Abstract
Arabidopsis thaliana has three genes encoding type I 3-ketoacyl-CoA thiolases (KAT1, KAT2, and KAT5), one of which (KAT5) is alternatively transcribed to produce both peroxisomal and cytosolic proteins. To evaluate the potential importance of these four gene products, their evolutionary history in plants and their expression patterns in Arabidopsis were investigated. Land plants as a whole have gene lineages corresponding to KAT2 and KAT5, implying conservation of distinct functions for these two genes. By contrast, analysis of synteny shows that KAT1 arose by duplication of the KAT2 locus. KAT1 is found in the Brassicaceae family, including in the genera Arabidopsis, Capsella, Thellungiella (=Eutrema) and Brassica, but not in the more distantly related Caricaceae (order Brassicales), or other plants. Gene expression analysis using qRT-PCR and β-glucuronidase reporter genes showed strong expression of KAT2 during germination and in many plant tissues throughout the life cycle, consistent with its observed dominant function in fatty acid β-oxidation. KAT1 was expressed very weakly while KAT5 was most strongly expressed during flower development and in seedlings after germination. Isoform-specific qRT-PCR analysis and promoter β-glucuronidase reporters revealed that the two splicing variants of KAT5 have similar expression profiles. Alternative splicing of KAT5 to produce cytosolic and peroxisomal proteins is specific to and ubiquitous in the Brassicaceae, and possibly had an earlier origin in the order Brassicales. This implies that an additional function for KAT5 arose between 43 and 115 mybp. We speculate that this KAT5 mutation was recruited for a cytosolic function in secondary metabolism.
Key words: 3-ketoacyl-CoA thiolase, β-oxidation, Brassicales, evolution, flowering, germination, peroxisome
Introduction
Germination and seedling establishment in oilseed species such as Arabidopsis thaliana require peroxisomal β-oxidation to degrade the seed storage lipids that fuel this stage of development (Eastmond and Graham, 2001; Graham, 2008). As well as its role during seed germination, β-oxidation is a significant pathway for the synthesis of the hormones jasmonic acid (JA) and indole-3-acetic acid (IAA) in plants (Baker et al., 2006), and is required for the turnover of fatty acids in plant cells during development and senescence (Yang and Ohlrogge, 2009). Three core enzymes catalyse peroxisomal β-oxidation: acyl-CoA oxidase (ACX), multifunctional protein (MFP, which can exhibit hydratase, dehydrogenase, epimerase and isomerase activities), and L-3-ketoacyl- CoA thiolase (KAT).
The ACX and MFP gene families, represented by six and two genes, respectively, have been extensively characterized in Arabidopsis (Graham, 2008; Arent et al., 2010). By contrast, of the three KAT genes, only the role of KAT2 has been investigated in detail (Hayashi et al., 1998; Germain et al., 2001; Footitt et al., 2007a). Thiolases of two types are distinguished. Type 1 enzymes (KAT: EC 2.3.1.16) are typically peroxisomal and catalyse the thiolysis of acetyl-CoA units from the thiol end of the fatty acyl-CoA during fatty acid catabolism. Type 2 enzymes (acetyl-CoA acetyltransferase or ACAT; EC 2.3.1.9) are typically cytosolic and are involved in acetoacetyl CoA synthesis in the mevalonate biosynthesis pathway. The three KAT genes in Arabidopsis were designated as KAT1 (At1g04710), KAT2 (At2g33150), and KAT5 (At5g48880) based on the chromosome on which they are located (Germain et al., 2001). Subsequent analysis has determined that while both KAT1 and KAT2 encode single peroxisome targeted proteins, KAT5 encodes the cytosolic KAT5.1 and the peroxisomal KAT5.2 isoforms. The KAT5.2px transcript differs from KAT5.1cyt in the 5’ region, with an additional exon encoding a peroxisome targeting signal type 2 (PTS2), and alternate transcription and translation start sites (Carrie et al., 2007).
Genes involved in lipid mobilization, including β-oxidation, the glyoxylate cycle, and gluconeogenesis, are expressed co-ordinately during early seedling growth in Arabidopsis, with transcript levels and enzyme activities peaking at 48h after the commencement of germination (Rylott et al., 2001). Of the three Arabidopsis KAT genes, expression of KAT2 is dominant during this stage of the life cycle, with KAT2 transcript levels far more abundant than KAT1 and KAT5 (Germain et al., 2001; Kamada et al., 2003). Genes of lipid mobilization decline in expression level in the late stages of seedling establishment, a process associated with peroxisome matrix remodelling as the function of the organelle changes from one primarily concerned with oil mobilisation to metabolism associated with photosynthesis (Rylott et al., 2001; Kamada et al., 2003; Pracharoenwattana and Smith, 2008; Lingard et al., 2009). There is growing evidence for the functional importance of β-oxidation in reproductive tissue and seed development (Richmond and Bleecker, 1999; Rylott et al., 2003, 2006; Chia et al., 2005; Footitt et al., 2007b; Schilmiller et al., 2007). Expression of the Arabidopsis KAT genes has been detected in reproductive tissue (Kamada et al., 2003), however, detailed analysis has not been reported, nor have KAT5.1cyt and KAT5.2px transcript expression patterns been distinguished. To characterize the function of members of the KAT gene family further, the phylogenetic relationships of the genes in sequenced plant genomes was investigated and a detailed analysis of their patterns of expression in Arabidopsis thaliana was conducted. Comparative genomics has highlighted the dynamic expansion, specialization, and contraction of plant genomes (Wang et al., 2011; Rutter et al., 2012) and evolution of the KAT gene family in the Brassicaceae provides an excellent and specific example of this.
Materials and methods
Bioinformatics
KAT protein sequences were retrieved from the collection of sequenced plant genomes at Phytozome v8.0 (http://www.phytozome.org) and as individually submitted sequences deposited at NCBI (http://www.ncbi.nlm.nih.gov). Thellungiella parvula (Eutrema parvulum) KAT sequences were obtained from the Compare Genomes site (http://genomevolution.org/CoGe/index.pl). Sequence visualization and manipulations were done using the Geneious (v5.4.6) package (Drummond et al., 2011). Multiple sequence alignment of full-length predicted protein sequences was made in Geneious using the MAFFT Alignment plug-in (Katoh et al., 2002) and phylogenetic analysis of this alignment was done using the PHYML plugin (Guindon and Gascuel, 2003) with the WAG substitution model and 1000 bootstrap replicates. Synteny analysis was done using the CoGe and SynMap (http://genomevolution.org/CoGe/index.pl) comparative genome analysis tools (Lyons et al., 2008).
