Abstract
Blond hair is a rare human phenotype found almost exclusively in Europe and Oceania. Here, we identify a cystine-to-arginine change at a highly conserved residue in tyrosinase-related protein 1 (TYRP1) as the single source of blond hair in Solomon Islanders. This missense mutation is predicted to impact catalytic activity of the protein and causes blond hair through a recessive mode of inheritance. The novel mutation is at a frequency of 26% in the Solomon Islands but is absent outside of Oceania and represents the largest genetic effect on a visible human phenotype reported to date. Our findings demonstrate that alleles of large effect reach appreciable frequencies in geographically isolated populations and underscore the importance of extending medical genomics to humans worldwide.
Human pigmentation varies considerably within and among populations and is a function of both variation in exposure to ultraviolet radiation (UVR) and the type and quantity of melanin produced in melanocytes and keratinocytes (1). While genome wide association (GWA) studies in European populations have yielded numerous insights into the genetic basis of pigmentation variation within Europeans (2), the relevance of these associations outside of Europe remains largely unexplored (but see (3)). Here, we focus on understanding the genetic basis of blond hair in the Solomon Islands, a population that breaks from the general trend of darker skin and hair pigmentation near the equator where there is higher UVR (1, 4). Strikingly, while individuals from the Solomon Islands and other locations in Oceania near the equator have both the darkest skin pigmentation outside of Africa, they also have the highest prevalence of blond hair (5–10%) outside of Europe (5). The earliest inhabitants of Near Oceania settled at least 40,000 years before present (YPB) and are thought to have remained in relative isolation on the archipelago for at least 25,000 years (6–9). There is clear evidence of at least one subsequent migration from Southeast Asia about 3,000–4,000 YBP, and waves of more recent gene flow from other islands in the Pacific and from Europe (10, 11). To understand whether sharing of this rare human phenotype is due to globally common genetic variants or arose independently, we investigated the genetic mechanism underlying hair color in Solomon Islanders.
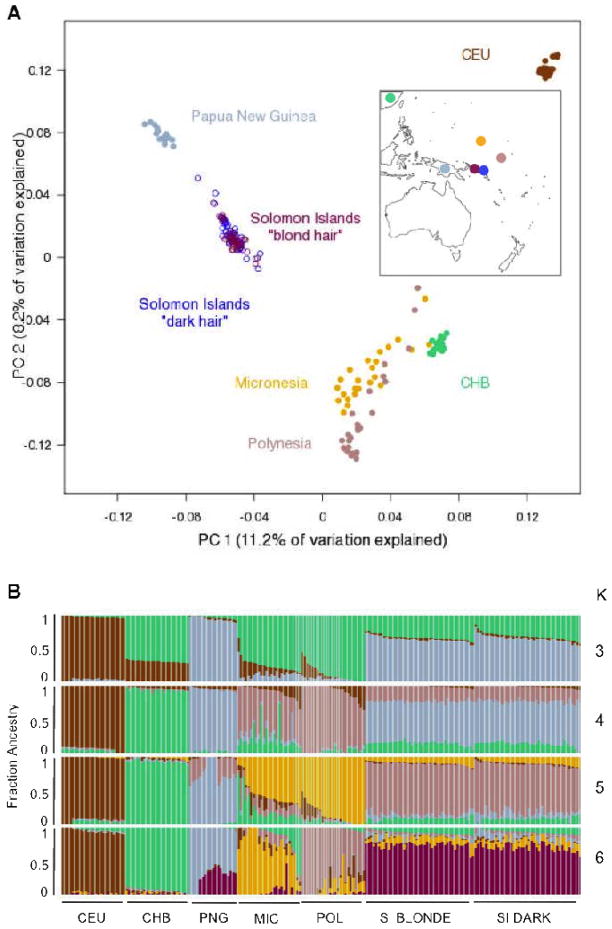
We measured skin and hair pigmentation in 1,209 Solomon Islanders and selected 85 individuals from the extreme 10% tails of the hair pigmentation distribution (43 blond and 42 dark-haired individuals) for genotyping with the Affymetrix 6.0 array (fig. S1–2 and table S1) (12). Estimates of global ancestry proportion showed that individuals from the Solomon Islands are genetically distinct from nearby populations (Fig. 1A and 1B), as has been previously observed (5). Further, no systematic differences in ancestry between blond and dark-haired Solomon Islanders were found suggesting that the presence of blond hair in the Solomon Islands is not due to recent gene flow from other populations (Fig. 1A, 1B and S3).
