeon SK, Kim KN, Ryu JJ, Kim MK. Effect of various implant coatings on biological responses in MG63 using cDNA microarray. J Oral Rehabil. 2006;33:368–379. doi: 10.1111/j.1365-2842.2005.01553.x. [DOI] [PubMed] [Google Scholar]
- Smith L, Lind MJ, Welham KJ, Cawkwell L. Cancer proteomics and its application to discovery of therapy response markers in human cancer. Cancer. 2006;107:232–241. doi: 10.1002/cncr.22000. [DOI] [PubMed] [Google Scholar]
- Chan E. Integrating transcriptomics and proteomics. Genom Proteom. 2006;6:20–26. [Google Scholar]
- Parri M, P Chiarugi. Rac and Rho GTPases in cancer cell motility control. Cell Commun Signal. 2010;8:23. doi: 10.1186/1478-811X-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson MF, Sahai E. The actin cytoskeleton in cancer cell motility. Clin Exp Metastasis. 2009;26:273–287. doi: 10.1007/s10585-008-9174-2. [DOI] [PubMed] [Google Scholar]
- MacKeigan JP, Clements CM, Lich JD, Pope RM, Hod Y, Ting JP. Proteomic profiling drug-induced apoptosis in non-small cell lung carcinoma: identification of RS/DJ-1 and RhoGDIalpha. Cancer Res. 2003;63:6928–6934. [PubMed] [Google Scholar]
- Zhang B, Zhang Y, Dagher MC, Shacter E. Rho GDP dissociation inhibitor protects cancer cells against drug-induced apoptosis. Cancer Res. 2005;65:6054–6062. doi: 10.1158/0008-5472.CAN-05-0175. [DOI] [PubMed] [Google Scholar]
- Zhao L, Wang H, Li J, Liu Y, Ding Y. Overexpression of Rho GDP-dissociation inhibitor alpha is associated with tumor progression and poor prognosis of colorectal cancer. J Proteome Res. 2008;7:3994–4003. doi: 10.1021/pr800271b. [DOI] [PubMed] [Google Scholar]
- Force T, Bonventre JV. Growth factors and mitogen-activated protein kinases. Hypertension. 1998;31:152–161. doi: 10.1161/01.hyp.31.1.152. [DOI] [PubMed] [Google Scholar]
- Park YJ, Ahn HJ, Kim YS, Cho Y, Joo DJ, Ju MK. Illumina-microarray analysis of mycophenolic acid-induced cell death in an insulin-producing cell line and primary rat islet cells: new insights into apoptotic pathways involved. Cell Signal. 2010;22:1773–1782. doi: 10.1016/j.cellsig.2010.07.005. [DOI] [PubMed] [Google Scholar]
- Qu MJ, Liu B, Qi YX, Jiang ZL. Role of Rac and Rho-GDI alpha in the frequency-dependent expression of h1-calponin in vascular smooth muscle cells under cyclic mechanical strain. Ann Biomed Eng. 2008;36:1481–1488. doi: 10.1007/s10439-008-9521-0. [DOI] [PubMed] [Google Scholar]
- Garcia-Mata R, Boulter E, Burridge K. The ‘invisible hand': regulation of RHO GTPases by RHOGDIs. Nat Rev Mol Cell Biol. 2011;12:493–504. doi: 10.1038/nrm3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovas A, Couchman JR. RhoGDI: multiple functions in the regulation of Rho family GTPase activities. Biochem J. 2005;390:1–9. doi: 10.1042/BJ20050104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamai T, Yamanishi T, Shirataki H, Takagi K, Asami H, Ito Y, et al. Overexpression of RhoA, Rac1, and Cdc42 GTPases is associated with progression in testicular cancer. Clin Cancer Res. 2004;10:4799–4805. doi: 10.1158/1078-0432.CCR-0436-03. [DOI] [PubMed] [Google Scholar]
- Bohil AB, Robertson BW, Cheney RE. Myosin-X is a molecular motor that functions in filopodia formation. Proc Natl Acad Sci USA. 2006;103:12411–12416. doi: 10.1073/pnas.0602443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh WI, Sudhaharan T, Lim KB, Sem KP, Lau CL, Ahmed S. Rif-mDia1 interaction is involved in filopodium formation independent of Cdc42 and Rac effectors. J Biol Chem. 2011;286:13681–13694. doi: 10.1074/jbc.M110.182683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobova F, Svitkina T. Arp2/3 complex is important for filopodia formation, growth cone motility, and neuritogenesis in neuronal cells. Mol Biol Cell. 2008;19:1561–1574. doi: 10.1091/mbc.E07-09-0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzelova K, Grebenova D, Pluskalova M, Kavan D, Halada P, Hrkal Z. Isoform-specific cleavage of 14-3-3 proteins in apoptotic JURL-MK1 cells. J Cell Biochem. 2009;106:673–681. doi: 10.1002/jcb.22061. [DOI] [PubMed] [Google Scholar]
- Brouillard F, Fritsch J, Edelman A, Ollero M. Contribution of proteomics to the study of the role of cytokeratins in disease and physiopathology. Proteom Clin Appl. 2008;2:264–285. doi: 10.1002/prca.200780018. [DOI] [PubMed] [Google Scholar]
- Zhang DH, Tai LK, Wong LL, Sethi SK, Koay ES. Proteomics of breast cancer: enhanced expression of cytokeratin19 in human epidermal growth factor receptor type 2 positive breast tumors. Proteomics. 2005;5:1797–1805. doi: 10.1002/pmic.200401069. [DOI] [PubMed] [Google Scholar]