GUS promoter reporter analysis
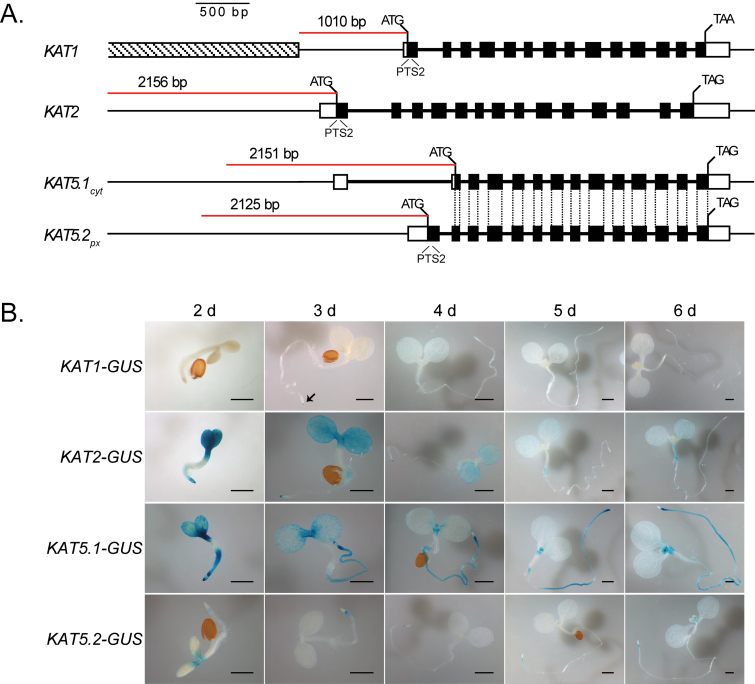
KAT gene promoter sequences, including the 5’-UTR and intergenic DNA upstream of the start codon, were amplified by PCR from wild-type Col-0 genomic DNA for Gateway cloning. Primer sequences are given in Supplementary Table S1 at JXB online. Amplified promoter fragments extended approximately 2kb upstream from the start codons (2156bp for the KAT2 promoter, 2151bp for KAT5.1cyt, and 2125bp for KAT5.2px) or until the adjacent gene (KAT1, 1010bp) as detailed in Figure 3A. Promoters were cloned into the GUS/GFP reporter plasmid pHGWFS7 (Karimi et al., 2002). Agrobacterium strain CV3130-C58 harbouring the pHGWFS7-KAT promoter vectors was used to transform Arabidopsis Col-0 plants using the floral dip method of Clough and Bent (1998). Transformed plants were screened for homozygosity over three generations by selection on hygromycin. Plants were grown for GUS staining under continuous light conditions to control for the diurnal regulation of promoter activity. Seedlings were grown on half-strength MS media (without sucrose). Flowers and siliques were removed from 6-week-old soil-grown plants and stained for GUS expression (Weigel and Glazebrook, 2002).
Fig. 3.
Activity of KAT:GUS promoter reporters during seedling establishment. (A) Thiolase gene structure and promoter regions used for reporter analysis. UTRs are indicated by white boxes, coding sequences by black boxes, introns by thick black lines, and upstream promoter regions by thin lines. The red line shows the region of promoter cloned upstream of the start codon. In the case of KAT1, the gene adjacent to the promoter is indicated by the shaded box. The common coding regions and the alternative start codons between KAT5.1cyt and KAT5.2px are indicated and emphasize the differences between the 5’ ends of the alternative transcripts. (B) GUS expression in Col-0 seedlings. Seeds of thiolase promoter-GUS reporter lines were stratified then seedlings grown in continuous light on half-strength MS media. Seedlings were stained for GUS activity from 2–6 d after transfer to the light. The length of the scale bar is 0.5mm.
Quantitative RT-PCR
Aerial tissue samples (about 100mg) were taken from 5-week-old Arabidopsis Col-0 plants grown in soil under continuous light conditions, and root samples were taken from hydroponically grown plants. Germinating seed samples utilized approximately 20mg of dry seed that had been spread on plates containing half-strength MS media then stratified for 48h before being transferred to continuous light. Samples were ground into a fine powder with a mortar and pestle pre-cooled with liquid nitrogen. Germinating seed RNA was extracted using the RNAqueous Kit with Plant RNA Isolation Aid (Ambion) and included LiCl precipitation. RNA from other plant tissues was isolated using an Aurum (Bio-Rad) kit. RNA was treated with Turbo DNA-free DNase (Ambion) and 1 µg used as the template for cDNA synthesis using the iScript cDNA Synthesis Kit (Bio-Rad). cDNAs were diluted 1/5 for quantitative PCR. Primer pairs for qRT-PCR that spanned introns were designed using Primer3. Due to the similarity of the KAT5.1cyt and KAT5.2px sequences, primers were designed in the respective 5’ UTRs and therefore could not bound introns. Primer sequences are listed in Supplementary Table S1 at JXB online.
qRT-PCR was performed on a Roche LC480. The reaction volume was 5 µl and included 1× LightCycler 480 SYBR Green I Master (Roche), 0.5 µl diluted cDNA, and 0.1 µl of 20 µM primers. Cycle conditions were: 95 °C for 10min; 45 cycles of 95 °C for 20 s, 60 °C for 20 s, and 72 °C for 20 s. Melt curve analysis of real-time PCR products was performed to verify amplification of a single product. Crossing point values were calculated under high confidence. Four biological replicates per tissue sample were examined, with at least two technical replicates of each real-time PCR. The average crossing point value of two technical replicates was used to calculate expression relative to an internal reference gene adjusted by primer efficiencies. Two reference genes were tested: ACT2 (At3g18780) and the clathrin adaptor complex subunit (CACS; At5g46630), identified by Czechowski et al. (2005) as a more suitable reference gene for transcript normalization. For analysis, CACS was used for normalization; the average relative expression and standard error for the four biological replicates are shown.