Figure 1.
PCA (A) and ADMIXTURE (B) plots demonstrating the genetic relationships between Solomon Islanders (blond and dark hair) genotyped in the present study and several other populations. In PC space, Solomon Islanders are found between New Guineans and Asians, which is consistent with their population history (7). Blond and dark-haired Solomon Islanders show no systematic differences in their ancestry suggesting that blond hair is unlikely to be due to gene flow from other populations (e.g. Europeans). (CEU = Europeans from HapMap; CHB = Han Chinese from HapMap; PNG = Papua New Guinea; MIC = Micronesians; POL = Polynesians; SI = Solomon Islanders).
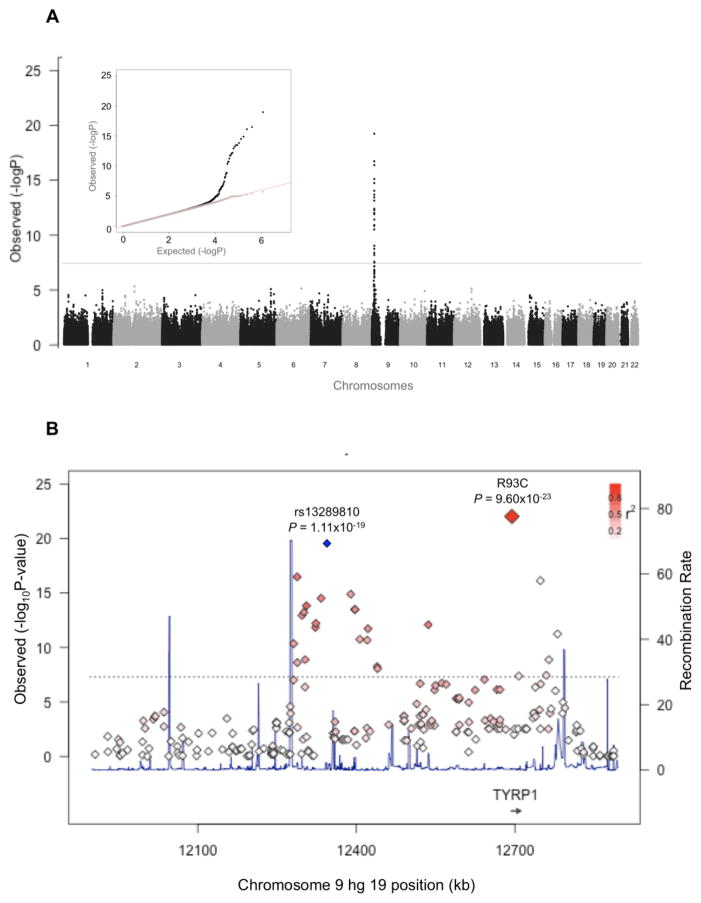
A case-control GWA study for hair color, comparing blond to dark-haired individuals, revealed a single strong association signal on chromosome 9 (Fig. 2A). The GWA signal was driven by 151 single nucleotide polymorphisms (SNPs) at 9p23 flanked at each side by a recombination hotspot (Fig. 2B). The SNP with the strongest association (rs13289810; odds ratio = 29.5±0.485 SE; P=1.11×10−19) had a frequency of 0.93 and 0.31 in blond -and dark-haired individuals, respectively. The mapping interval contained a single known candidate gene, tyrosinase-related protein 1 (TYRP1), which encodes a melanosomal enzyme involved in mammalian pigmentation (13).
Figure 2.
Mapping the blond hair locus in Solomon Islanders. (A) A Manhattan plot and QQ-plot (inset) of the association scores comparing blond and dark haired individuals. The grey line indicates the genome-wide threshold for statistical significance. The QQ-plot shows the distribution of p-values before (black points; λ=1.02) and after (grey points; λ=1.00) removal of the 151 SNPs in the associated 9p23 region. (B) A regional plot of the GWA including the R93C allele, where the signal for the top SNP on the GWA array (blue diamond) and R93C (large red diamond) are indicated. The degree of redness of all SNP’s in the region indicates linkage disequilibrium with R93C. Also shown is recombination rate (navy line) at the signal peak in a window of ± 500 kb around the top GWA SNP. Positions, recombination rates and gene annotations are according to NCBI build 37 (hg 19).
