Background: Dynamin proteins shape membranes by promoting membrane curvature, fission, and fusion.
Results: Cryo-EM demonstrates how the dynamin-related protein Mgm1 assembles onto and tethers membranes followed by nucleotide-dependent conformational changes.
Conclusion: Mgm1 may mediate mitochondrial fusion by bridging opposing membranes and undergoing structural transitions.
Significance: This study provides new mechanistic details of how dynamins may function as fusion molecules.
Keywords: Electron Microscopy (EM), GTPase, Lipid Structure, Membrane Fusion, Mitochondria, Cryo-electron Microscopy, Dynamin Proteins, Mitochondrial Dynamics, Optic Atrophy
Abstract
Cellular membrane remodeling events such as mitochondrial dynamics, vesicle budding, and cell division rely on the large GTPases of the dynamin superfamily. Dynamins have long been characterized as fission molecules; however, how they mediate membrane fusion is largely unknown. Here we have characterized by cryo-electron microscopy and in vitro liposome fusion assays how the mitochondrial dynamin Mgm1 may mediate membrane fusion. Using cryo-EM, we first demonstrate that the Mgm1 complex is able to tether opposing membranes to a gap of ∼15 nm, the size of mitochondrial cristae folds. We further show that the Mgm1 oligomer undergoes a dramatic GTP-dependent conformational change suggesting that s-Mgm1 interactions could overcome repelling forces at fusion sites and that ultrastructural changes could promote the fusion of opposing membranes. Together our findings provide mechanistic details of the two known in vivo functions of Mgm1, membrane fusion and cristae maintenance, and more generally shed light onto how dynamins may function as fusion proteins.
Introduction
Members of the dynamin superfamily are conserved in yeast, plants, and higher eukaryotes including humans. They share sequence homology, structural motifs, biochemical characteristics, and the ability to self-assemble into ordered structures and interact with cellular membranes. At the same time, they are implicated in diverse fundamental cellular processes involving membrane binding such as membrane fission and membrane fusion, plant cell plate formation, and chloroplast biogenesis (1, 2). A key question is, therefore, whether they all share a common mechanism of action. The mechanism of how dynamins mediate fission is largely evident from in vitro structural studies indicating membrane tubulation upon protein self-assembly and formation of highly ordered helical structures composed of repeated T-shaped dimers (3–10). The protein molecules dimerize via their G domain interface, and the dimers self-assemble into helical structures via the stalk (middle domain and GTPase effector domain (GED)) interface (11–14). Cryogenic-electron microscopy (cryo-EM)4 of dynamin and of Dnm1, which are involved in mitochondrial division, showed constriction of the lipid tubes in the presence of GTP and nonhydrolyzable analogs (6, 9, 15, 16) that is proposed to facilitate fission. In contrast to the detailed knowledge on the role of dynamin proteins in fission, exactly how the dynamin-related proteins (DRPs) of the mitochondrion promote fusion is unclear.
Mgm1 is a yeast DRP that has two important functions in the cell: mitochondrial membrane fusion and the formation and maintenance of cristae structures. The mechanism of action of the protein has yet to be determined, but requires two isoforms, an inner membrane-bound l-Mgm1 and an intermembrane space s-Mgm1; both isoforms contain a lipid-binding domain that is required for their in vivo function (17, 18). Recent studies showed that a functional GTPase domain is only required for s-Mgm1 function, and with their differential localizations along the mitochondrial inner membrane, both isoforms are proposed to have distinct roles (19). l-Mgm1 is proposed to serve a structural role, and s-Mgm1 is thought to use the energy of GTP binding and hydrolysis to drive the fusion reaction. In this study, we have focused on the possible mechanistic actions that s-Mgm1 may have by employing in vitro biochemical assays with purified proteins and liposomes as a model mitochondrial membrane. We demonstrate that s-Mgm1 can both tether membranes to likely support inner membrane cristae structures and also undergo a striking GTP-dependent conformational change that could promote the fusion of opposing membranes.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
s-Mgm1 was expressed and purified as described previously (20). In brief, cells were induced with 50 μm isopropyl-1-thio-β-d-galactopyranoside. Protein was purified by nickel-nitrilotriacetic acid resin and size exclusion chromatography and stored at −80 °C.
