Abstract
The ubiquitin–proteasome system targets selected proteins for degradation by the 26S proteasome. Rpn12 is an essential component of the 19S regulatory particle and plays a role in recruiting the extrinsic ubiquitin receptor Rpn10. In the present paper we report the crystal structure of Rpn12, a proteasomal PCI-domain-containing protein. The structure helps to define a core structural motif for the PCI domain and identifies potential sites through which Rpn12 might form protein–protein interactions. We demonstrate that mutating residues at one of these sites impairs Rpn12 binding to Rpn10 in vitro and reduces Rpn10 incorporation into proteasomes in vivo.
Keywords: PCI domain, proteasome, ubiquitin, X-ray crystallography
Abbreviations: cryo-EM, cryo-electron microscopy; CSN, COP9 signalosome; DUB, deubiquitylating enzyme; eIF3, eukaryotic initiation factor 3; ESRF, European Synchrotron Radiation Facility; HSQC, heteronuclear single-quantum coherence; MPN, Mpr1/Pad1 N-terminal; ORF, open reading frame; PCI, proteasome, COP9 signalosome, initiation factor 3; rmsd, root mean square deviation; RP, regulatory particle; TPR, tetratrico peptide repeat; UBL, ubiquitin-like; UIM, ubiquitin-interacting motif; vWA, von Willebrand factor A; WH, winged helix; WT, wild-type
INTRODUCTION
The ubiquitin-mediated protein degradation pathway, through its ability to selectively remove proteins from the cell, regulates diverse functions that include cell-cycle control, protein quality control and transcription regulation. The initial steps in the pathway generate proteins that are covalently tagged with a polyubiquitin chain that is then recognized by ubiquitin receptors of the 26S proteasome. This is a large complex composed of a 20S catalytic core particle and two 19S RPs (regulatory particles) that catalyses the final step in the pathway [1–4]. While the 20S particle composes a catalytic chamber for protein degradation, collectively the proteins that compose the 19S particle perform several proteasomal functions that include recognition of ubiquitylated substrates, cleavage of the polyubiquitin chain for ubiquitin recycling, control of access to the 20S proteolytic chamber, and substrate unfolding and subsequent translocation into the 20S core particle for degradation [1].
The 19S RP can be subdivided into base and lid subparticles and although its exact composition is dependent on cellular context, a set of core components can be identified. The base contains the AAA-ATPase subunits (Rpt1–6), and two large α-helical repeat proteins (Rpn1/2) that serve as a platform for binding UBL (ubiquitin-like)-domain-containing proteins [5–8]. The lid is composed of six PCI (proteasome, COP9 signalosome, initiation factor 3) repeat-containing proteins (Rpn3, 5–7, 9 and 12), Rpn8, Rpn15/Sem1 and the DUB (deubiquitylating enzyme) Rpn11 [1,9]. The ubiquitin receptors Rpn13 [10,11] and Rpn10 [12], which have also been considered as base subunits, are located peripherally in a distal part of the RP, sited above the Rpt4/5 and Rpt1/2 heterodimers respectively [13]. Beyond these subunits, the DUBs Ubp6/Usp14, and Uch37/UCHL5, and the RP chaperone Rpn14 are examples of proteasome-interacting proteins that frequently co-purify with it [1,2,4]. To date Rpn11 is the only lid subunit to which a catalytic function has been assigned, and it is unclear how the core subunits of the RP and the associated interacting proteins co-ordinate their activities to carry out their diverse functions.
A map of protein–protein interactions within the 19S lid has been derived from analysis of the lid subcomplex by MS under native conditions [14], combined with the results from yeast two-hybrid [15] and genetic [16,17] approaches. The map suggested a lid organized into two subcomplexes composed respectively of Rpn5, 6, 8, 9 and 11, and Rpn3, 7, 12 and Dss1/Sem1. Rpn10 was not detected in the proteasomes analysed by MS. Subsequent studies using either MS analysis [18] or protein cross-linking [19] with a series of Saccharomyces cerevisiae lid mutants confirmed the existence of these assemblies and proposed that the lid complex assembles from a core composed of Rpn5, 6, 8, 9 and 11 to which a module consisting of Rpn subunits 3 and 7 and Sem1 subsequently attaches, followed by the incorporation of Rpn12.
A more detailed picture of the lid architecture has been provided by the recent determination of the structure of the S. cerevisiae 19S RP [20] and of the Schizosaccharomyces pombe 26S proteasome [21] by electron microscopy methods. These structures have confirmed a number of the Rpn12 and Rpn10 protein interactions previously observed by other methods. The six PCI-repeat-containing subunits of the lid, including Rpn12, associate via their PCI domains into a conformationally flexible horseshoe-shaped structure. Rpn12 is at the edge of this structure and is flanked by Rpn3 on one side and is also observed to interact with the base subunit Rpn2 [21]. Interactions between Rpn12 and Rpn3, Rpt3 and Rpn8 have been detected by cross-linking [19], and between Rpn12 and Rpn3 and Rpt3 by yeast two-hybrid analysis [15,22]. Rpn10 also makes extensive contacts with surrounding subunits: the N-terminal vWA (von Willebrand factor A) domain binds to Rpn11 and Rpn9, whereas its C-terminal UIM (ubiquitin-interacting motif) is proposed to contact Rpt4 and Rpt5 [13,20,21]. Rpn10 has also been reported to bind to Rpn1 through an interaction stabilized by Rpn2 [8].
Although they do not contact each other in the electron microscopy reconstruction, Rpn12 and Rpn10 interact genetically [19,23], and the purified proteins bind to each other in vitro [23,24]. As well as being an integral proteasomal subunit, a substantial fraction of Rpn10 can also be isolated from the cytosol [23,25]. This Rpn10 fraction can engage the polyubiquitin receptors Rad23 and Dsk2 through an interaction between their respective UIM and UBL domains to regulate the accessibility of these receptors to the proteasome [26–28].
Translation initiation factor eIF3 (eukaryotic initiation factor 3) and the COP9 signalosome (termed CSN) resemble the 19S proteasome lid in that they also contain multiple subunits encoding PCI domains, together with subunits containing MPN (Mpr1/Pad1 N-terminal) domains [9,29]. eIF3 is a translation initiation factor that is essential for the interaction between the 43S pre-initiation complex and the mRNA transcript [30], whereas the CSN complex is an isopeptidase best characterized by its ability to de-NEDDylate the cullin subunit present in the CRL (Cullin-RING-Ligases) family of E3 ubiquitin ligases [31]. The arrangement of subunits in the 19S proteasome lid and CSN complexes is proposed to be similar and, by bioinformatic analysis, subunits corresponding to each of the six PCI- and two MPN-containing proteins in the 19S proteasome lid can be identified in the CSN complex [32]. Rpn12 consists of an N-terminal PCI domain followed by a C-terminal tail of unknown structure and corresponds to CSN8 and eIF3k.
In the present paper we report the structure of S. pombe Rpn12, a PCI-domain-containing protein from the 19S proteasome. We have identified potential conserved sites of Rpn12–protein interaction by sequence analysis and, using biophysical methods, we demonstrate that the introduction of mutations at two of these sites impairs Rpn12 binding to Rpn10 in vitro. Expression of one of these mutants in S. pombe results in reduced incorporation of Rpn10 into proteasomes and a corresponding accumulation of high-molecular-mass polyubiquitin conjugates.