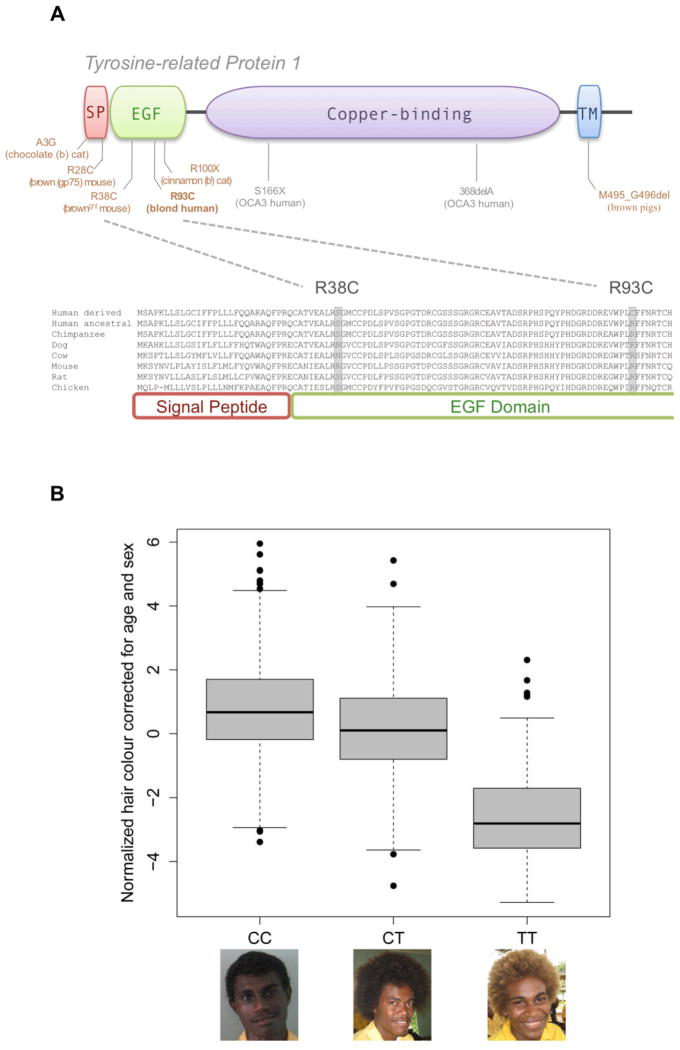
Resequencing of TYRP1 exons in 12 blond and 12 dark-haired Solomon Islanders detected only one polymorphism, a T to C transition at chr9:12,694,273 (GrCH37/hg19) which corresponds to a predicted arginine to cysteine mutation in exon 2 of TYRP1 at amino acid position 93. All 12 blond individuals were found to carry the TT genotype at this position and were thus homozygous for the cysteine residue, whereas dark-haired individuals were either heterozygous (CT genotype; N = 5) or homozygous (CC genotype; N = 7) for the ancestral arginine. The arginine residue is highly conserved (Fig. 3A) and the change to cysteine is predicted to be “probably damaging” with 0.999 posterior probability by the PolyPhen2 algorithm (14).
Figure 3.
Hair and skin pigmentation variants in TYRP1 (A) Top; a schema of the protein domains of TYRP1 (signal peptide (SI); epidermal growth factor (EGF), copper-binding and transmembrane (TM) domains) indicating variants in humans and other vertebrates that result in lightened skin and/or hair/coat (in brown) or albinism (in grey). Bottom; homology at the first 100 amino acids of TYRP1 in humans and other vertebrates. The location of the mouse brownlight mutation (R38C) and the Solomon Islands blond hair mutation (R93C) are given. (B) Boxplot showing the effect of the R93C genotypes on hair pigmentation in 921 Solomon Islanders.
To verify that 93C is the single causal allele driving the 9p23 GWA peak, the variant was genotyped directly in the discovery GWA panel and analyses repeated including R93C. R93C was more strongly associated with blond hair (P=9.60×10−23) than the top GWA SNP (Fig. 2B). Furthermore, analysis conditioning on the top GWA SNP (rs13289810) failed to remove all association from this region, as the residual signal for R93C still attained borderline genome-wide significance (P=3.11×10−7) (fig. S4A). Conversely, conditioning on R93C abolished the signal for association across this locus (fig. S4B), confirming a primary role for the coding R93C change.