Liposome Preparation
Liposomes were prepared from 1,2-dioleoyl-sn-glycero-3-phospho-l-serine and inner mitochondrial (IM) phospholipid composition as follows: 38% 1,2-dimyristoyl-sn-glycero-3-phosphocholine, 24% 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine, 16% l-α-phosphatidylinositol, 16% 1′,3′-bis[1,2-dimyristoyl-sn-glycero-3-phospho]-sn-glycerol, 4% 1,2-dimyristoyl-sn-glycero-3-phospho-l-serine, and 2% 1,2-dimyristoyl-sn-glycero-3-phosphate, as described previously (17).
Electron Microscopy
For cryo-EM, thin vitrified specimens were prepared at controlled temperatures and saturation and examined at cryogenic temperatures as described previously (21). Images were recorded at low-dose conditions to minimize beam exposure and electron beam radiation damage on an Ultrascan 1000 cooled CCD camera (Gatan). Negative stain was done as described previously (6, 10). Two-dimensional crystals were grown and manipulated as described previously (18). Images were recorded, developed, and scanned as described previously (22). Eighteen digitalized images of non-tilted crystals were processed by the 2dx image program (23) with 20 Å set as the upper resolution limit for the nonsymmetrized maps. Approximately 50 micrographs were analyzed for each set of experimental conditions in the structural transition analysis. Lattices were categorized into four groups according to their symmetry.
NBD-rhodamine Lipid Mixing Assay
Labeled liposomes containing 0.8% NBD and 0.8% rhodamine were prepared as described previously (24). In the lipid mixing assay, labeled and unlabeled liposomes were mixed at the ratio of 1:9 to obtain the final concentration of 50 μm liposomes. NBD signals were monitored by a spectrophotometer before and after proteins were added. The percentage of lipid mixing is calculated by 100×(I(t) − I(0))/(I(∞) − I(0)), with I(0) and I(∞) as residual and maximal NBD signals, respectively. Maximal level of NBD was obtained by lysing the liposomes with C12E8 detergent. the percentage of total lipid mixing was determined after 60 min.
RESULTS AND DISCUSSION
s-Mgm1 Assembles onto the Surface of Liposomes and Forms Protein Bridges to Promote Membrane Tethering
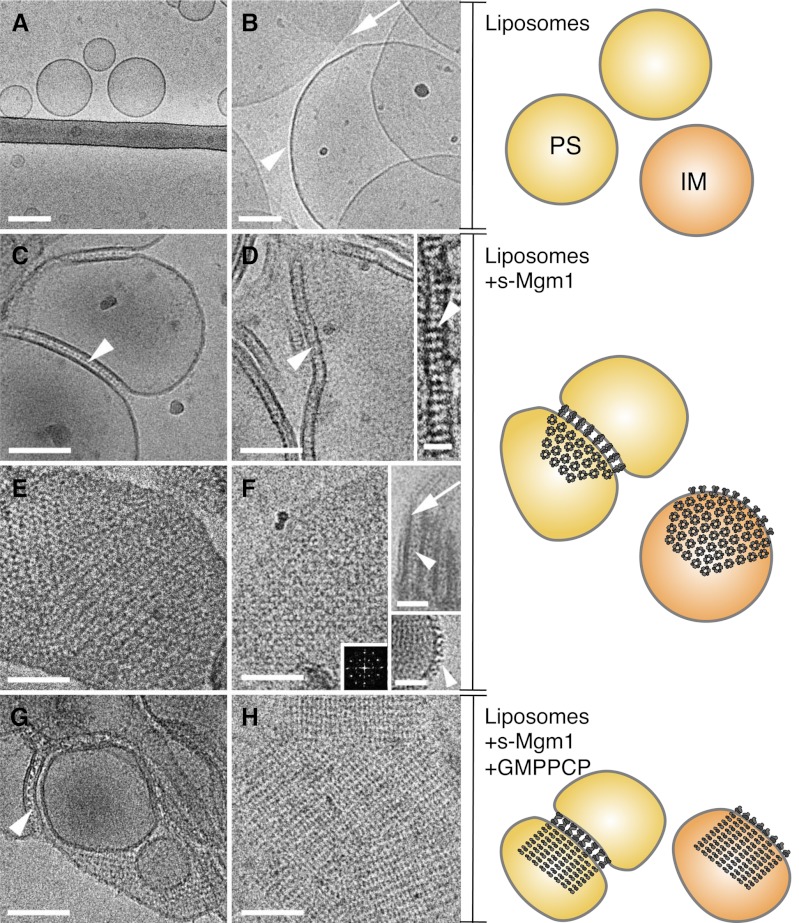
Using liposomes reflecting IM lipid composition, we have previously demonstrated by negative stain electron microscopy that s-Mgm1 assembled onto liposomes and promoted liposome aggregation (17). We have extended and refined our characterization of s-Mgm1-membrane associations. Cryo-EM analysis shows that s-Mgm1 self-assembles onto both phosphatidylserine (PS) liposomes (Fig. 1, C–E) and IM liposomes (Fig. 1F). Strikingly, images reveal liposome tethering by s-Mgm1 (Fig. 1, C, D, and F), through protein assembly onto both of the opposing membranes, creating protein bridges with a characteristic tethering distance of ∼15 