EXPERIMENTAL
Protein preparation
Full-length S. pombe Rpn12 repeatedly purified from recombinant Escherichia coli cells as three species suggesting that it is prone to degradation. Using limited proteolysis with subtilisin A followed by N-terminal sequencing and MS we identified three stable fragments truncated at residues 224, 228 and 250. Fragments 1–228, 1–250 and the full-length sequence were subcloned into pGEX6P-1, expressed in E. coli cells and purified by sequential affinity and size-exclusion chromatography. A detailed description of the Rpn12 purification procedure can be found in the Supplementary material (at http://www.BiochemJ.org/bj/448/bj4480055add.htm). Rpn12 mutants were prepared using the QuikChange® (Stratagene) method and verified by sequencing. The integrity of the mutant fold was verified by a comparison with the authentic protein using CD (Supplementary Figure S1 at http://www.BiochemJ.org/bj/448/bj4480055add.htm). Full-length Rpn10 was expressed and purified as described previously [24].
Rpn12 crystallization and structure determination
Rpn12 encoding residues 1–228 (Rpn12228) in HBS buffer [20 mM Hepes, 150 mM NaCl, 0.01% monothioglycerol and 0.02% sodium azide (pH 7.5)] was crystallized from a mother liquor solution containing 0.2 M sodium nitrate, 0.1 M Bis-Tris propane (pH 7.5) and 22.5% PEG [poly(ethylene) glycol]-3350. A native dataset was collected at the ESRF (European Synchrotron Radiation Facility, Grenoble, France) beamline ID14-2 to 1.6 Å (1 Å=0.1 nm) from a crystal cryoprotected by the addition of 25% glycerol and flash-frozen in liquid nitrogen. A second SAD dataset was collected to 1.9 Å at the ESRF beamline ID14-4 operating at 0.979 Å on a crystal grown from selenomethionine-derivatized protein and crystallized under similar conditions. Initial phase information was calculated from a SAD dataset (Table 1). The native and SAD datasets were integrated with MOSFLM [33] and scaled with SCALA [34]. For the structure calculation the SHELX suite was used [35]. Both datasets were prepared by SHELXC. The anomalous signal-to-noise (reported by SHELXC as d''/σ) dropped below 1.5 at 2.3 Å, therefore the maximal resolution for SHELXD to identify anomalous sites was limited to 2.4 Å. This search resulted in 14 anomalous sites with an occupancy above 0.3, consistent with 14 selenomethionine residues per asymmetric unit. On the basis of this substructure, SHELXE calculated phases for all reflections and optimized these by applying connectivity restraints and using solvent-flattening with a solvent content of 54%. This resulted in an electron-density map of sufficient quality for most of the molecule to be built. The model was refined by several rounds of alternating cycles of refinement in PHENIX.REFINE [36] and manual building in COOT [37].
Table 1. Rpn12 data collection and refinement statistics.
Values in parentheses are for the highest resolution shell. ![]() where Ih,j is the intensity of the jth observation of unique reflection h.
where Ih,j is the intensity of the jth observation of unique reflection h. ![]() where Foh and Fch are the observed and calculated structure factor amplitudes for reflection h. Rfree is equivalent to Rconv, but is calculated using a 5% disjoint set of reflections excluded from the maximum likelihood refinement stages.
where Foh and Fch are the observed and calculated structure factor amplitudes for reflection h. Rfree is equivalent to Rconv, but is calculated using a 5% disjoint set of reflections excluded from the maximum likelihood refinement stages. ![]() .
.
| Measurements | SAD | Native |
|---|---|---|
| Data collection | ||
| Wavelength (Å) beamline | 0.979 | 0.9685 |
| Beamline | ESRF ID14.EH4 | ESRF ID14.EH2 |
| Space group | P212121 | P212121 |
| Cell dimensions | ||
| a, b, c [Å] | 41.5, 91.3, 142.7 | 41.8, 91.4, 143.3 |
| α, β, γ [°] | 90, 90, 90 | 90, 90, 90 |
| Data quality | ||
| Resolution [Å] | 43.48–1.88 (1.98–1.88) | 42.34–1.59 (1.67–1.59) |
| Observations | 44716 | 74706 |
| Completeness [%] | 99.4 (99.3) | 99.7 (99.7) |
| Multiplicity | 7.0 (7.0) | 3.9 (3.9) |
| I/σ | 21.9 (4.1) | 17.1 (2.5) |
| Rsym | 0.057 (0.389) | 0.050 (0.497) |
| Rpim | 0.032 (0.170) | 0.030 (0.285) |
| Anomalous multiplicity | 3.7 (3.7) | |
| Solvent content (%) | 46 | 46 |
| Refinement | ||
| Resolution range [Å] | 40.11–1.59 | 40.11–1.59 |
| Number of reflections | 73328 | 73328 |
| Rconv/Rfree [%] | 18.80/22.48 | 18.80/22.48 |
| Average B factor [Å2] | 31.97 | 31.97 |
| Rmsd bonds (Å) | 0.015 | 0.015 |
| Rmsd angles (°) | 1.525 | 1.525 |
| Protein residues | 442 | 442 |
| Other atoms | four nitrate ions, 364 water, three glycerol and two monothioglycerol molecules. | four nitrate ions, 364 water, three glycerol and two monothioglycerol molecules. |
| Ramachandran outliers | 0 | 0 |
| Ramachandran favoured (%) | 99.1 | 99.1 |
NMR titrations
1H/15N HSQC (heteronuclear single-quantum coherence) titration samples were prepared at 100 μM concentration of 15N-labelled Rpn10. All NMR samples contained 5% 2H2O for deuterium locking and 0.5 mM DSS (4,4-dimethyl-4-silapentane-1-sulfonic acid) for chemical shift referencing. 1H/15N HSQC titrations were performed at 600 MHz on GE/Omega spectrometers with 102.4 and 30 ms acquisition times in the direct and indirect dimension respectively. Unlabelled Rpn12 proteins were added in increasing amounts up to a 5-fold molar excess. NMR data was processed using NMRPipe [38] and analysed using NMRView [39].
