Abstract
Members of the core pooids represent the most important crops in temperate zones including wheat, barley, and oats. Their importance as crops is largely due to the grain, particularly the storage capabilities of the endosperm. In this study, a comprehensive survey of grain morphology and endosperm organization in representatives of wild and cultivated species throughout the core pooids was performed. As sister to the core pooid tribes Poeae, Aveneae, Triticeae, and Bromeae within the Pooideae subfamily, Brachypodium provides a taxonomically relevant reference point. Using macroscopic, histological, and molecular analyses distinct patterns of grain tissue organization in these species, focusing on the peripheral and modified aleurone, are described. The results indicate that aleurone organization is correlated with conventional grain quality characters such as grain shape and starch content. In addition to morphological and organizational variation, expression patterns of candidate gene markers underpinning this variation were examined. Features commonly associated with grains are largely defined by analyses on lineages within the Triticeae and knowledge of grain structure may be skewed as a result of the focus on wheat and barley. Specifically, the data suggest that the modified aleurone is largely restricted to species in the Triticeae tribe.
Key words: Aleurone, Brachypodium, cereal grain, endosperm, monocot, temperate grasses
Introduction
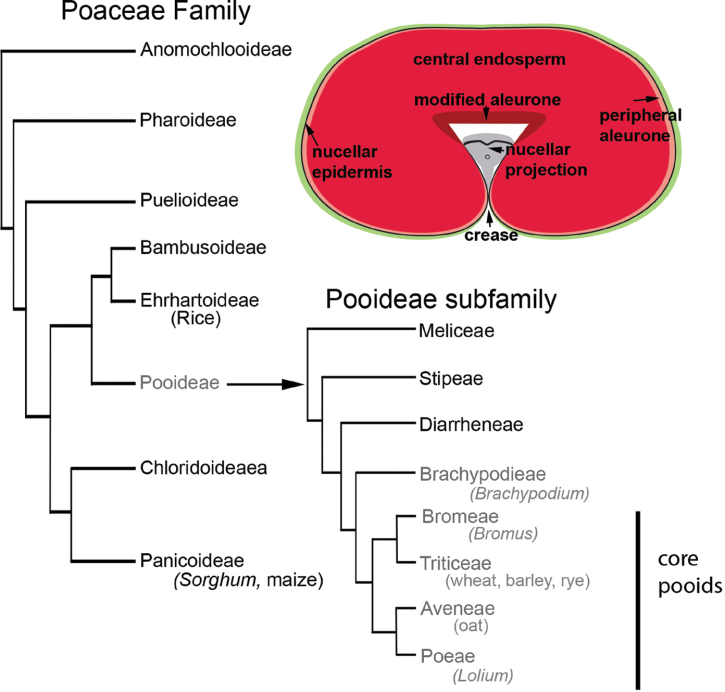
The grain (caryopsis) is a single-seeded fruit characteristic of the grasses. It is a composite organ with three genetically distinct compartments, the pericarp and associated maternal tissues, the embryo, and a prominent and persistent endosperm for which cereal species have been domesticated. The endosperm is rich in starch and protein and wheat grains remain one of the main sources in human nutrition. The ability of endosperm to store starch so proficiently is a character largely associated with monocot orders including the Poales. The family Poaceae within the Poales is synonymous with ‘The Grasses’ or ‘Grass Family’, and contains important grain crops such as wheat, rice, maize, and sorghum. Within the grass family, the Pooideae subfamily has diversified in cooler climates and contains the cereals of most value to the UK economy, wheat, barley, oats, and rye. In addition, it includes genera of important forage grasses such as Lolium (Hubbard, 1954), invasive weed genera such as Elymus and Bromus, and species such as Brachypodium distachyon that is a particularly amenable model with extensive genomic tools (Alves et al., 2009; Bevan et al., 2010; Vogel et al., 2010; Thole et al., 2010, Mur et al., 2011). Within the Pooideae, four tribes are termed the core pooids, Poeae, Aveneae, Triticeae, and Bromeae with the monogenic Brachypoideae as sister to these groups (Catalan et al., 1997; Jacobs et al., 2000; Doring et al., 2007; Vogel and Bragg, 2009).
Despite being a vital food source, grain development outside the key cultivated crop species has been less well characterized compared with other plant organs and relatively few regulatory genes have been identified (Sabelli and Larkins, 2009). While much work has been done on the evolution and development of such characters as inflorescence architecture in analyses across the cereal phylogeny that extend into uncultivated and wild relatives (Doebley et al., 1997; Vollbrecht et al., 2005; Preston and Kellogg, 2007), less is known about the evolution of grain form and function, particularly with regard to its tissue organization and the ability of the endosperm to store rich reserves.
In cultivated cereals and most grasses, the endosperm is the largest compartment in the grain. Endosperm development has certain features that seem well conserved (in studies to date). For example, early development progresses in the post-fertilization phase via the division of the central cell-derived triploid nucleus, whose descendants form a syncitial ring of nuclei around the central vacuole that divide and cellularize in a concerted sequence of anticlinal and periclinal cell divisions eventually to fill the central vacuole (Olsen, 2001; Costa et al., 2004; Wegel et al., 2005; Gubatz et al., 2007). The endosperm subsequently differentiates into functionally distinct subdomains, which can include the aleurone, modified aleurone or transfer layer, central starchy endosperm, and embryo-surrounding region endosperm. Temporal and spatial development can vary depending on the species and our studies also indicate that the number of subdomains is not fixed.
Maternal tissue organization also varies, affecting the possible routes by which sugars and amino acids are supplied to the developing endosperm. For example, the generally accepted conduit for maternal nutritional supplies in wheat grains is through the nucellar projection and modified aleurone (Wang et al., 1995; Zheng and Wang, 2011). Rye and barley share basic organization with wheat (Parker, 1981; Olsen et al., 1992). This differs in other grains such as rice with two pathways involved in the transport of nutrients within the developing caryopsis: one via a pathway analogous to the nucellar projection pathway of wheat and the other via the nucellar epidermis (Oparka and Gates, 1981). Rice grains have a vascular system that extends the length of the grain whereas the vascular tissue supplying the maize grain terminates at the junction of funiculus and ovule (as in Arabidopsis). Maize, in turn, has a pronounced transfer cell layer within the endosperm i.e., the basal endosperm transfer layer or BETL (Gomez et al., 2002; 2009; Costa et al., 2004). Rice lacks this basal layer of differentiated cells within its endosperm tissue and may utilize an alternative nucellar epidermis-mediated transport route, at least until the nucellar epidermis becomes compressed later in development. Even though different cereals have alternative transfer cells with functional homology, these may not be genetically homologous tissues in the evolutionary sense.
One of the key features in wheat domestication was the selection for larger and rounder grains (Glemin and Bataillon, 2009; Gegas et al., 2010). To be able to answer the question as to how a larger and rounder grain evolved, it is necessary to have a detailed understanding of grain morphology and anatomy in both cultivated and wild grasses. By identifying and characterizing differences and similarities at the structural and molecular levels, significant insights into the effects of domestication and cultivation on the grain’s development will be gained. Here, a comprehensive comparative survey of grain morphology and organization in the temperate grasses is described, focusing on the Pooideae subfamily and selected taxa therein (Fig. 1). The new model, Brachypodium distachyon, is used as a taxonomically-relevant reference point for the ‘core pooids’ encompassing wheat, barley, and oat. In addition, the Brachypodium genus has recently been re-assessed phylogenetically to provide relevant intra-genus candidates for comparison such as B. stacei and B. hybridum (Catalan et al., 2012). Selected taxa compose both crop and non-crop species and the features and criteria selected for analyses and comparison are centred on aspects of grain morphology relevant to their agronomic value and pertinent to their post-harvest processing requirements (Evers and Millar, 2002). This variation underpins the contrast between cultivated and non-crop species and features that have implications in nutritional profiles and post-harvest processing: crease, nucellar epidermis and projection, modified aleurone, peripheral aleurone, and central endosperm. The analyses reveal specific points of variation in grain structure at various levels.
Fig. 1.
Selected taxa based on published phylogenies; schematic of grain cross-section, based on wheat, indicating grain features in maternal and endosperm tissues analysed in this study.
Materials and methods
Seed material
Seed material for the majority of the wild species was obtained through specialist commercial seed suppliers (Herbiseed). For the cultivated and some wild crop species, seeds were obtained through the John Innes Centre germplasm resources centre. Seeds for B. hybridum and B. stacei were kindly supplied by the Doonan Laboratory (JIC).