Results
KAT2 and KAT5 isoforms are conserved in higher plants
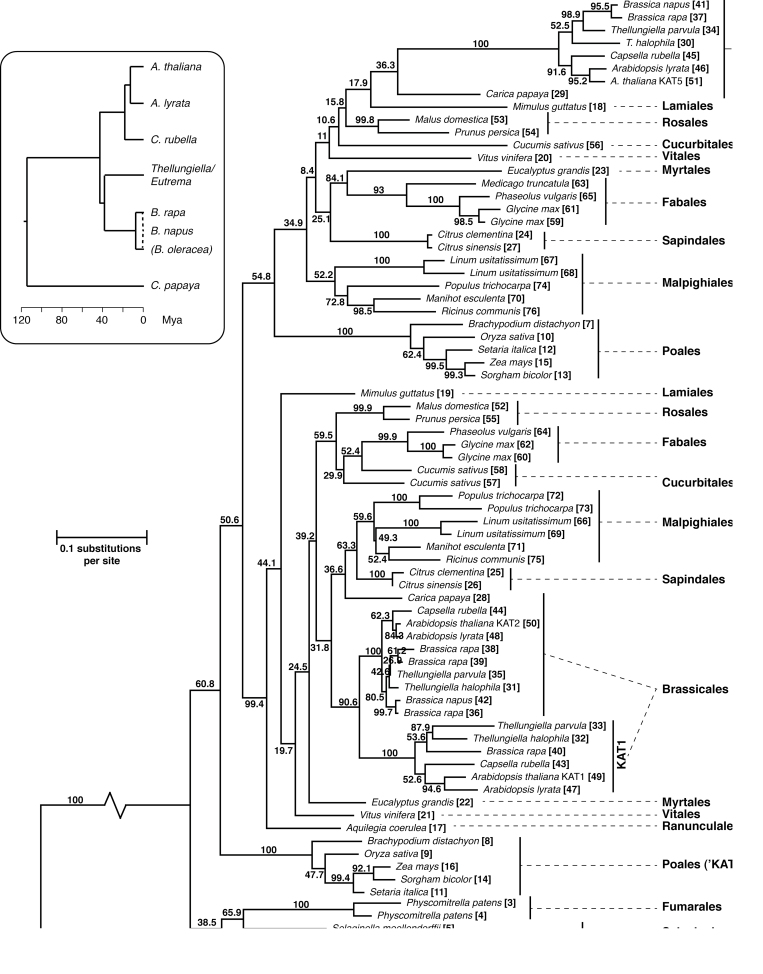
KAT protein sequences were obtained from Phytozome v8.0 to investigate the evolutionary history of the gene family. The Phytozome database includes genome sequences of 31 plant species that encompass major clades of plant (Viridiplantae) evolution, including green algae, mosses, spikemosses (lycophytes), monocot grasses, eudicots, and dicots. Interrogation of this database using Arabidopsis KAT2 (At3g33150) in a BLAST query yielded 82 protein sequences. This number was reduced to 71 hits (see Supplementary Table S2 at JXB online) after exclusion of proteins with obviously poor matches and partial alignments. KAT sequences from Brassica napus and Thellungiella parvula (which are not represented in Phytozome v8.0) were obtained from NCBI and CoGe, respectively, to yield a total of 76 KAT protein sequences from 33 species.
There was at least one KAT isoform encoded in each of the 33 genomes, with the majority (29/33) encoding two or more (see Supplementary Table S2 at JXB online). Notably, both green alga species (Volvox carteri and Chlamydomonas reinhardtii) only possessed a single gene. The exon–intron structure of the genes is highly conserved in land plants, with almost all of the KAT coding sequences from higher plants being assembled from 14 exons that span similar regions of the protein in each species (see Supplementary Table S2 at JXB online). In land plants, β-oxidation is a peroxisomal process and KAT proteins would thus be expected to localize to that organelle. Accordingly, 74/76 proteins possess a predicted peroxisome targeting signal 2 (PTS2) close to the N-terminus, the only exceptions being one of the two Selaginella moellendorffii isozymes (see Supplementary Table S2 at JXB online, Taxon #6) and C. reinhardtii, in which the KAT protein includes an incomplete PTS2 (RLAVLSRQF) (see Supplementary Table S2 at JXB online, Taxon #2). C. reinhardtii (and some other chlorophytes) have previously been noted to have peroxisomes that, although capable of PTS1 and PTS2-mediated import (Hayashi and Shinozaki, 2012), differ significantly from those in other plant lineages in lacking both catalase, which is found in their mitochondria, and glycolate oxidase, which is functionally replaced by a mitochondrial glycolate dehydrogenase (Kato et al., 1997; Atteia et al., 2009). In addition, a number of enzymes of β-oxidation have been identified in the C. reinhardtii mitochondrial proteome, including KAT, an acyl-CoA dehydrogenase (ACAD; EC 1.3.99.3), an enoyl-CoA hydratase (E-CoAH1; EC 4.2.1.17), but not acyl activating enzymes or acyl-CoA oxidases (Atteia et al., 2009).
The amino acid multiple sequence alignment of plant KAT protein sequences was analysed to generate a maximum likelihood phylogenetic tree (Fig. 1). Although many other plant thiolases are available via Genbank, the analysis was restricted to those from sequenced genomes available at Phytozome and CoGe so as to obtain the best picture of KAT diversity within and between species. The only exception to this was the inclusion of the two KAT sequences available from B. napus for which the full genome sequence is not yet available that were included due to their special relevance to evolution of the family in the Brassicales. The tree was rooted with the KATs from the green algae V. carteri and C. reinhardtii. KAT sequences of higher plants were divided into two relatively well-supported clades (Fig. 1). These clades were denoted KAT2 and KAT5 corresponding to the Arabidopsis KAT isozyme they contained. The taxonomic orders of the species are indicated on the tree for clarity. KAT proteins from the Poales formed two discrete clusters, one embedded in the KAT5 clade, and the second resolved basal to KATs of all angiosperms. We nominally label this later as KAT2 representatives of the Poales. Thus, of the 29 species of higher plants, 27 were represented by two KAT isoforms corresponding to KAT2 and KAT5. The two exceptions (Medicago truncatula and Aquilegia coerulea; see Supplementary Table S2 at JXB online) have only one annotated KAT gene. Whether the absence of another KAT gene from these genomes is genuine or due to incomplete sequencing or annotation remains to be determined. Interestingly, both moss genomes also encode two KAT proteins, but unlike those of higher plants these were not phylogenetically separated on the tree into KAT2 or KAT5 isoforms.
Fig. 1.
Phylogenetic analysis of the plant 3-ketoacyl-CoA thiolase (KAT) family proteins. Multiple sequence alignment of full-length KAT protein sequences predicted from sequenced plant genomes was made using MAFFT (Katoh et al., 2002) and phylogeny estimated using PHYML (Guindon and Gascuel, 2003). Numbers at the nodes are per cent bootstrap agreement from 1000 replicates. Major clades are named after similarity to Arabidopsis thaliana KAT2 (At2g33150) and KAT5 (At5g48880) isozymes and taxonomic order of the species is annotated to the right. The length of the branch connecting the ancestral (Volvocales) KATs has been shortened by 50%. Inset: The evolutionary history and timing of Brassicales species represented in the main tree, derived from Beilstein et al. (2010), Dassanayake et al. (2011), and Mun et al. (2009).
KAT1 is a duplication of KAT2 unique to the Brassicaceae lineage
The Brassicales appear to have experienced further duplication of KAT genes. The KAT2 cluster (Fig. 1) also contains six species (Arabidopsis thaliana, A. lyrata, Capsella rubella, Thellungiella halophila, T. parvula, and Brassica rapa) with a third KAT gene (KAT1) in addition to the KAT2 and KAT5 paralogues (see Supplementary Table S2 at JXB online). AtKAT1 does not appear to have orthologues other than in the Brassicaceae, suggesting that it is a duplication of KAT2 peculiar to that lineage. Indeed, there is significant synteny between the regions of A. thaliana chromosomes 1 and 2 surrounding the KAT1 and KAT2 genes. A fragment of about 2.2 Mbp on chromosome 2 shows nearly complete correspondence of genes with span of 1.3 Mbp on chromosome 1 (see Supplementary Fig. S1A at JXB online). It is likely that the duplication between chromosomes 1 and 2 was followed by a localized segmental inversion on one of the chromosomes, because about 170kb is inverted with respect to the rest of the duplicated region (see Supplementary Fig. S1B at JXB online). In contrast, there is no synteny between the AtKAT2 or AtKAT1 genomic regions and that of AtKAT5 (not shown).
