Background: Erythropoietin is required for erythrocyte production and stimulates erythroid gene expression including EPO-R.
Results: TAL1 induction promotes accessibility of EPO-R promoter to the GATA-1·TAL1·LMO2·LDB1 transcription activation complex to increase EPO-R expression.
Conclusion: Forced TAL1 expression increases EPO-R and erythropoietin hypersensitivity in erythroid progenitors.
Significance: Providing insight into the molecular link between TAL1 and erythropoietin activity.
Keywords: Erythropoeisis, Erythropoietin, Hematopoiesis, Transcription Factors, Transcription Regulation, TAL1, Excessive Erythrocytosis
Abstract
During erythropoiesis, erythropoietin stimulates induction of erythroid transcription factors that activate expression of erythroid genes including the erythropoietin receptor (EPO-R) that results in increased sensitivity to erythropoietin. DNA binding of the basic helix-loop-helix transcription factor, TAL1/SCL, is required for normal erythropoiesis. A link between elevated TAL1 and excessive erythrocytosis is suggested by erythroid progenitor cells from a patient that exhibits unusually high sensitivity to erythropoietin with concomitantly elevated TAL1 and EPO-R expression. We found that TAL1 regulates EPO-R expression mediated via three conserved E-box binding motifs (CAGCTG) in the EPO-R 5′ untranslated transcribed region. TAL1 increases association of the GATA-1·TAL1·LMO2·LDB1 transcription activation complex to the region that includes the transcription start site and the 5′ GATA and 3′ E-box motifs flanking the EPO-R transcription start site suggesting that TAL1 promotes accessibility of this region. Nucleosome shifting has been demonstrated to facilitate TAL1 but not GATA-1 binding to regulate target gene expression. Accordingly, we observed that with induced expression of EPO-R in hemotopoietic progenitor cells, nucleosome phasing shifts to increase the linker region containing the EPO-R transcription start site and TAL1 binds to the flanking 5′ GATA and 3′ E-box regions of the promoter. These data suggest that TAL1 binds to the EPO-R promoter to activate EPO-R expression and provides a potential link to elevated EPO-R expression leading to hypersensitivity to erythropoietin and the resultant excessive erythrocytosis.
Introduction
Erythropoietin (EPO)4 stimulates the proliferation, survival, and differentiation of erythroid progenitor cells. The bioavailability of EPO and the extent of EPO receptor (EPO-R) expression largely determine the erythropoietic response. EPO binding to its receptor on the surface of erythroid progenitor cells stimulates their proliferation and differentiation into mature red blood cells. The absence of EPO or EPO-R in mice is lethal in utero due to severe anemia (1). The EPO-R cytoplasmic region lacks intrinsic tyrosine kinase activity and is associated with the nonreceptor tyrosine kinase, JAK2. Binding of EPO to the EPO-R homodimer induces a conformational change of the two cytoplasmic tails bringing the associated JAK2 proteins into closer proximity, resulting in phosphorylation and activation of JAK2 and other downstream signal transduction pathways including STAT5 (2).
Increased EPO availability, EPO sensitivity, or EPO-related JAK2 signal transduction in erythroid progenitor cells stimulates erythropoiesis. Excessive erythropoiesis has been identified in individuals with increased production of EPO due to genetic mutations that disrupt the hypoxic regulation of EPO. These include mutations in proline hydroxylase or Von Hippel-Lindau (VHL) genes that mediate hypoxia-dependent stability of the α-subunit of hypoxia inducible factor, and a gain of function mutation in the hypoxia inducible factor-2α subunit (3–5). JAK2 mutations (JAK2V617F and other exon 12 mutations) that increase JAK2 activity and provide cytokine-independent growth of hematopoietic progenitor cell cultures have been detected in ∼95% of polycythemia vera patients (6, 7). The C-terminal EPO-R region acts as a negative growth-regulatory domain affecting receptor processing and degradation (8). EPO-R gene mutations resulting in truncation of this region have been identified in patients with familial polycythemia (9, 10).