To further evaluate its effects on hair pigmentation in Solomon Islanders, 93C was genotyped in 921 Solomon Islanders for whom we had measured hair pigmentation with spectrometry (fig. S5). Adjusting for age and geography, females had lighter hair than males (Student’s T test, P = 4.09×10−35), consistent with a previous study of hair color in Melanesians (5). Further, adjusting for sex and geography, hair color for 93C homozygotes (i.e. blonds) was not seen to darken significantly with age (R2 = 0.013, P = 0.187), whereas darkening was observed in heterozygotes (R2 = 0.026, P = 0.0017) and 93R homozygotes (R2 = 0.031, P = 0.0004). A recessive model provides the best fit for the association between 93C and hair color and a model including genotype, age and sex accounts for 46.4% of the variance in hair color on the Solomon Islands (P = 2.19×10−90; fig. S6 and table S3). There was evidence, however, that hair color differs between 93R homozygotes and heterozygotes (One-sided T test, P = 1.27×10−9), which suggests that 93C may have some co-dominant effect (Fig. 3B). Finally, we also assessed the effect of 93C on skin pigmentation in the Solomon Islands and found a modest additive effect accounting for 5.3% of variance in skin color (P = 4.56×10−8; fig. S7 and table S4).
The location and molecular alteration of the 93C mutation in TYRP1 suggests some intriguing possibilities for its function. TYRP1 encodes a key enzymatic step in melanin biosynthesis, giving rise to black/brown pigmentation (eumelanin), which occurs at high levels in individuals of African and Oceanic descent. Mutations in TYRP1, which is highly conserved in vertebrates, have been shown to lighten skin and/or hair pigmentation in several species including pigs, dogs, mice and zebra fish (Fig. 3A; 15, 16–19). In humans, previously discovered rare mutations leading to complete loss of function for TYRP1 are known to cause OCA3 or rufous albinism (20, 21). The 93C blond hair mutation in human TYRP1 resides in an epidermal growth factor (EGF)-like repeat near the N-terminus and shows similarity to the TYRP1 allele in the brownlight mouse (Fig. 3A). The brownlight mouse exhibits reduced TYRP1 stability and catalytic function resulting in decreased melanin content in mouse hair (19). Further, hair in these mice progressively lightens with age due to premature melanocyte death (19). Strikingly, like blond hair in Melanesians, the brownlight phenotype in the mouse is caused by an ARG→CYS substitution, located 55 amino acids upstream of R93C (R38C). R38C likely acts to interfere with normal disulfide bridges formed by the 15-Cys EGF repeat (Fig. 3A). It seems likely that the human 93C mutation confers a similar phenotype, namely premature melanocyte death that is exacerbated with age and melanogenic activity, resulting in blond hair that does not darken with age as observed in the Solomon Islands (Fig. 3B). However, it is also possible that the mouse R38C and human R93C mutations confer different effects on TYRP1 function, or that the R93C mutation is not disruptive to TYRP1 function, but rather is in linkage disequilibrium with non-coding functional genetic variant(s) within the ~500kb mapping interval.
The frequency of the 93C allele in the Solomon Islands is 0.26. To determine the worldwide distribution of the 93C allele, we genotyped the R93C SNP in 941 individuals from the 52 populations in the CEPH-HGDP (22). All 941 individuals were homozygous for 93R (table S4) and furthermore, the 93C allele was found to be absent from standard variant repositories (i.e. dbSNP, 1000 Genomes). Three 93C homozygous individuals were found in our cohort who reported that one of their parents came from outside the Solomon Islands: two individuals each with a parent from Fiji and one individual with a parent from New Guinea. Together, these observations suggest that the 93C allele is restricted to the Pacific and that the unique genetic mechanism causing blond hair in Solomon Islanders is shared with neighboring populations with the blond hair phenotype (5).
