Background: Phosphorylation of human DNA ligase I (hLigI) regulates its participation in DNA replication and repair.
Results: Identification of a single phosphorylation site that regulates the interaction of hLigI with replication factor C (RFC).
Conclusion: Interaction between hLigI and RFC is critical for DNA replication and repair.
Significance: We provide insights into the mechanism by which a single phosphorylation site regulates the cellular functions of hLigI.
Keywords: Cell Cycle, DNA Damage, DNA Repair, Mass Spectrometry (MS), Phosphorylation
Abstract
Human DNA ligase I (hLigI) joins Okazaki fragments during DNA replication and completes excision repair via interactions with proliferating cell nuclear antigen and replication factor C (RFC). Unlike proliferating cell nuclear antigen, the interaction with RFC is regulated by hLigI phosphorylation. To identity of the site(s) involved in this regulation, we analyzed phosphorylated hLigI purified from insect cells by mass spectrometry. These results suggested that serine 51 phosphorylation negatively regulates the interaction with RFC. Therefore, we constructed versions of hLigI in which serine 51 was replaced with either alanine (hLigI51A) to prevent phosphorylation or aspartic acid (hLigI51D) to mimic phosphorylation. hLigI51D but not hLigI51A was defective in binding to purified RFC and in associating with RFC in cell extracts. Although DNA synthesis and proliferation of hLigI-deficient cells expressing either hLig51A or hLig51 was reduced compared with cells expressing wild-type hLigI, cellular senescence was only observed in the cells expressing hLigI51D. Notably, these cells had increased levels of spontaneous DNA damage and phosphorylated CHK2. In addition, although expression of hLigI51A complemented the sensitivity of hLigI-deficient cells to a poly (ADP-ribose polymerase (PARP) inhibitor, expression of hLig151D did not, presumably because these cells are more dependent upon PARP-dependent repair pathways to repair the damage resulting from the abnormal DNA replication. Finally, neither expression of hLigI51D nor hLigI51A fully complemented the sensitivity of hLigI-deficient cells to DNA alkylation. Thus, phosphorylation of serine 51 on hLigI plays a critical role in regulating the interaction between hLigI and RFC, which is required for efficient DNA replication and repair.
Introduction
The human DNA ligase I (hLigI)2-deficient cell line 46BR.1G1 has a defect in joining Okazaki fragments and is sensitive to DNA damage, implicating this enzyme in DNA replication and excision repair (1, 2). In accord with its role in DNA replication, hLigI colocalizes with other DNA replication proteins in replication foci during the DNA synthesis phase of the cell cycle (3, 4). Surprisingly, LIG1 null mouse embryonic fibroblasts are viable suggesting that one of the other DNA ligases encoded by the LIG3 and LIG4 genes can substitute for hLigI in DNA replication (5, 6). Recent studies have shown that DNA ligase IIIα enables vertebrates to replicate their genome in the absence of hLigI (7).
Although the molecular mechanisms underlying the participation of hLigIIIα in DNA replication are not known, hLigI physically and functionally interacts with two DNA replication proteins, proliferating cell nuclear antigen (PCNA) and replication factor C (RFC) (2, 4, 8, 9). PCNA is a ring-shaped homotrimer that is loaded onto duplex DNA by RFC. At the replication fork, the topologically linked PCNA trimer serves as a processivity factor for the replicative DNA polymerases Pol δ and Pol ϵ and coordinates the activities of other replication proteins during Okazaki fragment processing and ligation (10). hLigI interacts with PCNA via an N-terminal PCNA binding motif (PCNA interaction protein (PIP) box) (2, 4, 8). This interaction is required for the recruitment of hLigI to replication foci in replicating cells and to correct the defects in DNA replication and repair in hLigI-deficient cell line 46BR.1G1 cells (2, 4).
Unlike PCNA binding, the interaction between hLigI and RFC is regulated by hLigI phosphorylation (11). Three cyclin-dependent kinase sites, serine 51, serine 76, and serine 91, and a casein kinase II site, serine 66, have been identified in hLigI (12, 13). These phosphorylation sites reside within an unstructured, proline-rich region between the N-terminal PIP box and the C-terminal catalytic domain. After phosphorylation of serine 91 at G1/S transition, human hLigI becomes increasingly phosphorylated during cell cycle progression (12). The serine 91 modification is required for the subsequent phosphorylation of serine 76 to generate the hyperphosphorylated form of hLigI in the G2 and M cell cycle phases (12). Serine 66 is dephosphorylated early in the G1 phase and then becomes progressively phosphorylated in S and G2/M, suggesting that the dephosphorylation establishes the prereplicative form of hLigI (13).
We have shown previously that replacement of the four phosphorylated serine residues, serine 51, serine 66, serine 76, and serine 91, with aspartic acid residues (4D) that mimic the charge of phosphorylated serine residues, abolishes the interaction with RFC (11). Furthermore, expression of both the 4D and the 4A version of hLigI in hLigI-deficient cell line 46BR.1G1 cells has a dominant negative effect on DNA replication and repair, indicating the important role of phosphorylation in regulating the cellular functions of hLigI (11, 14). Here we have identified a single serine residue, serine 51, among the four phosphorylated serines as the key residue that regulates the interaction with RFC and the participation of hLigI in DNA replication and repair.
EXPERIMENTAL PROCEDURES
Proteins
Wild-type hLigI was purified from baculovirus-infected insect cells as described previously (2, 15). In addition, cDNAs encoding FLAG-tagged versions of wild-type and mutant hLigI cDNAs were subcloned into the bacterial expression vector pRSF-Duet1, resulting in the presence of an additional His tag at the N terminus. Tagged versions of wild-type and mutant hLigI were purified from Escherichia coli extracts by SP-Sepharose Fastflow, Source Q, and Superdex 200 column chromatography. Recombinant human RFC was expressed in bacteria as described previously (16), with a modification of pET-hRfc1 in which a His tag was attached at its C terminus. The RFC complex was purified by SP-Sepharose FastFlow and HisTrap high performance column chromatography.