Fluorescence polarization measurements
Fluorescently labelled Rpn12 encoding residues 1–250 (Rpn12250) was prepared by incubating 100 μM Rpn12250 with a 10-fold molar excess of Oregon Green succinimidyl ester (Invitrogen) in bicarbonate/HCl (pH 8.5) buffer in a total reaction volume of 1 ml for several hours at 4°C. The reaction was stopped by the addition of a 10-fold molar excess of 2-mercaptoethanol and unreacted dye and labelled Rpn12250 were separated with concomitant buffer exchange into HBS [20 mM Hepes and 150 mM NaCl (pH 7.4)] by size-exclusion chromatography. Total fluorescence polarization (At) was measured for serially diluted full-length Rpn10 (0–370 μM) in HBS containing 100 nM labelled Rpn12250 [mutants and WT (wild-type)] in a total volume of 10 μl supplemented to 5 mM DTT (dithiothreitol) using a Pherastar FS platereader (BMG Labtech) and fluorescein optic module (excitation wavelength 485 nm/emission wavelength 520 nm). All samples were measured in black, low protein-binding, round-bottomed 384-well plates (Corning), and pre-incubated for 20 min at 25°C. Polarization due to non-specific binding (Ai was measured by adding a saturating concentration (500 μM) of unlabelled Rpn12250 to the buffer (as described in [40]). The background fluorescence level at each concentration was determined by setting up serial dilutions of 0–370 μM Rpn10 in HBS. Gain was calibrated so that a polarization value of 35 mP was recorded for the zero-point sample. The raw intensities were recorded with 200 flashes per point. Raw intensities for both channels were collected and corrected for background before calculating the measured anisotropy (Am). The binding curves were analysed in Prism 5 (GraphPad) using a single-site binding model. The polarization specific to the Rpn12250–Rpn10 interaction, AS, was calculated as described previously [40]. All values were measured in triplicate and are the average of at least two independent experiments.
Yeast methods and genetics
Standard molecular genetic methods and media were used [41]. All fission yeast strains were derived from the S. pombe WT heterothallic 972h− and 975h+ and are listed in Supplementary Table S1 (at http://www.BiochemJ.org/bj/448/bj4480055add.htm). The WT and K29A/Q76A Rpn12 mutant genes were each cloned into the pDUAL S. pombe expression vector and subsequently transformed into an Rpn12+ heterozygous diploid strain and the mts3-1 mutant allele [42]. For the insertion of the FLAG tag at the 3′ end of the Rpn1+ gene, the pFA6a-5FLAG-natMX6 vector was used (purchased from Addgene, [43]) and the FLAG tag epitope together with the antibiotic marker natMX6 were PCR-amplified and fused in-frame with the 3′ segment of the ORF (open reading frame) of the Rpn1+ gene. Rpn1–FLAG protein was then expressed under the control of its native promoter at its own genomic locus.
Spot assays
Cultures (5 ml) of Rpn12 WT and K29A/Q76A mutant S. pombe strains were grown overnight at 25°C in PMG medium supplemented with adenine (20 μg/ml) and nourseothicin (HKI, Jena) (100 μg/ml). The D (at 595 nm) of the cultures was adjusted to 0.5 and six 4-fold serial dilutions were made. Then, 5 μl of each dilution were spotted on to PMG agar plates and the plates were incubated at 20, 25, 30 and 36°C for 5 days.
Affinity purification of the 26S proteasome and Western blotting
26S proteasomes were isolated from both Rpn12 WT and mutant S. pombe tagged strains using a protocol modified from that described in [44]. A detailed description of the modified protocol can be found in the Supplementary material. Western blots were probed with anti-Pus1 (Rpn10) [23] and anti-FLAG (Sigma) antibodies and quantified with the ImageQuant software using an ImageQuant LAS 4000 CCD (charge-coupled device) camera system (GE Healthcare).
Stabilization of ubiquitin conjugates
Rpn12 WT and K29A/Q76A mutant together with a Rpn10Δ S. pombe strain were grown at the permissive temperature (25°C) to an D595 nm of 0.6. Half of the cells were harvested, washed with distilled water and frozen at −20°C. The remainder of the cells were then shifted to 36°C and incubated for an additional 6 h. Cell lysates were analysed by Western blotting using a polyclonal anti-ubiquitin antibody (Dako). Blots were also developed with an anti α-tubulin antibody as a loading control.
RESULTS
The structure of Rpn12
Full-length S. pombe Rpn12 repeatedly purified from recombinant E. coli cells as three species as seen by SDS/PAGE, suggesting that it is prone to degradation. Using limited proteolysis with subtilisin A followed by N-terminal sequencing and MS we identified three stable fragments truncated at residues 224, 228 and 250 (Supplementary Figure S2 at http://www.BiochemJ.org/bj/448/bj4480055add.htm). The crystal structure of Rpn12228 was solved by selenium SAD (single wavelength anomalous dispersion) and refined at 1.6 Å resolution (Figure 1 and Table 1). The crystals contained two molecules in the asymmetric unit linked via a disulfide bridge between their respective Cys178 residues. We presume that this covalent bond is a crystallization artefact as Rpn12228 is monomeric in solution as judged by size-exclusion chromatography. The symmetric interface between the chains is mediated by residues from their respective WH (winged helix) domains, including Val182, Tyr183 and Leu194. The model for chain A contains residues 1–222 and an N-terminal Gly-Pro-Leu-Gly-Ser cloning artefact; that for chain B contains residues 5–224. Both chains carry a monothioglycerol adduct on Cys65.
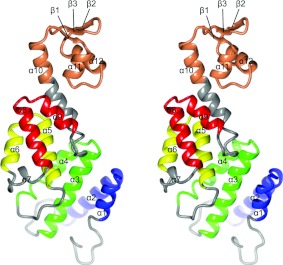
Figure 1. The structure of Rpn12.
A stereo view of Rpn12. Rpn12 (predominantly grey) consists of four TPR-like repeats coloured blue (7–35), green (44–82), yellow (91–117) and red (128–154) connected via a long helix (α10) to a WH domain (orange). Secondary structural elements are labelled.
The S. pombe Rpn12 structure consists of an N-terminal TPR (tetratrico peptide repeat)-like domain and a C-terminal WH domain. Unlike human Rpn12, the S. pombe Rpn12 sequence does not have a long N-terminal extension prior to the start of the PCI domain (Supplementary Figure S2). The TPR-like domain contains four repeats, each comprising two anti-parallel helices with a short helix (α7) inserted into the loop linking repeats 3 and 4. Together, the repeats form an elongated superhelix in which consecutive motifs are rotated by 20°. A long helix (α10), referred to as the ‘capping’ helix [45], packs against the last repeat and connects to the WH domain to give the molecule a banana-like shape, with convex and concave surfaces.
An alignment of diverse eukaryotic Rpn12 sequences defines 27 completely conserved residues (Supplementary Figure S2). Of these, 23 are included in the Rpn12228 structure, with the majority being hydrophobic, and playing structural roles. One cluster composed of conserved residues, Leu99, Phe111 and Leu115, together with Val130, Val133 and Leu134 forms the interface between helices α5, α6 and α8. The side chain of Phe70 and that of Phe163 at the start of α10 make a ring–face interaction as part of a hydrophobic cluster that also includes Leu97 and extends back to stabilize the packing of helices α3, α4 and α5. Trp212 and Leu195 form the nucleus of the hydrophobic core of the WH domain.
Glu55 is the only buried, conserved and charged residue. Its carboxylate group makes a hydrogen bond to the side-chain hydroxy group of Tyr81 that is also identical across species, presumably to correctly align α3 and α4. Similarly Arg172, although partially solvent-accessible, makes hydrogen bonds between its guanidinium group and the backbone carbonyl groups of Leu195 and Tyr196 to stabilize the start of the loop structure linking the two helices of the WH domain.