Grain preparation and physical measurements
Measurements of grain dimensions were collected from dehulled grains (with the exception of the barley species where it was not practical to remove the hull) using Clarke CM145 digital vernier callipers on 20 grain samples. Measurements for grain cross-sectional features were taken from images of dry mature grains cut transversely at the central point of the grain and observed using light microscopy as detailed below. Very brittle, fragile, and powdery grains were sometimes cut within a drop of 80% glycerol to reduce breakage. Sections prepared in this way were covered with a cover slip to reduce glare and imaged immediately to ensure features were not distorted by the uptake of moisture.
Light microscopy
External and macro-morphological analysis of grain features was performed using a Motic SMZ-168 dissecting microscope equipped with a Canon EOS 1000D digital camera. Images were collected using EOS utility software 2.4.0.1 and Canon Digital Photo professional. Bright and darkfield microscopy for observation and imaging of grain-stained sections was performed using a GX optical L3200 compound microscope equipped with a GT-vision GXCAM-5 5MP digital USB camera and GXCAPTURE software. Image analysis and measurement was performed using image-J software. For all cell size measurements the longest axis of the cell was recorded.
Scanning electron microscopy
Mature dry grains were imbibed overnight in distilled water. Imbibed grains were trimmed at the distal end to facilitate penetration of fixative into the tissue and transferred to freshly prepared FAA fixative (formaldehyde 3.7%, acetic acid 5%, ethanol 50%) Grains were exposed to three cycles of moderate vacuum (~500 mbar) to ensure penetration of fixative and fixed overnight at 4 °C with agitation. Fixed grains were transferred to 70% ethanol. Samples were dehydrated through a series of 80%, 90%, and 100% ethanol with 12–24h in each before critical point drying in a Bal-Tec 030 Critical Point Drier using CO2 and following manufacturer’s instructions. Samples were coated in gold using a Polaron SC7640 Sputter Coater for 90 s at ~2.0kV. Samples were analysed on a Hitachi S3000H scanning electron microscope equipped with digital image capture.
Vital staining, iodine staining and toluidine blue staining
For tetrazolium chloride (TZ) staining, thin sections were made by hand of living mature grains that had first been imbibed in distilled water overnight. Sections were taken as thinly as possible from the central point of the grain from three biological replicates. Freshly cut sections were immersed in 1ml of 0.5% TZ solution and incubated at 35 °C for 3–6h according to the intensity of staining. TZ solution was prepared fresh and tested before use to ensure a pH of 6.5–7 (optimal staining requires a close to neutral pH). Stained sections were mounted in 80% glycerol and photographed immediately using dissecting and compound microscopes. Evans Blue vital staining was performed as described previously (Young and Gallie, 1999; Opanowicz et al., 2011) with minor modification. Freshly cut sections were immersed in a 0.1% Evans Blue solution for 4min and then washed in several changes of distilled water for 3h with gentle agitation. Sections were mounted and photographed. For iodine staining, very thin, freshly prepared sections were immersed in 50% Lugol’s solution (Sigma L6146) for ~1min, briefly and gently washed in distilled water, and mounted in 80% glycerol. Histological analysis with Toluidine Blue was performed using 14 µm thick transverse central grain sections on glass slides that were fixed and sectioned as described for mRNA in situ hybridization, the cleared of wax in histo-clear II (National Diagnostics) baths with agitation before rehydration through 100%, 70%, and 50% ethanol series (10min each). Slides were immersed for 1min in 0.05% Toluidine blue in 0.1M phosphate buffer, pH 6.8, and then rinsed through several changes of deionized water. Slides were then allowed to air dry and permanently mounted in Entellan (Merck).
RNA isolation and RT-PCR
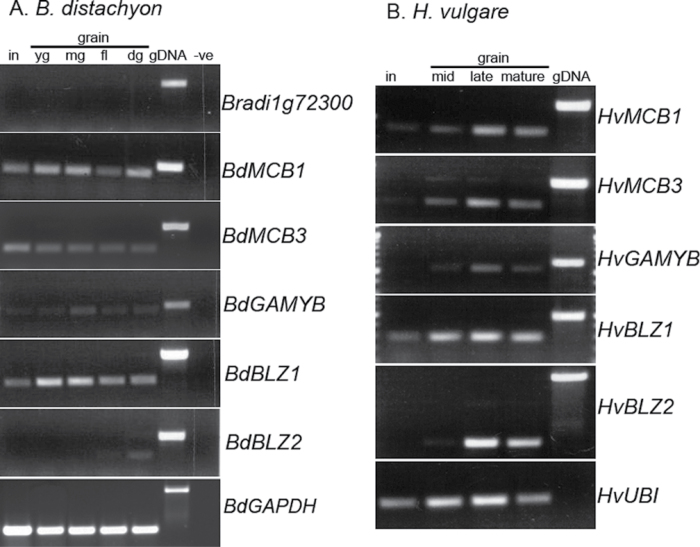
Grain samples for three biological replicates were collected according to grain size and distinct growth stages. For Brachypodium, the inflorescence stage comprises an entire spikelet prior to anthesis. Developing grains were then collected at four stages; a ‘young grain’ stage immediately after anthesis (~1 DAA), an early ‘mid-length’ stage (where grains are approximately halfway in proximal-distal development, ~5–6 DAA), a later developmental stage (as grains reached full proximal–distal development, correlating to ~8–10 DAA), and a fully developed stage (but before dessication, correlating to ~18–20 DAA). These developmental stages follow that of Opanowicz et al. (2011). For barley, the variety Optic was used and samples correlate to similar grain size stages. The inflorescence stage sample correlates to 2–3 entire florets from a central point in the spikelet collected at or very close to anthesis. Mid–early stage grains were typically halfway in proximal–distal development, whilst mid–late were fully developed in length but before the endosperm had begun to fill out appreciably. The mMature grain stage again comprised fully developed grains but prior to the onset of desiccation. Genomic DNA was extracted from leaf material using the Qiagen plant DNeasy mini kit. Total RNA was extracted using either the Trizol reagent (Invitrogen) or, for the more starch-rich mature grain tissue, the Spectrum plant total RNA extraction kit (Sigma) was used as per the manufacturer’s instructions. Approximately 300ng of DNase-treatded RNA was used in 10 µl cDNA synthesis reactions using BioscriptTM Reverse Transcriptase (Bioline) using the poly(T) primer 5’ GACTCGAGTCGACATCGA(T). PCR amplification was performed using 1 µl of cDNA template in 10 µl reactions with the following cycle: 94 °C for 5min, then 30 cycles of 94 °C for 30 s, 53 °C for 45 s, 72 °C for 1min followed by 72 °C for 6 m. Primers for gene expression comparisons in Brachypodium and barley are listed in Supplementary Table S2 at JXB online. Control primers for the former were BdGAPF and BdGAPR as described in Opanowicz et al. (2011). Barley control primers were HvUbiF 5’-ACTACAACATCCAGAAGGAG-3’ and HvUbiR 5’-TCGCGATAGGTAAAAGAGCAG-3’.
mRNA in situ hybridization
Grains were imbibed and fixed as above and transferred to 70% ethanol before automated fixation/dehydration/infiltration (Drea et al., 2005b). All slide processing and subsequent hybridization steps follow that described previously in Drea et al. (2005b). Probes for wheat ISH were as described in Drea et al. (2005a) with ID numbers 701993242 (PPDK; peripheral aleurone), 702006333 (oxidoreductase; modified aleurone), and 701994326 (gliadin; central endosperm).
Sequence analyses
Sequences were aligned using CLUSTAL_X (Thompson et al., 1997) and alignments refined by hand using BioEdit (Hall, 1999). Accession numbers for all sequences in alignments are list Supplementary Table S3 at JXB online.
Results
Distinct grain shape profiles can be defined
Twenty-eight species spanning the core pooids and Brachypodium comprising 12 genera were selected for analysis. On a very basic level the grains were grouped on the basis of shape determined by the ratios of simple size dimensions, length, width, and depth (see Supplementary Fig. S1 at JXB online; Table 1). High-yielding grains are marked by a round profile maximizing the accumulation of reserves in a confined space (see Supplementary Fig. S1 at JXB online; Fig. 2). However, within the Triticeae, and specifically the wheat grain, while the rounded grains may indicate a reserve-rich structure, the presence of a distinctive crease is a barrier to efficient milling. Though all the grains are curved around a vascular strand to form a crease, only the wheats, barley, and oat have a closed crease where the lobes of the grain make contact on the adaxial side of the grain.
Table 1.