nm (Fig. 1, D and F, insets). Interestingly, s-Mgm1 assembles onto liposomes independent of the presence of cardiolipin. This morphology of protein assembly is consistent both with the size of an s-Mgm1 dimer (18) and with the biological function of Mgm1 in mediating membrane fusion and possibly mitochondrial cristae maintenance. These data suggest that s-Mgm1 homo-complexes may act in trans like a zipper to tether the two membranes together to stabilize inner membrane folds and possibly facilitate fusion. Identifying this unique tethering activity of Mgm1 might explain the mechanistic requirement for Mgm1 in maintaining proper inner membrane cristae topology, a vital in vivo function for this protein.
FIGURE 1.
s-Mgm1 assembles onto and tethers liposomes. A and B, cryo-EM images of PS and IM liposomes, respectively. Arrowhead in B points to round liposome, and arrow points to the border of flat IM membrane. C and D, cryo-EM images show tethered PS liposomes and protein bridges (arrowheads). E and F, cryo-EM images show a crystalline protein array on PS (E) and IM (F) liposomes. The diffraction spots in F show the three-fold lattice symmetry. Arrowhead and arrows (E, upper inset) mark protein bridges and lipid bilayer. Lower inset shows T-shaped s-Mgm1 structures (arrowhead) on the outer liposome surface. G and H, cryo-EM images of crystalline arrays of s-Mgm1 in the presence of GMPPCP on PS (G) and IM (H) liposomes. Scale bar is 100 nm (A–H), 25 nm (inset in D), and 50 nm (insets in F). Schematics on the right summarize the observations from the cryo-EM images on the left. Yellow and orange circles represent PS and IM liposomes, respectively. At the periphery of the liposomes, a pair of light and dark gray structures represents the side view of an s-Mgm1 dimer. The ring of light and dark gray structures represents the top view of s-Mgm1 oligomers. Upon the addition of GMPPCP (from middle to bottom panels), the hexameric array of s-Mgm1 reorganizes into a square array.
Additionally, a highly ordered crystalline array of s-Mgm1 is frequently observed on the liposome surfaces (Fig. 1, E and F), and occasionally, perimeter structures resembling the T-shaped dimer of dynamin (Fig. 1F, inset). Crystalline assembly is more evident on IM liposomes due to the high content (16%) of cardiolipin, which has an inherent tendency to flatten as compared with PS liposomes (Fig. 1, A and B). Like dynamin, many DRPs, including Dnm1 and optic atrophy 1 (OPA1, the mammalian ortholog of Mgm1), form protein-lipid tubes (7–9, 16, 25). s-Mgm1 also transforms liposomes into protein-decorated tubes (Fig. 2, A–C), but infrequently. Instead, and uniquely, the majority of s-Mgm1 structures are tethered lipid bilayers and protein-lipid flat crystals, which agrees well with the suggested in vivo functions of this protein within mitochondria.