Comparison with eIF3k, CSN7 and Rpn6
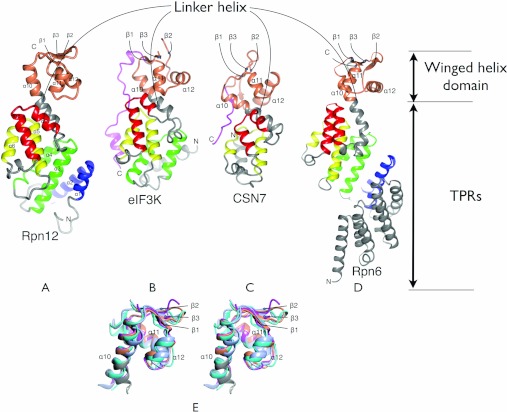
The PCI domains of eIF3k [46], CSN7 [47] and Rpn6 [48] have been structurally characterized, revealing a bi-partite fold in which a TPR-like-containing domain is associated with a WH domain. The TPR-like-containing domains from Rpn12 (Figure 2A), eIF3k (Figure 2B), CSN7 (Figure 2C) and Rpn6 (Figure 2D) vary in the number of helices, in the helix and linker lengths, and in the relative orientations of the helices to generate different tertiary folds. The WH domains, despite showing little sequence conservation (e.g. 13% identity for Rpn12 compared with eIF3k), are structurally well conserved. A superimposition of the four structures shows that the WH domains of CSN7, eIF3k and Rpn6 align to that of Rpn12 with an rmsd (root mean square deviation) of 1.83, 1.30 and 1.78 Å respectively over 50 equivalent residues (Figure 2E). A global superimposition of the structures excluding their respective WH domains shows that they align reasonably well over a longer region that includes two TPR-like repeats, the capping helix and the WH domain (Figure 2). Structural variability within this core manifests principally as variation in the relative disposition of the WH and TPR-like subdomains. These common elements are extended by one helix in CSN7, one TPR-like repeat in eIF3k, two TPR-like repeats in Rpn12 and five TPR-like repeats in Rpn6. In addition, eIF3k has an N-terminal leader helix that does not superimpose with either of the helices in the first Rpn12 repeat. This structural divergence extends into the first eIF3k TPR-like repeat: Rpn12 repeat 2 has a much longer first helix (α3), and eIF3k a longer loop linking the helices so that the second helices of each are displaced relative to the other (compare Figures 2A and 2B).
Figure 2. Structure of Rpn12 and comparison with other PCI-fold proteins.
Comparison of Rpn12 (A) with the complete PCI domains from eIF3K [PDB code 1RZ4 (B)], CSN7 [PDB code 3CHM (C)] and Rpn6 [PDB code 3TXN (D)]. The structures were superimposed on the basis of equivalent residues within the TPR-containing subdomains. The divergence in the direction that the linker helix takes in the different structures is apparent. The WH domain and TPRs that are common to Rpn12 are coloured according to the colours in Figure 1. C-terminal extensions of eIF3K and CSN7 are coloured magenta. Secondary structural elements of Rpn12 are labelled, and secondary structural elements of the WH domains of each of the other proteins are labelled according to their equivalent element in Rpn12. (E) A stereo view. Superimposition of the WH domains of Rpn12 (orange), eIF3k (magenta), CSN7 (cyan) and Rpn6 (light blue).
The structurally equivalent elements of the Rpn12, Rpn6, eIF3k and CSN7 TPR-like-containing and WH domains are scaffolded by a further common element, namely the linker helix between the TPR-like repeats and the WH domain. Towards its N-terminus this helix interacts intimately with the two preceding TPR-like repeats, and towards its C-terminal end it contributes to the three helical bundle of the WH domain. The structural homology of Rpn12, Rpn6, eIF3k and CSN7 is most apparent within this region, supporting a model in which the two TPR-like repeats, the linker helix and the WH domain constitute a common structural core of PCI domains.
The crystallized construct lacks 42 C-terminal residues of authentic S. pombe Rpn12. This sequence includes a well-conserved motif of 13 amino acids, attached via a linker of variable length (Supplementary Figure S2). In S. cerevisiae, mutations to Rpn12 Ile270 and Glu271 within this C-terminal motif disrupt its association respectively with the lid subparticle and the RP base [19]. The corresponding 32 C-terminal residues of Rpn6 are presumed to be disordered in the crystal structure and were also found to be proteolytically sensitive [48]. However, in eIF3k and CSN7 the corresponding C-terminal tails extend beyond the WH domain along the concave surface of the fold as unstructured peptides (Figure 2). Given the observed conservation of structure between eIF3k, CSN7 and Rpn12 and the conserved amphipathic nature of this tail motif, we speculate that the Rpn12 C-terminal tail may be conformationally flexible and adopt a similar extended structure in which the conserved C-terminal motif constitutes either an inter- or intra-molecular interaction motif.
Identification of potential sites of Rpn12–protein interaction
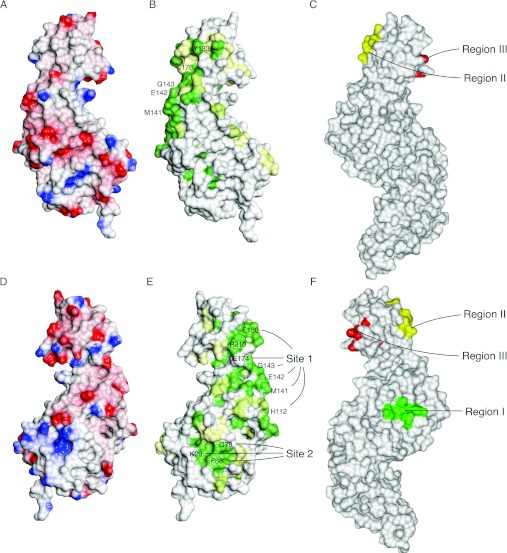
Rpn12 lacks extended hydrophobic surfaces that might suggest sites of protein–protein interaction (Figures 3A and 3D). There are, however, seven Rpn12 residues that are both solvent-exposed and identical across diverse eukaryotic species. Six of these residues (His112, Met141, Glu142, Gly143, Glu174 and Glu180), together with the majority of the highly conserved amino acids, are located at the end of the TPR-like domain on the convex side of Rpn12, suggestive of a protein-binding site (Site 1, Figures 3B and 3E). This patch involves α6 and α7 and extends into the WH domain via the long capping helix. Met141 and Glu142, within the Met-Glu-Gly motif at the end of α8, together with Glu174 in α10, further contribute to this conserved surface. Gly143, His112 and Glu180 appear to play structural roles in addition to their possible role in protein–protein interactions.
Figure 3. Rpn12 surface properties.
(A and D) Poisson–Boltzmann electrostatic potential mapped on the molecular surface. Positive and negative charge are shown in shades of blue and red respectively ranging from values of +0.5 kT/e to −0.5 kT/e. (B and E) Sequence conservation. Conserved surface residues were identified with CONSURF [51] and visualized using CCP4MG. The calculated conservation scores range from 1 (low) to 9 (high). Residues scoring <5 are coloured in white, those scoring 5–8 are highlighted with a gradient from white to green and those scoring >8 are in dark green. (C and F) View of Rpn6 in equivalent orientations, highlighting functionally important regions involved in intersubunit contacts. Peptide motifs at the heart of conserved Rpn6 patches are coloured green (Region I, residues 230–234), red (Region II, residues 366–370) and yellow (Region III, residues 340–343). (A–C) present the same view (similar to that in Figure 1), whereas (D–F) are rotated 180° around a vertical axis.