Summary of observed grain characters in mature selected species of Brachypodium and core pooids
| Species | Grain profile | Crease | Cavitya | Persistent nucellar epidermisa | Peripheral aleurone– cell layers | Modified aleurone –vital staina | Starch granulea |
|---|---|---|---|---|---|---|---|
| Brachypodium distachyon | Flat | Open | n | y | 1–4 irregular | y | Simple |
| Brachypodium stacei | Flat | Open | n | y | 1–4 irregular | y | nd |
| Bromus mollis | Flat | Open | n | y | 1 | y | Simple |
| Bromus sterilis | Flat | Open lobed | n | y | 3 | y~ | Simple |
| Elymus repens | Flat | Open | n | n | 1 | n~ | Bimodal |
| Hordeum vulgare | Round | Closed | y | n | 3 | n | Bimodal |
| Hordeum murineum | Round | Closed | y | n | 1 | nd | Bimodal |
| Triticum aestivum | Round | Closed | y | n | 1 | n | Bimodal |
| Triticum uratu | Narrow | Closed | y | n | 1 | n | Bimodal |
| Triticum speltoides | Flat | Closed | y | n | 1 | n~ | Bimodal |
| Triticu tauschii | Flat | Closed | y | n | 1 | n | Bimodal |
| Triticum dicoccoides | Round | Closed | y | n | 1 | n | Bimodal |
| Avena sativa | Round | Closed | y | n | 1 | y | Compound |
| Avena fatua | Round | Closed | y | n | 1 | y | Compound |
| Lolium perenne | Flat | Open | n | n | 1 | y | compound |
| Festuca pratensis | Flat | Open | n | n | 1 irregular | n~ | Compound/ simple |
a nd, not determined; n, not present; y, present; al, aleurone; ~ typical TZ staining pattern.
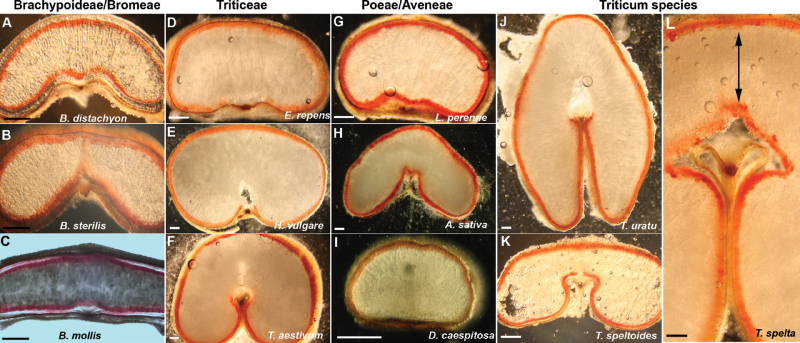
Fig. 2.
Vital staining of grains from several genera with tetrazolium chloride (TZ). Scale bars 400 µm. Double-ended arrow indicates bridge depth in spelt wheat.
Elymus repens, though also in the Triticeae, is exceptional in the openness of its adaxial side (Fig. 2d). With regard to this adaxial flatness it closely resembles the grains of Lolium, Festuca, and Dactylis in the Poeae (Fig. 2G). Within the Bromeae species, there was a noticeable difference between the mollis and diandrus/sterilis species. The latter species had a more obvious lobed profile reminiscent of the wheats, while mollis was starkly flat with barely any appearance of a crease (Fig. 2B, 2C). In addition, the diandrus and sterilis species are distinctively longer than mollis and, in fact, are longer than all the species examined (see Supplementary Fig. S1 at JXB online). The nucellar epidermis is persistent in Brachypodium and in Bromus genera only (Table 1). In the other grains sampled the nucellar epidermis was greatly reduced and compressed by maturity. A robust nucellar projection seems to persist only in the grains with the closed crease structure, the Triticeae (except Elymus) and Avena in the Poeae.
Though most grains with a closed crease are correspondingly round in shape, the narrow profile of diploid AA wheats such as T. uratu is a sharp contrast (Fig. 2J) to hexaploid wheats and to the flat T. speltoides (Fig. 2K). The narrow profile of uratu results in a very deep crease. The hexaploid spelt wheat, though round with a closed crease, has a narrow abaxial endosperm bridge domain (indicated by an arrow in Fig. 2L) and therefore a decidedly deep crease.
The organization of the aleurone layers varies
Our previous analyses revealed significant differences in endosperm development and differentiation in Brachypodium grain compared with wheat: for example, despite Brachypodium being a member of the Pooideae, the aleurone layer is not regionally differentiated into distinct peripheral and modified aleurone regions (Opanowicz et al., 2011). The modified aleurone layer, implicated as a major transfer tissue in wheat, is completely absent as judged by Evans Blue vital staining and expression analysis of the BdGLO1 (Bradi1g13040) gene (Opanowicz et al., 2011).
This analysis was extended to the species selected for this study and tetrazolium chloride (TZ) was used as a marker of vital tissues to ascertain if, as previously observed in hexaploid wheat and Brachypodium, differences in the viability of aleurone tissues in the mature grains could be identified. While all grains show that the peripheral aleurone layers are living at maturity, as is expected for these tissues, the assorted grains show variation in both the number of layers and in the regularity of their organization (Table 1; Fig. 3). Vital staining patterns in the adaxial domain where the modified aleurone would be expected to be located varied but, in Brachypodium and Bromus species, this region consistently stained with TZ and, like the peripheral aleurone, is living tissue at maturity (Fig. 2A–C). As previously shown using Evan’s Blue staining for dead tissues, no TZ staining was detected in hexaploid wheat modified aleurone (Fig. 2F). This was also the case in cultivated barley grains (Fig. 2E). However, there was some variation in the other Triticum genera tested, with speltoides consistently showing TZ staining in this region (Fig. 2K). Evans Blue staining was also performed which corroborated that the tissue in speltoides was live. While oat shows similar shape and organization as barley and wheat grains, living tissue was consistently detected in its modified aleurone domain (Fig. 2H).
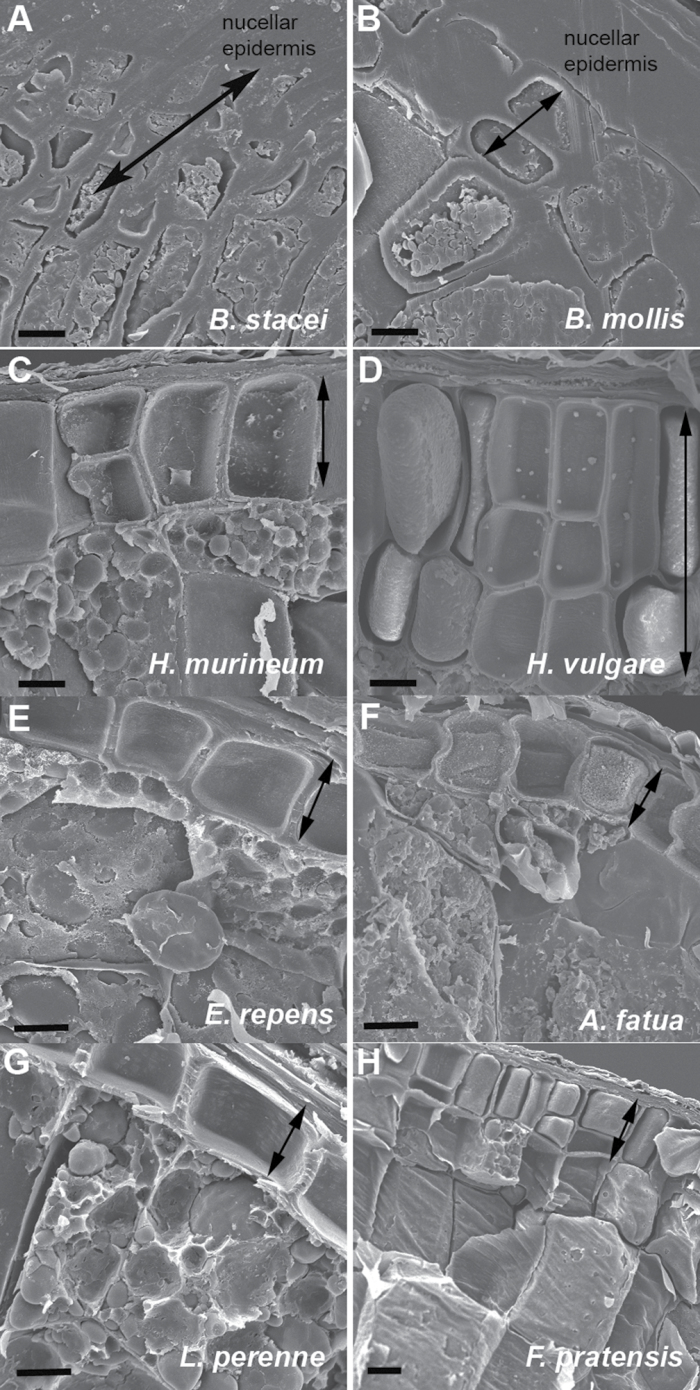
Fig. 3.
