Abstract
Three MADS-box genes were identified from a cDNA library derived from young flowers of Eucalyptus grandis W. Hill ex Maiden. The three egm genes are single-copy genes and are expressed almost exclusively in flowers. The egm1 and egm3 genes shared strongest homology with other plant MADS-box genes, which mediate between the floral meristem and the organ-identity genes. The egm3 gene was also expressed strongly in the receptacle or floral tube, which surrounds the carpels in the eucalypt flower and bears the sepals, petals, and numerous stamens. There appeared to be a group of genes in eucalypts with strong homology with the 3′ region of the egm1 gene. The egm2 gene was expressed in eucalypt petals and stamens and was most homologous to MADS-box genes, which belong to the globosa group of genes, which regulate organogenesis of the second and third floral whorls. The possible role of these three genes in eucalypt floral development is discussed.
The basic organization of the angiosperm flower is remarkably conserved among all species. The central generative organs (stamens and carpels) are always surrounded by perianth structures (sepals and petals). Alterations in the number, shape, size, color, and arrangement of these floral organs, usually adaptations to particular pollination strategies (Richards, 1986), have led to the evolution of an enormous range of floral structures. The diversification of patterns and structures in plants is attributable in part to evolution of the structure and regulation of genes that control morphogenesis (Doebley, 1993). In recent years there has been rapid expansion of our understanding of the genes controlling floral morphogenesis.
Genetic and molecular studies in Arabidopsis and snapdragon, among other plant species, have led to the identification of genes that direct floral development. Many of these genes contain a MADS-box domain, which is involved in DNA binding, and a K-box domain, which is involved in protein dimerization (Schwarz-Sommer et al., 1992; Davies et al., 1996). The MADS-box is highly conserved, about 56 aa long, and usually at the N terminus of the protein, whereas less homology is observed in the K-box (Ma et al., 1991; Pnueli et al., 1991). More than 12 MADS-box genes have been observed in angiosperm species, including Arabidopsis (Ma et al., 1991), maize (Mena et al., 1995), asparagus (Miller et al., 1995), and tomato (Pnueli et al., 1991). They have also been found in gymnosperms (Tandre et al., 1995; Mouradov et al., 1996, 1997). The function of many of these MADS-box genes is not known; however, most are almost exclusively expressed in flowers, suggesting that they are likely to play a role in flower development.
The MADS-box genes are likely to have played an important role in floral diversification because they regulate the major steps in floral morphogenesis. Phylogenetic analysis of the plant MADS-box genes has revealed that they are organized into distinct groups, the members of which share similar functional roles in flower development (Purugganan et al., 1995; Theissen and Saedler, 1995). MADS-box genes control floral meristem formation (Huijser et al., 1992; Mandel et al., 1992), floral organ formation (Sommer et al., 1990; Yanofsky et al., 1990; Angenent et al., 1992, 1993; Jack et al., 1992; Mandel et al., 1992; Tröbner et al., 1992; Bradley et al., 1993; Goto and Meyerowitz, 1994), and mediate between these meristem and organ-identity functions (Angenent et al., 1994; Pnueli et al., 1994). Molecular evolutionary analysis has revealed that the ancestral gene in each of these groups of genes arose before the appearance of the flowering plants (Purugganan et al., 1995).
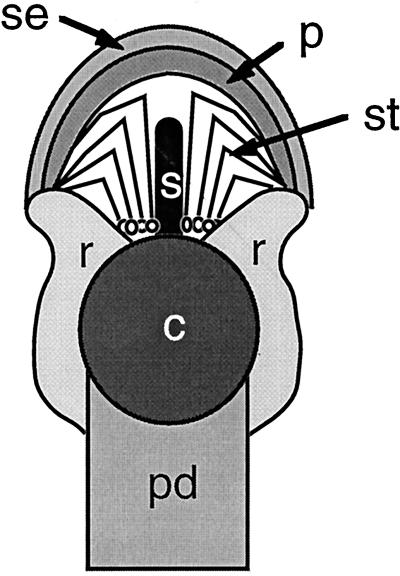
In eucalypts, flowers are generally arranged in a dichasium, which terminates in a single flower, the subtending bracts (deciduous) of which enclose all other flowers in the developing inflorescence. During flower development the sepal primordia of the first whorl (outermost) rapidly fuse to each other, as do the petal primordia of the second whorl. These two fused structures, separately or fused to each other, form a structure called the operculum, which covers and protects the developing reproductive structures until anthesis. In another modification, the calyx, corolla, and numerous stamens are borne on the summit of a floral tube, which surrounds and is fused to the wall of the inferior ovary (Beadle et al., 1972). Figure 1 shows the arrangement of floral organs in a typical eucalypt flower.
