Abstract
The Neurachninae is the only grass lineage known to contain C3, C4, and C3–C4 intermediate species, and as such has been suggested as a model system for studies of photosynthetic pathway evolution in the Poaceae; however, a lack of a robust phylogenetic framework has hindered this possibility. In this study, plastid and nuclear markers were used to reconstruct evolutionary relationships among Neurachninae species. In addition, photosynthetic types were determined with carbon isotope ratios, and genome sizes with flow cytometry. A high frequency of autopolyploidy was found in the Neurachninae, including in Neurachne munroi F.Muell. and Paraneurachne muelleri S.T.Blake, which independently evolved C4 photosynthesis. Phylogenetic analyses also showed that following their separate C4 origins, these two taxa exchanged a gene encoding the C4 form of phosphoenolpyruvate carboxylase. The C3–C4 intermediate Neurachne minor S.T.Blake is phylogenetically distinct from the two C4 lineages, indicating that intermediacy in this species evolved separately from transitional stages preceding C4 origins. The Neurachninae shows a substantial capacity to evolve new photosynthetic pathways repeatedly. Enablers of these transitions might include anatomical pre-conditions in the C3 ancestor, and frequent autopolyploidization. Transfer of key C4 genetic elements between independently evolved C4 taxa may have also facilitated a rapid adaptation of photosynthesis in these grasses that had to survive in the harsh climate appearing during the late Pliocene in Australia.
Keywords: C4 grass evolution, C4 photosynthesis, C3–C4 intermediate, grass phylogeny, lateral gene transfer, Neurachne, Paraneurachne, polyploidy
Introduction
Despite its complexity, the C4 photosynthetic pathway has evolved independently >62 times in flowering plants (Sage et al., 2011), thus constituting a striking example of convergent evolution. It is especially prevalent in grasses, where 22–24 distinct C4 lineages have been postulated in the PACMAD clade (Grass Phylogeny Working Group II, 2012). Such a clustering of C4 origins is also observed in other groups, with six independent lineages in the sedges (Cyperaceae) and 23 in the Caryophyllales (Sage et al., 2011). These patterns indicate that some plant groups have a higher propensity for C4 photosynthesis evolution, which may reflect ecological, genomic, and/or anatomical factors that facilitate the acquisition of novel traits (Sage, 2001; Marshall et al., 2007; McKown and Dengler, 2007; Christin et al., 2011; Edwards and Ogburn, 2012). Leading environmental factors promoting C4 evolution are low atmospheric CO2, heat, drought and salinity, often in combination (Sage et al., 2012). Anatomical factors include high vein density, which may be common in dry environments and certain taxonomic groups such as the grasses (Ehleringer et al., 1997; Sage et al., 2012).
The evolution of C4 photosynthesis is best studied using closely related taxa with different photosynthetic types, as in the eudicot groups Flaveria, Cleome, Molluginaceae, and Heliotropium (McKown et al., 2005; Marshall et al., 2007; Feodorova et al., 2010; Christin et al., 2011; Muhaidat et al., 2011). These groups, however, have limited utility for understanding the origins of the pathway in the grasses, where half of all C4 species occur. C4 grasses are the most successful group of C4 plants on Earth, dominating the 23% of global primary productivity attributable to C4 vegetation, and comprising the vast majority of C4 plants in agricultural use (Brown, 1999; Still et al., 2003). Efforts to engineer the C4 pathway into C3 crops to take advantage of the superior productivity of C4 photosynthesis have been directed towards the grasses rice and wheat (http://irri.org/c4rice); however, these efforts are hindered by the lack of a model group for studying C4 evolution in the Poaceae. Such a model group could be exploited to identify the genetic changes that occurred during C4 evolution, as well as elucidating the order in which the individual traits of the pathway were assembled. The major C4 lineages of grasses (e.g. Chloridoideae and Andropogoneae) are composed of numerous and ecologically successful C4 species, but lack C3–C4 intermediate species, and are only distantly related to C3 taxa (Christin et al., 2008; Grass Phylogeny Working Group II, 2012). Photosynthetic variation exists in some small groups of grasses, notably Steinchisma, which contains C3–C4 intermediates and C3 species, and Alloteropsis, which has both C3 and C4 taxa (Duvall et al., 2003; Ibrahim et al., 2009; Christin et al., 2010), but only one grass clade has been identified that contains C3, C4, and C3–C4 intermediate species. This is the Neurachne/Thyridolepis clade (in the subtribe Neurachninae; Morrone et al., 2012), a group of three genera and 11 species endemic to Australia (Blake, 1972; Macfarlane, 2007).
Within the Neurachninae, the genus Neurachne includes one C4 species (N. munroi), five C3 species, and one C3–C4 intermediate species, N. minor (Hattersley et al., 1982, 1986; Hattersley and Roksandic, 1983; Macfarlane, 2007). The monospecific genus Paraneurachne (P. muelleri) is C4, while the other genera (Thyridolepsis, Ancistrachne, Cleistochloa, and Calyptochloa) are C3. The clade belongs to the Panicoideae subfamily, which encompasses the vast majority of C4 grass lineages, and thus appears especially prone to transitions from C3 to C4 biochemistry (Grass Phylogeny Working Group II, 2012). Extensive work in the 1980s characterized the anatomy, biochemistry, and physiology of Neurachninae species (Hattersley et al., 1982, 1986; Hattersley and Roksandic, 1983; Hattersley and Stone, 1986; Moore and Edwards, 1989), indicating high potential for this group to serve as a model for C4 grass evolution. However, to make evolutionary inferences, it is necessary to have a well-resolved, species-level phylogeny. Such a phylogeny was not available for the Neurachninae, as only some members of the group have been analysed with a small number of molecular markers (Hudson et al., 1990; Christin et al., 2008; Grass Phylogeny Working Group II, 2012).
The objective of the present study was a reconstruction of the evolutionary history of the Neurachninae, with an emphasis on photosynthetic pathway evolution. Multiple accessions per species were sampled, and phylogenetic analyses of plastid as well as nuclear markers, photosynthetic pathway identification, and measures of genome size were included. This comparative approach established the phylogenetic relationships between Neurachninae species, and revealed the genome dynamics of the group. The outcomes also highlight the diversity of photosynthetic transitions in the Neurachninae, and provide a solid foundation for future studies aimed at elucidating the anatomical and molecular mechanisms underlying these transitions.
Materials and methods
Plant material
Live, field-collected individuals of Neurachninae species were sampled for genome size analyses using flow cytometry (Supplementary Table S1 available at JXB online), while carbon isotope ratios were determined using multiple herbarium samples of each species (Supplementary Appendix S1). Multiple individuals of each Neurachninae species were also sampled for phylogenetic analyses; these were herbarium specimens or plants collected from the field (Supplementary Tables S1, S2).
Carbon isotope ratios (δ13C)
Carbon isotope values were determined for all described Neurachne, Paraneurachne, and Thyridolepis species (Supplementary Appendix S1 at JXB online) by the University of California, Davis Stable Isotope Facility (http://stableisotopefacility.ucdavis.edu). Some of the samples were previously assayed for δ13C (Hattersley and Roksandic, 1983; Hattersley et al., 1986).
Determination of genome size
Nuclei were simultaneously released from fresh leaf material from a Neurachninae species and a calibration standard [Raphanus sativus cv. Saxa (2C DNA content = 1.11 pg; Doležel et al., 1992) or Lepidosperma gibsonii (2C DNA content = 0.56 pg; M. Wallace, unpublished results)] by chopping with a razor blade in cold buffer (Roberts, 2007). Samples were filtered, and the nuclei were stained with propidium iodide (Roberts, 2007) and analysed using a BD FACSCanto II flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA) with 488nm excitation and 585/424 band pass filter detection. Data were collected with BD FACSDIVA software (v. 5.0.2) and analysed using FlowJo v.7.6.3 software (Tree Star Inc., Ashland, OR, USA). At least 5000 nuclei per day were analysed from each individual on three separate days. The genome size of an individual was then calculated as the average of these three estimates. Genome size and standard deviation (SD) of conspecific homoploids (Supplementary Table S1 at JXB online) were calculated using the average genome sizes of the individual homoploids. Estimates were only included if the peak heights of the sample and standard were similar, and the coefficient of variation was <5%. Conversion of pg DNA to Mbp DNA followed Doležel et al. (2003).