SEM of selected grains showing peripheral aleurone and underlying central endosperm cells. The aleurone area in each species is indicated by the double-ended arrows. Scale bars (A, B, G, H) 10 µm; (C, D,E, F) 20 µm.
Endosperm cell size and cell wall thickness
Scanning electron microscopy (SEM) was performed on mature grains of representative species to determine cell wall thickness in the central endosperm compared with the aleurone layers. Conversely to wheat, B. distachyon central cell walls are comparable or slightly greater in thickness compared with the walls in the peripheral aleurone (Opanowicz et al., 2011; Table 2). In this study it was also found that, in Bromus, the central endosperm cell walls are comparable in thickness to the aleurone layer (Table 2; Fig. 3B; see Supplementary Table S1 at JXB online) or at least with a ratio greater than 0.7. In all other species examined, the aleurone cell walls are at least twice as thick as the central endosperm cell walls. However, only Brachypodium appeared to have the small central cell size throughout the endosperm and, in all other genera examined, the central endosperm cells were several times larger than the peripheral aleurone cells (Table 2; Fig. 2; see Supplementary Table. S1 at JXB online). This was particularly striking in Bromus mollis with a central cell length of up to 80 µm in some cases compared with an aleurone cell length as small as 10 µm. With such a flat grain there may only be 4–6 cells spanning the abaxial–adaxial axis (Fig. 3B). Barley seems to be the only grain having more than one regular layer of aleurone (Fig. 3D; Olsen et al., 1992) while only the Brachypodium species has an obviously disorganized cellular arrangement seen in both TZ staining and SEM images (and previously in gene expression patterns (Opanowicz et al., 2011)). SEM analysis indicated that Festuca pratensis aleurone cells are more irregular than those of other core pooids (Fig. 3H).
Table 2.
Measurements of cell wall thickness and cell size in central endosperm and peripheral aleurone of selected species. Measurements in µm with standard deviation and n=10.
| Species | Central wall | Aleurone wall | Central size | Aleurone size |
|---|---|---|---|---|
| B. distachyon | 2.73±0.42 | 2.07±0.51 | 27.5±4.68 | 18.23±4.29 |
| B. mollis | 2.41±0.67 | 3.06±1.04 | 59.22±6.23 | 12.59±1.65 |
| B. diandrus | 1.81±0.26 | 2.55±0.55 | 53.68±10.12 | 24.05±1.19 |
| E. repens | 0.41±0.14 | 1.08±0.20 | 45.31±9.53 | 17.43±4.45 |
| T. uratu | 0.39±0.07 | 0.79±0.14 | nda | 23.54±2.16 |
| T. speltoides | 0.31±0.04 | 1.31±0.11 | 78.15±9.27 | 19.89±2.62 |
| L. perenne | 0.37±0.17 | 1.42±0.34 | 79.81±33.56 | 16.59±2.64 |
| F. pratensis | 0.67±0.10 | 1.80±0.26 | 94.86±14.12 | 17.36±6.33 |
| A. fatua | 0.38±0.13 | 1.40±0.22 | 132.93±50. | 25.51±3.02 |
a nd, Not determined.
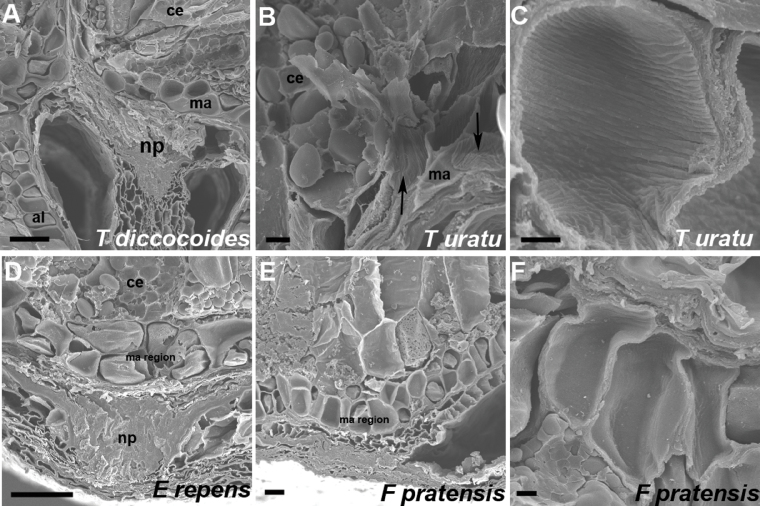
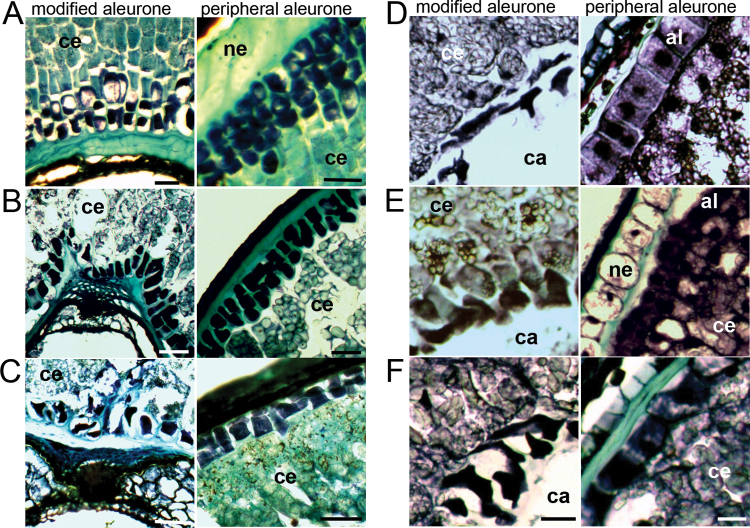
SEM can be used to examine some visual properties of the cell walls. Classic features of transfer cells, including the BETL of maize, include invaginations in the walls to increase surface area for transport. These cells are usually more irregularly shaped than the regular cuboidal cells of the peripheral aleurone. This was clearly visible in SEMs of cell walls of the modified aleurone of the wheat species observed, including those of primitive wheats (Fig. 4A–C). It was less obvious in grains with a flat profile such as E. repens, Lolium, and Festuca (Fig. 4D–F).
Fig. 4.
SEM of modified aleurone region in selected grains. ce, central endosperm; ma, modified aleurone; al, aleurone; np, nucellar projection. Arrows in (B) indicate internal surfaces of modified aleurone cells. Scale bars (A, D) 50 µm; (B, E) 10 µm; (C, F) 5 µm.
Molecular analyses of the grain’s aleurone layers between genera
Transcription factors are key specifiers of distinct cell and tissues types and, in grains, one of the few genes shown to specify particular tissues is ZmMRP-1, a determinant of the BETL layer in maize encoding an atypical single-repeat MYB protein (Gomez et al., 2009). No orthologues could be identified in temperate cereals (no sequence homology extended beyond the conserved MYB-domain of the protein). In addition, no orthologues were identified in rice, which lacks the BETL layer. This supports the idea that ZmMRP-1 is the key regulator specifying BETL cell fate but also suggests that, though functionally homologous, the modified aleurone of temperate cereals is genetically distinct and that it has evolved independently. The Brachypodium genome contains several single-repeat myb genes and Bradi1g72300 was identified as the closest in sequence homology to ZmMRP-1 focusing on the conserved VASHAQKYF domain (see Supplementary Fig. S2 at JXB online). However, expression of Bradi1g72300 was not detected in developing grain tissue by RT-PCR (Fig. 5A)—this is in contrast to the readily detectable expression of ZmMRP-1 described in developing maize endosperm (Gomez et al., 2002).
Fig. 5.
Expression analysis of Brachypodium orthologues of key aleurone/endosperm transcription factor genes in developing grain tissues. (A) RT-PCR in B. distachyon tissues and (B) RT-PCR in H. vulgare tissues. In, inflorescence; yg, young grain; mg, mid-length grain; fl, full-length grain; dg, dry grain; gDNA genomic DNA.
Other Brachypodium single-repeat MYB genes were identified and extracted from the genome showing homology to other published genes; ZmMybst1 in maize (Mercy et al., 2003) or HvMCBS3 in barley (Rubio-Somoza et al., 2006b ) and also to the HvMCB1 gene in barley (Table 3; see Supplementary Figs S2 and S5 at JXB online) which has been shown to be involved in aleurone development and in germinating grains (Rubio-Somoza et al., 2006a ). Orthologues of HvMCB1 and HvMCBS3 in Brachypodium are expressed in developing grain tissues (Fig. 5A) which compares with that reported in barley and maize. In addition to these single-repeat MYB genes, an R2R3 MYB gene, GAMYB, has been shown to play a role in peripheral aleurone function in germination. As has been shown in other species including barley (Gocal et al., 1999; Diaz et al., 2002; Gong and Bewley 2008), expression of the Brachypodium orthologue was detected in developing grain tissues (Table 3; Fig. 5A; a full amino acid alignment in shown in Supplementary Fig. S3 at JXB online).