Mass Spectrometry Analysis of hLigI Purified from Insect Cells
Aliquots of hLigI purified from insect cell (50 μg) protein were digested with either 1% (w/w) trypsin (Promega) or 1% (w/w) Glu-C (Roche) as described (17). In some cases, the tryptic digest was further purified using either TiO2- or gallium-immobilized resins to enrich for phosphopeptides. Peptides were analyzed by LC-MS/MS using either a LTQ or LTQ-Orbitrab mass spectrometer (ThermoElectron). MS/MS spectra were subsequently searched against a UniProtKB human protein database using both the SEQUEST and Mascot algorithms. To identify phosphorylation sites, differential modifications were used of +80 for serine, threonine, and tyrosine and −18 for serine and threonine (corresponding to dehydroalanine and 2-aminodehydrobutryic acid, respectively). Fully trypsin- or Glu-C-digested peptides with up to two missed cleavages and charge state-dependent cross-correlation scores = 2.5, 3.0, and 3.5 for 2+, 3+, and 4+ peptides, respectively, were considered as positive identification for further quantitative analyses. For Mascot search results, only peptides with a Mascot score larger than 25 were chosen. To further confirm the localization of the phosphorylated amino acid after visual inspection, a MS/MS spectrum corresponding to each phosphopeptide was extracted from the XCalibur raw file, and the location probability of phosphorylation site was calculated using Ascore.
Plasmids
A pVP-FLAG5 plasmid encoding an N-terminal FLAG-tagged version of hLigI (2) was mutated by site-directed mutagenesis using the QuikChange site-directed mutagenesis kit (Stratagene) according to the instructions of the manufacturer. The mutations altered the coding sequence so that serine residues 51, 66, 76, and 91 were replaced by either alanine (A) or aspartic acid (D) residues (51A, Ser-51 replaced by alanine; 51D, Ser-51 replaced by aspartic acid; 3A, Ser-66, 76, and 91 replaced by alanine; 3D, Ser-66, 76, and 91 replaced by aspartic acid; 4A, Ser-51, 66, 76, and 91 replaced by alanine; 4D, Ser-51, 66, 76, and 91 replaced by aspartic acid). After verification of the nucleotide sequence by DNA sequencing, N-terminal FLAG-tagged wild-type hLigI (WT), empty vector (Vector), and FLAG-tagged mutant hLigI cDNAs were subcloned into the mammalian expression vector pRC/RSV (Invitrogen).
Pull-down Assay
To prepare beads for the pull-down assay, 1 μg of purified wild-type or phosphomutants versions of hLigI were incubated with hLigI antibody (2) for 2 h at 4 °C in binding buffer (50 mm Tris-HCl (pH 7.5), 50 mm NaCl, 5% glycerol, 0.2% Nonidet P-40 containing a mixture of protease inhibitors). After the addition of 20 μl of a 50% suspension of protein A-Sepharose beads (Amersham Biosciences), incubation was continued for 2 h at 4 °C. Beads were collected, washed extensively with binding buffer, and then incubated with 1 μg of purified RFC complex in buffer 750 μl of binding buffer for 2 h at 4 °C. After collection and washing, the beads were resuspended in 20 μl SDS-PAGE sample buffer. Proteins eluted from the beads were detected by immunoblotting.
Generation of Cell Lines that Stably Express FLAG-tagged Versions of Wild-type and Mutant hLigI
Normal human embryonic kidney 293 cells and human DNA ligase I-deficient fibroblast 46BR.1G1 cells were transfected with pRC/RSV plasmids encoding the FLAG-tagged versions of wild-type and mutant hLigI using the using the FuGENE6 transfection reagent (Roche) and Lipofectamine transfection reagent (Invitrogen), respectively, according to the directions of the manufacturers. After selection for resistance to G418, the level of hLigI protein in individual clones was determined by immunoblotting with antibodies against hLigI (2) and the FLAG epitope (Sigma). Clones of the 293 cells that stably expressed tagged hLigI at similar levels were chosen for further analysis. Clones of the 46BR.1G1 that stably expressed tagged hLigI at levels similar to that of endogenous hLigI in wild-type simian virus 40-immortalized human fibroblasts were chosen for further analysis.
Immunoprecipitation
Cultures of 293 and derivatives expressing the FLAG-tagged versions of either wild-type or mutant hLigI cells (4 × 108 cells of each) were harvested and lysed in 1 ml of immunoprecipitation lysis buffer (10 mm Tris-HCl (pH 7.4), 100 mm NaCl, 2.5 mm MgCl2, and 0.5% Triton X-100) containing protease and phosphatase inhibitors. After centrifugation, the supernatant was incubated at 4 °C overnight with 100 μl of a 50% suspension of protein A-Sepharose beads (Amersham Biosciences) that had been preincubated with 2 μl of a mouse anti-FLAG M5 antibody. Beads were collected by centrifugation and then washed five times with immunoprecipitation lysis buffer. Proteins were eluted from the beads in 20 μl SDS-PAGE sample buffer, separated by SDS-PAGE, and then detected by immunoblotting.
Cell Proliferation and Replication
To measure cell growth rate, 104 cells were seeded in 60-mm dishes and then cultured at 37 °C in DMEM supplemented with 10% fetal bovine serum, 0.5 mg/ml of G418, 50 units of penicillin, and 50 μg of streptomycin. After 24, 48, 72, 96, and 120 h, cells were washed with phosphate-buffered saline (10 mm phosphate (pH 7.4), 137 mm NaCl, and 2.7 mm KCl), trypsinized, and then counted using a particle counter (Beckman Instruments).
To measure DNA synthesis, 105 cells were seeded in 60-mm dishes in DMEM containing 10% fetal bovine serum and 0.5 mg/ml G418. After 3 days, [methyl-3H] thymidine (40 to 60 Ci/mmol; GE Healthcare, Piscataway, NJ) was added (final concentration, 1 μCi/ml) and incubation was continued for 30 min. Cells were then washed with phosphate-buffered saline and resuspended in 0.5 m trichloroacetic acid. After extensive washing with 0.4 m trichloroacetic acid, acid-insoluble radioactivity was measured by liquid scintillation counting. To measure BrdU incorporation, we used the APC BrdU flow kit according to the instructions of the manufacturer (BD Biosciences). BrdU incorporation and cell cycle distribution was determined using a fluorescence-activated cell sorter in the Flow Cytometry Core of the University of Maryland Marlene and Stewart Greenebaum Cancer Center.
Cell Synchronization
Cells were synchronized in early S-phase with a double block of 2 mm thymidine (18) and at the G1/S border by growth for 24 h in 500 μm mimosine (12, 19, 20).
β-Galactosidase Staining
To detect expression of senescence-associated β-galactosidase (21), cells were fixed and stained using the senescence β-galactosidase staining kit (Cell Signaling Technology, Beverly, MA) according to the instructions of the manufacturer. Cell staining was visualized using a Nikon Eclipse TE200 microscope, and images were processed using Adobe Photoshop Elements (Adobe, San Jose, CA).