EPO stimulation up-regulates erythroid-specific transcription factor expression, such as TAL1/SCL, GATA-1, and EKLF, to promote erythroid differentiation (11). These transcription factors and their interacting coregulators including Friend of GATA-1 (FOG-1), Med1 for GATA-1, and lysine specific demethylase 1 (LSD1) for TAL1 are able to provide epigenetic modification of erythroid-specific target genes by recruiting histone acetyltransferases or histone deacetylases to, respectively, activate or repress gene expression (12, 13). For example, the basic helix-loop-helix (bHLH) transcription factor TAL1 is important for hematopoietic stem cell development, and TAL1 binding to E-box motifs (CANNTG) is required for erythroid maturation (14, 15). During erythroid differentiation TAL1 decreases association with the corepressors, mSin3A and ETO-2, with histone deacetylase, and transiently with LSD1; concomitantly, TAL1 acetylation by p300 increases resulting in increased DNA binding activity and selective activation of erythroid-specific genes (16–20). TAL1 associates with GATA-1, GATA-2, lim-only protein LMO2, and LDB1 in a multimeric complex to regulate erythroid transcription through GATA motifs as well as through E-box-GATA motifs separated by about one DNA helical turn (9 to 11 nucleotides) that are found in many erythroid regulatory elements (21–25).
EPO-R is expressed on early erythroid progenitor cells or BFU-E (burst forming unit erythroid) at a low level and is up-regulated with erythroid differentiation to CFU-Es (colony forming unit erythroid) that require EPO for survival (1). EPO induction of its own receptor is mediated in part by EPO induction of GATA-1, which can transactivate EPO-R (24, 26). The importance of TAL1 in erythroid differentiation is underscored by erythroid progenitor cell cultures from an individual with chronic isolated, excessive erythropoiesis (see below). These erythroid precursor cells exhibit EPO hypersensitivity and elevated EPO-R expression but normal GATA-1 and GATA-2 levels. We found abnormally high levels of TAL1 and increased TAL1 binding to EPO-R proximal promoter following EPO stimulation. We show that forced expression of EPO-R in human primary erythroid progenitor cells promotes EPO response via increased expression of GATA-1, TAL1, and EPO-R. Similarly, overexpression of TAL1 increases EPO-R expression and erythroid differentiation without affecting GATA-1 expression, whereas TAL1 knockdown decreases EPO-R expression and erythroid differentiation. TAL1 binds directly to the conserved E-box region downstream of the EPO-R transcription start site and regulates promoter activity. TAL1 regulation of EPO-R and the elevated induction of TAL1 and EPO-R in differentiating erythroid progenitor cell cultures from Patient A link to the observed EPO hypersensitivity in this case of chronic increase in erythropoiesis. In contrast, cultures of Jak2V617F erythroid progenitor cells show normal EPO-R and TAL1 levels.
EXPERIMENTAL PROCEDURES
Erythroid Cell Culture
Human primary erythroid progenitor cells were isolated from blood obtained from healthy volunteers through the National Institutes of Health (NIH) Department of Transfusion Medicine or from consenting patients and cultured as described (27). Informed consent was provided according to the Declaration of Helsinki. Briefly, buffy coats isolated from whole blood were diluted 1:1 with phosphate-buffered saline (PBS), pH 7.4, and gently layered on Ficoll-Hypaque (Sigma). They were centrifuged first to separate red blood cells, platelets, and plasma from the mononuclear cells, and then further centrifuged to wash the cells with PBS. Cells were then cultured 5–7 days in phase I medium containing α-minimal essential medium, 10% fetal bovine serum (FBS), 10% conditioned medium (from 5637 human bladder carcinoma cell line culture), 1.5 mm glutamine, 1 μg/ml of cyclosporin A, and antibiotics at 37 ºC with 5% CO2. To generate primary adult human erythroid progenitor cell (hAEPC) cultures, at the end of phase I, cells were then cultured in phase II medium containing EPO (1 unit/ml), α-minimal essential medium, 30% FBS, 1% bovine serum albumin, 10−6 m dexamethasone, 10−5 m β-mercapthoethanol, 0.3 mg/ml of human holotransferrin (Sigma), 10 ng/ml of stem cell factor, and antibiotics for up to 12 days. Viable cell counts were obtained using trypan blue exclusion. Benzidine staining was used for counting Hb containing cells. K562 cells were maintained in RPMI 1640 with 10% FBS. For other studies, CD34+ and CD133+ hematopoietic stem cells were isolated from apheresis products using immunomagnetic beads specific for CD34+ and CD133+ (Miltenyi CliniMacs system). For differentiation into CD36+ cells, the CD133+ cells were suspended in maintenance media prepared using α-minimal essential medium (for a final concentration of 10 mg/ml of BSA, 10 μg/ml of insulin, 200 μg/ml of holotransferrin and supplemented with 900 ng/ml of ferrous sulfate, 90 ng/ml of ferric nitrate, 10−6 m hydrocortisone, 100 ng/ml of stem cell factor, 5 ng/ml of IL-3, and 3 units/ml of EPO). The cells were incubated at 37 °C in a 5% CO2, 95% air atmosphere for 9–11 days, followed by FACS analysis for the presence of CD36+ cells.