Figure 1.
Diagrammatic median vertical section through a typical eucalypt flower before anthesis. c, Carpel; p, petal; pd, peduncle; r, receptacle; s, style; se, sepal; st, stamen. The fused sepals and fused petals, separately or fused together, form the operculum.
In this paper we report the isolation of three MADS-box genes from Eucalyptus grandis W. Hill ex Maiden, which are predominantly expressed in flowers. We compare the sequences of these genes with those of other plant MADS-box genes, and examine the expression of these genes in developing eucalypt flowers. Analysis of gene sequence and expression revealed similarities to other plant MADS-box genes with functional roles in floral development.
MATERIALS AND METHODS
Plant Material
Eucalyptus grandis W. Hill ex Maiden floral and foliar material was obtained on a number of occasions between May 1994 and December 1995, from a mature specimen growing at Lone Pine Koala Sanctuary, Fig Tree Pocket, Queensland, Australia. Roots were obtained from seedlings germinated aseptically and grown in a growth room for 1 month. Eucalyptus globulus Labill. flowers were collected in July 1996, by Peter Buxton from Australian Paper Plantations in Gippsland, Victoria. The large E. globulus flowers allowed easy dissection of floral organs, including the petals (inner operculum), stamens, carpels, receptacle, and style for northern-blot analysis. The sepals (outer operculum) had already abscised from these flowers.
Nucleic Acid Extraction
DNA was extracted from E. grandis leaves by the method described by Keil and Griffin (1994). RNA was extracted from various plant tissues using the method of Chang et al. (1993) with modifications. Three grams of tissue was ground in liquid nitrogen to a fine powder using a mortar and pestle, then added to 10 mL of extraction buffer (2% cetyl-trimethyl-ammonium bromide, 2% PVP 40, 100 mm Tris-HCl, pH 8.0, 25 mm EDTA, 2 m NaCl, 0.5 g/L spermidine, and 2% β-mercaptoethanol) at 65°C, and the tube was shaken thoroughly and incubated for 5 min at 65°C. The solutions were then extracted twice with an equal volume of chloroform, with centrifugation at 7500g for 15 min after each extraction. The supernatant from the second extraction was mixed with 0.25 volume of 10 m LiCl, and the RNA precipitated overnight at 4°C. RNA was pelleted by centrifugation at 7500g for 20 min, dissolved in 500 μL of SSTE (0.5% SDS, 1 m NaCl, 10 mm Tris-HCl, pH 8.0, and 1 mm EDTA), and then extracted with an equal volume of chloroform. The purified RNA was precipitated by addition of 2 volumes of ethanol and placed at −70°C for 1 h, pelleted by centrifugation at 10,000g in a microcentrifuge at 4°C, and the pellet was dried before dissolving in diethyl pyrocarbonate-treated water.
Library Construction and Screening
Poly(A+) RNA was purified using oligo(dT)25 Dynabeads, according to the manufacturer's methods (Dynal, Oslo, Norway), from RNA isolated from young E. grandis flowers, which were initiating formation of floral organ primordia. This poly(A+) RNA was used to construct a cDNA library using the Pharmacia Time Saver cDNA Synthesis kit and the cloning vector Lambda ExCell. Approximately 100,000 clones were screened at low stringency (5× SSC, 48°C) using, as a probe, a mixture of 32P-labeled MADS-box domain sequences that had been amplified from the tomato tdr1, tdr4, tdr5, and tdr8 genes using degenerate primers matching each end of the MADS-box.
Sequence Analysis
DNA sequences were determined using dye-terminator chemistry on an automated sequencer (model 377, ABI, Columbia, MD). All sequence analysis was performed using programs in the University of Wisconsin Genetics Computer Group package (Madison). Genetic distance analysis of nucleotide sequences from the MIK region (Purugganan et al., 1995) of several plant MADS-box genes were performed using the ClustalX program.