Amplification of plastid markers and ITS
Five plastid markers (ndhF, trnK/matK, rpoC2, rpl16, and trnLF) that have been used previously to investigate relationships among grasses (Duvall et al., 2003; Salariato et al., 2010; Grass Phylogeny Working Group II, 2012) were amplified from genomic DNA (gDNA) extracted from herbarium samples (MP FastDNA SPIN Kit and FastPrep Instrument; MP Biomedicals LLC, Solon, OH, USA). For ndhF and trnK/matK, previously published primers were used (Grass Phylogeny Working Group II, 2012), while primers for the three other markers were designed in the conserved regions of Panicoideae sequences downloaded from GenBank (Supplementary Table S3 at JXB online). The nuclear region encompassing the internal transcribed spacer 1, 5.8S rRNA, and internal transcribed spacer 2 (ITS) was amplified with the universal primers ITS4_rev and ITS5_for (Supplementary Table S3; White et al., 1990). However, in half the samples, these primers amplified endophytic fungal genes. A new forward primer, specific to grasses (ITS_grasses_for; Supplementary Table S3), was then used in combination with the ITS4 reverse primer. The plastid and ITS markers were amplified in overlapping fragments of ~300–700bp, with the protocol described in Grass Phylogeny Working Group II (2012). Single nucleotide polymorphisms were detected in most of the ITS sequences. These were coded as ambiguous characters in the phylogenetic analyses. All sequences have been submitted to GenBank (Supplementary Table S2).
Amplification of low-copy nuclear markers
As the gDNA extracted from herbarium specimens was too degraded to amplify low-copy nuclear markers, gDNA was isolated (DNeasy Plant Mini Kit, QIAGEN, Hilden, Germany) from selected accessions for which fresh material was available (Supplementary Table S4 at JXB online). Fragments of the genes encoding waxy and arodeh were amplified with previously designed primers (Christin et al., 2012b). The ppc-B2 lineage of phosphoenolpyruvate carboxylase (PEPC), which contains most C4-specific forms in grasses as well as non-C4 orthologues (Christin et al., 2007), was isolated in one fragment or two overlapping segments, using previously published primers (Christin et al., 2007, 2012b ). These three markers were isolated as described previously (Christin et al., 2012b ), but with an annealing temperature of 51 °C and an extension time of 2min. All sequences have been submitted to GenBank (Supplementary Table S4).
Phylogenetic analyses
The veracity of the sequence data was monitored throughout the analyses. The congruence between accessions of the same species and the lack of identical sequences for any of the plastid or nuclear markers, including the ppc-B2 genes, from different Neurachninae species indicated no cross-contamination or misidentification of the samples.
Sequences were aligned with MUSCLE (Edgar, 2004), and the alignments were manually refined. Outgroups were included based on previous work (Christin et al., 2012b ; Grass Phylogeny Working Group II, 2012). Best substitution models were determined by hierarchical likelihood ratio tests. In all cases, the general time reversible substitution model with a gamma shape parameter (GTR+G) was selected. Phylogenetic trees were obtained by Bayesian inference with MrBayes 3.2 (Ronquist and Huelsenbeck, 2003). Two analyses, each consisting of four parallel chains, were run for 7 000 000 generations after a burn-in period of 3 000 000, sampling a tree each 1000 generations. A consensus tree was computed on the 14 000 sampled trees. Phylogenetic trees were also inferred under a maximum likelihood (ML) criterion with the software PhyML (Guindon and Gascuel, 2003). Branch support of the ML trees was evaluated with 100 bootstrap pseudo-replications.
Phylogenetic trees were inferred for concatenated plastid data and ITS separately, and a further estimate of phylogeny was carried out by concatenating these two data sets. The three low-copy nuclear markers were analysed separately. For waxy, parts of the sequences that consisted of simple sequence repeats or short tandem repeats were difficult to align and were removed from the data set, resulting in the elimination of 52 aligned base pairs. To decrease the possibility of a bias due to convergent evolution (Christin et al., 2007, 2012a ), ppc-B2 phylogeny was also inferred from introns only. The significance of topological incongruence between the different markers was evaluated by Shimodaira–Hasegawa (S-H) tests with multiple-comparison correction (Shimodaira and Hasegawa, 1999), as implemented in baseml software (Yang, 2007).
Molecular dating
The divergence times of species within the Neurachninae were estimated with a molecular dating approach, which allows for rate variation among branches, following the recommendations from Rutschmann (2006), as described in Christin et al. (2008). Molecular dating requires time constraints set a priori on some nodes of the phylogeny as minimum or maximum ages. In the absence of a reliable fossil record for this group, the root of the Neurachninae was fixed to 11 Million years ago (Ma), a preliminary estimate obtained with a larger phylogeny, and without taking phytolith fossils into account (P.-A. Christin et al., unpublished). If phytoliths are taken into account, this node would probably be moved to ~20 Ma (P.-A.Christin et al., unpublished); however, as the same constraint was set for all markers, the relative ages can be compared among the different markers.
The topology inferred from combined plastid markers and ITS was used for molecular dating as it was better resolved and more complete than those of individual markers. However, the estimation of ages was based only on plastid markers to avoid strong variation in evolutionary rates due to differences between plastid and nuclear markers. Additional molecular dating analyses were carried out from the ITS, as well as each of the low-copy nuclear genes, with topologies compatible with the inferred species relationships. For these markers, an outgroup from outside the Neurachninae was added to the data set and used to estimate the first divergence time of the Neurachninae, but was removed during the molecular dating analyses.
Results
Photosynthetic pathway variation in the Neurachninae
Leaf δ13C assays confirmed previous photosynthetic pathway determinations in the Neurachninae (Hattersley and Roksandic, 1983; Hattersley et al., 1986; Macfarlane, 2007). Mean δ13C values near –13‰ in N. munroi and P. muelleri demonstrate that these species are C4 (Table 1). All other Neurachninae members, including the C3–C4 intermediate N. minor, showed δ13C values between –24‰ and –29‰, which are typical of C3 plants (Table 1).
Table 1.
Carbon isotope ratios (δ13C) and genome size data for Neurachninae species. Carbon isotope ratios are average values ±standard deviation (SD) determined from multiple herbarium samples of the same species (Supplementary Appendix S1 at JXB online). Photosynthetic pathway designations are shown based on these results and Hattersley et al. (1986) for the C3–C4 species N. minor. Genome size measurements ±SD were calculated from the average genome sizes of individual homoploids, which were measured from fresh leaf material on three consecutive days (see the Materials and Methods for further detail).