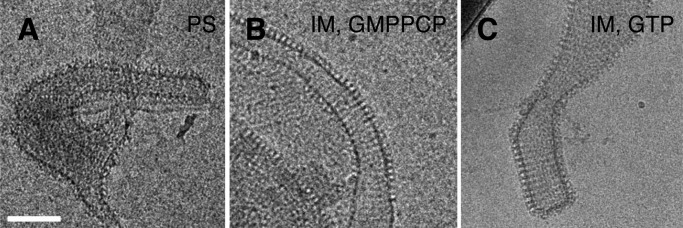
FIGURE 2.
s-Mgm1 can transform liposomes into protein-decorated tubes. Tubes are created with both PS (A) and IM (B and C) liposomes, without and with nucleotides as indicated. s-Mgm1 and lipid concentrations are 0.7 and 0.45 mg/ml respectively. Scale bar is 100 nm.
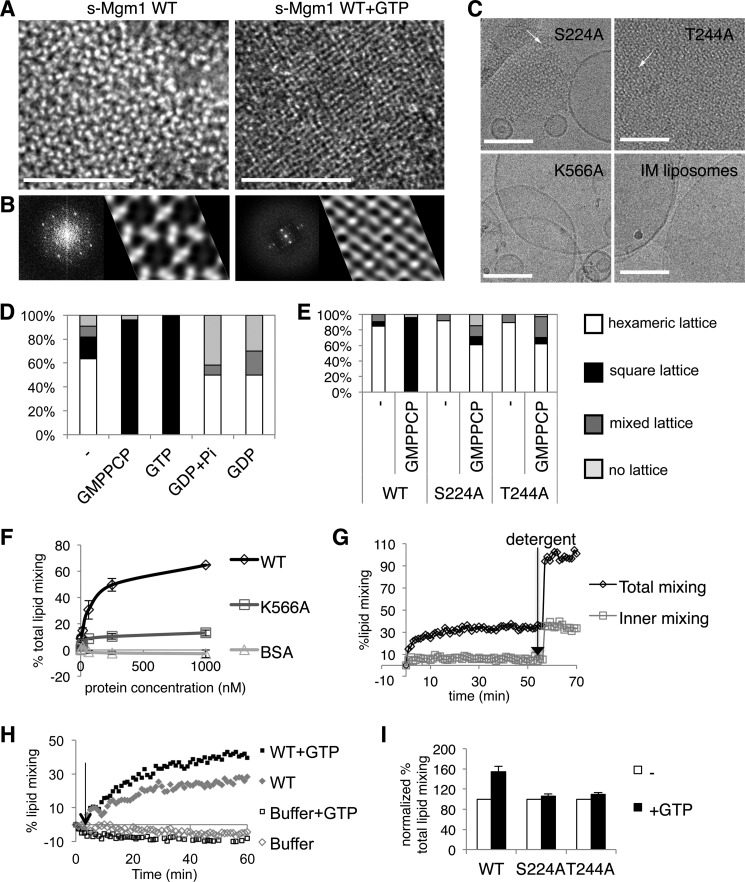
s-Mgm1 Lipid-bound Oligomers Undergo a Nucleotide-dependent Conformational Transition
Nucleotide-dependent conformational changes have been observed in several dynamin proteins (6, 15, 16). Hence, we investigated whether s-Mgm1 undergoes such structural transitions. EM images as well as two-dimensional crystallization analysis of the lattices by Fourier transformation and image processing showed a hexameric (flower-like) (26) three-fold symmetry lattice (Figs. 1F and 3, A and B, left panels), consistent with two previous studies (17, 18). A structural transition in the crystalline array to a square lattice is observed by the addition of 1 mm GTP and/or its nonhydrolyzable analog GMPPCP (Fig. 1, G and H, and 3, A and B, right panels) after allowing s-Mgm1 to bind to liposomes. Notably, this complete rearrangement of the lattice has not previously been reported for Mgm1, yet, importantly, it is very consistent with the fact that nucleotide binding induces transformations that promote the activity of other dynamin family members. The constriction in the planar packing of the protein-lipid lattice is clearly evident in both the EM images and the projection maps (Fig. 3B). We note that atlastin, a dynamin-like GTPase involved in endoplasmic reticulum morphology and fusion, has also been shown to tether membranes (27). Further, atlastin nucleotide-bound states suggest conformational changes upon GTPase activity (28). Thus, our data might suggest a new yet conserved mechanism for DRP-mediated fusion. Indeed, we directly show that Mgm1 membrane-bound oligomers undergo a GTP-dependent conformational change. We propose a model where this structural transition promotes a possible membrane deformation that can facilitate mitochondrial membrane fusion (Fig. 4).