These syntenic chromosome arrangements are also found in A. lyrata, T. parvula, and C. rubella around genes corresponding to KAT1 and KAT2 (not shown). BrKAT1 (B. rapa) is likely to have had the same origin at AtKAT1, but the situation is less clear because the Brassica lineage underwent whole genome triplication after the split from the Arabidopsis lineage (Mun et al., 2009). There are three clear orthologues of AtKAT2 in the B. rapa genome (Fig. 1; see Supplementary Table S2 at JXB online). Genomic regions around these B. rapa KAT2 genes display significant synteny with each other and with the AtKAT2 genomic region (see Supplementary Table S3 at JXB online). In addition, there are three other regions displaying synteny with these, only one of which has retained a KAT gene. This locus, accession Bra030586 (see Supplementary Table S2 and S3 at JXB online) groups with AtKAT1 in the phylogenetic analysis (Fig. 1). These latter three regions correspond to approximately the same genes duplicated between KAT2 and KAT1 in A. thaliana (see Supplementary Fig. S1B at JXB online). This evidence suggests that the duplication occurred before the Brassica–Arabidopsis split (and also before whole genome triplication in Brassica). It is also interesting to note that there are six regions in the B. rapa genome that display synteny with the AtKAT5 region (see Supplementary Table S3 at JXB online). All correspond to approximately the same span of the A. thaliana genome (~620 Mbp), but in B. rapa only one of the six regions has retained a KAT5 gene. There are no such regions of synteny between CpKAT2 and other genomic regions in Carica papaya, a more distantly related species in the order Brassicales. The separation of Brassicaceae, which have maintained a KAT1 orthologue, from Caricaceae, which have not, can be dated to between 43 and 115 mybp (see inset, Fig. 1); these ages thus provide the upper and lower bounds for the timing of the segmental duplication.
Dual targeting of KAT5 is conserved in Brassicales
AtKAT5 is alternately transcribed to produce two species of mRNA, KAT5.1cyt and KAT5.2px (Fig. 3A). These possess alternative start (Met) codons that direct the encoded proteins to the peroxisome and the cytosol, respectively (Carrie et al., 2007). Further investigation of the plant KAT gene family revealed that all KAT5 orthologues from the Brassicales (represented by two families in Phytozome; Brassicaceae and Caricaceae), but not other plant orders, have potential alternative start codons in the same relative positions (Fig. 2). Thus, A. thaliana, A. lyrata, Capsella rubella, T. halophila, T. parvula, B. rapa, and B. napus (Brassicaceae) and Carica papaya (Caricaceae) KAT5 orthologues putatively encode KAT5.1 and KAT5.2 isoforms. Examination of PASA assembled ESTs (via Phytozome v8.0) provided, where data were available, evidence for alternate transcription of KAT5 for each of these species (see Supplementary Table S2 at JXB online).
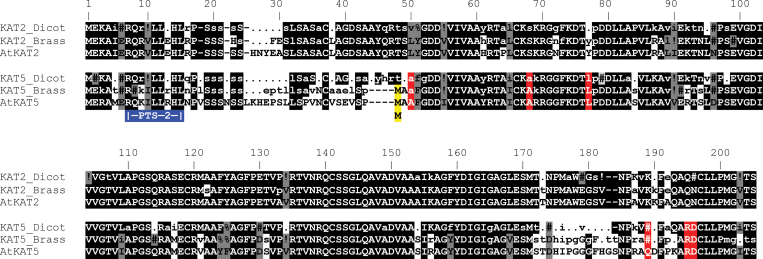
Fig. 2.
Alignment of dicotyledonous plant thiolase proteins. Consensus sequences were derived from four groups of sequences comprising dicotyledonous KAT2 or KAT5 and Brassicales KAT2 or KAT5 sequences. AtKAT2 and AtKAT5 are included as samples of specific sequences. Typical plant KAT proteins are about 460 amino acids; the first approximately 200 residues are shown. Highly conserved residues (present in >90% of sequences) in each of the four consensus sequences are shown in capital letters, while moderately conserved residues (present in 50–90% of sequences) in lower case. Non-conserved positions are depicted by a period (.) and gaps introduced into the alignment by a dash (-). Symbols used to indicate residues with strongly similar properties on the Gonnet PAM 250 matrix are: # (N, D, Q or E); % (F or Y);! (I or V). Residues conserved between the different consensus sequences are highlighted with black shading and grey shading indicates consensus sequence residues that are not identical but have similar properties. The red highlighting indicates residues that clearly distinguish KAT5. The blue box shows the PTS2 near the N-terminus of the proteins and the yellow highlighting the alternative initial Met that is conserved in all Brassicales KAT5 proteins.
Examination of alignments of plant thiolase proteins highlights differences between KAT2 and KAT5 and between KAT5s of Brassicales and other dicotyledonous species (Fig. 2). Groups of Brassicales KAT sequences that were identified as KAT2 or KAT5 in Fig. 1 were aligned separately using Multalin (http://multalin.toulouse.inra.fr/multalin/multalin.html). Similarly, other dicotyledonous plant KAT2 or KAT5 sequences were aligned (i.e. excluding those from Brassicales) and the four resulting consensus sequences compared (Fig. 2). After the conserved PTS2 (residues 7–15 in Fig. 2), all taxa have a linker that shows little conservation (residues 25–30). The sequence following this (residues 31–51) is highly conserved in all KAT2 proteins and less so in KAT5. Moreover, Brassicales KAT5s differ substantially from the KAT2s and the KAT5s of other dicots in this region and it includes the alternate Met described above and a four amino acid deletion relative to all other KAT proteins. Interestingly, although C. papaya (a basal Brassicales taxon) possesses the alternate Met in its KAT5 protein, it does not have the four amino acid deletion and the sequence surrounding the second Met is more similar to the typical KAT5 (and KAT2) consensus (not shown). Unfortunately, evidence for or against alternative transcription of CpKAT5 is not available because the EST collection is small and does not include the 5’ ends of the transcripts. A second region that distinguishes between KAT2 and KAT5 isoforms is found in residues 173–192. Here, KAT2 sequences are highly conserved and there is little conservation in the KAT5 sequences (Fig. 2).
Based on protein sequence similarity and EST data, we suggest that alternative transcription and protein localization of KAT5 isoforms is unique to the order Brassicales and originated with a mutation to make a second Met in a lineage ancestral to C. papaya (i.e. at least 115 mybp; Fig. 1) followed by further mutation and deletions between 115 and 43 mybp to yield the typical Brassicales KAT5 sequence.
Patterns of KAT gene expression by promoter::GUS reporters
Eleven KAT1-GUS, nine KAT2-GUS, ten KAT5.1-GUS, and ten KAT5.2-GUS independent transformants were obtained and screened for GUS activity. Lines giving GUS staining patterns representative of the group were selected for more detailed analysis. The promoter-reporter construct KAT5.2-GUS includes the KAT5 promoter up to the first start codon joined to the GUS reporter gene; KAT5.1-GUS includes the 5’ UTR and first exon of KAT5.2 and thus includes the full putative promoter that might be expected to drive independent transcription of KAT5.1cyt (Fig. 3A).