DNA and RNA Gel-Blot Analysis
Southern-blot analysis using standard methods was performed on 5-μg samples of DNA digested with EcoRI, HindIII, or BamHI. Northern-blot analysis was performed using standard methods on 10-μg samples of total RNA.
In Situ Hybridization
33P-labeled riboprobes were synthesized by T7 or SP6 RNA polymerase, using template plasmids linearized with restriction enzymes to enable synthesis of sense or antisense probes (minus the MADS-box) of each of the three E. grandis MADS-box genes.
Young flowers, roots, and leaves, germinated seedlings, and vegetative apices were fixed in 3.7% formaldehyde, dehydrated, cleared, and embedded in wax, and sections were prepared for in situ hybridization according to the method of Jack et al. (1992), with minor modifications. Histoclear (National Diagnostics, Atlanta, GA) was used instead of xylene throughout the method. Thick plant tissues were presectioned to a thickness of about 1 mm to aid infiltration of the fixative. Tissues were typically fixed for a period of 2.5 h with gentle agitation. Sections were baked onto slides coated in 3-aminopropyltriethoxysilane at 65°C for 18 h. The RNA probe was subjected to mild alkali hydrolysis by heating at 60°C for 1 h in 100 mm carbonate buffer (pH 10.2) before adding to 120 μL of hybridization buffer (Jack et al., 1992), and coated on each slide. Slides were incubated overnight at 42°C, then washed using the method of Jack et al. (1992). Slides were then coated in liquid emulsion and exposed for approximately 7 d. Slides were viewed under dark-field illumination, in which signals appeared as white grains against a dark background.
RESULTS
Sequence Analysis
Three different cDNAs (egm1, accession no. AF029975; egm2, accession no. AF029976; and egm3, accession no. AF029977) were isolated from an E. grandis cDNA library derived from young floral tissue as described in Methods. These cDNAs were sequenced and found to contain the MADS-box. The sequences for egm2 and egm3 each contained a full-length coding region and 5′ and 3′ untranslated sequences. The egm1 cDNA lacked 17 bases of the 5′ end of the coding region. The sequence of this portion of the egm1 gene was obtained by sequencing a genomic clone obtained from an E. grandis genomic library.
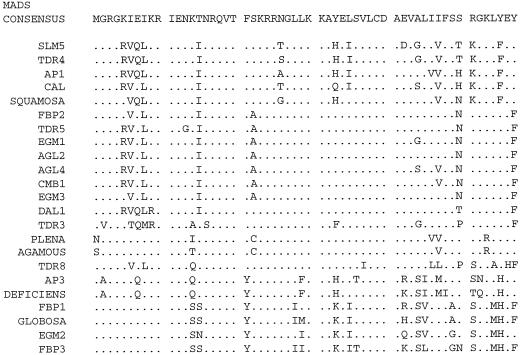
Figure 2 shows the alignment of the predicted protein sequences of the three egm genes with those of similar MADS-box genes from other plants. A predicted protein of 183 aa is encoded by egm1, which shares the strongest aa homology (85% identity over 170 aa) with the petunia FBP2 protein and the tomato TDR5 protein (83% identity over 173 aa). The cDNA egm2 encodes a predicted protein of 208 aa and shares 62% homology over 213 aa with the petunia FBP1 protein, and 60% homology over 215 aa with the snapdragon GLOBOSA protein. The cDNA egm3 encodes a predicted protein of 245 aa, which is most homologous to the Arabidopsis AGL4 protein (63% identity over 254 aa). An alignment of the MADS-box region of the three eucalypt genes with the MADS-box region of several closely related plant MADS-box genes is shown in Figure 3.
Figure 2.
A comparison of the deduced aa sequences of the cDNAs egm1 (a), egm2 (b), and egm3 (c), with similar sequences from other plant species. The MADS-box is underlined and the K-box is in italics. Dots indicate aa identical to the particular egm reference sequence, and dashes indicate gaps in the sequence alignment. Numbers indicate aa position in the egm sequences.
Figure 3.