FIGURE 3.
s-Mgm1 oligomeric arrays undergo a nucleotide-dependent structural transition that enhances s-Mgm1 membrane fusion activity. A, EM images of s-Mgm1 arrays on IM liposomes in the presence (right) and absence (left) of GTP. B, corresponding two-dimensional crystal analysis on protein crystalline arrays. Scale bar is 100 nm. Two-dimensional Fourier transform analysis and projection maps show the average patterns of s-Mgm1 arrays. C, cryo-EM images of GTPase mutants, S224A and T244A, and a lipid-binding mutant K566A. Upper panels: cryo-EM images of ordered protein arrays on IM liposome formed by S224A (left) and T244A (right). Arrows mark hexameric arrays. Lower panels: K566A does not assemble onto liposomes nor alter their morphology (compare left and right images). Scale bar is 100 nm. D, quantification of the nucleotide-dependent structural transition as categorized into four lattices, hexameric, square, mixed, or none. s-Mgm1-bound IM liposomes were incubated with buffer containing no nucleotide or 2 mm nucleotide as indicated. E, quantification of wild-type, S224A, and T244A s-Mgm1 assembly onto IM liposomes, categorized into four lattices. Analysis shows a nucleotide-induced transition from hexameric to square lattice. F, NBD-rhodamine assay showing the lipid mixing activity of Mgm1. Wild-type s-Mgm1 causes lipid mixing in a concentration-dependent manner, whereas the lipid mixing activity of the lipid-binding mutant K566A is significantly impaired. No activity is found for the control (bovine serum albumin, BSA). G, the lipid mixing of the inner leaflet only by s-Mgm1 was monitored by dithionite treatment. s-Mgm1 lipid mixing activity was monitored by the fusion of dithionite-treated and untreated labeled liposomes indicative of the mixing of the inner leaflet phospholipids only and total phospholipids, respectively. Detergent was added to determine the maximal NBD signals. H, GTP enhances Mgm1 lipid mixing activity. Arrow points to the 3-min point after the fusion has been initiated. GTP (1 mm) increases the total lipid mixing by wild-type s-Mgm1 (0.125 μm). I, the bar graph shows that GTP addition increased total lipid mixing induced by wild-type s-Mgm1 but not by the GTPase mutants S224A and T244A. Three separate experiments were performed. Basal levels of lipid mixing were normalized to 100%. The error bars represent standard deviations.
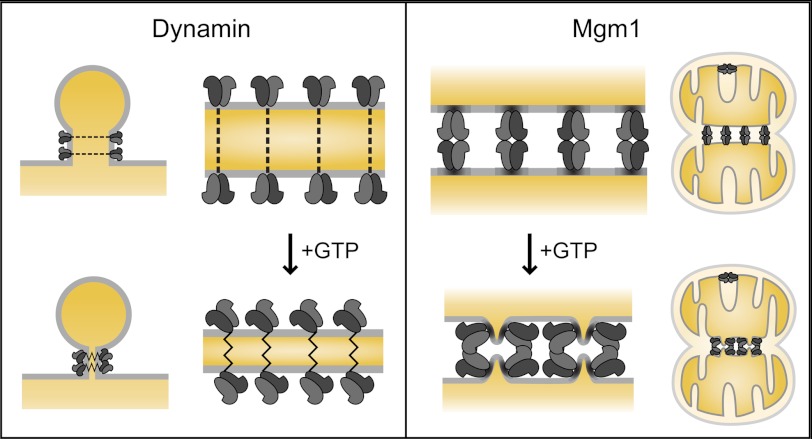
FIGURE 4.
Models of Mgm1-mediated membrane fusion. A model depicting the function of dynamin in fission (left), and s-Mgm1 in fusion (right). Both dynamin and Mgm1 shape membranes in vivo and in vitro. Dynamin dimers assemble into a helical collar, which upon GTPase activity constrict the underlying membrane to mediate fission. We propose that s-Mgm1 forms a homo-oligomeric complex in trans to create protein bridges and ordered lattices that tether opposing membranes to support mitochondrial inner membrane cristae structures, and to also undergo a GTP-induced transition to promote fusion.