| Species | δ13C ±SD | 2C ±SD (pg) | 2C (Mbp) | 1Cx ±SD (pg) | 2n | Ploidy |
|---|---|---|---|---|---|---|
| Neurachne alopecuroidea (C3) | −27.1±1.5 | 2.24±0.02 | 2190 | 0.56±0.01 | 36 | 4x |
| 2.77±0.05 | 2710 | 0.55±0.01 | 45a | 5x a | ||
| 3.33±0.04 | 3260 | 0.55±0.01 | 54 | 6x | ||
| 3.81 | 3730 | 0.54 | 63a | 7x a | ||
| N. annularis (C3) | −24.8±0 | 1.30±0.01 | 1270 | 0.65±0.01 | 18 | 2x |
| N. lanigera (C3) | −28.4±1.2 | 0.98±0.01 | 958 | 0.49±0.01 | 18 | 2x |
| ND | ND | ND | 36 | 4x | ||
| N. minor (C3–C4) | −27.1±1.0 | 2.76 | 2700 | 0.69 | 36 | 4x |
| N. munroi (C4) | −12.7±1.0 | ND | ND | ND | 18 | 2x |
| 3.61 | 3530 | 0.60 | 36 | 4x | ||
| ND | ND | ND | 54 | 6x | ||
| N. queenslandica (C3) | −27.1±0.4 | ND | ND | ND | 54 | 6x |
| N. tenuifolia (C3) | −25.7±0.5 | 1.48 | 1450 | 0.74 | 18 | 2x |
| Paraneurachne muelleri (C4) | −13.3±0.4 | 3.97±0.03 | 3880 | 0.99±0.01 | 36 | 4x |
| Thyridolepis mitchelliana (C3) | −26.6±1.4 | 1.32±0.01 | 1290 | 0.66±0.01 | 18 | 2x |
| T. multiculmis (C3) | −26.3±1.8 | 2.50±0.02 | 2450 | 0.63±0.01 | 36 | 4x |
| T. xerophila (C3) | −27.6±0.6 | ND | ND | ND | 18 | 2x |
ND, not determined.
a Ploidy and chromosome numbers are inferred from the flow cytometry results of this study; other values are from Prendergast and Hattersley (1985) and Macfarlane (2007).
Neurachninae genome size analyses
The 2C DNA estimates varied 4-fold from 0.98±0.01 pg DNA for the diploid N. lanigera S.T.Blake to 3.97±0.04 pg DNA for the tetraploid P. muelleri (Table 1). These values are well within the range observed in the Poaceae (Bennett and Leitch, 2010, Plant DNA C-values database, http://data.kew.org/cvalues/), and agree with previous estimates from the Neurachninae based on 2C values or chromosome counts (Prendergast and Hattersley, 1985; Morgan and Westoby, 2005; Macfarlane, 2007).
Intraspecific ploidy variation was found in N. alopecuroidea R.Br. (4x, 5x, 6x, and 7x), and although multiple ploidy levels have been observed previously for N. lanigera and N. munroi (Prendergast and Hattersley, 1985), only diploid and tetraploid forms of these species, respectively, were detected in this study.
Phylogenetic relationships among the Neurachninae
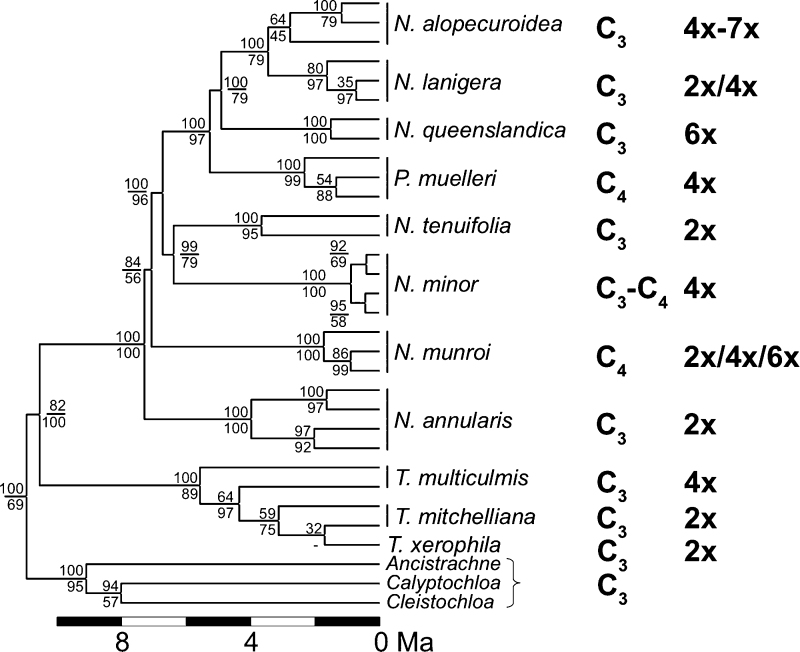
All plastid and nuclear markers showed that Neurachne and Paraneurachne species formed a strongly supported monophyletic clade, which was sister to Thyridolepis spp., and the monospecific Paraneurachne was nested inside Neurachne (Supplementary Figs S1–S4 at JXB online). The original name of this species, Neurachne muelleri Hack. (Hackel, 1895), should thus be resurrected.
Based on plastid markers, each species of the Neurachne/ Paraneurachne group was strongly supported as monophyletic, with the exception of N. lanigera, which was poorly resolved with respect to N. alopecuroidea (Supplementary Fig. S1 at JXB online). The two C4 taxa (N. munroi and P. muelleri) were not closely related, and the C3–C4 intermediate N. minor was not sister to either of the C4 taxa (Supplementary Fig. S1).
The topology of the phylogeny inferred from ITS (Supplementary Fig. S2 at JXB online) was almost identical to that from plastid markers. The only exception was N. queenslandica S.T.Blake, which was positioned as sister to N. alopecuroidea and not P. muelleri. Forcing N. queenslandica to be sister to N. alopecuroidea in the phylogeny inferred from plastid markers led to a significant decrease of likelihood (S-H test, difference of log-likelihoods= –55.801, P < 0.001), as did forcing it to be sister to P. muelleri in the ITS phylogeny (S-H test, difference of log-likelihoods= –35.332, P < 0.005). This incongruence between the plastid and ITS inferred phylogenies thus appears real and is symptomatic of incomplete lineage sorting or reticulate evolution. Since N. queenslandica is hexaploid (Prendergast and Hattersley, 1985), an allopolyploid origin is likely, with the female parent related to the P. muelleri lineage and the male parent to the N. alopecuroidea/N. lanigera lineage (assuming maternal chloroplast inheritance). Neurachne queenslandica, therefore, may be a natural hybrid between C3 and C4 parents, although further support for this hypothesis is required as C4 photosynthesis may have evolved in P. muelleri after the hybridization event.
The phylogeny inferred from combined plastid and ITS markers (plastid+ITS) was strongly resolved (Fig. 1), and was thus considered representative of the species relationships. In this phylogeny, the monophyly of all species was strongly supported, with the exception of Thyridolepis taxa.
Fig. 1.
Phylogeny of the Neurachninae, photosynthetic types, and ploidy levels. The tree was obtained through Bayesian inference using the plastid markers ndhF, trnK/matK, trnLF, rpl16, and rpoC2, and the genomic region encoding the internal transcribed spacer 1, 5.8S rRNA, and internal transcribed spacer 2 (ITS). Bayesian support values and bootstrap values are indicated above and below branches, respectively. The tree was calibrated and branch lengths are proportional to divergence times, in million years ago (Ma). Photosynthetic types are indicated on the right, as are ploidy levels. Multiple branches to the same species denote different genes or alleles.
The relationships between species inferred from arodeh (Supplementary Fig. S3 at JXB online) and waxy (Supplementary Fig. S4) were poorly supported, but nevertheless compatible with the plastid+ITS phylogeny. Forcing the topologies to that deduced from plastid+ITS did not significantly decrease the likelihood (S-H test, difference of log-likelihoods = –2.786 and –0.822, respectively, P=0.257 and 0.269, respectively). All arodeh and waxy sequences isolated from the same species were monophyletic, with the exception of the sequences encoding these markers isolated from T. multiculmis S.T.Blake. Two sequences encoding both waxy and arodeh were isolated from this species, and in both cases one sequence was more similar to that from T. mitchelliana S.T.Blake than it was to the other sequence isolated from T. multiculmis.
Evolutionary history of ppc-B2 in the Neurachninae
The coding sequences encompassing exons 5–10 (1786bp; 270 parsimoniously informative sites within the Neurachninae) of the Neurachninae ppc-B2 genes were placed in a larger data set encompassing many Panicoideae ppc-B2 sequences (Christin et al., 2012b ). In this inferred phylogeny, the Neurachninae sequences were positioned near those of affiliated C3 Paniceae, and were monophyletic, with the exception of one Ancistrachne sequence, which was positioned outside the Neurachninae and was therefore excluded from further analyses.