Table 3.
Brachypodium orthologues of aleurone/endosperm regulator genes
| Gene | Grass species | Brachypodium orthologue | Reference |
|---|---|---|---|
| ZmMRP-1 | Zea mays | Bradi1g72300 (closest hit) | Gomez et al., 2002, 2009 |
| HvMCB1 | Hordeum vulgare | Bradi3g33440 | Rubio-Somoza et al., 2006a |
| ZmMybst1/HvMCBS3 | Zea mays, Hordeum vulgare | Bradi3g33400 | Mercy et al., 2003, Rubio-Somoza et al., 2006b |
| GAMYB | Hordeum vulgare, Lolium tenulatum | Bradi2g53010 | Diaz et al., 2002 |
| BLZ1 | Hordeum vulgare | Bradi1g05480 | Vicente-Carbajosa et al., 1998 |
| BLZ2 /Opaque2/TaSPA | Hordeum vulgare/Zea mays/ Triticum aestivum | Bradi1g55450 | Onate et al., 1999 |
| Cr4 | Zea mays, Hordeum vulgare | Bradi1g30430 | Becraft et al., 1996 |
| Dek1 | Zea mays, Hordeum vulgare | Bradi3g53020 | Lid et al., 2002 |
| Sal1 | Zea mays, Hordeum vulgare | Bradi1g30430 | Shen et al., 2003 |
Orthologues of other key endosperm regulators such as the Opaque2/BLZ1/2 bZIP genes are present in Brachypodium and indeed throughout the Poaceae. Cognate orthologues in Brachypodium for BLZ1 and BLZ2 (Opaque2) were identified (Table 3; full amino acid alignments are shown in Supplementary Fig. S4 at JXB online) and RT-PCR performed on developing grain samples (Fig. 5A). BdBLZ1 is expressed in leaf tissue as was found in barley and other species (Vicente-Carbajosa et al., 1998). Strikingly, expression of the BLZ2/O2 orthologue (Bradi1g55450) was detected at very low levels in mature grains (Fig. 5A). This contrasting expression pattern was verified by performing RT-PCR on developing barley grains with primers for the corresponding orthologues (Fig. 5B) and HvBLZ2 showed high expression in maturing grains as expected based on previous northern analysis (Onate et al., 1999).
In addition to transcription factors, other key determinants of peripheral aleurone specification have been identified in maize: Cr4, Dek1, and Sal1 (Becraft et al., 1996; Lid et al., 2002; Shen et al., 2003). Single orthologues of these genes were identified in the Brachypodium genome (Table 3) and RT-PCR detected expression in all Brachypodium samples with a slight decrease in the older grain samples (see Supplementary Fig. S6 at JXB online;).
Histological and molecular analyses of the grain’s aleurone layers
It was hypothesized that a highly differentiated and regularly organized peripheral aleurone was correlated to whether the species was wild or cultivated so that a distinctive well-differentiated aleurone was selected for, perhaps by conferring more efficient storage properties on the grain or enhancing seed germination. SEM and vital staining suggested that a selection of both wild and cultivated grains had distinctive aleurone layers (Figs 3, 4).
Further histological analysis using toluidine blue was performed on a selection of the grains to delineate and compare the aleurone layers. Toluidine blue will stain nucleic acids and cell walls but not callose or starch (O’Brien et al., 1964), though starch granules are visible in background contrast. Figure 6A–C shows staining patterns for B. sylvaticum, B. sterilis, and E.repens. The irregularity of the Brachypodium peripheral aleurone, particularly in terms of the unfixed number of layers, is again apparent and also striking is the similarity in staining density between the two aleurone cell types in all these species. The delineation between aleurone and central endosperm domains is very clear in all cases. Within the wheat species specifically, in Fig. 6D–F including T. uratu, tauschii, and speltoides, there is dense staining of the contents of the modified aleurone cells (which are not regularly cuboidal as are the peripheral aleurone cells) while, in the peripheral aleurone cells, the densely stained nuclei can be distinguished from the surrounding cytoplasm. In all cases, the aleurone layers display a distinct organization to the central endosperm, though it was noted in speltoides (Fig. 6F) that the dense staining pattern was visible in both aleurones as in the species in Fig. 6A–C. In tauschii it was observed that the nucellar epidermis persisted as the grain reached maturity (Fig. 6E) and the cells of the peripheral aleuone were small and more difficult to distinguish from the central endosperm.
Fig. 6.
Toluidine blue staining of grains focusing on the aleurone cells (modified and peripheral) juxtaposed with the central endosperm and endosperm cavity. (A) Brachypodium (sylvaticum). (B) Bromus (sterilis). (C) Elymus (repens). (D–F) Triticum (uratu, tauschii, speltoides). ne, Nucellar epidermis; ce, central endosperm; al, aleurone; ca, cavity. Scale bars (A, B, D, E, F) 20 µm; (C) 50 µm.
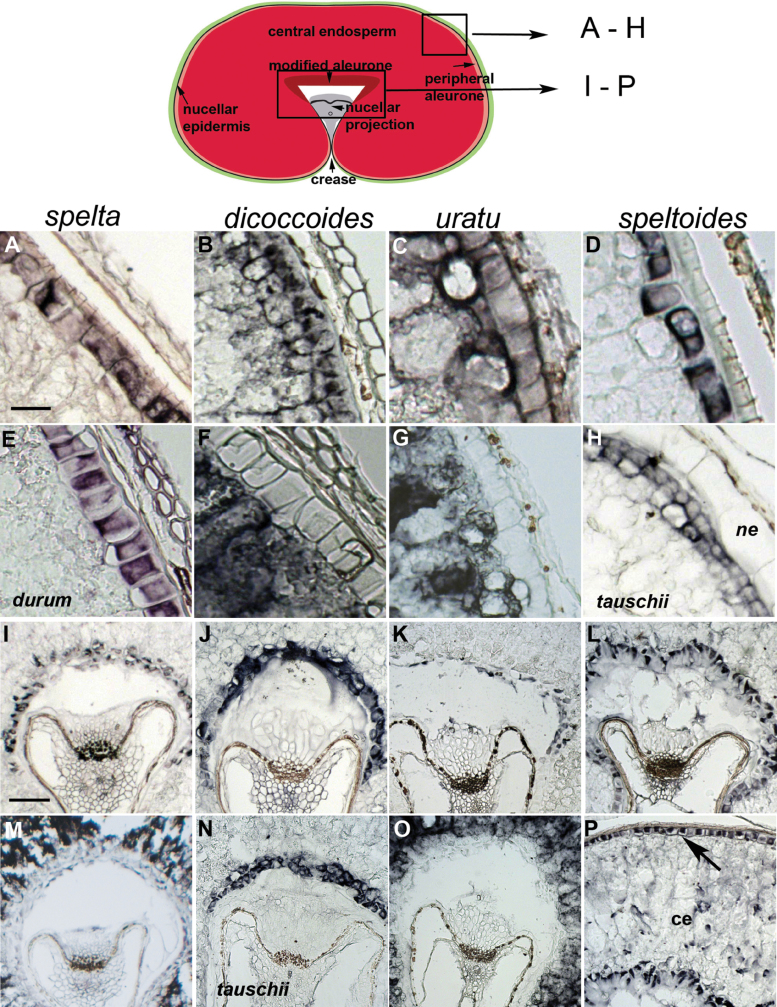
The distinction of peripheral and modified aleurone was investigated further at the molecular level. Wheat provides a useful system for exploring both primitive/uncultivated and cultivated/domesticated species within one genus, Triticum. In addition, the species within the genus represent all the shape classifications, flat, round, and narrow. Genes previously shown to have expression patterns specific for the peripheral aleurone, the modified aleurone, and the central endosperm (excluding the aleurone tissues) in wheat encoding pyruvate orthophosphate dikinase (PPDK), an iron/ascorbate oxidoreductase and a gliadin storage protein (Drea et al., 2005a ) were selected and DIG-labelled RNA probes were used in mRNA ISH on cultivated and primitive/uncultivated wheat species to identify if the level of specificity of gene expression in endosperm domains was preserved in both (Fig. 7). While all the wheat grains had general expression patterns similar to those observed in hexaploid bread wheat some subtle variations were noted. In dicoccoides and uratu the peripheral aleurone markers were also detected in the subaleurone (Fig. 7B, 7C). This was in contrast to the discretely-localized aleurone transcripts in hexaploid spelta, tetraploid durum (Fig, 7E), and the other diploid species, speltoides and tauschii (Fig. 7D, 7H) when hybridized with the same aleurone marker. However, when using the central endosperm probe, which excludes the aleurone layers, the expression extended to a distinct aleurone border in both dicoccoides and uratu (Fig. 7F, 7G) which was also apparent in histological sections (Fig. 6D). The expression of the modified aleurone marker in uratu was weak and it was difficult to discern the extent of the modified aleurone region (Fig. 7K). However, when probed with the central endosperm marker, the expected ‘exclusion zone’ for the modified aleurone region was visible (Fig. 7O). It was noted that in histological (Fig. 6D) and in situ analysis the cells in the modified aleurone layer of uratu sometimes appeared damaged or depleted which may account for some loss of signal intensity in the ISH analysis. Interestingly, in T. speltoides the markers for the peripheral and modified aleurones were expressed in both domains simultaneously (Fig. 7L, 7P) which could reflect the similar histological staining patterns observed in these layers (Fig. 6F).