Immunoblotting
Cell extracts were prepared as described previously (11). Protein concentration was determined using the method of Bradford (22). Protein in whole cell extracts (20 μg) were detected by immunoblotting after separation by SDS-PAGE using the following primary antibodies: hLigI (2); phospho-histone H2AX (Ser-139) (clone JBW301) from Upstate Biotechnology; FLAG M5 from Sigma; anti-phospho-Chk1 (Ser-354), phospho-Chk2 (Thr-68), p53, and phospho-p53 (Ser-15) from Cell Signaling Technology; RFC1 p140 from Novus; and Chk1, Chk2, PCNA, and p16 from Santa Cruz Biotechnology.
Measurement of DNA Damage in Cells
To measure DNA damage and cell cycle distribution, 46BR.1G1 and derivatives expressing recombinant human hLigI (106 of each) were grown in 10-cm plates overnight and synchronized at the G1/S border as described above and then released. After 1 h, phosphorylated H2AX was labeled with Alexa Fluor 488 mouse anti-γH2AX (pS139) according to the APC BrdU flow kit instructions (BD Biosciences) (23). Cells with a γH2AX intensity higher than 1000 were counted as γH2AX-positive cells. The extent of DNA damage in individual cells at different cell cycle stages was determined by fluorescence-activated cell sorting in the Flow Cytometry Core of the University of Maryland Marlene and Stewart Greenebaum Cancer Center.
Cell Survival Assay
Derivatives of the 46BR.1G1 cell line (1 × 104 cells) were plated in 10-cm plates in DMEM supplemented with 10% fetal bovine serum and G418 (500 μg/ml). Various concentrations of methyl methanesulfonate (MMS) and the PARP inhibitor (NU1025) were added to the medium, and then growth was continued in drug-containing media for 10 days. After cells were washed twice with phosphate-buffered saline and then fixed with 100% methanol, they were stained with crystal violet solution, washed with water, and air-dried. Colonies were counted by using Quantity One 4.5.
Statistical Analysis
Data represent the mean ± S.D. from at least three independent experiments. Statistical comparisons were performed with Student's two-tailed paired t test and analysis of variance. Values of p < 0.01 were considered statistically significant.
RESULTS
Identification of Phosphorylated Residues in hLigI Purified from Insect Cells and Role of Serine 51 in the Interaction of hLigI with RFC p140 and Association with the RFC Complex
We showed previously that phosphorylated hLigI purified after expression in insect cells exhibited more efficient binding to RFC p140 than unmodified hLigI purified after expression in E. coli but that replacement of the four phosphorylated serine residues identified by the Montecucco laboratory (12, 13) with aspartic acid to mimic phosphorylation in unmodified hLigI abolished binding to RFC (11). Therefore, we determined the sites of phosphorylation in wild-type hLigI purified from insect cells by nanospray liquid chromatography-tandem mass spectrometry. Following proteolytic digestion, we identified peptides encompassing about 90% of the 919 amino acids of hLigI (Fig. 1) and eight phosphorylated amino acids within these peptides (Fig. 1 and supplemental Table S1). These included peptides containing three of the four serine residues, Ser-66, 76, and 91 (supplemental Fig. S1, A–C), identified by the Montecucco laboratory (12, 13), that were among the most abundant phosphopeptides detected by spectral counting (supplemental Table S1) and phosphorylated residues identified in recent analyses of the human phosphoproteome (24–30).
FIGURE 1.
Amino acid sequences and phosphorylated amino acids identified LC-MS/MS after proteolytic digestion of hLigI purified from insect cells. The amino acid sequence of human DNA ligase I is shown (UniprotKB Entry P18858, DNLI1_HUMAN). Sequenced peptides and phosphorylated amino acids are indicated in boldface and with an encircled P, respectively.
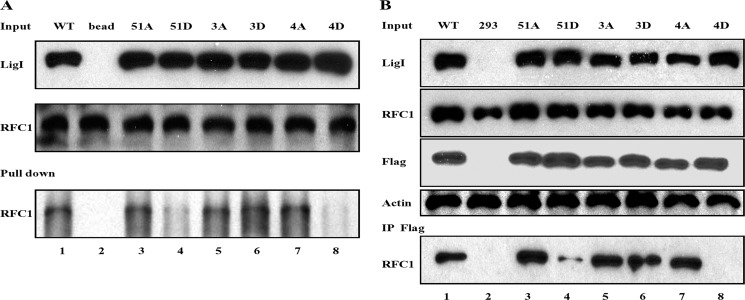
Although a peptide containing phosphorylated Ser-51 was not detected, we did identify a non-phosphorylated tryptic peptide, AALKEWNGVVSESDSPVK, containing unmodified Ser-51 (supplemental Fig. S2A) and a phosphorylated version of this peptide in which Ser-49 was phosphorylated (supplemental Fig. S2B). Because the 4D version of hLigI fails to bind to RFC (11), the absence of phosphorylated Ser-51 in hLigI purified from insect cells suggested that phosphorylation of Ser-51 may play a key role in negatively regulating the interaction with RFC p140. To test this idea, we constructed mutant versions of hLigI in which Ser51 was replaced with either aspartic acid (51D) to mimic phosphorylation or alanine (51A) and mutant versions in which Ser-51 was retained but Ser-66, 76, and 91 were each replaced with either aspartic acid (3D) or alanine (3A). After purification from E. coli, we measured the DNA joining activity of wild-type hLigI and the phosphorylation site mutant versions and their binding to the RFC complex. As expected (11), the amino acid changes in the non-catalytic N-terminal region of hLigI had no detectable effect on the ligation of nicked DNA (data not shown). In accord with our previous study, the binding of hLigI, in which all four serine residues were replaced with aspartic acid (4D), was greatly reduced compared with wild-type hLigI and the 4A version (Fig. 2A, compare lanes 1, 7, and 8). Notably, the 51D mutant exhibited a similar reduction in binding (Fig. 2A, lane 4). In contrast, neither replacement of Ser-51 with alanine nor the replacement of Ser-66, 76, and 91 with either alanine or aspartic acid significantly impacted the binding of hLigI to the RFC complex (Fig. 2A). Thus, we conclude that the presence of a negatively charged amino acid that mimics phosphorylation of Ser-51 disrupts the binding of hLigI to RFC.
FIGURE 2.