Inside the Neurachninae, despite a lack of resolution of the deeper nodes, the phylogenetic relationships deduced from ppc-B2 sequences were almost identical to those deduced from other markers (Supplementary Fig. S5 at JXB online). The only exceptions were the sequences from P. muelleri and N. munroi. Three groups of highly divergent ppc-B2 sequences were retrieved from P. muelleri. One of these groups was supported as sister to N. alopecuroidea/N. lanigera ppc-B2 sequences, as expected based on other markers (Fig. 1). The exact positions of the other two clusters of P. muelleri ppc-B2 sequences, however, were not resolved with confidence, although one of them is strongly supported as sister to sequences from N. munroi (Supplementary Fig. S5, subclade highlighted in red). The close relationship between N. munroi ppc-B2 sequences and some ppc-B2 sequences from P. muelleri was strongly supported even when a phylogeny was inferred from the five introns only (774 aligned base pairs, including 129 parsimoniously informative sites within the Neurachninae; Supplementary Fig. S6, subclade highlighted in red).
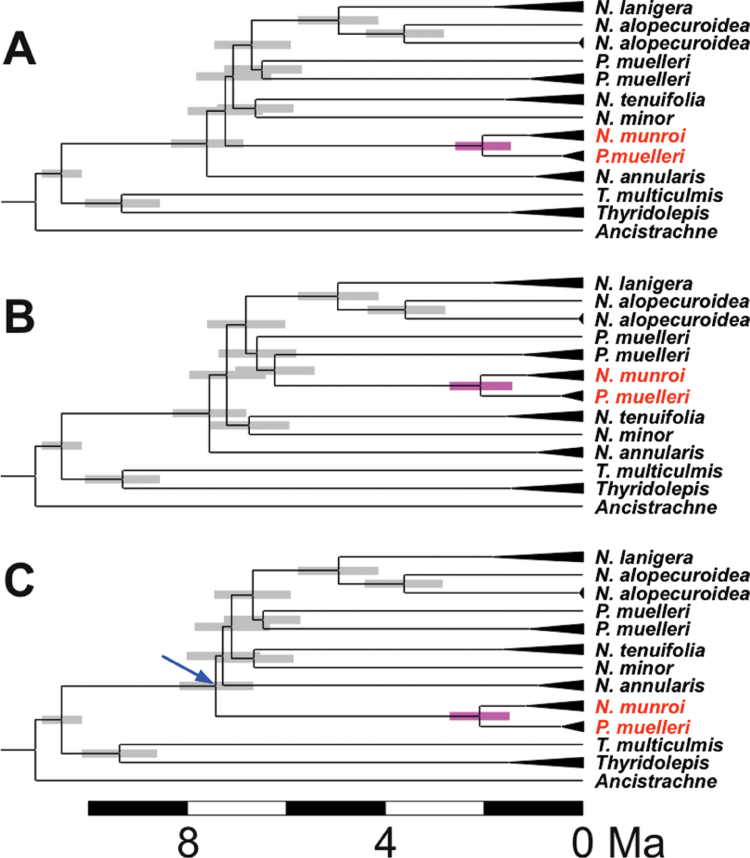
Different hypotheses reconciling ppc-B2 phylogeny with those inferred from other markers were evaluated using the sequence data (Fig. 2). Forcing the monophyly of all P. muelleri ppc-B2 sequences and placing them as expected for the species, and placing the N. munroi sequences in the predicted species position—the hypothesis of gene transmission that followed species genealogy—very strongly decreased the likelihood of the data (S-H test, difference of log-likelihoods= –246.516, P < 0.0001). Thus the close relationship between some of the P. muelleri ppc-B2 sequences and the N. munroi ppc-B2 sequences is unambiguous. A scenario in which a gene transfer occurred from N. munroi to P. muelleri involved placing the closely related N. munroi and P. muelleri sequences as expected for N. munroi, and the other P. muelleri sequences as expected for this species (Fig. 2A). This treatment of the data did not significantly affect the likelihood (S-H test, difference of log-likelihoods= –5.542, P=0.621). The alternative hypothesis, a gene transfer from P. muelleri to N. munroi, was also examined, and involved moving all N. munroi and P. muelleri ppc-B2 sequences to the position expected for P. muelleri (Fig. 2B). Again, no significant effect on the likelihood was seen (S-H test, difference of log-likelihoods= –6.799, P=0.588). The hypothesis that the data set might encompass different paralogues that appeared through gene duplication before the diversification of Neurachne/Paraneurachne was also tested. This implied placing P. muelleri sequences as sister to all other Paraneurachne/Neurachne sequences (Fig. 2C), which did not significantly decrease the likelihood (S-H test, difference of log-likelihoods= –9.391, P=0.485). This hypothesis, however, is not supported by the molecular dating analyses, which indicate that the divergence of the closely related N. munroi and P. muelleri ppc-B2 genes occurred long after the initial diversification of Neurachne/Paraneurachne (Fig. 2C). Finally, it was considered whether the phylogenetic analyses might be biased by convergent amino acid changes in these two C4 species (Christin et al., 2007, 2012a ). However, topological tests based only on introns yielded identical results, ruling out such a scenario. The data, therefore, show that at least some Neurachninae ppc-B2 genes were not transmitted following the species genealogy, but the direction of transmission, N. munroi to P. muelleri or vice versa, cannot be determined with confidence from the current data.
Fig. 2.
Different evolutionary scenarios for ppc-B2, and divergence times within the Neurachninae. The inferred chronograms are represented for the different statistically plausible hypotheses: (A) gene transfer from N. munroi to P. muelleri; (B) gene transfer from P. muelleri to N. munroi; and (C) ancient duplication resulting in different paralogues being compared. Branch lengths are proportional to divergence times, in million years ago (Ma). Sequences with low divergence and belonging to the same species are compressed. Putative C4-specific forms are highlighted in red. Confidence intervals of age estimates are represented by boxes for interspecific nodes. Purple boxes represent putative lateral gene transfer events. The blue arrow points to the ancient gene duplication postulated for hypothesis C.
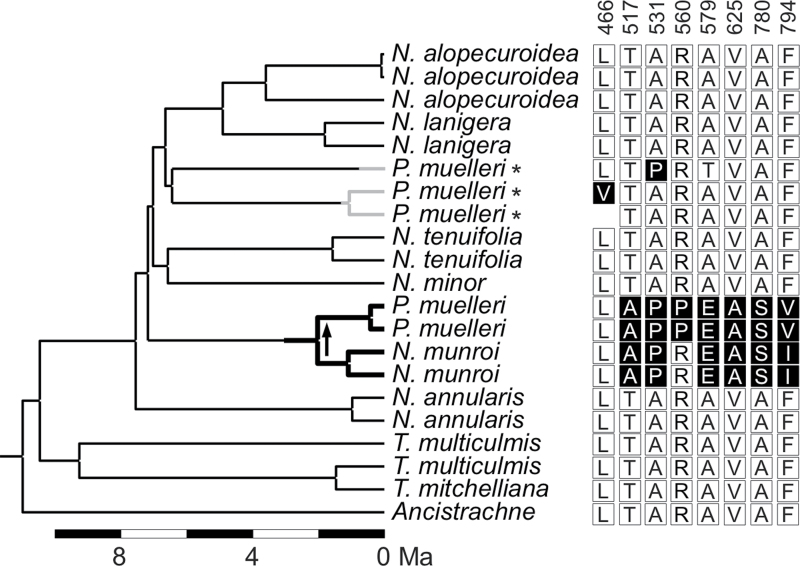
Residues at positions putatively selected for C4-specific function (Christin et al., 2007) were compared among the predicted amino acid sequences of the Neurachninae ppc-B2 sequences (Fig. 3). Of the 12 positions with the highest probability of having been positively selected in grasses (Christin et al., 2007), four were conserved in all members of the Neurachninae, with amino acid residues characteristic of non-C4 ppc-B2 proteins (positions 577, 637, 761, and 807). At the other positions, the predicted amino acid sequences encoded by the closely related N. munroi and P. muelleri ppc-B2 genes presented residues that characterize independently evolved C4 ppc-B2 genes (Fig. 3), with a serine at position 780 that was shown to be a major determinant of C4-specific PEPC activity (Bläsing et al., 2000; Svensson et al., 2003). The other P. muelleri ppc-B2 sequences were predicted to encode some C4-characteristic residues (Fig. 3), but also contained several stop codons and deletions altering the reading frame, and are very probably non-functional pseudogenes.