The frequency of the 93C allele in Solomon Islanders relative to other populations and its robust association with a visible human phenotype suggest that the 93C allele may have been driven to intermediate frequency by positive selection. The TYRP1 locus was evaluated for evidence of past positive selection using statistics based on allele frequency differentiation and long-range linkage disequilibrium (12). No strong evidence of recent selection at the TYRP1 locus was found in our sample from the Solomon Islands (fig. S8–S10). This result should be interpreted with caution, however, since our sample has relatively low power to detect selection on variants that follow a recessive mode of inheritance (23, 24). Moreover, even if the 93C allele was driven to its present frequency by selection, it remains a possibility that a different phenotype was targeted by selection and that blond hair is the result of phenotypic hitchhiking (25). For example, skin pigmentation may have been targeted by selection as we find that the 93C allele is associated with a minor reduction in skin pigmentation (table S3). This locus does show evidence of selection in Europeans, but it remains unclear what functional variant(s) may have been the target of selection (26, 27).
Regardless of the mechanisms responsible for the intermediate frequency of the 93C allele in Melanesia, this study realizes some of the postulated benefits of mapping complex human traits in population isolates (28); namely, the strong influence of genetic drift (and/or local positive selection) in such populations means that, while many recessive and/or rare alleles will be lost, some have a chance to rise to an appreciable frequency. Thus, it is expected (and demonstrated in recent studies) that population isolates may harbor alleles at appreciable frequency that influence complex traits and that are otherwise rare or absent in other populations (29–31). In the present case, the 93C TYRP1 variant accounts for ~46% of the variation in hair color in Solomon Islanders, which to our knowledge is the strongest reported effect on a visible human phenotype attributable to a common polymorphism. More generally, this work strongly supports the growing notion that population-specific variation is important in accounting for the heritable variance in clinically relevant phenotypes, and underscores the importance of enabling medical genomics in diverse worldwide populations.
Supplementary Material
Acknowledgments
We are deeply grateful to the people of the Solomon Islands for their participation in this study. We thank Itsik Pe’er, Marcus Stoffel, David Altshuler, Jan Breslow, Jeffery Friedman and the Kosrae consortium for access to genetic data. We thank Graham Coop and Greg Barsh for helpful comments. We thank Claudia Wegener for technical assistance. Funded by a Wenner-Gren Foundation for Anthropological Research grant to SM, MRC Centre for Causal Analyses in Translational Epidemiology grant RD1634 to NJT and the Max Planck Society.
References
- 1.Jablonski NG, Chaplin G. Human skin pigmentation as an adaptation to UV radiation. Proceedings of the National Academy of Sciences. 2010 May 11;107:8962. doi: 10.1073/pnas.0914628107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sturm RA. Molecular genetics of human pigmentation diversity. Hum Mol Genet. 2009 Apr 15;18:R9. doi: 10.1093/hmg/ddp003. [DOI] [PubMed] [Google Scholar]
- 3.Stokowski RP, et al. A Genomewide Association Study of Skin Pigmentation in a South Asian Population. Am J Hum Genet. 2007 Dec 1;81:1119. doi: 10.1086/522235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barsh GS. What Controls Variation in Human Skin Color? PLoS Biol. 2003;1:e27. doi: 10.1371/journal.pbio.0000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Norton HL, et al. Skin and hair pigmentation variation in Island Melanesia. Am J Phys Anthropol. 2006 Jun;130:254. doi: 10.1002/ajpa.20343. [DOI] [PubMed] [Google Scholar]
- 6.Friedlaender JS, et al. The genetic structure of Pacific Islanders. PLoS Genet. 2008 Jan;4:e19. doi: 10.1371/journal.pgen.0040019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kayser M, et al. Melanesian and Asian Origins of Polynesians: mtDNA and Y Chromosome Gradients Across the Pacific. Mol Biol Evol. 2006 Nov 1;23:2234. doi: 10.1093/molbev/msl093. [DOI] [PubMed] [Google Scholar]
- 8.Reich D, et al. Denisova admixture and the first modern human dispersals into southeast Asia and oceania. American journal of human genetics. 2011 Oct 7;89:516. doi: 10.1016/j.ajhg.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rasmussen M, et al. An Aboriginal Australian genome reveals separate human dispersals into Asia. Science. 2011 Oct 7;334:94. doi: 10.1126/science.1211177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kayser M. The Human Genetic History of Oceania: Near and Remote Views of Dispersal. Current Biology. 2010;20:R194. doi: 10.1016/j.cub.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 11.Delfin F, et al. Bridging Near and Remote Oceania: MtDNA and NRY Variation in the Solomon Islands. Molecular Biology and Evolution. 2011 Jul 18; doi: 10.1093/molbev/msr186. in press. [DOI] [PubMed] [Google Scholar]
- 12.Materials and methods are available as supporting material on Science Online.