Quantitative analysis of the structural transition from hexameric to square lattice suggested a GTP-bound state because it was observed with GMPPCP and GTP (Fig. 3D). To directly test this, we found that the GTPase mutants S224A and T244A predominantly created the hexameric lattice, demonstrating that they retain the basal ability to assemble onto the lipid bilayer (Fig. 3, C and E). However, they could not undergo this GTP-dependent structural transition and displayed the same hexameric lattice with and without nucleotides (Fig. 3E), further demonstrating that nucleotide binding and/or hydrolysis is the driving force behind the lattice transformation.
To investigate the functional significance of the GTP-dependent structural transitions, we monitored potential s-Mgm1 membrane fusion activity using a well characterized NBD-rhodamine lipid mixing assay (24, 29). We showed that s-Mgm1 promoted IM lipid mixing in a dose-dependent manner independently of nucleotide (Fig. 3F). Furthermore, we confirmed that this lipid mixing activity is dependent on the lipid binding activity as the lipid-binding mutant K566A was significantly impaired (Fig. 3F). Importantly, no spontaneous mixing of liposomes was found using BSA as control (Fig. 3F). Further, we fond by dithionite treatment of liposomes that the lipid mixing we were monitoring was that of the outer leaflet as s-Mgm1 does not induce lipid mixing of the inner leaflet of the liposome bilayer (Fig. 3G).
Although nucleotide was not absolutely required for s-Mgm1-mediated lipid mixing shown in this NBD-rhodamine assay, the addition of GTP enhances the rate and amount of this lipid mixing. After IM liposome fusion was induced by the addition of s-Mgm1 for 3 min, 1 mm GTP was added (indicated in Fig. 3H by an arrow). In comparison with the sample that has only added buffer (gray diamond), the initial rate of fusion and the total lipid mixing was higher by the addition of GTP (black square) (Fig. 3H). We repeated the experiments with the nucleotide mutants S224A and T244A and monitored the total lipid mixing after 60 min of reaction. Importantly, GTP was unable to stimulate the basal rate of lipid mixing with the nucleotide mutants S224A and T244A as compared with WT s-Mgm1 (Fig. 3I). Therefore, these data suggest that GTP binding and hydrolysis enhance liposome fusion likely by inducing conformational changes, as we have demonstrated by the cryo-EM images.
Although the GTP-induced structural transition is reminiscent of dynamins, the mechanical forces that would be transferred to the membrane would likely be quite distinct, suggesting a key mechanistic difference in how these related proteins function. Dynamin and s-Mgm1 GTPase activity is highly stimulated in the presence of lipid (17, 18, 30–32), suggesting that these proteins use the hydrolysis energy to apply force on the membrane while performing their biological activity: membrane fusion by s-Mgm1 and fission by dynamin. However, their different biological activity reflects the unique structures they form in vivo and in vitro. Dynamin shapes liposomes in vitro into helical tubes, with a diameter similar to that of endocytic buds. GTP hydrolysis induces conformational changes that generate force, which constricts and twists the underlying membrane, decreasing the distance between the bilayers as a step toward fission (6, 13–15, 27, 28) (Fig. 4). Our data point to Mgm1 as an unconventional dynamin-like protein, uniquely acting to bridge the bilayers of opposing membranes and anchor them at a fixed distance, to both support mitochondrial inner membrane cristae structures and promote the fusion of opposing membranes. In addition, we propose that the GTP-dependent conformational transitions and GTP hydrolysis could constrict or deform the membrane to promote membrane fusion (Fig. 4).
Given that Mgm1/OPA1 is an important human disease gene, our demonstration of its biophysical properties will direct future studies to reveal mechanisms of disease. Specifically, identifying and characterizing the protein/protein interfaces that both stabilize the lattice work and allow for the gross nucleotide-dependent conformational changes will further reveal its in vivo function. In addition, future studies will need to incorporate the long isoform of Mgm1 to fully understand the breadth of the in vivo activities of Mgm1. These studies will help to uncover the mechanistic roles of other membrane-fusing proteins in the cell and the unique biophysical properties required to regulate membrane dynamics.