Fig. 3.
Evolutionary history of ppc-B2 in the Neurachninae. The calibrated phylogeny represents one of the hypotheses compatible with the species relationships, which assumes a gene transfer from N. munroi to P. muelleri (see Fig. 2A). Branch lengths are proportional to divergence times, in million years ago (Ma). Branches leading to the putative C4-specific genes are shown in bold. The black arrow indicates a putative horizontal transfer. Pseudogenes are indicated by asterisks and grey branches. Amino acids at the positions hypothesized to have been under C4-specific selection and that were variable inside the Neurachninae are shown on the right. C4-characteristic residues are highlighted in black.
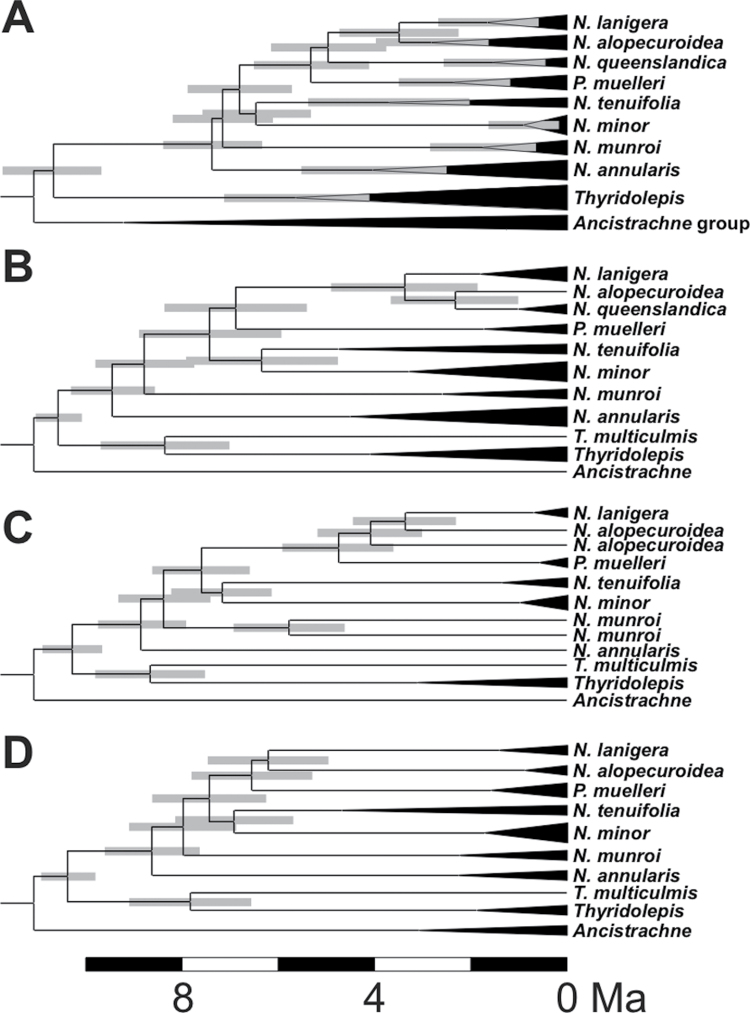
Molecular dating of the Neurachninae
The confidence intervals of estimated species divergence times based on plastid and nuclear markers overlapped (Fig. 4). Within most species, the divergence time between nuclear sequences overlaps with the crown age of the species. However, N. munroi arodeh sequences are estimated to have diverged long before the different N. munroi accessions. The occurrence of this pattern on a single gene can be explained by gene duplication or polyploidy followed by the loss of some gene copies. In addition, with multiple nuclear markers, the divergence of the different T. multiculmis sequences is estimated before the divergence of the different Thyridolepis species. This pattern, restricted to the tetraploid T. multiculmis, suggests an allopolyploid origin, with parents inside Thyridolepis, one of which was either not sampled or is extinct. The different hypotheses for the evolutionary history of ppc-B2 described above yielded similar age estimates (Fig. 2).
Fig. 4.
Divergence times for members of the Neurachninae estimated from different markers. The inferred chronograms are represented for (A) plastid markers, (B) ITS; (C) arodeh, and (D) waxy. Branch lengths are proportional to divergence times, in million years ago (Ma). For the plastid markers, the main taxa are compressed. For nuclear markers, sequences with low divergence and belonging to the same species are compressed. Confidence intervals of age estimates are represented by grey boxes for interspecific nodes in all trees, and species divergence in the plastid tree only.
Discussion
Evolutionary origins of C4 biochemistry in the Neurachninae
The C4 and C3–C4 members of the Neurachninae are not sister species, but are separated in both plastid- and nuclear marker-based phylogenies by C3 taxa (Fig. 1; Supplementary Figs S1–S4 at JXB online). In other groups containing species demonstrating different photosynthetic types, and for which robust phylogenies exist, some C3–C4 species are directly sister to C4 taxa as in Flaveria (McKown et al., 2005), Cleome (Feodorova et al., 2010), Heliotropium (Hilger and Diane, 2003), and the Molluginaceae (Christin et al., 2011). This pattern suggests that the photosynthetic variation observed in extant taxa reflects a gradual transition from C3 to C3–C4 and finally to C4 photosynthesis. However, several C3–C4 taxa are not closely related to any C4 species (Sage et al., 2011), and many groups encompass a mixture of C3, C3–C4, and C4 taxa, including the Neurachninae, but also other Flaveria (McKown et al., 2005) and Molluginaceae (Christin et al., 2011) lineages, sedges (Roalson et al., 2010), and the Chenopodiaceae (Sage et al., 2011).
The clustering of taxa using different photosynthetic pathways that is observed in the phylogenetic trees of the Neurachninae and other groups suggests a complex evolutionary history, with either multiple transitions from C3 to C3–C4 or C4 photosynthesis, multiple losses of an ancestral C4 or C3–C4 type, or a combination of both (Christin et al., 2010). A simple scenario of a single C4 origin followed by reversals to the C3 (or C3–C4) state can be ruled out for the Neurachninae by the analyses of genes encoding PEPC. During the evolution of this key C4 pathway enzyme, mutations occurred in the genes that resulted in modifications to the catalytic and regulatory properties of the encoded proteins, allowing them to function optimally in C4 photosynthesis (Dong et al., 1998; Bläsing et al., 2000). These mutations often occur at identical positions in the PEPC coding sequences of distantly related species (Christin et al., 2007; Besnard et al., 2009). Several amino acid changes characteristic of C4-specific PEPC were predicted from the ppc-B2 genes of C4 Neurachninae, including the key C4 determinant alanine to serine mutation at position 780 (Fig. 3). If the C4-specific properties of PEPC appeared before the diversification of the Neurachninae, the ppc-B2 genes of non-C4 taxa most probably would have maintained codons for some C4-characteristic amino acids. However, the ppc-B2 genes of the C3 and C3–C4 Neurachninae do not encode C4-specific amino acids, and are indistinguishable at these positions from species known never to have been in a C4 state (Fig. 3), strongly suggesting that the ppc-B2 genes of these non-C4 taxa did not descend from genes encoding enzymes that were C4 optimized.