Interaction of hLigI phosphomutants with RFC. A, purified RFC complex (1 μg) was incubated with wild-type hLigI and the indicated phosphomutants (1 μg of each) prior to immunoprecipitation with hLigI antibody (2). Lane 1, wild type hLigI; lane 2, no protein; lane 3, hLig 51A; lane 4, hLigI51D; lane 5, hLigI3A; lane 6, hLigI3D; lane 7, hLigI4A; lane 8, hLigI4D. hLigI (LigI) and RFC p140 (RFC1) were detected by immunoblotting. Upper panel, 10% of the hLigI input. Center panel, 10% of RFC input. Bottom panel, immunoprecipitated RFC p140. B, immunoprecipitation of extracts from 293 cells (4 × 108 cells) expressing the following: lane 1, FLAG-tagged wild-type hLigI; lane 2, no FLAG-tagged protein; lane 3, FLAG-tagged hLigI51A; lane 4, FLAG-tagged hLigI51D; lane 5, FLAG-tagged hLigI3A; lane 6, FLAG-tagged hLigI3D; lane 7, FLAG-tagged hLigI4A; lane 8, FLAG-tagged hLigI4D. The levels of endogenous and FLAG-tagged hLigI (LigI and FLAG), β-actin (Actin), and RFC p140 (RFC1) in the extracts (Input, 5%) were determined by immunoblotting with the indicated antibodies. The extracts were incubated with the FLAG antibody (IP FLAG), and the immunoprecipitates were probed for RFC p140 (RFC1) by immunoblotting.
To examine the effect of replacing Ser-51 with either alanine or aspartic acid in hLigI on the association with the RFC complex in cell extracts, we isolated derivatives of 293 cells that stably express similar levels of FLAG-tagged wild-type hLigI and phosphorylation site mutant versions (Fig. 2B). These experiments were carried out in 293 cells because the tagged versions of hLigI were expressed at higher levels (about 10-fold higher than endogenous) and more stably in these cells compared with hLigI-deficient 46BR.1G1 cells. In accord with the results of the protein-protein interactions studies, less RFC p140 was coimmunoprecipitated with the 51D and 4D versions of hLigI compared with wild-type hLigI and the 51A and 4A versions (Fig. 2B). The 3A and, more notably, the 3D version of hLigI also had no detectable defect in the association with RFC p140 (Fig. 2B). Together, these results show that the replacement of serine 51 with aspartic acid is sufficient to disrupt the association of hLigI with RFC in cell extracts.
Effect of Ser-51 Phosphorylation on Cell Proliferation and DNA Replication
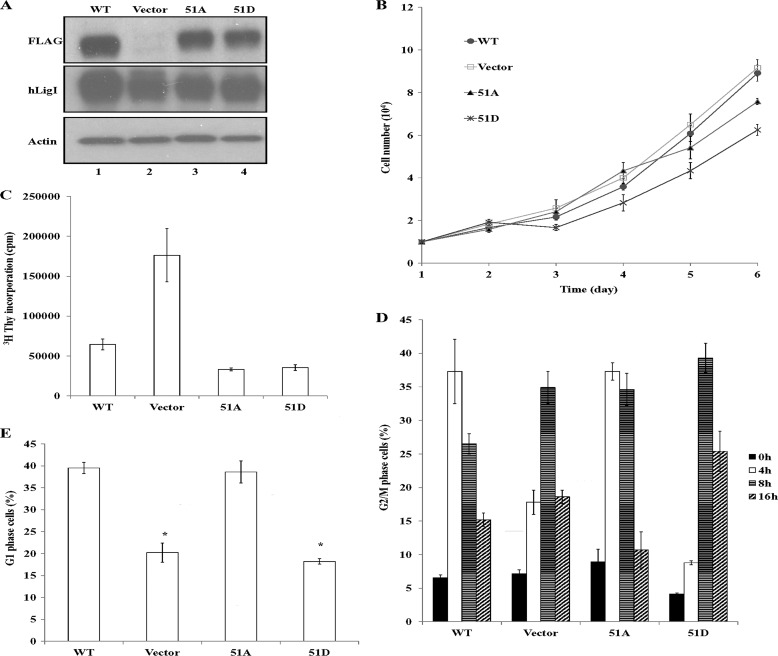
Next we asked whether replacing serine 51 of hLigI with either alanine or aspartic acid disrupted the ability of hLigI to complement the replication defect of hLigI-deficient 46BR.1G1 cells. Derivatives of 46BR.1G1 cells that stably express similar levels of FLAG-tagged versions of wild-type hLigI and the Ser51A and Ser51D mutants compared with endogenous hLigI were isolated (Fig. 3A). Expression of the mutant versions of hLigI resulted in a moderate reduction in the rate of proliferation, with the Ser51D mutant having a slightly greater effect than the Ser51A mutant (Fig. 3B). As expected (11), DNA synthesis activity was significantly higher in parental hLigI-deficient 46BR.1G1 cells compared with a derivative expressing wild-type hLigI (Fig. 3C). In contrast, expression of both the Ser51A and Ser51D versions resulted in significantly lowers levels of DNA synthesis activity than in cells expressing wild-type hLigI (Fig. 3C).
FIGURE 3.
Expression of hLigI51A and hLigI51D in hLigI-deficient cells and effects on proliferation and DNA synthesis. A, extracts (20 μg) from 46BR.1G1 cells that stably express the following: lane 1, FLAG-tagged wild-type hLigI; lane 2, no FLAG-tagged protein; lane 3, FLAG-tagged hLigI51A; lane 4, FLAG-tagged hLigI51D. The levels of endogenous and FLAG-tagged hLigI (LigI and FLAG) and β-actin (Actin) in the extracts were determined by immunoblotting with the indicated antibodies. B, growth of derivatives of 46BR.1G1 cells either transfected with the empty vector (□) or expressing FLAG-tagged wild-type hLigI (●), hLigI51A (▴) or hLigI51D (cross). The graph shows data compiled from three independent experiments, with the error bars indicating mean ± S.E. C, the indicated 46BR.1G1 derivatives were seeded in duplicate into 60-mm dishes (105 cells/dish). After 3 days, [3H]thymidine was added for 30 min. Incorporation of [3H]thymidine into DNA was measured by liquid scintillation counting. The graphs shows data compiled from two independent experiments, with the error bars indicating mean ± S.E. D and E, the indicated 46BR.1G1 derivatives were synchronized in early S phase as described under “Experimental Procedures” and then harvested at 0, 4, 8, and 12 h after release. The percentage of G2/M cells at each time point (D) and the percentage of G1 cells after 8 h (E) were determined by FACS. The graph shows data compiled from three independent experiments, with the error bars indicating S.D. *, p < 0.001 compared with cells expressing FLAG-tagged wild-type hLigI.