A second smaller solvent-exposed conserved patch (Site 2) is located between TPRs 1 and 2 and is composed of residues Leu25, Lys29, Phe38 and Gln76 (Figure 3E). Phe38 makes a face-to-face packing interaction across a crystallographic symmetry axis, supporting the hypothesis that this patch might be an Rpn12–protein interaction site.
The determination of the structure of Rpn6 has allowed the use of conservation mapping to propose likely sites of intermolecular interaction in another PCI-containing component of the RP [48]. This analysis identified three surfaces (regions I–III, Figures 3C and 3F) that might mediate protein–protein contacts, and a further set of conserved charged residues that might also be involved in inter-subunit interactions. Of these patches, region III, together with a conserved but structurally flexible C-terminal helix, was shown by mutagenesis and pull-down assay to mediate binding to Rpn7. By docking the Rpn6 structure into a 9.1Å resolution cryo-EM (cryo-electron microscopy) reconstruction, it was also possible to propose the participation of region I and the positively charged cluster in contacts with Rpt6 and Pre8 respectively. Region II corresponds approximately to Site 1 of Rpn12, whereas the extended Region I of Rpn6 spans the amino acids that constitute Site 2 in Rpn12.
The interaction between Rpn12 and Rpn10
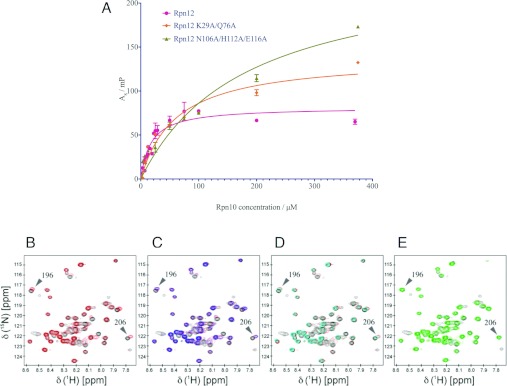
Rpn10 is composed of an N-terminal vWA domain and a C-terminal tail that encodes one (in yeast) or two (in higher eukaryotes) helical UIMs. We have shown by NMR that the interaction between Rpn12 and Rpn10 is mediated by residues from both the vWA domain and the UIM of Rpn10, suggesting an extended interaction between the two proteins [24]. In order to further characterize this interaction, three Rpn12 mutants were constructed by changing conserved surface-exposed residues to alanine. Lys29 and Gln76 contribute to Site 2, whereas Asn106, His112 and Glu116 contribute to Site 1. Tyr183 is at the end of the capping helix (Figure 3B). In order to measure the affinity of authentic and mutant Rpn12 proteins for Rpn10, fluorescence polarization of fluorescently labelled Rpn12250 was measured in the presence of increasing concentrations of full-length Rpn10 (Figure 4A). Under these conditions, the interaction between Rpn12250 and Rpn10 displays a Kd of 17.8±2.7 μM. In contrast, the affinities between Rpn10 and the Rpn12250K29A/Q76A double and Rpn12250N106A/H112A/E116A triple mutants were respectively approximately 3.5- (61.1±9.3 μM) and 10- (182±35 μM) fold lower, suggesting that the residues mutated in these proteins contribute to the Rpn10-binding site.
Figure 4. Characterization of the interaction between Rpn12 and Rpn10.
(A) Fluorescence polarization measurements. Binding of Rpn10 to Rpn12 (magenta), Rpn12 N106A/H112A/E116A (green) and Rpn12K29A/Q76A (orange). (B–E) NMR titrations of Rpn10 and Rpn12. The spectrum of 1H15N Rpn10 is shown in each panel (black) and overlaid with the spectra collected in the presence of a 5:1 excess of ligands. (B) Rpn12 (red), (C) Rpn12Y183A (blue) and Rpn12 (red), (D) Rpn12N106A/H112A/E116A (cyan) and Rpn12 (red), and (E) Rpn12K29A/Q76A (green). Rpn10 residues 196 and 206 in the linker sequence are labelled.
To confirm this result, we exploited the assigned Rpn10 1H15N HSQC spectrum [24]. WT Rpn12250 and each mutant were added in 5-fold excess to 15N-labelled Rpn10. Binding of Rpn12250 to Rpn10 results in the formation of a 48 kDa complex that precludes unambiguous identification of the residues that form the binding site of Rpn12 on Rpn10. However, peaks assigned to residues around the linker sequence between the Rpn10 vWA domain and UIM undergo concentration-dependent chemical-shift changes that report on the Rpn10–Rpn12 interaction.
Using this approach, the chemical-shift changes that accompanied addition of the Rpn12250Y183A mutant were identical with WT (Figures 4B and 4C). Addition of Rpn12250N106A/H112A/E116A caused reduced binding-induced chemical-shift changes, indicating a decrease in the affinity compared with the WT protein (Figure 4D). Rpn12250K29A/Q76A showed significantly decreased binding to Rpn10, as the 1H15N Rpn10 spectrum in the presence of the mutant was identical with that in its absence (Figure 4E). Taken together, the biophysical and structural analyses suggest two possible Rpn10-binding sites on Rpn12. The first (Site 1) includes residues Asn106, His112 and Glu116. The second (Site 2) is centred near Gln76 and Lys29 and probably includes Phe38. However, by analogy with the related structures of eIF3k and CSN7, Site 1 is hypothesized to possibly interact with a C-terminal extension of Rpn12 that was included in the fragment used for biophysical measurements, but not in the crystallized construct, so that Site 1 mutations might be affecting Rpn10 binding indirectly. We therefore continued our further functional analysis using the Rpn12 Site 2 mutant.
Proteasomes incorporating mutant Rpn12 protein rescue an rpn12+ deletion strain but are impaired for Rpn10 binding
To test in vivo whether Lys29 and Gln76 residues are important for the interaction of Rpn12 with the proteasome, the Site 2 mutant as well as the authentic version of the rpn12+ gene were cloned and expressed from the S. pombe expression vector pDUAL [49]. We first tested whether this mutant could rescue the temperature-sensitive phenotype of the mts3-1 strain, which carries a point mutation in the essential rpn12+ gene [42]. It was found that the mutant version rescued as well as WT. We further assessed the Lys29/Gln76 mutant by testing its ability to function under more stringent conditions and rescue a complete deletion of rpn12+. The plasmids containing the authentic and mutant Rpn12 gene were each transformed into a heterozygous rpn12+ diploid deletion strain where one of the authentic ORFs was replaced with the ura4+ selectable marker [42]. Strains were sporulated and both the deletion and expression plasmid were selected for by looking for haploid cells that were prototrophic for uracil and leucine. As shown in Figure 5(A), the mutant version rescued a complete deletion of the rpn12+ gene as well as WT. We conclude from these results that the K29A and Q76A mutations did not significantly affect the essential function(s) of the Rpn12 subunit.