Fig. 7.
ISH of domain-specific endosperm markers on cultivated and primitive (uncultivated) wheats. (A–E, H) peripheral aleurone marker; (I–L, N, P) modified aleurone marker; (F, G, M, O) central endosperm marker. Species are arranged vertically as labelled except where indicated. ne, Nucellar epidermis; ce, central endosperm. Scale bars (A–H) 50 µm, (I–P) 200 µm.
Discussion
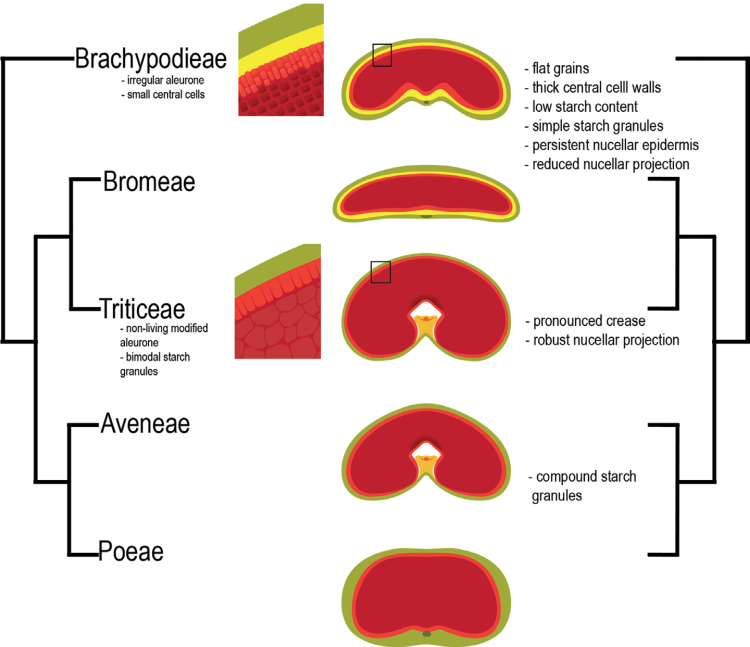
Brachypodium distachyon was used as the reference point for surveying several characters of grain morphology and endosperm organization in the core pooideae, Bromeae, Aveneae, Triticeae, and Poeae. As a sister group to this important section of the Poaceae family and a model species for the temperate grasses—with all the tools that accompany that status (Mur et al., 2011)—it is valuable to analyse and assess its features compared with neighbouring taxonomic groups both wild and cultivated. A summary of the trends and key features are summarized in Table 1 and schematically in Fig. 8.
Fig. 8.
Schematic diagram summarizing trends in grain characteristics in terms of basic phylogeny. NOT to scale. Green, pericarp; yellow, nucellar tissues; light red, aleurone; red, central endosperm.
Ultimately, the final properties of the grain are a consequence of the developmental processes that generate its tissue organization. What we know about grain development and endosperm organization has largely been restricted to cultivated grains such as rice, maize, barley, and wheat until Brachypodium became established as a reference species. This left some gaps in what is known about grain form evolution both under the artificial selective pressures of domestication and independent of it. While it was not unexpected that there are many features of grain development and composition in Brachypodium that are significantly different from wheat (Guillon et al., 2011; Opanowicz et al., 2011), the spectrum of that variation in the context of pooid phylogeny has not been addressed from the aspect of the grain’s internal features.
A significant observation is the restriction of a distinctive modified aleurone and prominent crease to the Triticeae. Transfer cells have a unique architecture that perfectly mediate their central role in nutrient transfer (Offler et al., 2003). The transfer cells in the seeds and grains of cotton and Sorghum have been shown to have implications in grain size and yield (Pugh et al., 2010; Wang et al., 2012). The apparent lack of a typical modified aleurone and robust nucellar projection—the nucellar projection is also rich in transfer characteristics (Zheng and Wang, 2011)—in genera such as Brachypodium may help explain its flat and starch-poor grains. Though the Triticeae species examined have aleurone layers that are readily identified, our spatial molecular analyses using mRNA ISH with markers previously shown to be layer-specific (Drea et al., 2005b ; Fig. 7) indicated that the aleurone layers of some species (speltoides, uratu, and dicoccoides) may be less definitively differentiated though the central endosperm marker patterns were more tightly delineated. This is something that requires further analyis to assess the implications for grain evolution and development.
Both Brachypodium and Bromus genera possess a persistent nucellar epidermis and a central endosperm cell wall thickness comparable with the aleurone layer. This is in contrast to wheat (Opanowicz et al., 2011) and indeed to the Triticeae in general. Within the Bromus genus there was a noticeable difference between the mollis and diandrus/sterilis species. The latter species had a more obvious lobed profile reminiscent of the wheats, while mollis was starkly flat with barely any appearance of a crease. All the core pooids seem to have the ability to generate large starch reserves except for the Bromus species examined, though Bromus starch reserves are generally greater than the Brachypodium species (see Supplementary Fig. S7 at JXB online). In discussing differences such as these, however, the selective forces of domestication need to be taken into account. Generally, this feature of low-starch reserves is linked to more irregular aleurone layers, a persistent nucellar epidermis, and lack of a distinct crease. In addition, we have found that the cells of the central endosperm in B. mollis are significantly bigger than those of its peripheral aleurone layer which contrasts with Brachypodium. Vital staining patterns in the adaxial domain where the modified aleurone would be expected to be located varied: in B. distachyon and B. mollis species, this region consistently stained with tetrazolium (TZ) indicating a living modified aleurone at maturity while, in B. sterilis staining in this region was absent, similar to the pattern in cultivated wheat or barley. These dramatic variations within Bromus may reflect its complex taxonomy (Smith, 1970; Oja and Jaaska, 1998).
Transcription factors (TFs) have been shown to be key players in the genetic differences distinguishing wild and cultivated species (Doebley et al., 2006) and of ten domestication genes cloned so far in grasses (rice, maize, wheat, and barley), eight are TFs (Doebley et al., 2006; Glemin and Bataillon, 2009) and four of these affect grain characters such as colour, grain shattering/threshability, and size – the others affect inflorescence architecture and growth habits that can consequently affect grain properties. Molecular evolution analyses using these genes has revealed much about the evolution of grass growth and morphology (Doebley et al., 1997; Vollbrecht et al., 2005) and transcription factors are recognized as having an important role to play in advances in agricultural biotechnology (Century et al., 2008). These domestication transcription factors were generally identified using QTL approaches, eventually narrowing down contributory loci to individual genes. Other forward genetics approaches (mutant screens) identified genes such as opaque-2 in maize as key regulators of storage protein gene expression (Hartings et al., 1989), and several TFs have been identified in the aleurone of barley where they regulate storage protein genes and genes responding to GA at germination (Rubio-Somoza et al., 2006a , b ). However, there are few TFs that have been shown to be specific determinants of early grain development and endosperm differentiation in temperate grasses. One of the few genes shown to specify particular tissues in the grain is ZmMRP-1, a determinant of the BETL layer in maize (Gomez et al., 2009). The absence/presence of key genes (or paralogues) in genomes or variation in the expression pattern in one species compared with another, can be a powerful indicator of what the genetic bases for pronounced morphological and developmental differences are. The lack of expression of the Brachypodium ZmMRP-1 and BLZ2/Opaque2 orthologues (as determined by sequence analysis and RT-PCR) could be molecular genetic underpinnings of the differences between Brachypodium and barley and maize. BLZ genes are regulators of prolamin storage protein genes in barley, rice, and maize and polymorphisms in maize and wheat have been shown to have huge implications in grain composition and quality (Onate et al., 1999; Onodera et al., 2001; Gibbon and Larkins, 2005; Ravel et al., 2009). Given that the storage protein profile of Brachypodium is enriched for globulins rather than prolamins (Laudencia-Chingcuanco and Vensel, 2008; Larre et al., 2010) this variation in expression of BLZ2 may not be unexpected.