Quantification of Gene Expression
Total cellular RNA was extracted using RNeasy (Qiagen, Valencia, CA). Synthesis of first-strand cDNA from 1 μg of total RNA was performed using Moloney murine leukemia virus reverse transcriptase (RT) and oligo(dT)16 (Applied Biosystems, Foster City, CA). Quantitative real-time RT-PCR was performed as previously described using a 7900HT Fast Real-time PCR (Applied Biosystems) and gene-specific primers and TaqMan probes for EPO-R, GATA-1, GATA-2, TAL1, EKLF, β-globin, γ-globin, and β-actin (11). Serial dilutions of plasmids containing the cDNA of interest were used to create standard curves.
Colony Assay
Mononuclear cells (2 × 105) from peripheral blood were cultured in 35-mm plastic tissue culture dishes in methylcellulose medium (Stemcell number 04434) and incubated at 37 ºC, in 5% CO2, with ≥95% humidity for 14–16 days. CFU-E and BFU-E were identified by their red color via a microscope with a ×4 objective for CFU-E (×40 magnification), and a ×2 objective for BFU-E (×20 magnification).
Transfection of Expression Vectors
After 3 days of EPO exposure, erythroid progenitor cells were transfected with an expression plasmid containing mouse Epo-R (expression vector PIRES2-EGFP-mEpoR), TAL1 (expression vector PIRES2-EGFP-TAL1), TAL1 antisense cDNA, mouse Eklf (mEklf) cDNA, or no plasmid using the Amaxa Nucleofector for CD34+ cells (Lonza, Walkersville, MD). Cells were analyzed on the days indicated following EPO stimulation. For stable TAL1 overexpression in K562 erythroid cells, K562 were co-transfected with a TAL1 expression vector PIRES2-EGFP-TAL1 and RSVneo. Cells were seeded at low dilution in media containing G418. K562/TAL1 colonies were isolated and propagated in suspension culture and the expression level of TAL1, EPO-R, and other erythroid genes determined.
Reporter Gene Analysis
A human EPO-R reporter gene construct (EPOR) extending 5′ 194 and 3′ 120 bp from the transcription start site was linked to the firefly luciferase reporter gene (26). ΔEPOR with deletion of the conserved E-box region was constructed similarly with the promoter fragment extending only 26 bp 3′. Δ3EPR was constructed from EPOR by mutation of each of the three E-box motifs. Luciferase plasmids were co-transfected with a Renilla luciferase construct (pRL-TK) into K562 cells with Lipofectamine 2000 (Invitrogen). Cells were harvested after 48 h and luciferase activity was determined by the Dual Luciferase Assay System (Promega, Madison, WI). A promoterless construct containing only the 5′ untranslated transcribed region of EPO-R linked to luciferase was used as a negative control.
Chromatin Immunoprecipitation (ChIP)
Analysis was carried out in K562 cells without and with TAL1 overexpression (K562/TAL1) as previously described (28). In brief, isolated 1% formaldehyde cross-linked cells were lysed (lysis buffer: 1% SDS, 10 mm EDTA, 50 mm Tris, pH 8.1) and sonicated on ice. DNA was sheared to an average length of around 500 bp. The sonicated cell supernatant was diluted 10-fold in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris−HCl, 167 mm NaCl, pH 8.1). Chromatin was precleared using protein G-agarose (Millipore, Billerica, MA) and incubated at 4 °C overnight with specific antibodies or nonspecific IgG (control). Immunoprecipitates were recovered with protein G-agarose (4 °C for 2 h), followed by low-speed centrifugation. Washed pellets were reverse cross-linked. DNA was phenol chloroform/isoamyl alcohol (25:24:1) extracted, ethanol precipitated, and used for PCR analysis.