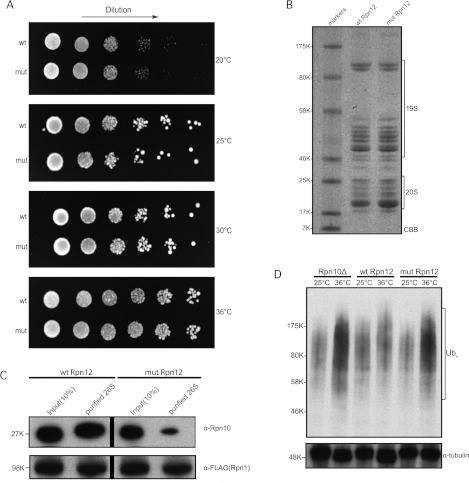
Figure 5. Proteasomes containing the Rpn12K29A/Q76A mutant show decreased incorporation of Rpn10.
(A) Rpn12K29A/Q76A rescues Rpn12Δ S. pombe cells as well as WT Rpn12. Strains were cultured overnight at 25°C and 4-fold serial dilutions were spotted on to PMG plates and incubated at 20, 25, 30 and 36°C. Images were taken after 5 days. (B) SDS/PAGE (4–12% gradient) of the 26S proteasome preparations from Rpn12Δ S. pombe expressing WT Rpn12 (wt Rpn12) and Rpn12K29A/Q76A mutant (mut Rpn12) respectively. 19S regulatory and 20S core subunits are indicated. (C) Rpn10 incorporation into the proteasome is reduced in Rpn12Δ fission yeast cells expressing the mutant Rpn12 subunit. Top panel: Western blotting of proteasomes isolated from the indicated strains using an anti-Rpn10 antibody. Bottom panel: Western blotting using an antibody against the FLAG epitope indicating equal loading of cell extract and isolated proteasomes. Vertical bars denote grouping of samples from different parts of the same gel. (D) Ubiquitin conjugates are accumulated in Rpn12Δ fission yeast cells expressing the mutant Rpn12 protein. Rpn10Δ, WT Rpn12 and mutant Rpn12 strains were grown to exponential phase at the permissive temperature (25°C). Half of each culture was then shifted to the restrictive temperature (36°C) for 6 h. Cell extracts were prepared and analysed by Western blotting for the accumulation of polyubiquitin conjugates using an anti-ubiquitin antibody (top panel). The blot was also probed with an α-tubulin antibody as a loading control (bottom panel). For all gels, the molecular mass in kDa is indicated. CBB, Coomassie Brilliant Blue; Ub, ubiquitin.
Although, the Rpn12 mutant was able to rescue the lethal phenotype of the rpn12+ deletion, we postulated that it could still effect the incorporation of the Rpn10 subunit into the proteasome in vivo, given that the rpn10+ gene does not encode an essential subunit. To test this, we used the rpn12+deletion strains carrying the Site 2 K29A/Q76A mutant and WT expression plasmids shown in Figure 5(A) and tagged the rpn1+ gene encoding another proteasomal subunit with a FLAG epitope. We then affinity-purified the proteasome from these strains (Figure 5B) and assayed the incorporation of Rpn10 in the complex by Western blot analysis using an anti-Rpn10 antibody [23]. As shown in Figure 5(C), the amount of Rpn10 protein present in proteasomes isolated from the mutant strain was significantly reduced. When digitally quantified, a 12-fold difference was calculated for the amount of Rpn10 between the WT and mutant Rpn12-derived 26S preparations.
In light of the misincorporation of Rpn10 into the proteasome complex, we then tested whether the K29A/Q76A Rpn12 mutant was phenotypically similar to an Rpn10-null deletion and shows an accumulation of polyubiquitin conjugates. Western blot analysis of cell extracts from the Rpn12 mutant strain did indeed show an accumulation of ubiquitylated proteins, suggesting impaired proteasomal function presumably as a consequence of the reduced incorporation of Rpn10 in the proteasome (Figure 5D) [23]. Taken together, these results show that a significant function of Rpn12 is to recruit Rpn10 to the proteasome.
DISCUSSION
Biochemical and biophysical studies have demonstrated that Rpn12 is essential for proteasome integrity and deletion of the gene in S. cerevisiae [50] and S. pombe [42] is lethal. Further genetic dissection of the function of Rpn12 revealed that the temperature-sensitive S. pombe mts3-1 allele encodes a version of Rpn12 that is truncated at residue 197 and that this allele is synthetically lethal with an Rpn10 deletion [23]. The Rpn12 structure reveals that this allele removes the WH domain and the C-terminal tail and helps to confirm the importance of these substructures for Rpn12 function and for proteasome integrity. The synthetic lethality observed in the mts3-1Δrpn10 strain points to a significant functional link between the two proteins. On the basis of the observed activity of the Rpn12K29A/Q76A mutant in vivo we would propose that this link is a requirement for the presence of Rpn12 as a prerequisite for successful incorporation of Rpn10 into the proteasome. This conclusion is in agreement with recent models for the assembly of the 19S lid complex which propose that inclusion of Rpn12 is the last step before the binding of Rpn10 [18,19].
Our in vitro experiments support a direct interaction between Rpn12 and Rpn10 that is not apparent in the current cryo-EM structures of the 19S RP, which present a snapshot of the RP in which Rpn2 binds to Rpn12 through the site on Rpn12 that we have found to bind to Rpn10 (F. Foerster, personal communication). To reconcile our data with these structures, we have considered three possible models. In the first model, conformational changes during a cycle of substrate turnover expose the Rpn10-binding site on Rpn12 to facilitate Rpn10 binding and hence substrate recruitment and/or processing. In this model, Rpn2 may provide the functional as well as the structural link between the activities and locations of Rpn12 and Rpn10: binding of a ubiquitylated substrate–Rpn10 complex to Rpn1 may trigger the hypothetical reconfiguration of the Rpn2–Rpn12 subcomplex.
In a second model, the site that we have identified acts as a co-receptor for the cytoplasmic pool of Rpn10, rather than the stably integrated Rpn10 subunit. Docking of cytoplasmic Rpn10, possibly as a ternary Rpn10–UBA/UBL receptor–substrate complex to Rpn1 may elicit a conformational change or relocation of Rpn2 to allow Rpn10 to bind to Rpn12.
In a third model, the interaction that we have observed between Rpn12 and Rpn10 reflects the propensity of both molecules to form protein–protein interactions, rather than their participation in a contact in an authentic functional state of the RP. The micromolar affinity that we have measured is typical of protein–protein interactions that contribute to the structure and stability of large multisubunit complexes, but can also occur for non-cognate protein–protein interactions. Recent cryo-EM maps show that the surface of Rpn12 defined by Lys29/Gln76 is occupied by a helix from Rpn2 in one conformational state of the RP (Supplementary Figure S3 at http://www.BiochemJ.org/bj/448/bj4480055add.htm; F. Foerster, personal communication). We have previously reported that the helical UIM of Rpn10 binds to Rpn12 [24]. Whether our biophysical measurements are reporting on an opportunistic interaction of the Rpn10 UIM with a site that normally accommodates an helix from Rpn2, and the functional significance of any potential alternative helical interactions remain to be determined. In this model, it is necessary to invoke an indirect effect of mutating Rpn12 Lys29 and Gln76 on the association of Rpn10 to the proteasome. This effect could arise through the mutations interfering with the function of Rpn12 subsequent to its integration into the proteasome, making it incompatible with the subsequent integration of Rpn10.