The C4-specific properties of the PEPC used by N. munroi and P. muelleri evolved after the divergence from non-C4 Neurachninae, but the C4 PEPC genes of these two species strongly grouped together in the phylogeny, even when only introns were considered, arguing against genetic convergence as a phylogenetic bias (Supplementary Figs S5, S6 at JXB online). The divergence of these genes occurred long after the split of N. munroi and the lineage leading to P. muelleri (Fig. 2), excluding paralogy as an explanation for this grouping. Therefore, the transmission of these C4-optimized genes did not follow the species genealogy. A lateral transfer might have occurred via allopolyploidization, but this is unlikely given that none of the other nuclear markers isolated from P. muelleri showed affinities with those of N. munroi. It is more likely that the transmission occurred via hybridization, followed by backcrossing and introgression into the population, or alternatively through a lateral gene transfer event, as shown for the grass Alloteropsis (Christin et al., 2012b ), and suggested for the sedge Eleocharis (Besnard et al., 2009). Although the direction of the ppc-B2 gene transfer is not known yet, a transfer from N. munroi to P. muelleri is more consistent with the present data. In some regions of Australia, N. munroi and P. muelleri co-occur (Prendergast and Hattersley, 1985; Supplementary Fig. S7), providing opportunities for a gene transfer that might have been favoured by natural selection. Some of the native P. muelleri ppc-B2 copies encode residues at positions 466 and 531 that generally characterize C4-specific PEPC (Besnard et al., 2009), which suggests that these genes previously fulfilled the C4 function in P. muelleri and underwent a few changes toward C4 optimization of the encoded enzyme. The acquisition of ppc-B2 genes from N. munroi that were more C4 optimized would have been advantageous, leading to a replacement of the vertically acquired P. muelleri genes, and their subsequent pseudogenization (Fig. 3).
Anatomical enablers of photosynthetic transitions in the Neurachninae
The optimization of C4 biochemistry is hypothesized to be one of the final steps in the evolution of C4 plants (Sage et al., 2012), with the initial steps involving the reduction of the mesophyll cell to bundle sheath cell ratio, the enhancement of bundle sheath organelles, and the appearance of a photorespiratory CO2 pump, which characterizes nearly all known C3–C4 intermediates (Hylton et al., 1988; Griffiths, 1989; Hattersley and Watson, 1992; Sage et al., 2012). The C4 P. muelleri and N. munroi and the C3–C4 N. minor have very similar foliar anatomies, with short interveinal distances, low mesophyll to bundle sheath tissue volume, and high numbers of bundle sheath organelles, typical of C4 taxa (Hattersley et al., 1982, 1986; Brown and Hattersley, 1989). The C3 taxa that separate them in the phylogeny have a comparatively larger interveinal distance and smaller bundle sheath cells (Hattersley et al., 1982, 1986; Macfarlane, 2007), although the interveinal distance tends to be shorter than that of typical C3 taxa in N. queenslandica and N. annularis Macfarlane, and especially in N. tenuifolia S.T.Blake, where additional minor bundles reduce the average interveinal distance (Hattersley et al., 1982; Macfarlane, 2007). C3 species in the Neurachne/Thyridolepis clade have conspicuous mestome sheaths, the tissue that has been recruited for carbon reduction in C3–C4 and C4 Neurachninae species (Dengler et al., 1985). Indeed, while this tissue is usually devoid of organelles in C3 taxa, it contains some chloroplasts and a high number of mitochondria in these C3 Neurachninae (Hattersley et al., 1982, 1986; Hattersley and Roksandic, 1983). The presence of these organelles in the mestome sheath may have facilitated the evolution of a photorespiratory CO2 pump in the Neurachninae, enabling the subsequent transition to a complete C4 pathway (Sage et al., 2012). As has been hypothesized for other anatomical characters in some eudicot lineages with multiple C4 origins (Marshall et al., 2007; McKown and Dengler, 2007; Christin et al., 2011), the high organelle content of C3 Neurachninae mestome sheaths may have contributed to the multiple origins of biochemical carbon-concentrating mechanisms in this grass group.
Recurrent polyploidizations in the Neurachninae as diversification facilitators?
The distribution of ploidy levels in the species phylogeny (Fig. 1) suggests either multiple polyploidy events or a few followed by a return to a diploid state (diploidization). The latter suggestion can be disregarded because it would have led to a high chromosome number in the newly formed diploids, which is not observed in the Neurachninae (Table 1). In addition, multiple polyploidizations are supported by the existence of intraspecific ploidy variation in several taxa (Table 1; Prendergast and Hattersley, 1985). The data therefore suggest that seven independent polyploidization events have occurred in the Neurachne/Thyridolepis clade, one per polyploid species (Fig. 1).
The analyses of markers from multiple genomes identified T. multiculmis and N. queenslandica as probable allotetraploids, the latter possibly representing a hybrid from a C3 and a C4 parent. In contrast, the tetraploid C3–C4 intermediate N. minor does not appear to be the result of an allopolyploidization between C3 and C4 Neurachninae species, or at least not between known members, as the sequences from N. minor clustered together in the phylogenies at the same position for all plastid and nuclear markers (Fig. 1; Supplementary Figs S1–S4 at JXB online). With leaf anatomy similar to that of the C4 N. munroi (Hattersley et al., 1982), and a typically C3 isotopic value (Table 1; Hattersley and Roksandic, 1983; Hattersley et al., 1986), consistent with minimal or limited C4 cycle activity (Hattersley and Stone, 1986; Hattersley et al., 1986; von Caemmerer, 1992), N. minor may represent a transitional stage toward an additional origin of full C4 photosynthesis in the Neurachninae. As for N. minor, all the plastid and nuclear marker sequences belonging to the polyploid N. alopecuroidea, N. lanigera, and N. munroi clustered together in the phylogenies (Fig. 1; Supplementary Figs S1–S6), suggesting that the dominant mechanism for increased chromosome numbers in this grass group was autopolyploidization.
The two C4 taxa and the C3–C4 intermediate species of the Neurachninae are all polyploids of putatively recent origin (Fig. 1), and autopolyploidization might have allowed photosynthetic diversification by providing additional copies of genes encoding enzymes that fulfilled housekeeping functions in the C3 ancestors. A relaxation of selection due to genetic redundancy has indeed been suggested to facilitate the recruitment of enzymes into novel photosynthetic functions (Monson, 2003; Wang et al., 2009); however, as yet no empirical evidence exists to support this hypothesis, and comparison of distantly related grass genomes questioned the importance of gene duplication in the evolution of the C4 syndrome (Williams et al., 2012). The apparent correlation of autopolyploidy and derived photosynthetic types in the Neurachninae offers an opportunity to test the link between genetic redundancy and photosynthetic diversification within a sound phylogenetic framework.
Conclusion
The Neurachninae have long been viewed as a potential model for understanding the evolution of C4 photosynthesis in the grasses due to natural photosynthetic variation in the group; however, evolutionary inferences were hampered by the lack of a phylogenetic framework. Using multiple markers from the chloroplast and nuclear genomes, it has been shown that C4 biochemistry has evolved more than once in this grass group, and may have been facilitated by anatomical enablers, a high frequency of autopolyploidization, and transfer of C4-optimized genes between related taxa. Molecular dating indicates that these events most probably occurred during the late Pliocene, an epoch of relatively low atmospheric CO2 (Gerhart and Ward, 2010), when the Australian continent became drier (Bowler, 1976). The present-day distribution of extant Neurachninae (Prendergast and Hattersley, 1985) indicates that the group appeared in the red sand soils and stony rises of semi-arid west-central Australia. These observations are consistent with recent models of C4 evolution that hypothesize that climate deterioration in the past 5 million years selected for a series of traits that predisposed taxa to evolve novel photosynthetic mechanisms to deal with the combined effects of severe stress and low atmospheric CO2 (Sage et al., 2012).
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Neurachninae phylogeny inferred from plastid markers.