To determine whether alterations in the cell cycle may contribute to the reduced DNA synthesis in the cells expressing the hLigI phosphomutants, we measured cell cycle distribution in asynchronous cell populations by FACS but did not observe any major differences between 46BR.1G1 cells and derivatives expressing wild-type and mutant versions of hLigI (data not shown). Therefore, we synchronized cells in early S phase using a double thymidine block (12) and followed progression of the cells through the cell cycle after release from the block. hLigI-deficient 46BR.1G1 cells and a derivative expressing the 51D mutant version of hLigI exhibited a delay in reaching G2/M (Fig. 3D) and the G1 phase of the next cell cycle (Fig. 3E) compared with 46BR.1G1 cells expressing either wild-type hLigI or the Ser51A mutant. Because we did not observe an increased fraction of S phase cells in the asynchronous cultures, the delay in cell cycle progression observed in the hLigI-deficient 46BR.1G1 cells and the derivative expressing the 51D mutant may be due to defects in DNA replication that are exacerbated by the synchronization conditions, resulting in delayed recovery from the cell cycle block.
Expression of the hLigI Ser51D Mutant Affects Cell Morphology and Induces Cellular Senescence
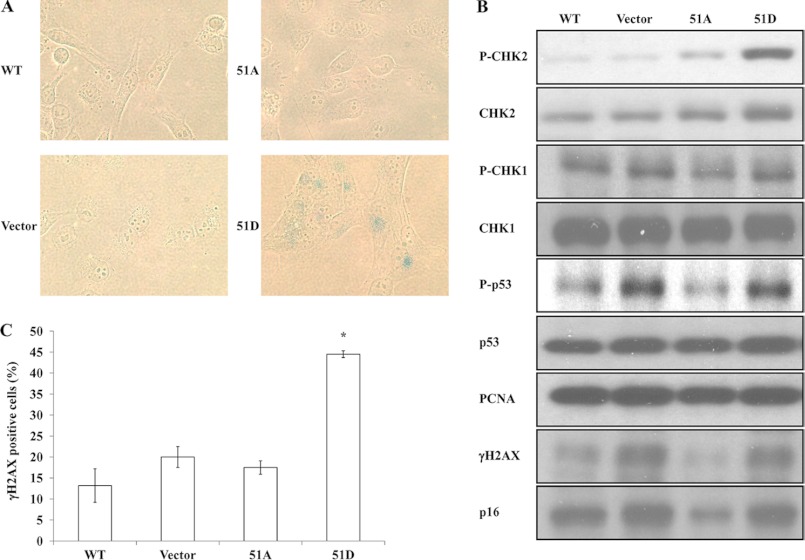
Because expression of either the 4A or 4D version of hLigI in 46BR.1G1 cells induced cellular senescence (11), we examined the morphology and expression of senescence-associated β-galactosidase in 46BR.1G1 cells expressing the Ser51A and Ser51D mutant versions of hLigI. Cells were plated and then monitored over a 5-day period. Expression of either wild-type hLigI or the Ser51A version had no detectable effect on the morphology of 46BR.1G1 cells (Fig. 4A). In contrast, larger cells with a flattened appearance were observed in the culture of 46BR.1G1 cells expressing the Ser51D mutant version of hLigI (Fig. 4A). The fraction of cells with this altered appearance increased over time, reaching more than 70% after 5 days. Unlike 46BR.1G1 cells and derivatives expressing either wild-type hLigI or the Ser51A version, β-galactosidase activity was detectable in 46BR.1G1 cells expressing the Ser51D version of hLigI (Fig. 4A), with the expression of β-galactosidase correlating with the appearance of cells with altered morphology.
FIGURE 4.
Expression of hLigI51D in hLigI-deficient cells induces cellular senescence and DNA damage. A, cultures of 46BR.1G1 cells either transfected with the empty vector (Vector) or expressing FLAG-tagged wild-type hLigI, hLigI51A (51A), or hLigI51D (51D) were fixed and stained for expression of β-galactosidase (magnification ×40). B, CHK1 and phosphorylated CHK1 (P-CHK1), CHK2 and phosphorylated CHK2 (p-CHK2), p53 and phosphorylated p53 (P-p53), PCNA, p16 and phosphorylated H2AX (γH2AX) were detected in whole cell extracts (20 μg) from the indicated derivatives of 46BR.1G1 cells by immunoblotting. C, the indicated 46BR.1G1 derivatives were synchronized at the G1/S transition as described under “Experimental Procedures” and then harvested 1 h after release. Cells with a phosphorylated H2AX (γH2AX) intensity higher than 1000, determined by FACS, were counted as γH2AX-positive cells. The graph shows the percentage of γH2AX-positive cells determined in three independent experiments, with the error bars indicating S.D. *, p < 0.001 compared with cells expressing FLAG-tagged wild-type hLigI.
Because DNA damage can induce cellular senescence (31) and the replication defect in hLigI-deficient 46BR.1G1 cells leads to the accumulation of DNA single- and double-strand breaks and activation of ATM (14), we asked whether DNA damage generated as a consequence of abnormal DNA replication was the cause of the senescence in the 46BR.1G1 cells expressing the Ser51D version of hLigI. Both the parental 46BR.1G1 cells and the derivative expressing the Ser51D version had elevated levels of phosphorylated H2AX, an indicator of DNA double strand breaks (32), phosphorylated p53, a downstream target of the ATM pathway, and the cyclin-dependent kinase inhibitor p16, which is expressed at high levels in senescent cells (33, 34), compared with 46BR.1G1 cells expressing either wild-type hLig1 or the Ser51A version (Fig. 4B). Although these results show that DNA damage response remains activated in hLigI-deficient cells expressing the Ser51D version of hLigI, they do not explain why the cells expressing the hLigI 51D mutant undergo senescence. Notably, these cells, but not the parental 46BR.1G1 cells, have an elevated level of phosphorylated Chk2 (Fig. 4B). This prompted us to ask whether expression of the Ser51D version of hLigI exacerbates the replication defect in hLigI-deficient 46BR.1G1 cells. To provide support for this idea, the level of phosphorylated H2AX was measured in cells that were synchronized at different stages of the cell cycle and then released. In cultures synchronized at the G1/S transition with mimosine and then released (12), there were significantly higher levels of phosphorylated H2AX in cells expressing hLigI Ser51 compared with the parental 46BR.1G1 cells and derivatives expressing either wild-type hLigI or the Ser51A mutant version (Fig. 4C). No significant differences were observed in cultures synchronized either in early S by double thymidine block or at G2/M with nocodazole (data not shown).