A comparison of the deduced aa sequence of the MADS-box domain of the egm genes with the MADS-box domain of other plant genes. Dots indicate aa identical to the consensus sequence.
Genomic Organization of the egm Genes
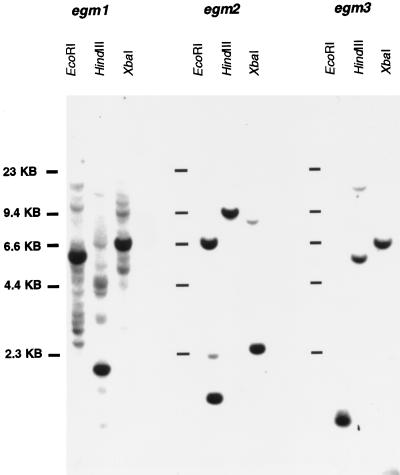
To determine the number of copies of the respective egm genes in the eucalypt genome, DNA gel-blot analysis was performed on E. grandis genomic DNA digested with various restriction enzymes. Figure 4 shows genomic DNA hybridizing to 3′ probes specific for each of the three egm genes. In all three blots, single, strongly hybridizing bands were observed under high-stringency conditions, indicating that each of the three egm genes occurs as a single copy in the eucalypt genome. In the egm1 blots, the weakly hybridizing bands indicated the presence of a number of genes with significant homology to the 3′ region of the egm1 sequence outside the MADS-box. Hybridizations with the 5′ region of all of the egm genes revealed a large number of bands, indicating that there are a large number of MADS-box genes in the eucalypt genome (results not shown).
Figure 4.
DNA gel-blot analysis of the egm genes. DNA isolated from E. grandis was digested with EcoRI, HindIII, or Xbal and hybridized with the egm gene minus the MADS-box under high stringency. Each lane contains 5 μg of digested genomic DNA.
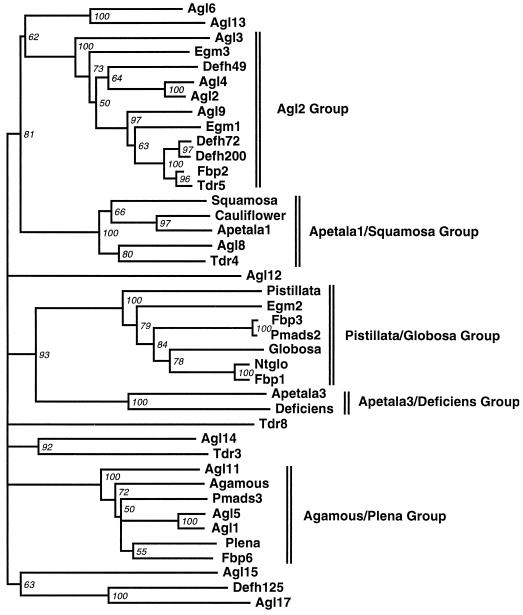
Figure 5 shows a phylogenetic tree of plant MADS-box genes based on analysis of the MIK region. Both egm1 and egm3 were included in the Agl2 group. Some of the genes in this group are involved in interactions between the meristem and the organ-identity genes. The egm2 gene was included in the Pistillata/globosa group, which, together with the Ap3/deficiens group of genes, is involved in regulating the development of petals and stamens.
Figure 5.
Phylogenetic tree of several dicot MADS-box genes and the three eucalypt egm genes based on genetic distance analysis of the MIK region. The lengths of the horizontal branches are proportional to the number of base substitutions. Numbers next to the nodes indicate bootstrap values from 100 replicates. Nodes with <50% bootstrap support are collapsed.