Western Blotting
Erythroid progenitor cells from normal volunteers and Patient A were washed twice with cold PBS and lysed in RIPA buffer (50 mm Tris-HCl, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, 0.25% sodium deoxycholate and protease inhibitors). Proteins were resolved on 4–12% NuPAGE BisTris gels (Invitrogen), transferred onto nitrocellulose membranes, incubated with EPO-R antibody (M-20), TAL1 antibody, or anti-β-actin antibody (Santa Cruz Biotechnology, Santa Cruz, CA) followed by horseradish-peroxidase (HRP)-coupled secondary antibodies, and developed by enhanced chemiluminescence (ECL) (GE Healthcare).
DNA Sequencing and Southern Blotting
DNA was isolated directly from erythroid progenitor cell cultures from Patient A. Primer pairs were designed to PCR amplify and sequence the erythroid 1a promoter, +19 enhancer, and +51 enhancer regions of the TAL1 gene. Sequencing results were analyzed using the BLASTn tool. Genomic DNA isolated from immortalized B-cells from Patient A was digested by restriction enzyme and analyzed for TAL1 rearrangements by Southern blotting (supplemental Fig. 1A). DNA primers specific for the interstitial deletion between the TAL1 and SIL genes were synthesized and also used for genomic analysis (29). Restriction enzyme digestion and direct DNA sequencing confirmed the identity of the PCR product.
EPO-R RNA-seq and EPO-R Nucleosome Phasing Analysis
EPO-R RNA-seq was performed as described (30). Briefly, mRNAs isolated from CD34+ and CD36+ cells from a healthy donor were converted to cDNA using standard protocols. The cDNAs were fragmented to 100−200 bp using sonication, followed by end repair and Solexa adaptor ligation. The products were sequenced on an Illumina GAII system according to established procedures. Nucleosome phasing was performed using Solexa Sequencing technology as described (31).
RESULTS
Erythrocytosis Linked to Elevated EPO-R and TAL1 Expression in Erythroid Progenitor Cells
We identified a patient with unusually high hemoglobin of 22 g/dl upon emergency hospitalization for a brain hemorrhage. Patient A, an adult male, had normal white blood cell count, differential white blood cell count and platelet count were normal. Mean corpuscular volume and red cell morphology were normal. No vascular abnormality aside from the brain hemorrhage and no enlargement of liver or spleen were found. The skin of the patient was a gray-purple color at presentation but returned to normal when hemoglobin was decreased to the normal range. EPO level, pO2, oxygen saturation, p50, and hemoglobin electrophoresis were normal. Patient A was emergently treated with phlebotomy to lower his hemoglobin to the normal range and he recovered from all neurologic defects. The elevated erythrocytosis appeared to be inherited as the patient's father and daughter had high hemoglobin. Sequencing of genomic DNA from Patient A did not reveal any mutations in the erythropoietin receptor or in other genes associated with isolated erythrocytosis such as VHL, PDH-2, hypoxia inducible factor-2α, Jak2V617F, exon 12 mutations of the JAK2 gene, and hemochromatosis.
The hAEPC cultures derived from CD34+ cells from Patient A required EPO for colony formation but exhibited unusually large erythroid colonies. Erythroid progenitor cells exhibited hypersensitivity to EPO with increased induction of EPO-R and β-globin expression compared with the control hAEPC culture (Fig. 1, A and B). In contrast, hAEPC cultures from patients with the Jak2V617F mutation exhibit increased numbers of early erythroid progenitor cells even in the absence of EPO (32). Furthermore, EPO stimulated induction of EPO-R and β-globin expression in Jak2V617F cultures did not exhibit the increased level observed in erythroid progenitor cell cultures from Patient A (Fig. 1B). The increased level of EPO-R expression provides a possible explanation for the hypersensitivity to EPO during erythroid differentiation of hematopoietic progenitor cells from Patient A. The colony assays of the patient progenitor cells showed an increase in CFU-E but not BFU-E (Fig. 1C), consistent with the increased proliferative response with EPO stimulation.
FIGURE 1.