Figure S2. Neurachninae phylogeny inferred from the ITS region.
Figure S3. Phylogenetic relationships among arodeh sequences.
Figure S4. Phylogenetic relationships among waxy sequences.
(Figure S5. Phylogenetic relationships among ppc-B2 sequences.
Figure S6. Phylogenetic relationships among ppc-B2 sequences deduced from intron sequences.
Figure S7. Distribution of Neurachne spp. and Paraneurachne muelleri in Australia.
Table S1. Neurachne, Paraneurachne, and Thyridolepis samples used for genome size analyses.
Table S2. Plastid and ITS sequences from Neurachne, Paraneurachne, and Thyridolepis species used in phylogenetic analyses.
Table S3. Primers used in this study.
Table S4. Neurachninae sampled for low-copy number nuclear markers.
Appendix S1. Herbarium vouchers for specimens of Neurachne, Paraneurachne, and Thyridolepis sampled for carbon isotope assays.
Supplementary Material
Acknowledgements
The authors acknowledge the facilities, scientific, and technical assistance of the Australian Microscopy and Microanalysis Research Facility at the Centre for Microscopy, Characterisation and Analysis, The University of Western Australia, a facility funded by the University, State, and Commonwealth Governments. The authors thank J. Doležel (Institute of Experimental Botany, Olomouc, Czech Republic) for supplying seeds of Raphanus sativus cv. Saxa. Parts of this work resulted from discussions at the workshop ‘Using Functional Genomics to Harness Adaptive Traits in Australian Native Plants’, held at the University of Western Australia, 1–5 November 2010. We thank D. Albrecht, Northern Territory Herbarium, Alice Springs, for supplying seeds of Neurachne tenuifolia and Paraneurachne muelleri. This work was supported by the Marie Curie IOF 252568 fellowship to PAC. Funding provided by the Western Australia Department of Environment and Conservation, and the University of Western Australia for collection of living Neurachninae species by the Wiluna 7 (TDM, ML, RFS, HC, P. Finnegan, J. Cheeseman, and B. Chapman), and subsequently by the Kookynie 3 (TDM, ML, and P. Finnegan) is gratefully acknowledged.
References
- Besnard G, Muasya AM, Russier F, Roalson EH, Salamin N, Christin PA. 2009. Phylogenomics of C4 photosynthesis in sedges (Cypercaeae): multiple appearances and genetic convergence Molecular Biology and Evolution 26 1909–1919 [DOI] [PubMed] [Google Scholar]
- Blake ST. Neurachne and its allies (Gramineae) Contributions from the Queensland Herbarium No. 13 1972 [Google Scholar]
- Bläsing OE, Westhoff P, Svensson P. 2000. Evolution of C4 phosphoenolpyruvate carboxylase in Flaveria, a conserved serine residue in the carboxyl-terminal part of the enzyme is a major determinant for C4-specific characteristics Journal of Biological Chemistry 275 27917–27923 [DOI] [PubMed] [Google Scholar]
- Bowler JM. 1976. Aridity in Australia: age, origins and expression in Aeolian landforms and sediments Earth-Science Reviews 12 279–310 [Google Scholar]
- Brown RH. 1999. Agronomic implications of C4 photosynthesis. In: Sage RF, Monson RK, eds.C4plant biology San Diego, CA: Academic Press; 473–517 [Google Scholar]
- Brown RH, Hattersley PW. 1989. Leaf anatomy of C3–C4 species as related to evolution of C4 photosynthesis Plant Physiology 91 1543–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin PA, Besnard G, Edwards EJ, Salamin N. 2012. a Effect of genetic convergence on phylogenetic inference Molecular Phylogenetics and Evolution 62 921–927 [DOI] [PubMed] [Google Scholar]
- Christin PA, Besnard G, Samaritani E, Duvall MR, Hodkinson TR, Savolainen V, Salamin N. 2008. Oligocene CO2 decline promoted C4 photosynthesis in grasses Current Biology 18 37–43 [DOI] [PubMed] [Google Scholar]
- Christin PA, Edwards EJ, Besnard G, Boxall SF, Gregory R, Kellogg EA, Hartwell J, Osborne CP. 2012. b Adaptative evolution of C4 photosynthesis through recurrent lateral gene transfer Current Biology 22 445–449 [DOI] [PubMed] [Google Scholar]
- Christin PA, Freckleton RP, Osborne CP. 2010. Can phylogenetics identify C4 origins and reversals? Trends in Ecology and Evolution 25 403–409 [DOI] [PubMed] [Google Scholar]
- Christin PA, Sage TL, Edwards EJ, Ogburn RM, Khoshravesh R, Sage RF. 2011. Complex evolutionary transitions and the significance of C3–C4 intermediate forms of photosynthesis in Molluginaceae Evolution 65 643–660 [DOI] [PubMed] [Google Scholar]
- Christin PA, Salamin N, Savolainen V, Duvall MR, Besnard G. 2007. C4 photosynthesis evolved in grasses via parallel adaptive genetic changes Current Biology 17 1241–1247 [DOI] [PubMed] [Google Scholar]
- Dengler NG, Dengler RE, Hattersley PW. 1985. Differing ontogenetic origins of PCR (‘Kranz’) sheaths in leaf blades of C4 grasses (Poaceae) American Journal of Botany 72 284–302 [Google Scholar]
- Doležel J, Bartoš J, Voglmayr H, Greilhuber J. 2003. Nuclear DNA content and genome size of trout and human Cytometry Part A 51A 127–128 [DOI] [PubMed] [Google Scholar]
- Doležel J, Sgorbati S, Lucretti S. 1992. Comparison of three DNA fluorochromes for flow cytometric estimation of nuclear DNA content in plants Physiologia Plantarum 85 625–631 [Google Scholar]
- Dong L, Masuda T, Kawamura T, Hata S, Izui K. 1998. Cloning, expression, and characterization of a root-form phosphoenolpyruvate carboxylase from Zea mays: comparison with the C4-form enzyme Plant and Cell Physiology 39 865–873 [DOI] [PubMed] [Google Scholar]
- Duvall MR, Saar DE, Grayburn WS, Holbrook GP. 2003. Complex transitions between C3 and C4 photosynthesis during the evolution of Paniceae: a phylogenetic case study emphasizing the position of Steinchisma hians (Poaceae), a C3–C4 intermediate International Journal of Plant Sciences 164 949–958 [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput Nucleic Acids Research 32 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards EJ, Ogburn RM. 2012. Angiosperm responses to a low-CO2 world: CAM and C4 photosynthesis as parallel evolutionary trajectories International Journal of Plant Science 173, in press [Google Scholar]
- Ehleringer JR, Cerling TE, Helliker BR. 1997. C4 photosynthesis, atmospheric CO2, and climate Oecologia 112 285–299 [DOI] [PubMed] [Google Scholar]
- Feodorova TA, Voznesenskaya EV, Edwards GE, Roalson EH. 2010. Biogeographic patterns of diversification and the origins of C4 in Cleome (Cleomaceae) Systematic Botany 35 811–826 [Google Scholar]
- Gerhart LM, Ward JK. 2010. Plant responses to low [CO2] of the past New Phytologist 188 674–695 [DOI] [PubMed] [Google Scholar]
- Grass Phylogeny Working Group II 2012. New grass phylogeny resolves deep evolutionary relationships and discovers C4 origins New Phytologist 193 304–312 [DOI] [PubMed] [Google Scholar]
- Griffiths H. 1989. Carbon dioxide concentrating mechanisms in relation to the evolution of CAM in vascular epiphytes. In: Luttge U, ed. Ecological studies 76. Vascular plants as epiphytes Berlin: Springer-Verlag; 42–86 [Google Scholar]
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood Systematic Biology 52 696–704 [DOI] [PubMed] [Google Scholar]