Expression of the egm Genes in Eucalypts
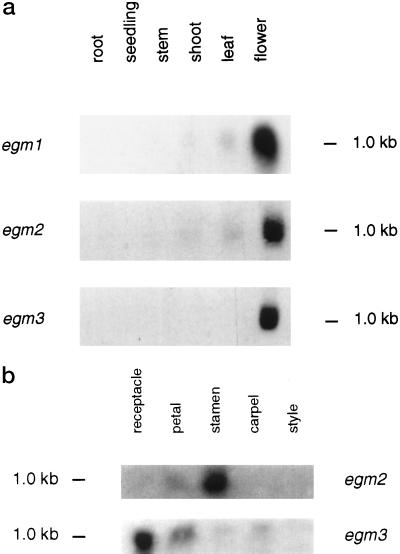
To determine the pattern of expression of the egm genes, northern blots of total RNA from a range of tissues were probed with each of the three genes. As shown in Figure 6a, all three genes were strongly expressed in floral tissue. Expression of egm3 was virtually exclusive to floral tissue, since in overexposed northern blots only a very weak signal was detected in nonfloral tissue. Weak expression of the egm1 gene, and to a lesser extent the egm2 gene, was detected in vegetative tissue, including vegetative shoots, leaves, stems, and seedlings. The expression pattern of the egm1 gene may also include expression of some of the other genes sharing homology to the egm1 gene as indicated by Southern-blot analysis.
Figure 6.
Analysis of the expression of the egm genes in different eucalypt tissues. a, RNA gel-blot analysis of egm1, egm 2, and egm3 expression in roots, seedlings, stems, shoots, leaves, and flowers. b, RNA gel-blot analysis of egm2 and egm3 expression in floral receptacles, petals, stamens, carpels, and styles. The blots contained 10 μg of total RNA and were hybridized with 3′ regions of egm1, egm2, and egm3 genes. The lengths of each of the three egm messages are estimated at 1.1 kb.
Further analysis of egm2 and egm3 expression within floral organs was performed on total RNA isolated from flowers of E. globulus. These flowers are large, allowing easier dissection of floral organs and collection of sufficient material for RNA extraction. Unfortunately, RNA from the sepals (outer operculum) could not be collected because they are shed early in floral development, when the floral organs are difficult to dissect. As shown in Figure 6b, a strong egm2 signal was detected in stamens, and a weaker signal was observed in petals. No expression was detected in the receptacle, carpels, and the style. The expression of egm3 was strongest in the receptacle, moderate in petals, carpels, and stamens, and absent in the style.
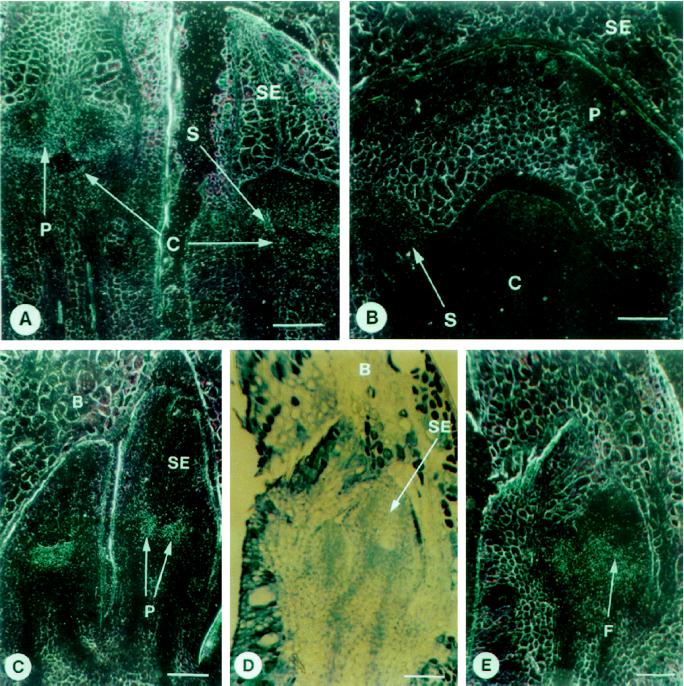
In situ hybridization was used to localize the expression of the egm genes in eucalypt floral tissue. In sections probed with the egm1 probe, a signal was detected in the primordia of developing petals, stamens, and carpels (Fig. 7A). Within the carpels at later stages of development, expression was strongest in the developing ovules. The egm2 gene was expressed in the petals and stamens at the primordial stages (Fig. 7C), and weakly in these organs during later stages as the flower enlarged (Fig. 7B). Expression of egm3 was detected in very young flowers in the floral meristem and the base of the sepals (Fig. 7, D and E), but was not detected later in development. Failure to detect expression of the three egm genes later in floral development is not consistent with the RNA-blot analysis. We have been unable to detect in situ expression of several other eucalypt genes in mature flowers, which we have shown by northern-blot analysis to be strongly expressed in these tissues.