Effect of Ser-51 Phosphorylation on Cellular Sensitivity to DNA Damage
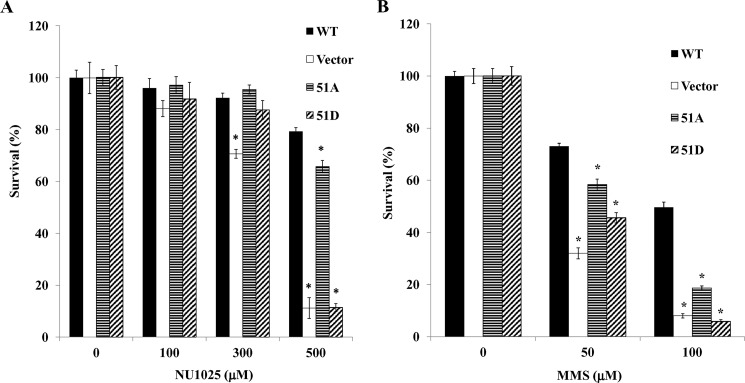
hLigI-deficient 46BR.1G1 cells are hypersensitive to 3 aminobenzamide, an inhibitor of poly(ADP-ribose) polymerases (35). Although PARP-1 does not appear to participate in the processing and joining of Okazaki fragments under normal circumstances (36), it may be involved in the repair of DNA damage resulting from inefficient joining of Okazaki fragments in hLigI-deficient cells. In support of this idea, expression of hLigI and the Ser51A derivative in 46BR.1G1 cells markedly increased resistance to the PARP inhibitor NU1025 compared with the parental cells, whereas expression of the Ser51D derivative did not (Fig. 5A). 46BR.1G1 cells are also hypersensitive to simple DNA alkylating agents such as MMS because of a defect in long patch base excision repair (2). Both the Ser51A and the Ser 51D versions of hLigI increased the resistance of 46BR.1G1 cells to MMS but not to the same extent as wild-type hLigI (Fig. 5B).
FIGURE 5.
Effect of expression of hLigI51A and hLigI51D on the sensitivity of hLigI-deficient cells to a PARP inhibitor and MMS. Cultures of 46BR.1G1 cells either transfected with the empty vector (Vector) or expressing FLAG-tagged wild-type hLigI, hLigI51A (51A), or hLigI51D (51D) were grown in the presence of the indicated concentrations of the PARP inhibitor NU1025 (A) and the DNA alkylating agent MMS as described under “Experimental Procedures” (B). After 10 days, colonies were stained with crystal violet solution and then counted by using Quantity One 4.5. The graph shows data compiled from three independent experiments, with the error bars indicating S.D. *, p < 0.001) compared with cells expressing FLAG-tagged wild-type hLigI.
DISCUSSION
Human and mouse cells with either reduced or no LigI activity have a defect in joining Okazaki fragments during DNA replication (1, 2, 5, 6, 37). In addition to interactions with PCNA and RFC (2, 4, 9), the participation of hLigI in DNA replication is mediated by phosphorylation (11–14). hLigI is phosphorylated at multiple sites, with these modifications accumulating during cell cycle progression to yield a hyperphosphorylated form in M phase (12, 13). To date, studies have focused on three serine residues phosphorylated by Cdks: Ser-51, Ser-76, and Ser-91, and Ser-66 phosphorylated by casein kinase II (12, 13). Interestingly, expression of mutant versions of hLigI, in which all four of these residues are substituted with either alanine to prevent phosphorylation or aspartic acid to mimic phosphorylation, has a dominant negative effect on the proliferation and DNA damage sensitivity of hLigI-deficient 46BR.1G1 cells (11, 14). Although the phosphorylation status of the Ser-66, 76, and 91 residues at different cell cycle stages and the dependence of Ser-76 phosphorylation upon prior phosphorylation of Ser-91 have been described (12, 13), the role of Ser-51 phosphorylation has not been elucidated. Here we have shown that posttranslational modification of Ser-51 plays a critical role in regulating the interaction of hLigI with RFC and the participation of hLigI in replication and repair.
Since hLigI is phosphorylated in S phase (12, 13), it appears likely that its participation in DNA replication is regulated by phosphorylation at specific sites. We showed previously that phosphorylated hLigI purified from insect cells bound more efficiently to RFC p140 than unmodified hLigI purified from E. coli, whereas replacement of the phosphorylated serine residues 51, 66, 76, and 91, with aspartic acid to mimic phosphorylation, disrupted the interaction of unmodified hLigI with RFC (11). In contrast, these different species of hLigI did not differ in their ability to interact with PCNA (11). To gain insights into the role of phosphorylation in regulating the interaction of hLigI with RFC, we identified the phosphorylated residues in hLigI purified from insect cells. Although Ser-66, Ser-76, and Ser-91 were phosphorylated, Ser-51 was not, suggesting that phosphorylation of Ser-51 may negatively regulate the interaction with RFC. In support of this model, replacement of Ser-51 alone with aspartic acid had a similar effect in terms of reducing the in vitro interaction of hLigI with RFC p140 to replacing all four serine residues with aspartic acid. However, replacement of serines 66, 76, and 91 with aspartic acid to mimic the phosphorylation state of insect cell-expressed hLigI had no significant effect on the binding of unmodified hLigI to RFC p140. These results were reiterated in immunoprecipitation assays with extracts from cells expressing the mutant versions of hLigI. With the caveat that aspartic acid may not mimic phosphoserine in some contexts, it appears that phosphorylation of Ser-51 disrupts the binding of hLigI to RFC and that phosphorylation at sites other than Ser-51, 66, 76, and 91 enhance the binding of insect cell-expressed hLigI to RFC. Although phosphorylation of Ser-91 by a Cdk at the G1/S transition is necessary for the subsequent phosphorylation of Ser-76 and the appearance of hyperphosphorylated hLigI in M phase cells, preventing Ser-51 phosphorylation did not result in altered mobility of hLigI (12). Thus, phosphorylation of Ser-51 does not appear to be required for phosphorylation of Ser-76 and Ser-91. Instead, we suggest that Ser-51 phosphorylation most likely occurs when replication has been completed and contributes to the disassembly of the replication fork and/or replication factories.