- Hackel E. Neurachne muelleri n. sp. Österreichische Botanische Zeitschrift. 1895;45:329. [Google Scholar]
- Hattersley PW, Roksandic Z. 1983. δ13C values of C3 and C4 species of Australian Neurachne and its allies (Poaceae) Australian Journal of Botany 31 317–321 [Google Scholar]
- Hattersley PW, Stone NE. 1986. Photosynthetic enzyme activities in the C3–C4 intermediate Neurachne minor S.T. Blake (Poaceae) Australian Journal of Plant Physiology 13 399–408 [Google Scholar]
- Hattersley PW, Watson L. 1992. Diversification of photosynthesis. In: Chapman GP, ed. Grass evolution and domestication Cambridge, UK: Cambridge University Press; 38–116 [Google Scholar]
- Hattersley PW, Watson L, Johnston CR. 1982. Remarkable leaf anatomical variations in Neurachne and its allies (Poaceae) in relation to C3 and C4 photosynthesis Botanical Journal of the Linnean Society 84 265–272 [Google Scholar]
- Hattersley PW, Wong SC, Perry S, Roksandic Z. 1986. Comparative ultrastructure and gas exchange characteristics of the C3–C4 intermediate Neurachne minor S. T. Blake (Poaceae) Plant, Cell and Environment 9 217–233 [Google Scholar]
- Hilger HH, Diane N. 2003. A systematic analysis of Heliotropiaceae (Boraginales) based on trnL and ITS1 sequence data Botanische Jahrbücher 125 19–51 [DOI] [PubMed] [Google Scholar]
- Hudson GS, Mahon JD, Anderson PA, Gibbs MJ, Badger MR, Andrews TJ, Whitfeld PR. 1990. Comparisons of rbcL genes for the large subunit of ribulose-bisphosphate carboxylase from closely related C3 and C4 taxa Journal of Biological Chemistry 265 808–814 [PubMed] [Google Scholar]
- Hylton CM, Rawsthorne S, Smith AM, Jones A, Woolhouse HW. 1988. Glycine decarboxylase is confined to the bundle-sheath cells of leaves of C3–C4 intermediate species Planta 175 452–459 [DOI] [PubMed] [Google Scholar]
- Ibrahim DG, Burke R, Ripley BS, Osborne CP. 2009. A molecular phylogeny of the genus Alloteropsis (Panicoideae, Poaceae) suggests an evolutionary reversion from C4 to C3 photosynthesis Annals of Botany 103 127–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane TD. 2007. A new species of Neurachne (Poaceae) from Western Australia Nuytsia 17 215–222 [Google Scholar]
- Marshall DM, Muhaidat R, Brown NJ, Liu Z, Stanley S, Griffiths H, Sage RF, Hibberd JM. 2007. Cleome, a genus closely related to Arabidopsis, contains species spanning a developmental progression from C3 to C4 photosynthesis The Plant Journal 51 886–896 [DOI] [PubMed] [Google Scholar]
- McKown AD, Dengler NG. 2007. Key innovations in the evolution of Kranz anatomy and C4 vein pattern in Flaveria (Asteraceae) American Journal of Botany 94 382–399 [DOI] [PubMed] [Google Scholar]
- McKown AD, Moncalvo JM, Dengler NG. 2005. Phylogeny of Flaveria (Asteraceae) and inference of C4 photosynthesis evolution American Journal of Botany 92 1911–1928 [DOI] [PubMed] [Google Scholar]
- Monson RK. 2003. Gene duplication, neofunctionalization, and the evolution of C4 photosynthesis International Journal of Plant Science 164 S43–S54 [Google Scholar]
- Moore BD, Edwards GE. 1989. Metabolism of 14CO2 by leaves of different types of Neurachne species Plant Science 60 155–161 [Google Scholar]
- Morgan HD, Westoby M. 2005. The relationship between nuclear DNA content and leaf strategy in seed plants Annals of Botany 96 1321–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrone O, Aagesen L, Scataglini MA, Salariato DL, Denham SS, Chemisquy MA, Sede SM, Giussani LM, Kellogg EA, Zuloaga FO. 2012. Phylogeny of the Paniceae (Poaceae: Panicoideae): integrating plastid DNA sequences and morphology into a new classification Cladistics 28 333–356 [DOI] [PubMed] [Google Scholar]
- Muhaidat R, Sage TL, Frohlich MW, Dengler NG, Sage RF. 2011. Characterization of C3–C4 intermediate species in the genus Heliotropium L. (Boraginaceae): anatomy, ultrastructure and enzyme activity Plant, Cell and Environment 34 1723–1736 [DOI] [PubMed] [Google Scholar]
- Prendergast HDV, Hattersley PW. 1985. Distribution and cytology of Australian Neurachne and its allies (Poaceae), a group containing C3, C4 and C3–C4 intermediate species Australian Journal of Botany 33 317–336 [Google Scholar]
- Roalson EH, Hinchliff CE, Trevisan R, da Silva CRM. 2010. Phylogenetic relationships in Eleocharis (Cyperaceae): C4 photosynthesis origins and patterns of diversification in the spikerushes Systematic Botany 35 257–271 [Google Scholar]
- Roberts AV. 2007. The use of bead beating to prepare suspensions of nuclei for flow cytometry from fresh leaves, herbarium leaves, petals and pollen Cytometry Part A 71 1039–1044 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models Bioinformatics 19 1572–1574 [DOI] [PubMed] [Google Scholar]
- Rutschmann F. 2006. Molecular dating of phylogenetic trees: a brief review of current methods that estimate divergence times Diversity and Distribution 12 35–48 [Google Scholar]
- Sage RF. 2001. Environmental and evolutionary preconditions for the origin and diversification of the C4 photosynthetic syndrome Plant Biology 3 202–213 [Google Scholar]
- Sage RF, Christin PA, Edwards EJ. 2011. The C4 plant lineages of planet Earth Journal of Experimental Botany 62 3155–3169 [DOI] [PubMed] [Google Scholar]
- Sage RF, Sage TL, Kocacinar F. 2012. Photorespiration and the evolution of C4 photosynthesis Annual Review of Plant Biology 63 19–47 [DOI] [PubMed] [Google Scholar]
- Salariato DL, Zuloaga FO, Giussani LM, Morrone O. 2010. Molecular phylogeny of the subtribe Melinidinae (Poaceae: Panicoideae: Paniceae) and evolutionary trends in the homogenization of inflorescences Molecular Phylogenetics and Evolution 56 355–369 [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. 1999. Multiple comparisons of log-likelihoods with applications to phylogenetic inference Molecular Biology and Evolution 16 1114–1116 [Google Scholar]
- Still CJ, Berry JA, Collatz GJ, DeFries RS. 2003. Global distribution of C3 and C4 vegetation: carbon cycle implications Global Biogeochemical Cycles 17 1006–1030 [Google Scholar]
- Svensson P, Bläsing OE, Westhoff P. 2003. Evolution of C4 phosphoenolpyruvate carboxylase Archives of Biochemistry and Biophysics 414 180–188 [DOI] [PubMed] [Google Scholar]
- von Caemmerer S. 1992. Carbon isotope discrimination in C3–C4 intermediates Plant, Cell and Environment 15 1063–1072 [Google Scholar]
- Wang X, Gowik U, Tang H, Bowers JE, Westhoff P, Paterson AH. Comparative genomic analysis of C4 photosynthetic pathway evolution in grasses. Genome Biology. 2009;10:R68. doi: 10.1186/gb-2009-10-6-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor JW. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, eds. PCR protocols: a guide to methods and applications New York: Academic Press; 315–322 [Google Scholar]
- Williams BP, Aubry S, Hibberd JM. 2012. Molecular evolution of genes recruited into C4 photosynthesis Trends in Plant Science 17 213–220 [DOI] [PubMed] [Google Scholar]
- Yang ZH. 2007. PAML 4: phylogenetic analysis by maximum likelihood Molecular Biology and Evolution 24 1586–1591 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.