Figure 7.
RNA in situ-hybridization analysis of egm2 and egm3 expression in eucalypt flowers. B, Bract; C, carpel; F, floral meristem; P, petal; SE, sepal; S, stamen. Bar = 100 μm. A, Dark-field view of a longitudinal section through two E. grandis flowers, with developing petal, stamen, and carpel primordia hybridized to the egm1 gene probe. B, Dark-field view of a longitudinal section through an E. grandis flower initiating stamen primordia, hybridized to the egm2 gene probe. C, Dark-field view of a longitudinal section through two E. grandis flowers initiating petal primordia, hybridized to the egm2 gene probe. D, Bright-field view of a longitudinal section through two young E. grandis flowers with developing sepal primordia. E, Dark-field view of the section in D hybridized to the egm3 gene probe.
DISCUSSION
In this study we report the isolation and characterization of three MADS-box genes from E. grandis. The eucalypt genome, in common with the genomes of other plants (Ma et al., 1991; Pnueli et al., 1991; Mena et al., 1995; Miller et al., 1995; Tandre et al., 1995), contains a family of MADS-box genes. All three eucalypt genes are expressed predominantly in flowers, like most other identified MADS-box genes. Similarities between the eucalypt genes and other plant MADS-box genes were identified by sequence and expression analysis to provide clues to the part these genes play in eucalypt flower development.
The predicted protein of the egm1 gene shares strong homology with the petunia fbp2 (Angenent et al., 1994) and tomato tdr5 (Pnueli et al., 1994) genes in the Agl2 group. Both the fbp2 and tdr5 genes are strongly expressed in the inner three whorls (petals, stamens, and carpels) of the flower. Functional analysis of these genes revealed that they are required for determination of the three inner whorls of the flower in each species (Angenent et al., 1994; Pnueli et al., 1994). Incomplete homeotic transformation of all three whorls is observed in plants lacking the function of these genes. Analysis of the egm1 gene in floral development is complicated by the occurrence of a small family of genes in eucalypts that share significant homology to egm1 outside of the MADS-box region. RNA gel-blot analysis revealed that expression of egm1 is likely to be almost exclusive to flowers. In situ analysis with an egm1 probe revealed expression in primordia of petals, stamens, and carpels, and in the ovules within the developing carpels; however, these results may also depict expression of egm1 homologs.
The predicted protein of the egm2 gene shares strong homology with the petunia fbp1 and snapdragon globosa genes. The genes fbp1 (Angenent et al., 1993) and globosa (Tröbner et al., 1992) have been shown to function as organ-identity genes specifying second- and third-whorl petal and stamen organogenesis. They function as class B genes according to the A, B, and C model, which describes the way in which the combinatorial expression of homeotic genes specifies floral organ identity. Both the fbp1 and globosa genes are expressed in petals and stamen primordia, and more strongly during their development. Expression of egm2 was detected only in petals and stamens of eucalypt flowers. RNA-blot analysis revealed much stronger expression of egm2 in stamens than in petals. Expression in the petals may have been low because the flowers sampled were close to anthesis, when the operculum (fused petals) is shed from the flower. It is also possible that high expression of the egm2 gene is associated with the formation of the large number of stamens typically found in eucalypt flowers.
Like egm1, the egm3 gene shares closest homology to the Agl2 group of genes. Its expression in all four floral whorls and in the receptacle suggests functional divergence from egm1. In situ analysis revealed expression in the floral meristem and in the sepal primordia. As the sepals developed, egm3 expression was restricted to the base of the sepals. In eucalypts the receptacle is modified into a floral tube that surrounds and sometimes protrudes above the carpels. Sepals, petals, and stamens are borne on the apex of the receptacle. These results suggest that the floral tube is initiated from the floral meristem, and may be derived from the base of the developing sepals. It would be interesting to determine the function of the egm3 gene in the development of the floral tube in eucalypt flowers.
Attempting to determine gene function on the basis of similarities in gene sequence or expression pattern, particularly with regulatory proteins (Gruenberg et al., 1992), can be misleading. Despite this, recent phylogenetic analysis of plant MADS-box genes has revealed that the gene family sorts into groups of genes that share a similar functional role in flower development (Purugganan et al., 1995; Theissen and Saedler, 1995). The members of these groups also share similar expression patterns and sequence homology. Our analysis of the MADS-box gene family is in agreement with this work.