- Oshima RG, Baribault H, Caulin C. Oncogenic regulation and function of keratins 8 and 18. Cancer Metastasis Rev. 1996;15:445–471. doi: 10.1007/BF00054012. [DOI] [PubMed] [Google Scholar]
- Chang GC, Liu KJ, Hsieh CL, Hu TS, Charoenfuprasert S, Liu HK, et al. Identification of alpha-enolase as an autoantigen in lung cancer: its overexpression is associated with clinical outcomes. Clin Cancer Res. 2006;12:5746–5754. doi: 10.1158/1078-0432.CCR-06-0324. [DOI] [PubMed] [Google Scholar]
- Tsai ST, Chien IH, Shen WH, Kuo YZ, Jin YT, Wong TY, et al. ENO1, a potential prognostic head and neck cancer marker, promotes transformation partly via chemokine CCL20 induction. Eur J Cancer. 2010;46:1712–1723. doi: 10.1016/j.ejca.2010.03.018. [DOI] [PubMed] [Google Scholar]
- Mazurek S, Boschek CB, Hugo F, Eigenbrodt E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin Cancer Biol. 2005;15:300–308. doi: 10.1016/j.semcancer.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Chen G, Gharib TG, Wang H, Huang CC, Kuick R, Thomas DG, et al. Protein profiles associated with survival in lung adenocarcinoma. Proc Natl Acad Sci USA. 2003;100:13537–13542. doi: 10.1073/pnas.2233850100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Xiao Z, Chen Z, Zhang X, Li J, Wu X, et al. Proteome analysis of human lung squamous carcinoma. Proteomics. 2006;6:547–558. doi: 10.1002/pmic.200500256. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zhang H, Xu A, Li N, Liu J, Liu C, et al. Elevation of serum l-lactate dehydrogenase B correlated with the clinical stage of lung cancer. Lung Cancer. 2006;54:95–102. doi: 10.1016/j.lungcan.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Nagakubo D, Taira T, Kitaura H, Ikeda M, Tamai K, Iguchi-Ariga SM, et al. DJ-1, a novel oncogene which transforms mouse NIH3T3 cells in cooperation with ras. Biochem Biophys Res Commun. 1997;231:509–513. doi: 10.1006/bbrc.1997.6132. [DOI] [PubMed] [Google Scholar]
- Kim RH, Peters M, Jang Y, Shi W, Pintilie M, Fletcher GC, et al. DJ-1, a novel regulator of the tumor suppressor PTEN. Cancer Cell. 2005;7:263–273. doi: 10.1016/j.ccr.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Hod Y. Differential control of apoptosis by DJ-1 in prostate benign and cancer cells. J Cell Biochem. 2004;92:1221–1233. doi: 10.1002/jcb.20159. [DOI] [PubMed] [Google Scholar]
- Gu L, Cui T, Fan C, Zhao H, Zhao C, Lu L, et al. Involvement of ERK1/2 signaling pathway in DJ-1-induced neuroprotection against oxidative stress. Biochem Biophys Res Commun. 2009;383:469–474. doi: 10.1016/j.bbrc.2009.04.037. [DOI] [PubMed] [Google Scholar]
- He X, Zheng Z, Li J, Ben Q, Liu J, Zhang J, et al. DJ-1 promotes invasion and metastasis of pancreatic cancer cells by activating SRC/ERK/uPA. Carcinogenesis. 2012;33:555–562. doi: 10.1093/carcin/bgs002. [DOI] [PubMed] [Google Scholar]
- Kumar S, Singh BK, Kalra N, Kumar V, Kumar A, Prasad AK, et al. Novel thiocoumarins as inhibitors of TNF-alpha induced ICAM-1 expression on human umbilical vein endothelial cells (HUVECs) and microsomal lipid peroxidation. Bioorg Med Chem. 2005;13:1605–1613. doi: 10.1016/j.bmc.2004.12.013. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra R, Dubey R, Saini N. Gene expression profiling indicate role of ER stress in miR-23a∼27a∼24-2 cluster induced apoptosis in HEK293T cells. RNA Biol. 2011;8:648–664. doi: 10.4161/rna.8.4.15583. [DOI] [PubMed] [Google Scholar]
- Dubey R, Chhabra R, Saini N. Small interfering RNA against transcription factor STAT6 leads to increased cholesterol synthesis in lung cancer cell lines. PLoS One. 2011;6:e28509. doi: 10.1371/journal.pone.0028509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales-Cadenas R, Carmona-Saez P, Vazquez M, Vicente C, Yang X, Tirado F, et al. GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res. 2009;37 (Web Server issue:W317–W322. doi: 10.1093/nar/gkp416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona-Saez P, Chagoyen M, Tirado F, Carazo JM, Pascual-Montano A. GENECODIS: a web-based tool for finding significant concurrent annotations in gene lists. Genome Biol. 2007;8:R3. doi: 10.1186/gb-2007-8-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel A, Prasad AK, Parmar VS, Ghosh B, Saini N. 7,8-Dihydroxy-4-methylcoumarin induces apoptosis of human lung adenocarcinoma cells by ROS-independent mitochondrial pathway through partial inhibition of ERK/MAPK signaling. FEBS Lett. 2007;581:2447–2454. doi: 10.1016/j.febslet.2007.04.052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.