- 13.del Marmol V, Beermann F. Tyrosinase and related proteins in mammalian pigmentation. FEBS Letters. 1996;381:165. doi: 10.1016/0014-5793(96)00109-3. [DOI] [PubMed] [Google Scholar]
- 14.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Meth. 2010;7:248. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ren J, et al. A 6-bp deletion in the TYRP1 gene causes the brown colouration phenotype in Chinese indigenous pigs. Heredity. 2011;106:862. doi: 10.1038/hdy.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braasch I, Liedtke D, Volff J-N, Schartl M. Pigmentary function and evolution of tyrp1 gene duplicates in fish. Pigment Cell & Melanoma Research. 2009;22:839. doi: 10.1111/j.1755-148X.2009.00614.x. [DOI] [PubMed] [Google Scholar]
- 17.Schmutz SM, Berryere TG, Goldfinch AD. TYRP1 and MC1R genotypes and their effects on coat color in dogs. Mammalian Genome. 2002;13:380. doi: 10.1007/s00335-001-2147-2. [DOI] [PubMed] [Google Scholar]
- 18.Javerzat S, Jackson IJ. White-based brown (Tyrp1B-w) is a dominant mutation causing reduced hair pigmentation owing to a chromosomal inversion. Mammalian Genome. 1998;9:469. doi: 10.1007/s003359900798. [DOI] [PubMed] [Google Scholar]
- 19.Johnson R, Jackson IJ. Light is a dominant mouse mutation resulting in premature cell death. Nat Genet. 1992 Jun;1:226. doi: 10.1038/ng0692-226. [DOI] [PubMed] [Google Scholar]
- 20.Boissy RE, et al. Mutation in and lack of expression of tyrosinase-related protein-1 (TRP-1) in melanocytes from an individual with brown oculocutaneous albinism: a new subtype of albinism classified as “OCA3”. Am JHum Genet. 1996 Jun;58:1145. [PMC free article] [PubMed] [Google Scholar]
- 21.Manga P, et al. Rufous Oculocutaneous Albinism in Southern African Blacks Is Caused by Mutations in the TYRP1 Gene. The American Journal of Human Genetics. 1997;61:1095. doi: 10.1086/301603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cann HM, et al. A human genome diversity cell line panel. Science. 2002 Apr 12;296:261. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- 23.Teshima KM, Przeworski M. Directional Positive Selection on an Allele of Arbitrary Dominance. Genetics. 2006 Jan 1;172:713. doi: 10.1534/genetics.105.044065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Przeworski M, Coop G, Wall JD. The Signature of Positive Selection on Standing Genetic Variation. Evolution. 2005;59:2312. [PubMed] [Google Scholar]
- 25.Barton NH. Genetic hitchhiking. Philos Trans R Soc Lond B Biol Sci. 2000 Nov 29;355:1553. doi: 10.1098/rstb.2000.0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sulem P, et al. Two newly identified genetic determinants of pigmentation in Europeans. Nat Genet. 2008;40:835. doi: 10.1038/ng.160. [DOI] [PubMed] [Google Scholar]
- 27.Sulem P, et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat Genet. 2007 Dec;39:1443. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- 28.Kristiansson K, Naukkarinen J, Peltonen L. Isolated populations and complex disease gene identification. Genome Biology. 2008;9:109. doi: 10.1186/gb-2008-9-8-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holm H, et al. A rare variant in MYH6 is associated with high risk of sick sinus syndrome. Nature genetics. 2011 Apr;43:316. doi: 10.1038/ng.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Acuna-Alonzo V, et al. A functional ABCA1 gene variant is associated with low HDL-cholesterol levels and shows evidence of positive selection in Native Americans. Hum Mol Genet. 2010 Jul 15;19:2877. doi: 10.1093/hmg/ddq173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kenny EE, et al. Systematic haplotype analysis resolves a complex plasma plant sterol locus on the Micronesian Island of Kosrae. Proc Natl Acad Sci U S A. 2009 Aug 18;106:13886. doi: 10.1073/pnas.0907336106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.