Taken together, the first two models share the hypothesis that binding of ubiquitylated substrates to the proteasome results in a rearrangement to the structure of the RP that affects interactions made by Rpn12. In addition to delineating an important element in the 19S protein–protein interaction network, by significantly depleting proteasome incorporation of Rpn10, the Rpn12K29A/Q76A mutant strain also provides a tool to dissect the roles of Rpn10 as a proteasomal and cytoplasmic ubiquitin-binding protein.
Online data
AUTHOR CONTRIBUTION
Rpn12 preparation, mutagenesis and crystallization was carried out by Jonas Boehringer assisted by Christina Khoudian and Dominique Smith. Sample preparation for NMR and subsequent characterization was carried out by Christiane Riedinger. Konstantinos Paraskevopoulos carried out the yeast genetics and biochemical characterization. Jonas Boehringer and Eachan Johnson carried out the fluorescence spectroscopy. Edward Lowe and Jonas Boehringer collected the X-ray data and determined the Rpn12 structure. Jonas Boehringer, Christiane Riedinger, Konstantinos Paraskevopoulos, Martin Noble, Colin Gordon and Jane Endicott designed the experiments, interpreted the results and wrote the paper.
ACKNOWLEDGEMENTS
We thank the staff at the ESRF and Diamond synchrotrons for providing excellent facilities. We thank J. Boyd and N. Soffe for NMR facilities, A. Willis, J. Nettleship and D. Staunton for protein analysis, H. Waller for collecting the CD spectra, I. Taylor for technical assistance and our colleagues, J. Dean, Vakonakis, R. Gilbert, N. Solcan and J. McDonnell for assistance and useful discussions. We extend our thanks to Dr Friedrich Foerster (Max Planck Institute of Biochemistry, Martinsried, Germany) and members of the Baumeister group for generously sharing their results prior to publication.
FUNDING
This work was supported by the Medical Research Council [grant number G0700053], the RUBICON EU network of excellence, the Biotechnology and Biological Sciences Research Council and The Wellcome Trust [grant number 080823/Z/06/Z].
References
- 1.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marques A. J., Palanimurugan R., Matias A. C., Ramos P. C., Dohmen R. J. Catalytic mechanism and assembly of the proteasome. Chem. Rev. 2009;109:1509–1536. doi: 10.1021/cr8004857. [DOI] [PubMed] [Google Scholar]
- 3.Voges D., Zwickl P., Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 4.Bedford L., Paine S., Sheppard P. W., Mayer R. J., Roelofs J. Assembly, structure, and function of the 26S proteasome. Trends Cell Biol. 2010;20:391–401. doi: 10.1016/j.tcb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elsasser S., Gali R. R., Schwickart M., Larsen C. N., Leggett D. S., Muller B., Feng M. T., Tubing F., Dittmar G. A., Finley D. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat. Cell Biol. 2002;4:725–730. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- 6.Saeki Y., Sone T., Toh-e A., Yokosawa H. Identification of ubiquitin-like protein-binding subunits of the 26S proteasome. Biochem. Biophys. Res. Commun. 2002;296:813–819. doi: 10.1016/s0006-291x(02)02002-8. [DOI] [PubMed] [Google Scholar]
- 7.Gomez T. A., Kolawa N., Gee M., Sweredoski M. J., Deshaies R. J. Identification of a functional docking site in the Rpn1 LRR domain for the UBA-UBL domain protein Ddi1. BMC Biol. 2011;9:33. doi: 10.1186/1741-7007-9-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenzweig R., Bronner V., Zhang D., Fushman D., Glickman M. H. Rpn1 and Rpn2 coordinate ubiquitin processing factors at proteasome. J. Biol. Chem. 2012;287:14659–14671. doi: 10.1074/jbc.M111.316323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pick E., Hofmann K., Glickman M. H. PCI complexes: beyond the proteasome, CSN, and eIF3 Troika. Mol. Cell. 2009;35:260–264. doi: 10.1016/j.molcel.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 10.Husnjak K., Elsasser S., Zhang N., Chen X., Randles L., Shi Y., Hofmann K., Walters K. J., Finley D., Dikic I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X., Lee B. H., Finley D., Walters K. J. Structure of proteasome ubiquitin receptor hRpn13 and its activation by the scaffolding protein hRpn2. Mol. Cell. 2010;38:404–415. doi: 10.1016/j.molcel.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deveraux Q., Ustrell V., Pickart C., Rechsteiner M. A 26 S protease subunit that binds ubiquitin conjugates. J. Biol. Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- 13.Sakata E., Bohn S., Mihalache O., Kiss P., Beck F., Nagy I., Nickell S., Tanaka K., Saeki Y., Forster F., Baumeister W. Localization of the proteasomal ubiquitin receptors Rpn10 and Rpn13 by electron cryomicroscopy. Proc. Natl. Acad. Sci. U.S.A. 2012;109:1479–1484. doi: 10.1073/pnas.1119394109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharon M., Taverner T., Ambroggio X. I., Deshaies R. J., Robinson C. V. Structural organization of the 19S proteasome lid: insights from MS of intact complexes. PLoS Biol. 2006;4:e267. doi: 10.1371/journal.pbio.0040267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu H., Reis N., Lee Y., Glickman M. H., Vierstra R. D. Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. EMBO J. 2001;20:7096–7107. doi: 10.1093/emboj/20.24.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isono E., Saeki Y., Yokosawa H., Toh E. A. Rpn7 is required for the structural integrity of the 26S proteasome of Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:27168–27176. doi: 10.1074/jbc.M314231200. [DOI] [PubMed] [Google Scholar]
- 17.Isono E., Saito N., Kamata N., Saeki Y., Toh E. A. Functional analysis of Rpn6p, a lid component of the 26 S proteasome, using temperature-sensitive rpn6 mutants of the yeast Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:6537–6547. doi: 10.1074/jbc.M409364200. [DOI] [PubMed] [Google Scholar]
- 18.Fukunaga K., Kudo T., Toh E. A., Tanaka K., Saeki Y. Dissection of the assembly pathway of the proteasome lid in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2010;396:1048–1053. doi: 10.1016/j.bbrc.2010.05.061. [DOI] [PubMed] [Google Scholar]