In addition to transcription factors, other key determinants of peripheral aleurone specification have been identified in maize: Cr4, Dek1, and Sal1 (Becraft et al., 1996; Lid et al., 2002; Shen et al., 2003). These genes encode a receptor kinase, a calpain protease, and a vacuolar sorting protein, respectively. In a barley mutant with reduced aleurone layers, des5, these genes, particularly HvCr4, were found to be significantly reduced in expression in the grains (Olsen et al., 2008). In this study, significant differences in gene expression for Brachypodium orthologues were not detected.
In the context of a sister group to the core pooids, Brachypodium was used as a reference point for a detailed analyses of grain characters at several levels macro-, microscopic, physiological, and molecular. A spectrum of specific character variation is described across the phylogenetic groups, groups that include wild, primitive, and cultivated species, as well as forage grass species that are under different selective forces less focused on grain characters. In addition, preliminary evidence is provided of gene expression differences underpinning this variation. A cross-species comparative analysis provides a firm basis on which to interpret molecular data derived from various groups and contribute generally to our knowledge of grain form evolution. By aligning a grain survey focused on agriculturally relevant traits in the grain alongside the established phylogenies, not only are evolutionary patterns in grain structure and organization within the Pooideae revealed but also candidate species for informative genomic/transcriptomic comparisons are identified.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. S1. Graph of basic dimensions (length, width, and depth) of all grain species surveyed.
Supplementary Fig. S2. Amino acid alignment of the myb domain in the single repeat MYB genes from Brachypodium, barley, and wheat.
Supplementary Fig. S3. Amino acid alignment of GAMYB orthologues.
Supplementary Fig. S4. Amino acid alignment of BLZ orthologues.
Supplementary Fig. S5. Amino acid alignment of R1MYB orthologues.
Supplementary Fig. S6. RT-PCR of BdCR4, BdSAL1, and BdDEK.
Supplementary Fig. S7. Iodine staining of grain sections to compare levels of starch accumulation.
Supplementary Table S1. Grain characters of each of the 28 species examined.
Supplementary Table S2. Primers used in RT-PCR.
Supplementary Table S3. Accession numbers of all sequences used in alignments.
Acknowledgements
The authors wish to thank John Doona and Kay Trafford for helpful discussions on the manuscript, Mike Ambrose at the John Innes Centre for providing grain material, and Stefan Hyman and Natalie Allcock for help with SEM. SD is funded by a BBSRC Research Grant and The Leverhulme Trust. There are no conflicts of interest to declare.
References
- Albani D, Hammond-Kosack M, Smith C, Conlan S, Colot V, Holdsworth M, Bevan MW. 1997. The wheat transcriptional activator SPA: a seed-specific bZIP protein that recognizes the GCN4-like motif in the bifactorial endosperm box of prolamin genes The Plant Cell Online 9 171–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves SC, Worland B, Thole V, Snape JW, Bevan MW, Vain P. 2009. A protocol for Agrobacterium-mediated transformation of Brachypodium distachyon community standard line Bd21 Nature Protocols 4 638–649 [DOI] [PubMed] [Google Scholar]
- Becraft PW, Stinard PS, McCarty DR. 1996. CRINKLY4, A TNFR-like receptor kinase involved in maize epidermal differentiation Science 273 1406–1409 [DOI] [PubMed] [Google Scholar]
- Bevan MW, Garvin DF, Vogel JP. 2010. Brachypodium distachyon genomics for sustainable food and fuel production Curent Opinion in Biotechnology 21 211–217 [DOI] [PubMed] [Google Scholar]
- Catalan P, Kellogg EA, Olmstead RG. 1997. Phylogeny of Poaceae subfamily Pooideae based on chloroplast ndhF gene sequences Molecular Phylogenetics and Evolution 8 150–166 [DOI] [PubMed] [Google Scholar]
- Catalan P, Muller J, Hasterok R, Jenkins G, Mur LA, Langdon T, Betekhtin A, Siwinska D, Pimentel M, Lopez-Alvarez D. 2012. Evolution and taxonomic split of the model grass Brachypodium distachyon Annals of Botany 109 385–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century K, Reuber TL, Ratcliffe OJ. 2008. Regulating the regulators: the future prospects for transcription-factor-based agricultural biotechnology products Plant Physiology 147 20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa LM, Gutierrez-Marcos JF, Dickinson HG. 2004. More than a yolk: the short life and complex times of the plant endosperm Trends in Plant Science 9 507–514 [DOI] [PubMed] [Google Scholar]
- Diaz I, Vicente-Carbajosa J, Abraham Z, Martinez M, Isabel-La Moneda I, Carbonero P. 2002. The GAMYB protein from barley interacts with the DOF transcription factor BPBF and activates endosperm-specific genes during seed development The Plant Journal 29 453–464 [DOI] [PubMed] [Google Scholar]
- Doebley JF, Gaut BS, Smith BD. 2006. The molecular genetics of crop domestication Cell 127 1309–1321 [DOI] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L. 1997. The evolution of apical dominance in maize Nature 386 485–488 [DOI] [PubMed] [Google Scholar]
- Doring E, Schneider J, Hilu KW, Roser M. 2007. Phylogenetic relationships in the Aveneae/Poeae complex (Pooideae, Poaceae) Kew Bulletin 62 407–424 [Google Scholar]
- Drea S, Corsar J, Crawford B, Shaw P, Dolan L, Doonan JH.2005bA streamlined method for systematic, high resolution in situ analysis of mRNA distribution in plants Plant Methods 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drea S, Leader DJ, Arnold BC, Shaw P, Dolan L, Doonan JH. 2005. a Systematic spatial analysis of gene expression during wheat caryopsis development The Plant Cell 17 2172–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers T, Millar S. 2002. Cereal grain structure and development: some implications for quality Journal of Cereal Science 36 261–284 [Google Scholar]
- Gegas VC, Nazari A, Griffiths S, Simmonds J, Fish L, Orford S, Sayers L, Doonan JH, Snape JW. 2010. A genetic framework for grain size and shape variation in wheat The Plant Cell 22 1046–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbon BC, Larkins BA. 2005. Molecular genetic approaches to developing quality protein maize Trends in Genetics 21 227–233 [DOI] [PubMed] [Google Scholar]
- Glemin S, Bataillon T. 2009. A comparative view of the evolution of grasses under domestication New Phytologist 183 273–290 [DOI] [PubMed] [Google Scholar]
- Gocal GF, Poole AT, Gubler F, Watts RJ, Blundell C, King RW. 1999. Long-day up-regulation of a GAMYB gene during Lolium temulentum inflorescence formation Plant Physiology 119 1271–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez E, Royo J, Guo Y, Thompson R, Hueros G. 2002. Establishment of cereal endosperm expression domains: identification and properties of a maize transfer cell-specific transcription factor, ZmMRP-1 The Plant Cell 14 599–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez E, Royo J, Muniz LM, Sellam O, Paul W, Gerentes D, Barrero C, Lopez M, Perez P, Hueros G. 2009. The maize transcription factor myb-related protein-1 is a key regulator of the differentiation of transfer cells The Plant Cell 21 2022–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong X, Bewley JD. 2008. A GAMYB-like gene in tomato and its expression during seed germination Planta 228 563–572 [DOI] [PubMed] [Google Scholar]
- Gubatz S, Dercksen VJ, Bruss C, Weschke W, Wobus U. 2007. Analysis of barley (Hordeum vulgare) grain development using three-dimensional digital models The Plant Journal 52 779–790 [DOI] [PubMed] [Google Scholar]
- Guillon F, Bouchet B, Jamme F, Robert P, Quemener B, Barron C, Larre C, Dumas P, Saulnier L. 2011. Brachypodium distachyon grain: characterization of endosperm cell walls Journal of Experimental Botany 62 1001–1015 [DOI] [PubMed] [Google Scholar]
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows Nucleic Acids Symposium Series 41 95–98 [Google Scholar]