Expression of both the 51A and 51D version of hLigI in hLigI-deficient 46BR.1G1 cells reduced DNA synthesis and the rate of cell proliferation albeit to a lesser extent than the 4A and 4D versions of hLigI. We had shown previously that expression of either the 4A or 4D version of hLigI induced 46BR.1G1 cells to undergo senescence (11). A similar effect was observed with the Ser51D but not the Ser51A version of hLigI. Like the cells expressing the 4D version of hLigI (14), elevated levels of phospho-Chk2 but not phospho-Chk1 were present in the cells expressing the Ser51D version of hLigI. Notably, cells expressing the 51D version of hLigI contained higher levels of γH2AX than the parental hLigI-deficient 46BR.1G1 cells, indicating that these cells have higher levels of replication-dependent DNA damage. It is likely that these increased levels of DNA damage activate the ATM-Chk2 pathway (14), leading to cellular senescence. We suggest that the high levels of replication-dependent DNA damage underlie the sensitivity of 46BR.1G1 cells and the derivative expressing hLigI Ser51D to PARP inhibitors, as it is likely that the DNA damage generated by abnormal DNA replication is repaired by PARP-dependent DNA repair pathways. Similarly, we envision that the abnormalities in DNA replication and the resultant DNA damage in 46BR.1G1 cells and the derivative expressing hLigI Ser51D contribute to the delayed progression through the cell cycle after synchronization in early S phase by double thymidine block. Because the 4D version of hLigI is targeted to replication foci (14), it is likely that the hLig51D version is recruited to the replication machinery by virtue of its interaction with PCNA but is unable to functionally interact with RFC. This may cause DNA damage by disrupting Okazaki fragment processing and ligation and/or hindering the ability of DNA ligase IIIα to substitute for hLigI during DNA replication (7).
In addition to the defect in joining Okazaki fragments, hLigI-deficient 46BR.1G1 cells are hypersensitive to simple DNA alkylating agents (2). Expression of either the Ser51A or the Ser51D version of hLigI only partially corrected the DNA damage sensitivity, indicating that posttranslational modification of Ser-51 contributes to the repair of DNA alkylation damage. In contrast, expression of the 4A and 4D versions of hLigI increased the sensitivity of 46BR.1G1 cells to DNA alkylation (11), indicating that other phosphorylation sites are involved in dictating the participation of hLigI in DNA repair. In summary, we have shown that posttranslation modification of Ser-51 regulates the interaction of hLigI with RFC and that this interaction plays a key role in coordinating Okazaki fragment processing and ligation during DNA replication and contributes to the DNA repair functions of hLigI.
Acknowledgments
We thank Drs. Rassool, Song, and Vijayakumar for reagents.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 GM 57479 and P01 CA 92584 (to A. E. T.) and R01 AG25323 (to A. Y.).

This article contains supplemental Fig. S1 and S2 and Table S1.
- hLigI
- human DNA ligase I
- PCNA
- proliferating cell nuclear antigen
- RFC
- replication factor C
- γH2AX
- phosphorylated histone H2AX.
REFERENCES
- 1. Barnes D. E., Tomkinson A. E., Lehmann A. R., Webster A. D., Lindahl T. (1992) Mutations in the DNA ligase I gene of an individual with immunodeficiencies and cellular hypersensitivity to DNA-damaging agents. Cell 69, 495–503 [DOI] [PubMed] [Google Scholar]
- 2. Levin D. S., McKenna A. E., Motycka T. A., Matsumoto Y., Tomkinson A. E. (2000) Interaction between PCNA and DNA ligase I is critical for joining of Okazaki fragments and long-patch base-excision repair. Curr. Biol. 10, 919–922 [DOI] [PubMed] [Google Scholar]
- 3. Lasko D. D., Tomkinson A. E., Lindahl T. (1990) Mammalian DNA ligases. Biosynthesis and intracellular localization of DNA ligase I. J. Biol. Chem. 265, 12618–12622 [PubMed] [Google Scholar]
- 4. Montecucco A., Rossi R., Levin D. S., Gary R., Park M. S., Motycka T. A., Ciarrocchi G., Villa A., Biamonti G., Tomkinson A. E. (1998) DNA ligase I is recruited to sites of DNA replication by an interaction with proliferating cell nuclear antigen. Identification of a common targeting mechanism for the assembly of replication factories. EMBO J. 17, 3786–3795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bentley D., Selfridge J., Millar J. K., Samuel K., Hole N., Ansell J. D., Melton D. W. (1996) DNA ligase I is required for fetal liver erythropoiesis but is not essential for mammalian cell viability. Nat. Genet. 13, 489–491 [DOI] [PubMed] [Google Scholar]
- 6. Bentley D. J., Harrison C., Ketchen A. M., Redhead N. J., Samuel K., Waterfall M., Ansell J. D., Melton D. W. (2002) DNA ligase I null mouse cells show normal DNA repair activity but altered DNA replication and reduced genome stability. J. Cell Sci. 115, 1551–1561 [DOI] [PubMed] [Google Scholar]
- 7. Arakawa H., Bednar T., Wang M., Paul K., Mladenov E., Bencsik-Theilen A. A., Iliakis G. (2012) Functional redundancy between DNA ligases I and III in DNA replication in vertebrate cells. Nucleic Acids Res. 40, 2599–2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Levin D. S., Bai W., Yao N., O'Donnell M., Tomkinson A. E. (1997) An interaction between DNA ligase I and proliferating cell nuclear antigen. Implications for Okazaki fragment synthesis and joining. Proc. Natl. Acad. Sci. U.S.A. 94, 12863–12868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Levin D. S., Vijayakumar S., Liu X., Bermudez V. P., Hurwitz J., Tomkinson A. E. (2004) A conserved interaction between the replicative clamp loader and DNA ligase in eukaryotes. Implications for Okazaki fragment joining. J. Biol. Chem. 279, 55196–55201 [DOI] [PubMed] [Google Scholar]
- 10. Waga S., Stillman B. (1998) The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67, 721–751 [DOI] [PubMed] [Google Scholar]
- 11. Vijayakumar S., Dziegielewska B., Levin D. S., Song W., Yin J., Yang A., Matsumoto Y., Bermudez V. P., Hurwitz J., Tomkinson A. E. (2009) Phosphorylation of human DNA ligase I regulates its interaction with replication factor C and its participation in DNA replication and DNA repair. Mol. Cell. Biol. 29, 2042–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ferrari G., Rossi R., Arosio D., Vindigni A., Biamonti G., Montecucco A. (2003) Cell cycle-dependent phosphorylation of human DNA ligase I at the cyclin-dependent kinase sites. J. Biol. Chem. 278, 37761–37767 [DOI] [PubMed] [Google Scholar]