Footnotes
- cryo-EM
- cryogenic-electron microscopy
- DRP
- dynamin-related proteins
- PS
- phosphatidylserine
- IM
- inner mitochondrial membrane
- NBD
- 12-(N-methyl-N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl))
- GMPPCP
- guanylyl β,γ-methylenediphosphonate.
REFERENCES
- 1. Danino D., Hinshaw J. E. (2001) Dynamin family of mechanoenzymes. Curr. Opin. Cell Biol. 13, 454–460 [DOI] [PubMed] [Google Scholar]
- 2. Praefcke G. J., McMahon H. T. (2004) The dynamin superfamily: universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 5, 133–147 [DOI] [PubMed] [Google Scholar]
- 3. Takei K., Haucke V., Slepnev V., Farsad K., Salazar M., Chen H., De Camilli P. (1998) Generation of coated intermediates of clathrin-mediated endocytosis on protein-free liposomes. Cell 94, 131–141 [DOI] [PubMed] [Google Scholar]
- 4. Sweitzer S. M., Hinshaw J. E. (1998) Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell 93, 1021–1029 [DOI] [PubMed] [Google Scholar]
- 5. Stowell M. H., Marks B., Wigge P., McMahon H. T. (1999) Nucleotide-dependent conformational changes in dynamin: evidence for a mechanochemical molecular spring. Nat. Cell Biol. 1, 27–32 [DOI] [PubMed] [Google Scholar]
- 6. Danino D., Moon K. H., Hinshaw J. E. (2004) Rapid constriction of lipid bilayers by the mechanochemical enzyme dynamin. J. Struct. Biol. 147, 259–267 [DOI] [PubMed] [Google Scholar]
- 7. Yoon Y., Pitts K. R., McNiven M. A. (2001) Mammalian dynamin-like protein DLP1 tubulates membranes. Mol. Biol. Cell 12, 2894–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ban T., Heymann J. A., Song Z., Hinshaw J. E., Chan D. C. (2010) OPA1 disease alleles causing dominant optic atrophy have defects in cardiolipin-stimulated GTP hydrolysis and membrane tubulation. Hum. Mol. Genet. 19, 2113–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mears J. A., Ray P., Hinshaw J. E. (2007) A corkscrew model for dynamin constriction. Structure 15, 1190–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. von der Malsburg A., Abutbul-Ionita I., Haller O., Kochs G., Danino D. (2011) Stalk domain of the dynamin-like MxA GTPase protein mediates membrane binding and liposome tubulation via the unstructured L4 loop. J. Biol. Chem. 286, 37858–37865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Low H. H., Sachse C., Amos L. A., Löwe J. (2009) Structure of a bacterial dynamin-like protein lipid tube provides a mechanism for assembly and membrane curving. Cell 139, 1342–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Faelber K., Posor Y., Gao S., Held M., Roske Y., Schulze D., Haucke V., Noé F., Daumke O. (2011) Crystal structure of nucleotide-free dynamin. Nature 477, 556–560 [DOI] [PubMed] [Google Scholar]
- 13. Ford M. G., Jenni S., Nunnari J. (2011) The crystal structure of dynamin. Nature 477, 561–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chappie J. S., Mears J. A., Fang S., Leonard M., Schmid S. L., Milligan R. A., Hinshaw J. E., Dyda F. (2011) A pseudoatomic model of the dynamin polymer identifies a hydrolysis-dependent powerstroke. Cell 147, 209–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang P., Hinshaw J. E. (2001) Three-dimensional reconstruction of dynamin in the constricted state. Nat. Cell Biol. 3, 922–926 [DOI] [PubMed] [Google Scholar]