- Hartings H, Maddaloni M, Lazzaroni N, Di Fonzo N, Motto M, Salamini F, Thompson R. 1989. The O2 gene which regulates zein deposition in maize endosperm encodes a protein with structural homologies to transcriptional activators EMBO Journal 8 2795–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard CE. 1954. Grasses: London, UK: Pelican Books; [Google Scholar]
- Jacobs SWL, Everett J, Wilson KL, Morrison D. Grasses: systematics and evolution. Melbourne, Australia: CSIRO Publishing; 2000. [Google Scholar]
- Larre C, Pennick B, Bouchet V, Lollier O, Tranquet S, Denery-Papini S, Guillon F, Rogniaux H. 2010. Brachypodium distachyon grain: identification and subcellular localization of storage proteins Journal of Experimental Botany 61 1771–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudencia-Chingcuanco DL, Vensel WH. 2008. Globulins are the main seed storage proteins in Brachypodium distachyon Theoretical and Applied Genetics 117 555–563 [DOI] [PubMed] [Google Scholar]
- Lid SE, Gruis D, Jung R, Lorentzen JA, Ananiev E, Chamberlin M, Niu X, Meeley R, Nichols S, Olsen OA. 2002. The defective kernel 1 (dek1) gene required for aleurone cell development in the endosperm of maize grains encodes a membrane protein of the calpain gene superfamily Proceedings of the National Academy of Sciences USA 99 5460–5465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercy IS, Meeley RB, Nichols SE, Olsen OA. 2003. Zea mays ZmMybst1 cDNA, encodes a single Myb-repeat protein with the VASHAQKYF motif Journal of Experimental Botany 54 1117–1119 [DOI] [PubMed] [Google Scholar]
- Mur LA, Allainguillaume J, Catalan P, Hasterok R, Jenkins G, Lesniewska K, Thomas I, Vogel J. 2011. Exploiting the Brachypodium tool box in cereal and grass research New Phytologist 191 334–347 [DOI] [PubMed] [Google Scholar]
- O’Brien TP, Feder N, McCully ME. 1964. Polychromatic staining of plant cell walls by toluidine blue O Protoplasma 59 368–373 [Google Scholar]
- Offler CE, McCurdy DW, Patrick JW, Talbot MJ. 2003. Transfer cells: cells specialized for a special purpose Annual Review of Plant Biology 54 431–454 [DOI] [PubMed] [Google Scholar]
- Oja T, Jaaska V. 1998. Molecular phylogeny and reticulate origins of the polyploid bromus species from section genea (poaceae) Annales Botanici Fennici 35 123–130 [DOI] [PubMed] [Google Scholar]
- Olsen LT, Divon HH, Al R, Fosnes K, Lid SE, Opsahl-Sorteberg H-G. 2008. The defective seed5 (des5) mutant: effects on barley seed development and HvDek1, HvCr4, and HvSal1 gene regulation Journal of Experimental Botany 59 3753–3765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen OA. 2001. Endosperm development: cellularization and cell fate specification Annual Review of Plant Physiology and Plant Molecular Biology 52 233–267 [DOI] [PubMed] [Google Scholar]
- Olsen OA, Potter RH, Kalla R. 1992. Histo-differentiation and molecular biology of developing cereal endosperm Seed Science Research 2 117–131 [Google Scholar]
- Onate L, Vicente-Carbajosa J, Lara P, Diaz I, Carbonero P. 1999. Barley BLZ2, a seed-specific bZIP protein that interacts with BLZ1 in vivo and activates transcription from the GCN4-like motif of B-hordein promoters in barley endosperm Journal of Biological Chemistry 274 9175–9182 [DOI] [PubMed] [Google Scholar]
- Onodera Y, Suzuki A, Wu CY, Washida H, Takaiwa F. 2001. A rice functional transcriptional activator, RISBZ1, responsible for endosperm-specific expression of storage protein genes through GCN4 motif Journal of Biological Chemistry 276 14139–14152 [DOI] [PubMed] [Google Scholar]
- Opanowicz M, Hands P, Betts D, Parker ML, Toole GA, Mills EN, Doonan JH, Drea S. 2011. Endosperm development in Brachypodium distachyon 62 735–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oparka KJ, Gates P. 1981. Transport of assimilates in the developing caryopsis of rice (Oryza sativa L.) Planta 152 388–396 [DOI] [PubMed] [Google Scholar]
- Parker ML. 1981. The structure of mature rye endosperm Annals of Botany 47 181–186 [Google Scholar]
- Preston JC, Kellogg EA. 2007. Conservation and divergence of APETALA1/FRUITFULL-like gene function in grasses: evidence from gene expression analyses The Plant Journal 52 69–81 [DOI] [PubMed] [Google Scholar]
- Pugh DA, Offler CE, Talbot MJ, Ruan Y-L. 2010. Evidence for the role of transfer cells in the evolutionary increase in seed and fiber biomass yield in cotton Molecular Plant 3 1075–1086 [DOI] [PubMed] [Google Scholar]
- Ravel C, Martre P, Romeuf I, Dardevet M, El-Malki R, Bordes J, Duchateau N, Brunel D, Balfourier F, Charmet G. 2009. Nucleotide polymorphism in the wheat transcriptional activator Spa influences its pattern of expression and has pleiotropic effects on grain protein composition, dough viscoelasticity, and grain hardness Plant Physiology 151 2133–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Somoza I, Martinez M, Abraham Z, Diaz I, Carbonero P. 2006. b Ternary complex formation between HvMYBS3 and other factors involved in transcriptional control in barley seeds The Plant Journal 47 269–281 [DOI] [PubMed] [Google Scholar]
- Rubio-Somoza I, Martinez M, Diaz I, Carbonero P. 2006. a HvMCB1, a R1MYB transcription factor from barley with antagonistic regulatory functions during seed development and germination The Plant Journal 45 17–30 [DOI] [PubMed] [Google Scholar]
- Sabelli PA, Larkins BA. 2009. The development of endosperm in grasses Plant Physiology 149 14–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Li C, Min Z, Meeley RB, Tarczynski MC, Olsen OA. 2003. sal1 determines the number of aleurone cell layers in maize endosperm and encodes a class E vacuolar sorting protein Proceedings of the National Academy of Sciences USA 100 6552–6557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PM. 1970. Taxonomy and nomenclature of the brome-grasses (Bromus L. s.l.) Notes RBG Edinburgh 30 361–375 [Google Scholar]
- Thole V, Worland B, Wright J, Bevan MW, Vain P. 2010. Distribution and characterization of more than 1000 T-DNA tags in the genome of Brachypodium distachyon community standard line Bd21 Plant BiotechnologyJournal 8 734–747 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools Nucleic Acids Research 25 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Carbajosa J, Onate L, Lara P, Diaz I, Carbonero P. 1998. Barley BLZ1: a bZIP transcriptional activator that interacts with endosperm-specific gene promoters The Plant Journal 13 629–640 [DOI] [PubMed] [Google Scholar]
- Vogel J, Bragg J. 2009. Brachypodium distachyon, a new model for the Triticeae. In: Feuillet C, Muehlbauer GJ, eds. Genetics and genomics of the Triticeae New York: USA: Springer Science; 427–449 [Google Scholar]
- Vogel JP, Garvin DF, Mockler TC, et al. 2010. Genome sequencing and analysis of the model grass Brachypodium distachyon Nature 463 763–768 [DOI] [PubMed] [Google Scholar]
- Vollbrecht E, Springer PS, Goh L, Buckler ESt, Martienssen R. 2005. Architecture of floral branch systems in maize and related grasses Nature 436 1119–1126 [DOI] [PubMed] [Google Scholar]
- Wang HH, Wang Z, Wang F, Gu YJ, Liu Z. 2012. Development of basal endosperm transfer cells in Sorghum bicolor (L.) Moench and its relationship with caryopsis growth Protoplasma 249 309–321 [DOI] [PubMed] [Google Scholar]
- Wang HL, Patrick JW, Offler CE, Wang XD. 1995. The cellular pathway of photosynthate transfer in the developing wheat grain. III. A structural analysis and physiological studies of the pathway from the endosperm cavity to the starchy endosperm Plant, Cell and Environment 18 389–407 [Google Scholar]
- Wegel E, Pilling E, Calder G, Drea S, Doonan J, Dolan L, Shaw P. 2005. Three-dimensional modelling of wheat endosperm development New Phytologist 168 253–262 [DOI] [PubMed] [Google Scholar]
- Young TE, Gallie DR. 1999. Analysis of programmed cell death in wheat endosperm reveals differences in endosperm development between cereals Plant Molecular Biology 39 915–926 [DOI] [PubMed] [Google Scholar]
- Zheng Y, Wang Z. 2011. Contrast observation and investigation of wheat endosperm transfer cells and nucellar projection transfer cells Plant Cell Reports 30 1281–1288 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.