The three egm genes clustered with genes with which they shared strongest overall sequence and expression pattern homology. This provides a clue to the part these genes may play in eucalypt flower development. The results suggest that the egm1 gene may function in mediating between the floral meristem and the organ-identity genes in a manner similar to the fbp2 and tdr5 genes in petunia and tomato, respectively. The egm2 gene is likely to be a homeotic B function gene regulating organogenesis of the petals and stamens in eucalypt flowers in a manner similar to the globosa gene in snapdragon (Tröbner et al., 1992). The strong expression of egm3 in the floral receptacle provides an interesting lead for further investigations to elucidate its function. Further analysis, either by heterologous complementation of floral mutants or by gene suppression in eucalypts, may assist in the determination of the function of these three MADS-box genes in eucalypt floral morphogenesis.
ACKNOWLEDGMENTS
The authors thank Peter Buxton for assistance with collection of plant material, Albert Ng and Adrian Steele for assistance with computer analysis, and Corinna Lange for critical reading and preparation of the manuscript.
Abbreviation:
- aa
amino acid(s)
LITERATURE CITED
- Angenent GC, Busscher M, Franken J, Mol JNM, van Tunen AJ. Differential expression of two MADS-box genes in wild-type and mutant petunia flowers. Plant Cell. 1992;4:983–993. doi: 10.1105/tpc.4.8.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angenent GC, Franken J, Busscher M, Colombo L, van Tunen AJ (1993) Petal and stamen formation in petunia is regulated by the homeotic gene fbp1. Plant J 4: 101–112 [DOI] [PubMed]
- Angenent GC, Franken J, Busscher M, Weiss D, van Tunen AJ. Co-suppression of the petunia homeotic gene fbp2affects the identity of the generative meristem. Plant J. 1994;5:33–44. doi: 10.1046/j.1365-313x.1994.5010033.x. [DOI] [PubMed] [Google Scholar]
- Beadle NCW, Evans OD, Carolin RC (1972) Flora of the Sydney Region. Reed, Sydney, Australia
- Bradley D, Carpenter R, Sommer H, Hartley N, Coen E. Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of Antirrhinum. Cell. 1993;72:85–95. doi: 10.1016/0092-8674(93)90052-r. [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11: 113–116
- Davies B, Egea-Cortines M, de Andrade Silva E, Saedler H, Sommer H. Multiple interactions amongst floral homeotic MADS-box proteins. EMBO J. 1996;15:4330–4343. [PMC free article] [PubMed] [Google Scholar]
- Doebley J. Genetics, development and plant evolution. Curr Opin Genet Dev. 1993;3:865–872. doi: 10.1016/0959-437x(93)90006-b. [DOI] [PubMed] [Google Scholar]
- Goto K, Meyerowitz EM (1994) Function and regulation of the Arabidopsis floral homeotic gene Pistillata. Genes Dev 8: 1548–1560 [DOI] [PubMed]
- Gruenberg DA, Natesan S, Alexandre C, Gilman MZ. Human and Drosophilahomeodomain proteins that enhance the DNA binding activity of serum response factor. Science. 1992;257:1089–1095. doi: 10.1126/science.257.5073.1089. [DOI] [PubMed] [Google Scholar]
- Huijser P, Klein J, Lonnig W-E, Meijer H, Saedler H, Sommer H (1992) Bracteomania, an inflorescence anomaly, is caused by the loss of function of the MADS-box gene squamosa in Antirrhinum majus. EMBO J 11: 1239–1249 [DOI] [PMC free article] [PubMed]
- Jack T, Brockman LL, Meyerowitz EM. The homeotic gene APETALA3 of Arabidopsis thalianaencodes a MADS-box and is expressed in petals and stamens. Cell. 1992;68:683–697. doi: 10.1016/0092-8674(92)90144-2. [DOI] [PubMed] [Google Scholar]
- Keil M, Griffin AR (1994) Use of random amplified polymorphic DNA (RAPD) markers in the discrimination and verification of genotypes in Eucalyptus. Theor Appl Genet 89: 442–456 [DOI] [PubMed]