- 19.Tomko R. J., Jr, Hochstrasser M. Incorporation of the Rpn12 subunit couples completion of proteasome regulatory particle lid assembly to lid-base joining. Mol. Cell. 2011;44:907–917. doi: 10.1016/j.molcel.2011.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lander G. C., Estrin E., Matyskiela M. E., Bashore C., Nogales E., Martin A. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lasker K., Forster F., Bohn S., Walzthoeni T., Villa E., Unverdorben P., Beck F., Aebersold R., Sali A., Baumeister W. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc. Natl. Acad. Sci. U.S.A. 2012;109:1380–1387. doi: 10.1073/pnas.1120559109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen C., Huang C., Chen S., Liang J., Lin W., Ke G., Zhang H., Wang B., Huang J., Han Z., et al. Subunit-subunit interactions in the human 26S proteasome. Proteomics. 2008;8:508–520. doi: 10.1002/pmic.200700588. [DOI] [PubMed] [Google Scholar]
- 23.Wilkinson C. R., Ferrell K., Penney M., Wallace M., Dubiel W., Gordon C. Analysis of a gene encoding Rpn10 of the fission yeast proteasome reveals that the polyubiquitin-binding site of this subunit is essential when Rpn12/Mts3 activity is compromised. J. Biol. Chem. 2000;275:15182–15192. doi: 10.1074/jbc.275.20.15182. [DOI] [PubMed] [Google Scholar]
- 24.Riedinger C., Boehringer J., Trempe J. F., Lowe E. D., Brown N. R., Gehring K., Noble M. E., Gordon C., Endicott J. A. Structure of Rpn10 and its interactions with polyubiquitin chains and the proteasome subunit Rpn12. J. Biol. Chem. 2010;285:33992–34003. doi: 10.1074/jbc.M110.134510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu H., Sadis S., Rubin D. M., Glickman M., van Nocker S., Finley D., Vierstra R. D. Multiubiquitin chain binding and protein degradation are mediated by distinct domains within the 26 S proteasome subunit Mcb1. J. Biol. Chem. 1998;273:1970–1981. doi: 10.1074/jbc.273.4.1970. [DOI] [PubMed] [Google Scholar]
- 26.Matiuhin Y., Kirkpatrick D. S., Ziv I., Kim W., Dakshinamurthy A., Kleifeld O., Gygi S. P., Reis N., Glickman M. H. Extraproteasomal Rpn10 restricts access of the polyubiquitin-binding protein Dsk2 to proteasome. Mol. Cell. 2008;32:415–425. doi: 10.1016/j.molcel.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang D., Chen T., Ziv I., Rosenzweig R., Matiuhin Y., Bronner V., Glickman M. H., Fushman D. Together, Rpn10 and Dsk2 can serve as a polyubiquitin chain-length sensor. Mol. Cell. 2009;36:1018–1033. doi: 10.1016/j.molcel.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lipinszki Z., Kovacs L., Deak P., Udvardy A. Ubiquitylation of Drosophila p54/Rpn10/S5a regulates its interaction with the UBA-UBL polyubiquitin receptors. Biochemistry. 2012;51:2461–2470. doi: 10.1021/bi3001006. [DOI] [PubMed] [Google Scholar]
- 29.Kim T., Hofmann K., von Arnim A. G., Chamovitz D. A. PCI complexes: pretty complex interactions in diverse signaling pathways. Trends Plant Sci. 2001;6:379–386. doi: 10.1016/s1360-1385(01)02015-5. [DOI] [PubMed] [Google Scholar]
- 30.Silvera D., Formenti S. C., Schneider R. J. Translational control in cancer. Nat. Rev. Cancer. 2010;10:254–266. doi: 10.1038/nrc2824. [DOI] [PubMed] [Google Scholar]
- 31.Wei N., Serino G., Deng X. W. The COP9 signalosome: more than a protease. Trends Biochem. Sci. 2008;33:592–600. doi: 10.1016/j.tibs.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 32.Scheel H., Hofmann K. Prediction of a common structural scaffold for proteasome lid, COP9-signalosome and eIF3 complexes. BMC Bioinf. 2005;6:71. doi: 10.1186/1471-2105-6-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leslie A. G. W. Recent changes to the MOSFLM package for processing film and image plate data. Protein Crystallogr. 1992;26:27–33. [Google Scholar]
- 34.Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 35.Sheldrick G. M. A short history of SHELX. Acta Crystallogr. Sect. A Found. Crystallogr. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 36.Adams P. D., Afonine P. V., Bunkoczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 39.Johnson B. A., Blevins R. A. NMRView: a computer program for the visualisation and analysis of NMR data. J. Biomol. NMR. 1994;5:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 40.Rossi A. M., Taylor C. W. Analysis of protein-ligand interactions by fluorescence polarization. Nat. Protoc. 2011;6:365–387. doi: 10.1038/nprot.2011.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moreno S., Klar A., Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 42.Gordon C., McGurk G., Wallace M., Hastie N. D. A conditional lethal mutant in the fission yeast 26 S protease subunit mts3+ is defective in metaphase to anaphase transition. J. Biol. Chem. 1996;271:5704–5711. doi: 10.1074/jbc.271.10.5704. [DOI] [PubMed] [Google Scholar]
- 43.Noguchi C., Garabedian M. V., Malik M., Noguchi E. A vector system for genomic FLAG epitope-tagging in Schizosaccharomyces pombe. Biotechnol. J. 2008;3:1280–1285. doi: 10.1002/biot.200800140. [DOI] [PubMed] [Google Scholar]
- 44.Verma R., Chen S., Feldman R., Schieltz D., Yates J., Dohmen J., Deshaies R. J. Proteasomal proteomics: identification of nucleotide-sensitive proteasomeinteracting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell. 2000;11:3425–3439. doi: 10.1091/mbc.11.10.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.D’Andrea L. D., Regan L. TPR proteins: the versatile helix. Trends Biochem. Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 46.Wei Z., Zhang P., Zhou Z., Cheng Z., Wan M., Gong W. Crystal structure of human eIF3k, the first structure of eIF3 subunits. J. Biol. Chem. 2004;279:34983–34990. doi: 10.1074/jbc.M405158200. [DOI] [PubMed] [Google Scholar]
- 47.Dessau M., Halimi Y., Erez T., Chomsky-Hecht O., Chamovitz D. A., Hirsch J. A. The Arabidopsis COP9 signalosome subunit 7 is a model PCI domain protein with subdomains involved in COP9 signalosome assembly. Plant Cell. 2008;20:2815–2834. doi: 10.1105/tpc.107.053801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pathare G. R., Nagy I., Bohn S., Unverdorben P., Hubert A., Korner R., Nickell S., Lasker K., Sali A., Tamura T., et al. The proteasomal subunit Rpn6 is a molecular clamp holding the core and regulatory subcomplexes together. Proc. Natl. Acad. Sci. U.S.A. 2012;109:149–154. doi: 10.1073/pnas.1117648108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matsuyama A., Shirai A., Yashiroda Y., Kamata A., Horinouchi S., Yoshida M. pDUAL, a multipurpose, multicopy vector capable of chromosomal integration in fission yeast. Yeast. 2004;21:1289–1305. doi: 10.1002/yea.1181. [DOI] [PubMed] [Google Scholar]
- 50.Nisogi H., Kominami K., Tanaka K., Toh E. A. A new essential gene of Saccharomyces cerevisiae, a defect in it may result in instability of nucleus. Exp. Cell Res. 1992;200:48–57. doi: 10.1016/s0014-4827(05)80070-9. [DOI] [PubMed] [Google Scholar]
- 51.Landau M., Mayrose I., Rosenberg Y., Glaser F., Martz E., Pupko T., Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.