- 16. Mears J. A., Lackner L. L., Fang S., Ingerman E., Nunnari J., Hinshaw J. E. (2011) Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat. Struct. Mol. Biol. 18, 20–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rujiviphat J., Meglei G., Rubinstein J. L., McQuibban G. A. (2009) Phospholipid association is essential for dynamin-related protein Mgm1 to function in mitochondrial membrane fusion. J. Biol. Chem. 284, 28682–28686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. DeVay R. M., Dominguez-Ramirez L., Lackner L. L., Hoppins S., Stahlberg H., Nunnari J. (2009) Coassembly of Mgm1 isoforms requires cardiolipin and mediates mitochondrial inner membrane fusion. J. Cell Biol. 186, 793–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zick M., Duvezin-Caubet S., Schäfer A., Vogel F., Neupert W., Reichert A. S. (2009) Distinct roles of the two isoforms of the dynamin-like GTPase Mgm1 in mitochondrial fusion. FEBS Lett. 583, 2237–2243 [DOI] [PubMed] [Google Scholar]
- 20. Meglei G., McQuibban G. A. (2009) The dynamin-related protein Mgm1p assembles into oligomers and hydrolyzes GTP to function in mitochondrial membrane fusion. Biochemistry 48, 1774–1784 [DOI] [PubMed] [Google Scholar]
- 21. Danino D., Bernheim-Groswasser A., Talmon Y. (2001) Digital cryogenic transmission electron microscopy: an advanced tool for direct imaging of complex fluids. Colloids Surf. A Physicochem. Eng. Asp. 183, 113–122 [Google Scholar]
- 22. Bueler S. A., Rubinstein J. L. (2008) Location of subunit d in the peripheral stalk of the ATP synthase from Saccharomyces cerevisiae. Biochemistry 47, 11804–11810 [DOI] [PubMed] [Google Scholar]
- 23. Gipson B., Zeng X., Zhang Z. Y., Stahlberg H. (2007) 2dx—user-friendly image processing for 2D crystals. J. Struct. Biol. 157, 64–72 [DOI] [PubMed] [Google Scholar]
- 24. Marsden H. R., Tomatsu I., Kros A. (2011) Model systems for membrane fusion. Chem. Soc. Rev. 40, 1572–1585 [DOI] [PubMed] [Google Scholar]
- 25. Ingerman E., Perkins E. M., Marino M., Mears J. A., McCaffery J. M., Hinshaw J. E., Nunnari J. (2005) Dnm1 forms spirals that are structurally tailored to fit mitochondria. J. Cell Biol. 170, 1021–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Avinoam O., Fridman K., Valansi C., Abutbul I., Zeev-Ben-Mordehai T., Maurer U. E., Sapir A., Danino D., Grünewald K., White J. M., Podbilewicz B. (2011) Conserved eukaryotic fusogens can fuse viral envelopes to cells. Science 332, 589–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Roux A., Uyhazi K., Frost A., De Camilli P. (2006) GTP-dependent twisting of dynamin implicates constriction and tension in membrane fission. Nature 441, 528–531 [DOI] [PubMed] [Google Scholar]
- 28. Lenz M., Morlot S., Roux A. (2009) Mechanical requirements for membrane fission: common facts from various examples. FEBS Lett. 583, 3839–3846 [DOI] [PubMed] [Google Scholar]
- 29. Weidberg H., Shpilka T., Shvets E., Abada A., Shimron F., Elazar Z. (2011) LC3 and GATE-16 N termini mediate membrane fusion processes required for autophagosome biogenesis. Dev. Cell 20, 444–454 [DOI] [PubMed] [Google Scholar]
- 30. Sever S., Damke H., Schmid S. L. (2000) Garrotes, springs, ratchets, and whips: putting dynamin models to the test. Traffic 1, 385–392 [DOI] [PubMed] [Google Scholar]
- 31. Marks B., Stowell M. H., Vallis Y., Mills I. G., Gibson A., Hopkins C. R., McMahon H. T. (2001) GTPase activity of dynamin and resulting conformation change are essential for endocytosis. Nature 410, 231–235 [DOI] [PubMed] [Google Scholar]
- 32. He B., Yu X., Margolis M., Liu X., Leng X., Etzion Y., Zheng F., Lu N., Quiocho F. A., Danino D., Zhou Z. (2010) Live-cell imaging in Caenorhabditis elegans reveals the distinct roles of dynamin self-assembly and guanosine triphosphate hydrolysis in the removal of apoptotic cells. Mol. Biol. Cell 21, 610–629 [DOI] [PMC free article] [PubMed] [Google Scholar]