- 13. Rossi R., Villa A., Negri C., Scovassi I., Ciarrocchi G., Biamonti G., Montecucco A. (1999) The replication factory targeting sequence/PCNA-binding site is required in G(1) to control the phosphorylation status of DNA ligase I. EMBO J. 18, 5745–5754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Soza S., Leva V., Vago R., Ferrari G., Mazzini G., Biamonti G., Montecucco A. (2009) DNA ligase I deficiency leads to replication-dependent DNA damage and impacts cell morphology without blocking cell cycle progression. Mol. Cell. Biol. 29, 2032–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang Y. C., Burkhart W. A., Mackey Z. B., Moyer M. B., Ramos W., Husain I., Chen J., Besterman J. M., Tomkinson A. E. (1994) Mammalian DNA ligase II is highly homologous with vaccinia DNA ligase. Identification of the DNA ligase II active site for enzyme-adenylate formation. J. Biol. Chem. 269, 31923–31928 [PubMed] [Google Scholar]
- 16. Fazlieva R., Spittle C. S., Morrissey D., Hayashi H., Yan H., Matsumoto Y. (2009) Proofreading exonuclease activity of human DNA polymerase delta and its effects on lesion-bypass DNA synthesis. Nucleic Acids Res. 37, 2854–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cripps D., Thomas S. N., Jeng Y., Yang F., Davies P., Yang A. J. (2006) Alzheimer disease-specific conformation of hyperphosphorylated paired helical filament-τ is polyubiquitinated through Lys-48, Lys-11, and Lys-6 ubiquitin conjugation. J. Biol. Chem. 281, 10825–10838 [DOI] [PubMed] [Google Scholar]
- 18. Stein J. L. (1989) Cell Synchronization (Baserga R., ed.) pp. 133–137, IRL Press, Oxford, UK [Google Scholar]
- 19. Krude T. (1999) Mimosine arrests proliferating human cells before onset of DNA replication in a dose-dependent manner. Exp. Cell Res. 247, 148–159 [DOI] [PubMed] [Google Scholar]
- 20. Song W., Levin D. S., Varkey J., Post S., Bermudez V. P., Hurwitz J., Tomkinson A. E. (2007) A conserved physical and functional interaction between the cell cycle checkpoint clamp loader and DNA ligase I of eukaryotes. J. Biol. Chem. 282, 22721–22730 [DOI] [PubMed] [Google Scholar]
- 21. Dimri G. P., Lee X., Basile G., Acosta M., Scott G., Roskelley C., Medrano E. E., Linskens M., Rubelj I., Pereira-Smith O. (1995) A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. U.S.A. 92, 9363–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 23. Porcedda P., Turinetto V., Brusco A., Cavalieri S., Lantelme E., Orlando L., Ricardi U., Amoroso A., Gregori D., Giachino C. (2008) A rapid flow cytometry test based on histone H2AX phosphorylation for the sensitive and specific diagnosis of ataxia telangiectasia. Cytometry A 73, 508–516 [DOI] [PubMed] [Google Scholar]
- 24. Hsu P. P., Kang S. A., Rameseder J., Zhang Y., Ottina K. A., Lim D., Peterson T. R., Choi Y., Gray N. S., Yaffe M. B., Marto J. A., Sabatini D. M. (2011) The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 332, 1317–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rigbolt K. T., Prokhorova T. A., Akimov V., Henningsen J., Johansen P. T., Kratchmarova I., Kassem M., Mann M., Olsen J. V., Blagoev B. (2011) System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci. Signal. 4, rs3. [DOI] [PubMed] [Google Scholar]
- 26. Olsen J. V., Vermeulen M., Santamaria A., Kumar C., Miller M. L., Jensen L. J., Gnad F., Cox J., Jensen T. S., Nigg E. A., Brunak S., Mann M. (2010) Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3, ra3. [DOI] [PubMed] [Google Scholar]
- 27. Iliuk A. B., Martin V. A., Alicie B. M., Geahlen R. L., Tao W. A. (2010) In-depth analyses of kinase-dependent tyrosine phosphoproteomes based on metal ion-functionalized soluble nanopolymers. Mol. Cell. Proteomics 9, 2162–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen R. Q., Yang Q. K., Lu B. W., Yi W., Cantin G., Chen Y. L., Fearns C., Yates J. R., 3rd, Lee J. D. (2009) CDC25B mediates rapamycin-induced oncogenic responses in cancer cells. Cancer Res. 69, 2663–2668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang Y. T., Tsai C. F., Hong T. C., Tsou C. C., Lin P. Y., Pan S. H., Hong T. M., Yang P. C., Sung T. Y., Hsu W. L., Chen Y. J. (2010) An informatics-assisted label-free quantitation strategy that depicts phosphoproteomic profiles in lung cancer cell invasion. J. Proteome Res. 9, 5582–5597 [DOI] [PubMed] [Google Scholar]
- 30. Christensen G. L., Kelstrup C. D., Lyngsø C., Sarwar U., Bøgebo R., Sheikh S. P., Gammeltoft S., Olsen J. V., Hansen J. L. (2010) Quantitative phosphoproteomics dissection of seven-transmembrane receptor signaling using full and biased agonists. Mol. Cell Proteomics 9, 1540–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wahl G. M., Carr A. M. (2001) The evolution of diverse biological responses to DNA damage. Insights from yeast and p53. Nat. Cell Biol. 3, E277–286 [DOI] [PubMed] [Google Scholar]
- 32. Löbrich M., Shibata A., Beucher A., Fisher A., Ensminger M., Goodarzi A. A., Barton O., Jeggo P. A. (2010) γH2AX foci analysis for monitoring DNA double-strand break repair. Strengths, limitations, and optimization. Cell Cycle 9, 662–669 [DOI] [PubMed] [Google Scholar]
- 33. Serrano M., Lin A. W., McCurrach M. E., Beach D., Lowe S. W. (1997) Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88, 593–602 [DOI] [PubMed] [Google Scholar]
- 34. Di Micco R., Fumagalli M., Cicalese A., Piccinin S., Gasparini P., Luise C., Schurra C., Garre' M., Nuciforo P. G., Bensimon A., Maestro R., Pelicci P. G., d'Adda di Fagagna F. (2006) Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444, 638–642 [DOI] [PubMed] [Google Scholar]
- 35. Lehmann A. R., Willis A. E., Broughton B. C., James M. R., Steingrimsdottir H., Harcourt S. A., Arlett C. F., Lindahl T. (1988) Relation between the human fibroblast strain 46BR and cell lines representative of Bloom's syndrome. Cancer Res. 48, 6343–6347 [PubMed] [Google Scholar]
- 36. Balakrishnan L., Bambara R. A. (2011) Eukaryotic lagging strand DNA replication employs a multi-pathway mechanism that protects genome integrity. J. Biol. Chem. 286, 6865–6870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Harrison C., Ketchen A. M., Redhead N. J., O'Sullivan M. J., Melton D. W. (2002) Replication failure, genome instability, and increased cancer susceptibility in mice with a point mutation in the DNA ligase I gene. Cancer Res. 62, 4065–4074 [PubMed] [Google Scholar]