GUS expression in 2–6-d-old seedlings indicated low levels of expression for KAT1-GUS and KAT5.2-GUS, with higher levels for KAT2-GUS and KAT5.1-GUS (Fig. 3B). In KAT1-GUS lines, activity was nearly absent but some GUS staining was observed in the root tip (indicated by the arrow in Fig. 3B). KAT2-GUS was expressed strongly in the cotyledons and hypocotyls, as well as in the root tip, with this activity diminishing significantly by day four. KAT5.1-GUS expression was observed in the cotyledons, but appeared to be stronger near the apical meristem and in the root at day two. Expression in apical parts and the root tip diminished significantly during the five days analysed, but was maintained in the rest of the root. KAT5.1-GUS activity was also observed in the new leaves forming at the meristem. KAT5.2-GUS had lower expression levels with some staining of the petioles and also just behind the root tip during the first three days of germination. Seedlings grown for 16 d on MS medium were also stained for GUS (see Supplementary Fig. S2 at JXB online). Evidence for promoter activity was not seen in KAT1-GUS lines. KAT2-GUS and KAT5.1-GUS were expressed in the roots, petioles, and new leaves of the rosette and KAT5.2-GUS promoter activity was mostly limited to petioles (see Supplementary Fig. S2at JXB online). Collectively, these results suggest that there are genomic elements in the region between the two KAT5 start codons that contribute significantly to transcription of the gene and that potentially drive expression of the isoforms to different levels.
KAT2 and KAT5 show differential expression patterns in reproductive tissue
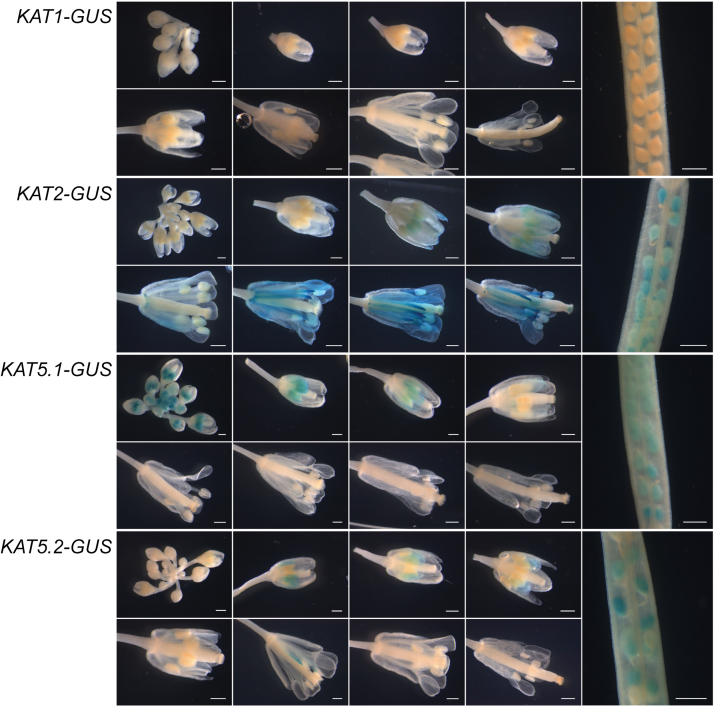
In flowers, as in seedlings, KAT1-GUS promoter expression was essentially absent at all stages (Fig. 4). KAT2-GUS expression was absent during early flower development, and was first observed at early stages in young anthers and petals, corresponding to stage 12 of flower development. As the flowers matured, strong staining was observed in the anther filament and petals as well as the tip of the gynoecium and developing ovules. KAT5.1-GUS reported activity from the earliest stages of flower development, primarily in anthers, but was not present beyond stage 12 of flower development. Staining in KAT5.2-GUS lines was seen during the middle stages (~9–12) of flower development and was largely absent at the later stages. Promoter analysis in silique tissue indicated activity in the developing seeds for KAT2-GUS, KAT5.1-GUS, and KAT5.2-GUS, but not for the KAT1-GUS lines (Fig. 4).
Fig. 4.
KAT:GUS promoter reporter activity in flowers and siliques. Flowers and siliques were removed from soil-grown KAT:GUS reporter plants at various stages of flower and silique development, and stained for GUS activity. The length of the scale bar is 0.5mm.
Publicly available Arabidopsis thiolase gene expression data retrieved from the BAR eFP browser (http://bar.utoronto.ca/) (Winter et al., 2007) broadly agrees with the experimental data from promoter analysis (see Supplementary Fig. S3 at JXB online). KAT1 transcript levels were uniformly low. KAT2 transcript abundance vastly exceeded KAT1 and KAT5, with expression highest in germinating seeds (and also in dry seed and senescent leaves; see Supplementary Fig. S3A, B at JXB online). In flowers and developing seed, KAT2 expression was initially relatively low, and increased in maternal tissues from about stage 12 of flower development. KAT2 expression increases appreciably in the style once developing seeds have passed the torpedo stage (see Supplementary Fig. S3C, D, E at JXB online). As seen in the GUS reporter lines, KAT5 transcript abundance follows a complementary distribution to KAT2, with expression highest during early stages of flower and seed development (see Supplementary Fig. S3C, D, F at JXB online). In terms of the total abundance of transcript, KAT5 is substantially lower in most tissues (see Supplementary Fig. S3A, B at JXB online). However, KAT5 expression level matches KAT2 in early flower and seed development, after which KAT2 transcript abundance increases several-fold while KAT5 decreases substantially (see Supplementary Fig. S3D at JXB online).
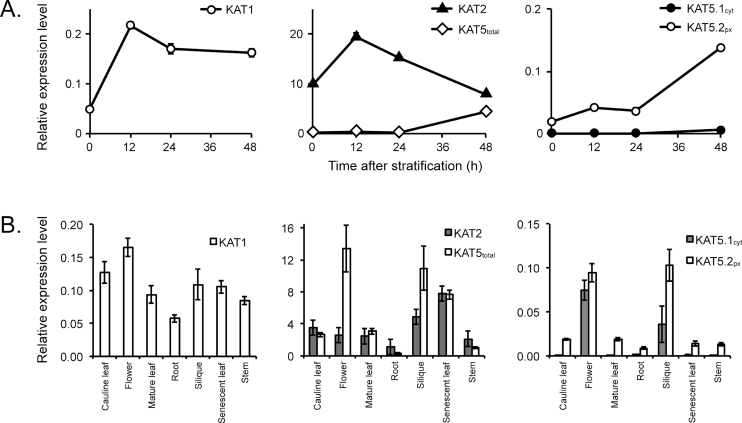
As data derived from microarray analysis does not allow differentiation between KAT5.1cyt and KAT5.2px transcripts, qRT-PCR was used to detail the microarray expression data further (Fig. 5). Firstly, in germinating seeds (Fig. 5A), the KAT2 expression level was high and spiked 12h after stratification but decreased again by the time seeds began to germinate at 48h. KAT1 expression was much lower, but followed a similar pattern to KAT2. KAT5 expression was examined by use of primers located towards the 3’ end of the transcript, which assessed total transcript level (KAT5total), and by two alternative primer pairs at the 5’ end that were able to discriminate KAT5.1cyt and KAT5.2px. KAT5total levels, initially very low, began to rise significantly by 48h when seeds were germinating. KAT5.2px followed a similar pattern to KAT5total and KAT5.1cyt remained barely detectable during the first 48h of germination (Fig. 5A). In tissues of mature plants, KAT1 expression was low and relatively uniform (Fig. 5B). KAT2 and KAT5total were expressed at comparable levels in most tissues, although KAT5total expression exhibited significantly higher peaks in flowers and siliques. KAT5.2px transcript was expressed at low levels in most plant tissues, with substantially higher expression in flowers and siliques, and KAT5.1cyt was expressed exclusively in the flowers and siliques (Fig. 5B). Together with the GUS reporter and microarray data, these observations suggest a role for both KAT2 and KAT5 in fertility, in both flowers and siliques, with a role for KAT5 primarily during early flower and embryo development and KAT2 during seed filling and maturation.