- Ma H, Yanofsky ME, Meyerowitz EM. AGL1-AGL6, an Arabidopsisgene family with similarity to floral homeotic and transcription factor genes. Genes Dev. 1991;5:484–495. doi: 10.1101/gad.5.3.484. [DOI] [PubMed] [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky ME. Molecular characterisation of the Arabidopsis floral homeotic gene APETALA1. Nature. 1992;360:273–277. doi: 10.1038/360273a0. [DOI] [PubMed] [Google Scholar]
- Mena M, Mandel MA, Lerner DR, Yanofsky MF, Schmidt RJ. A characterisation of the MADS-box gene family in maize. Plant J. 1995;8:845–854. doi: 10.1046/j.1365-313x.1995.8060845.x. [DOI] [PubMed] [Google Scholar]
- Miller HG, Kocher TD, Loy B (1995) New MADS-box domains in Asparagus officinalis L. Sex Plant Reprod 8: 318–320
- Mouradov A, Glassick T, Teasdale R. Isolation and characterization of a new MADS-box cDNA from Pinus radiata (accession no. U76726) (PGR 97-027) Plant Physiol. 1997;113:664. [Google Scholar]
- Mouradov A, Glassick T, Vivian-Smith A, Teasdale R. Isolation of a MADS-box gene family from Pinus radiata (accession nos. U42399 and U42400) (PGR 96-002) Plant Physiol. 1996;110:1047. [Google Scholar]
- Pnueli L, Abu-Abeid M, Zamir D, Nacken W, Schwarz-Sommer Z, Lifschitz E (1991) The MADS-box gene family in tomato: temporal expression during floral development, conserved secondary structures and homology with homeotic genes from Antirrhinum and Arabidopsis. Plant J 1: 255–266 [PubMed]
- Pnueli L, Hareven D, Broday L, Lifschitz E. The TM5 [tdr5] MADS-box gene mediates organ differentiation in the three inner whorls of tomato flowers. Plant Cell. 1994;6:175–186. doi: 10.1105/tpc.6.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purugganan MD, Rounsley SD, Schmidt RJ, Yanofsky ME. Molecular evolution of flower development: diversification of the plant MADS-box regulatory gene family. Genetics. 1995;140:345–356. doi: 10.1093/genetics/140.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards AJ. Plant Breeding Systems. Winchester, MA: Allen and Unwin; 1986. [Google Scholar]
- Schwarz-Sommer Z, Hue I, Huijser R, Flor RJ, Hansen R, Tetens E, Lönnig W, Saedler H, Sommer H. Characterisation of the Antirrhinum floral homeotic MADS-box gene deficiens: evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J. 1992;11:251–263. doi: 10.1002/j.1460-2075.1992.tb05048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer H, Beltran J-R, Huijser R, Pape H, Lönnig W-E, Saedler H, Schwarz-Sommer Z. Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: the protein shows homology to transcription factors. EMBO J. 1990;9:605–613. doi: 10.1002/j.1460-2075.1990.tb08152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandre K, Albert VA, Sundas A, Engström P. Conifer homologues to genes that control floral development in angiosperms. Plant Mol Biol. 1995;27:69–78. doi: 10.1007/BF00019179. [DOI] [PubMed] [Google Scholar]
- Theissen G, Saedler H. MADS-box genes in plant ontogeny and phylogeny: Haeckel's ‘biogenetic law’ revisited. Curr Opin Genet Dev. 1995;5:628–639. doi: 10.1016/0959-437x(95)80032-8. [DOI] [PubMed] [Google Scholar]
- Tröbner W, Ramirez L, Motte P, Hue I, Huijser P, Lönnig W-E, Saedler H, Sommer H, Schwarz-Sommer Z. Globosa: a homeotic gene which interacts with deficiens in the control of Antirrhinumfloral organogenesis. EMBO J. 1992;11:4693–4704. doi: 10.1002/j.1460-2075.1992.tb05574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky ME, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM. The protein encoded by the Arabidopsis homeotic gene agamousresembles transcription factors. Nature. 1990;346:35–39. doi: 10.1038/346035a0. [DOI] [PubMed] [Google Scholar]