Fig. 5.
Quantitative RT-PCR analysis of KAT transcript abundance in Arabidopsis thaliana. (A) KAT transcript abundance during germination. (B) Relative expression of KAT transcripts in different plant organs. The abundance of KAT1, KAT2, KAT5total, KAT5.1cyt. and KAT5.2px transcripts was measured and normalized to reference gene (CACS) transcript levels. Error bars represent standard error from the mean (n=4).
Discussion
Evolution of KAT genes
Ancient Viridiplantae taxa such as green algae have a single KAT gene, whereas almost all sequenced land plant genomes (mosses and higher plants) possess at least two KAT genes. Phylogenetic analysis of predicted KAT protein sequences suggests that duplication of KAT genes may have been associated with the adoption of a land habitat and that two KAT genes have been selectively maintained ever since. In angiosperms the encoded proteins cluster into two groups that may be defined by their similarity to A. thaliana KAT2 and KAT5. Where a genome encodes three or more KAT proteins, the extra isozymes cluster with the respective KAT2 or KAT5 from that species suggesting that they are more recent lineage-specific duplications (Fig. 1). For example, in the KAT2 clade, Populus trichocarpa, Linum usitatissimum, and Glycine max show duplicates clustered together, and there are three closely related Brassica rapa KAT2 proteins. KAT1 in the Brassicaceae appears to be an older duplication in that it was found in six species from four Brassicaceae genera (Arabidopsis, Capsella, Thellungiella, and Brassica) and there is extensive synteny between genomic sequences surrounding KAT1 and KAT2 in these species (see Supplementary Fig. S1 and Table S3 at JXB online). Despite its relatively high rate of divergence (branch lengths between KAT1s in Fig. 1 are long compared with those between KAT2s in the Brassicales), and very low levels of expression (Figs 3–5; see Supplementary Figs S2 and S3 at JXB online), KAT1 has persisted in the Brassicaceae for at least 43 million years suggesting a selective advantage in maintaining this gene.
Roles for thiolases during the plant life cycle
In A. thaliana, both KAT2 and KAT5 are expressed in distinct (although not mutually exclusive) patterns. KAT2 is dominant during seed germination and its expression is co-ordinated with that of other β-oxidation genes, including MFP2, PXA1, ACX1, LACS6, and LACS7 (Fulda et al., 2004), during the first 2–3 d of seedling establishment when the bulk of storage lipid breakdown occurs. Indeed, the kat2-1 mutant can germinate, but does not establish unless supplied with sugar in the medium (Germain et al., 2001). KAT2 expression is high throughout the life cycle with other peaks of expression during later stages of flower and seed development, and in senescence. By comparison, KAT5 expression is relatively minor during germination and strongest in young flowers and early in seed development.
Strong expression of KAT2 and KAT5 in inflorescences and siliques (Figs 4, 45; see Supplementary Fig. S3 at JXB online) and compromised reproductive success in mutants of core β-oxidation genes including kat2, cts, acx1 acx5, and lacs6 lacs7 (Footitt et al., 2007a, b; Schilmiller et al., 2007) suggests a role for β-oxidation in reproduction. β-oxidation potentially contributes to reproductive success via its roles in fatty-acid turnover and/or synthesis of the hormones JA and IAA. Auxin can be synthesized via tryptophan-dependent and -independent pathways, and can also be derived from stored forms; either IAA conjugates (amino acid, sugar or peptide), or from indole-3-butyric acid (IBA) via one cycle of β-oxidation (Baker et al., 2006). Similarly, JA biosynthesis involves the precursor, 3-oxo-2-(2’-[Z]-pentenyl)-cyclopentane-1-octanoic acid (OPC:8), undergoing three cycles of β-oxidation to produce JA (Baker et al., 2006).
Many mutants disrupted in JA synthesis or signalling (including acx1 acx5) display male sterility, usually in the form of defective pollen development, that can be rescued by exogenously supplied JA (Schilmiller et al., 2007). kat2 mutants, however, are not sterile, presumably due to compensation of KAT function by KAT5 (Afitlhile et al., 2005). KAT2 appears to be the dominant thiolase in IBA metabolism: kat2 mutant seedlings have an IBA-resistant phenotype while kat1 and kat5 knockouts do not (Wiszniewski et al., 2009). It is likely that β-oxidation of IBA to IAA contributes significantly to levels of free IAA in seedlings (Strader et al., 2011). In flowers, IAA generated in the stamens controls flower development by promoting anther growth and suppressing petal development (Aloni et al., 2006). Anther elongation in cts and kat2-1 mutants is inhibited, but it can be rescued by exogenous supply of 1-naphthaleneacetic acid (NAA; a synthetic auxin) or IAA (but not by JA) (Footitt et al., 2007b). High KAT2 expression in anther filaments and KAT5 expression in anthers (Fig. 4) is consistent with an essential role for β-oxidation-mediated hormone metabolism in flower development.
During fertilization, pollen tube growth is extremely rapid and lipids may provide some of the energy required to drive this growth. Lipid bodies and peroxisomes accumulate in developing and mature Arabidopsis (Kuang and Musgrave, 1996) and olive (Rodríguez-García et al., 2003) pollen. They then diminish in number during pollen tube growth. In vitro germination tests of pollen from kat2-1 and cts mutants revealed reduced germination frequency and shorter pollen tube length, but these could be rescued by exogenous sucrose supply, suggesting lipid degradation via β-oxidation may contribute to the growth of germinating pollen tubes in vivo (Footitt et al., 2007b).
β-oxidation is also implicated during seed maturation. While fatty acid accumulation in developing embryos originates maternally as photosynthetically fixed carbon, β-oxidation of stored oil may provide respiratory substrates following the breaking of the trophic connection with maternal tissue (Baud et al., 2002). In fact, a 10% decrease in seed oil content has been observed to occur in B. napus embryos late in their development (Eastmond and Rawsthorne, 2000) and β-oxidation, the glyoxylate cycle, and gluconeogenesis are all active in developing embryos (Eastmond and Graham, 2001; Chia et al., 2005). A lower respiration rate in kat2-1 mutant ovules compared with the wild type suggests that β-oxidation is important for carbon flow into sugars via gluconeogenesis and respiration (Footitt et al., 2007a). This is corroborated by strong expression of KAT2 and KAT5 in silique tissue (Figs 4, 45; see Supplementary Fig. S3 at JXB online) implying a role for both thiolases in reproductive success.
A specific function for thiolases in Brassicales?
The apparent coincident origin and maintenance in the Brassicales (but not other plant orders) of a third KAT gene (KAT1; Fig. 1) and a KAT5 orthologue that is alternatively transcribed to produce cytosol- and peroxisome-targeted proteins (Fig. 2) raises the question as to whether these events were related and facilitated a specific function for thiolases in Brassicales. The first product of the phenylpropanoid pathway, trans-cinnamic acid, represents a branch point to either flavonoids or benzenoid metabolism (Boatright et al., 2004). Intriguingly, KAT5 coexpresses with genes of flavonoid metabolism (Carrie et al., 2007). Benzoic acid (BA) synthesis from trans-cinnamic acid can occur cytosolically or in peroxisomes. In the first case, hydration is followed by retro-aldol cleavage (to release an acetic acid molecule) and dehydrogenation of benzaldehyde to yield BA. BA can then be activated to form benzoyl-CoA. In peroxisomes, CoA-activated trans-cinnamic acid undergoes one cycle of β-oxidation: hydration followed by dehydrogenation and KAT-mediated thiolysis to yield acetyl-CoA and benzoyl-CoA. Both of these pathways have been shown in petunia to contribute to the plant BA pool (Boatright et al., 2004). Mutations that reduce thiolase activity in flowers also result in reduced BA or benzoyl-CoA accumulation. For example, silencing of the Petunia hybrida PhKAT1 results in a significant decrease in BA and volatile benzenoid compound production in petunia petals (Moerkercke et al., 2009). Arabidopsis chy1 knockout mutants exhibit greatly reduced KAT activity (Lange et al., 2004) and chy1 mutant seeds are deficient in BA and BA-containing glucosinolates (benzoyloxyglucosinolates) (Ibdah and Pichersky, 2009). As glucosinolate synthesis, including benzyl- and benzoyloxy-glucosinolates, is essentially a cytosolic process, a cytosolic KAT5 may have been co-opted in the Brassicales for benzenoid metabolism. We suggest that future research should address KAT function in secondary metabolism including benzoyloxyglucosinolate and flavonoid synthesis.
The complement, structure, and expression of three KAT genes in the Brassicaceae has survived genome expansion and contraction in the lineage and persisted through millions of years. In the same time span, plant metabolism has undergone some major changes including the evolution of C4 photosynthesis. This implies that KAT gene specialization has a fundamentally important function.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. S1. Synteny of AtKAT2 and AtKAT1 chromosomal regions.
Supplementary Fig. S2. Activity of KAT promoter reporters visualised by GUS staining in 16-d-old seedlings.
Supplementary Fig. S3. KAT transcript abundance in Arabidopsis tissues.
Supplementary Table S1. Primers used in this study.
Supplementary Table S2. Species and gene identifiers used in the phylogenetic tree.
Supplementary Table S3. Synteny of Arabidopsis thaliana KAT genomic regions in the Brassica rapa genome.
Acknowledgements
This work was supported by the Australian Research Council [Grant numbers FF0457721 and CE0561495]; the Western Australian Government’s Centre of Excellence Program; and an Australian Postgraduate Award to AAGW.
References
- Afitlhile M, Fukushige H, Nishimura M, Hildebrand D. 2005. A defect in glyoxysomal fatty acid β-oxidation reduces jasmonic acid accumulation in Arabidopsis Plant Physiology and Biochemistry 43 603–609 [DOI] [PubMed] [Google Scholar]
- Aloni R, Aloni E, Langhans M, Ullrich CI. 2006. Role of auxin in regulating Arabidopsis flower development Planta 223 315–328 [DOI] [PubMed] [Google Scholar]
- Arent S, Christensen CE, Pye VE, Norgaard A, Henriksen A. 2010. The multifunctional protein in peroxisomal β-oxidation: structure and substrate specificity of the Arabidopsis thaliana protein MFP2 Journal of Biological Chemistry 285 24066–24077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atteia A Adrait A Brugiere S et al. 2009. A proteomic survey of Chlamydomonas reinhardtii mitochondria sheds new light on the metabolic plasticity of the organelle and on the nature of the α-proteobacterial mitochondrial ancestor Molecular Biology and Evolution 26 1533–1548 [DOI] [PubMed] [Google Scholar]
- Baker A, Graham IA, Holdsworth M, Smith SM, Theodoulou FL. 2006. Chewing the fat: β-oxidation in signalling and development Trends in Plant Science 11 124–132 [DOI] [PubMed] [Google Scholar]
- Baud S, Boutin J-P, Miquel M, Lepiniec L, Rochat C. 2002. An integrated overview of seed development in Arabidopsis thaliana ecotype WS Plant Physiology and Biochemistry 40 151–160 [Google Scholar]
- Beilstein MA, Nagalingum NS, Clements MD, Manchester SR, Mathews S. 2010. Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana Proceedings of the National Academy of Sciences USA 107 18724–18728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boatright J, Negre F, Chen X, Kish CM, Wood B, Peel G, Orlova I, Gang D, Rhodes D, Dudareva N. 2004. Understanding in vivo benzenoid metabolism in petunia petal tissue Plant Physiology 135 1993–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrie C, Murcha MW, Millar AH, Smith SM, Whelan J. 2007. Nine 3-ketoacyl-CoA thiolases (KATs) and acetoacetyl-CoA thiolases (ACATs) encoded by five genes in Arabidopsis thaliana are targeted either to peroxisomes or cytosol but not to mitochondria Plant Molecular Biology 63 97–108 [DOI] [PubMed] [Google Scholar]
- Chia TYP, Pike MJ, Rawsthorne S. 2005. Storage oil breakdown during embryo development of Brassica napus (L.) Journal of Experimental Botany 56 1285–1296 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana The Plant Journal 16 735–743 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W-R. 2005. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis Plant Physiology 139 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassanayake M Oh DH Haas JS et al. 2011. The genome of the extremophile crucifer Thellungiella parvula Nature Genetics 43 913–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ Ashton B Buxton S et al. 2011. Geneious v5.4. Available from http://www.geneious.com/
- Eastmond PJ, Graham IA. 2001. Re-examining the role of the glyoxylate cycle in oilseeds Trends in Plant Science 6 72–78 [DOI] [PubMed] [Google Scholar]
- Eastmond PJ, Rawsthorne S. 2000. Coordinate changes in carbon partitioning and plastidial metabolism during the development of oilseed rape embryos Plant Physiology 122 767–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Footitt S, Cornah JE, Pracharoenwattana I, Bryce JH, Smith SM. 2007a The Arabidopsis 3-ketoacyl-CoA thiolase-2 (kat2-1) mutant exhibits increased flowering but reduced reproductive success Journal of Experimental Botany 58 2959–2968 [DOI] [PubMed] [Google Scholar]
- Footitt S, Dietrich D, Fait A, Fernie AR, Holdsworth MJ, Baker A, Theodoulou FL. 2007. b The COMATOSE ATP-binding cassette transporter is required for full fertility in Arabidopsis Plant Physiology 144 1467–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulda M, Schnurr J, Abbadi A, Heinz E, Browse J. 2004. Peroxisomal acyl-CoA synthetase activity is essential for seedling development in Arabidopsis thaliana The Plant Cell 16 394–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain V, Rylott EL, Larson TR, Sherson SM, Bechtold N, Carde JP, Bryce JH, Graham IA, Smith SM. 2001. Requirement for 3-ketoacyl-CoA thiolase-2 in peroxisome development, fatty acid β-oxidation and breakdown of triacylglycerol in lipid bodies of Arabidopsis seedlings The Plant Journal 28 1–12 [DOI] [PubMed] [Google Scholar]
- Graham IA. 2008. Seed storage oil mobilization Annual Review of Plant Biology 59 115–142 [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood Systematic Biology 52 696–704 [DOI] [PubMed] [Google Scholar]
- Hayashi M, Toriyama K, Kondo M, Nishimura M. 1998. 2,4-Dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid beta-oxidation The Plant Cell 10 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Shinozaki A. 2012. Visualization of microbodies in Chlamydomonas reinhardtii Journal of Plant Research 125 579–586 [DOI] [PubMed] [Google Scholar]
- Ibdah M, Pichersky E. 2009. Arabidopsis Chy1 null mutants are deficient in benzoic acid-containing glucosinolates in the seeds Plant Biology 11 574–581 [DOI] [PubMed] [Google Scholar]
- Kamada T, Nito K, Hayashi H, Mano S, Hayashi M, Nishimura M. 2003. Functional differentiation of peroxisomes revealed by expression profiles of peroxisomal genes in Arabidopsis thaliana Plant and Cell Physiology 44 1275–1289 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. 2002. GATEWAY vectors for Agrobacterium-mediated plant transformation Trends in Plant Science 7 193–195 [DOI] [PubMed] [Google Scholar]
- Kato J, Yamahara T, Tanaka K, Takio S, Satoh T. 1997. Characterization of catalase from green algae Chlamydomonas reinhardtii Journal of Plant Physiology 151 262–268 [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform Nucleic Acids Research 30 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang A, Musgrave ME. 1996. Dynamics of vegetative cytoplasm during generative cell formation and pollen maturation in Arabidopsis thaliana Protoplasma 194 81–90 [DOI] [PubMed] [Google Scholar]
- Lange PR, Eastmond PJ, Madagan K, Graham IA. 2004. An Arabidopsis mutant disrupted in valine catabolism is also compromised in peroxisomal fatty acid β-oxidation FEBS Letters 571 147–153 [DOI] [PubMed] [Google Scholar]
- Lingard MJ, Monroe-Augustus M, Bartel B. 2009. Peroxisome-associated matrix protein degradation in Arabidopsis Proceedings of the National Academy of Sciences USA 106 4561–4566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons E Pedersen B Kane J et al. 2008. Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with rosids Plant Physiology 148 1772–1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerkercke AV, Schauvinhold I, Pichersky E, Haring MA, Schuurink RC. 2009. A plant thiolase involved in benzoic acid biosynthesis and volatile benzenoid production The Plant Journal 60 292–302 [DOI] [PubMed] [Google Scholar]
- Mun JH, Kwon SJ, Yang TJ. et al. Genome-wide comparative analysis of the Brassica rapa gene space reveals genome shrinkage and differential loss of duplicated genes after whole genome triplication. Genome Biology. 2009;10:R111. doi: 10.1186/gb-2009-10-10-r111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pracharoenwattana I, Smith SM. 2008. When is a peroxisome not a peroxisome? Trends in Plant Science 13 522–525 [DOI] [PubMed] [Google Scholar]
- Richmond TA, Bleecker AB. 1999. A defect in β-oxidation causes abnormal inflorescence development in Arabidopsis The Plant Cell 11 1911–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-García MI, M’Rani-Alaoui M, Fernández MC. 2003. Behavior of storage lipids during development and germination of olive (Olea europaea L.) pollen Protoplasma 221 237–244 [DOI] [PubMed] [Google Scholar]
- Rutter MT, Cross KV, Van Woert PA. 2012. Birth, death and subfunctionalization in the Arabidopsis genome Trends in Plant Science 17 204–212 [DOI] [PubMed] [Google Scholar]
- Rylott EL, Eastmond PJ, Gilday AD, Slocombe SP, Larson TR, Baker A, Graham IA. 2006. The Arabidopsis thaliana multifunctional protein gene (MFP2) of peroxisomal β-oxidation is essential for seedling establishment The Plant Journal 45 930–941 [DOI] [PubMed] [Google Scholar]
- Rylott EL, Hooks MA, Graham IA. 2001. Co-ordinate regulation of genes involved in storage lipid mobilization in Arabidopsis thaliana Biochemical Society Transactions 29 283–287 [DOI] [PubMed] [Google Scholar]
- Rylott EL, Rogers CA, Gilday AD, Edgell T, Larson TR, Graham IA. 2003. Arabidopsis mutants in short- and medium-chain acyl-CoA oxidase activities accumulate acyl-CoAs and reveal that fatty acid β-oxidation is essential for embryo development Journal of Biological Chemistry 278 21370–21377 [DOI] [PubMed] [Google Scholar]
- Schilmiller AL, Koo AJK, Howe GA. 2007. Functional diversification of acyl-coenzyme A oxidases in jasmonic acid biosynthesis and action Plant Physiology 143 812–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Wheeler DL, Christensen SE, Berens JC, Cohen JD, Rampey RA, Bartel B. 2011. Multiple facets of Arabidopsis seedling development require indole-3-butyric acid-derived auxin The Plant Cell 23 984–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X Wang H Wang J et al. 2011. The genome of the mesopolyploid crop species Brassica rapa Nature Genetics 43 1035–1039 [DOI] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J. Arabidopsis: a laboratory manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2002. [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiszniewski AAG, Zhou W, Smith SM, Bussell JD. 2009. Identification of two Arabidopsis genes encoding a peroxisomal oxidoreductase-like protein and an acyl-CoA synthetase-like protein that are required for responses to pro-auxins Plant Molecular Biology 69 503–515 [DOI] [PubMed] [Google Scholar]
- Yang Z, Ohlrogge JB. 2009. Turnover of fatty acids during natural senescence of Arabidopsis, Brachypodium, and switchgrass and in Arabidopsis β-oxidation mutants Plant Physiology 150 1981–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.