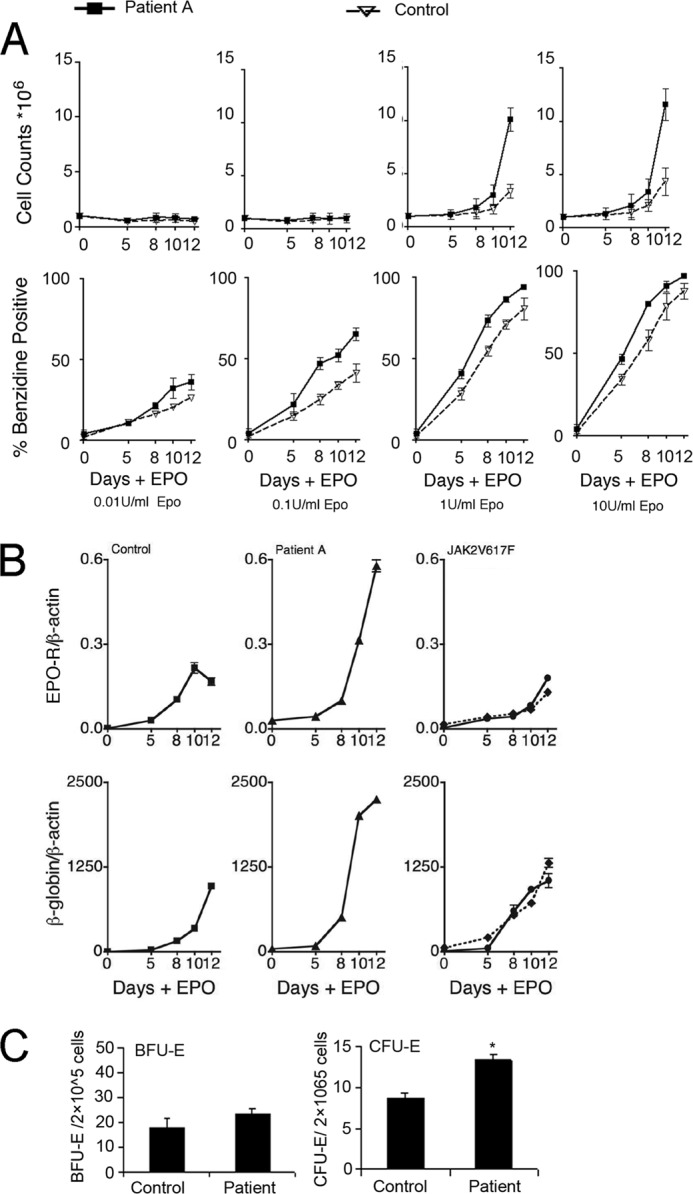
Gene expression in human primary erythroid progenitor cells. A, cell proliferation (top) and % benzidine positive cells (bottom) following 12 days of treatment with EPO in primary erythroid progenitor cell cultures from Patient A with excessive erythrocytosis (solid line) compared with normal control (dashed line) treated with 0.01 to 10 units/ml of EPO as indicated. Data represent mean ± S.D. B, expression of EPO-R and β-globin are shown for corresponding cultures treated with 1 unit/ml of EPO and corresponding cultures from Patient B (solid line) and Patient C (dashed line) with the JAK2V617F mutation associated with polycythemia vera (right) and treated with 1 unit/ml of EPO. C, colony assay of BFU-E and CFU-E in peripheral blood of patient and control in culture. Data represent mean ± S.D. (n = 3), and * indicates p ≤ 0.05.
EPO-R promoter is transactivated by GATA-1, which requires the GATA-1 binding motif for high-level EPO-R expression. However, quantification of GATA-1 expression in EPO-stimulated hAEPC cultures from Patient A revealed that EPO induction of GATA-1, as well as down-regulation of GATA-2, was similar for cultures from Patient A and for normal control (Fig. 2A). Erythroid progenitor cells from Patient A did show increased induction of EKLF in response to EPO stimulation, consistent with the increased induction of β-globin expression compared with control or Jak2V617F cultures (Fig. 2B). However, overexpression of mEklf in control hAEPC did not increase the EPO-R mRNA level, but did increase erythroid differentiation, determined by benzidine staining, and decreased progenitor cell number (supplemental Fig. 1B).
FIGURE 2.
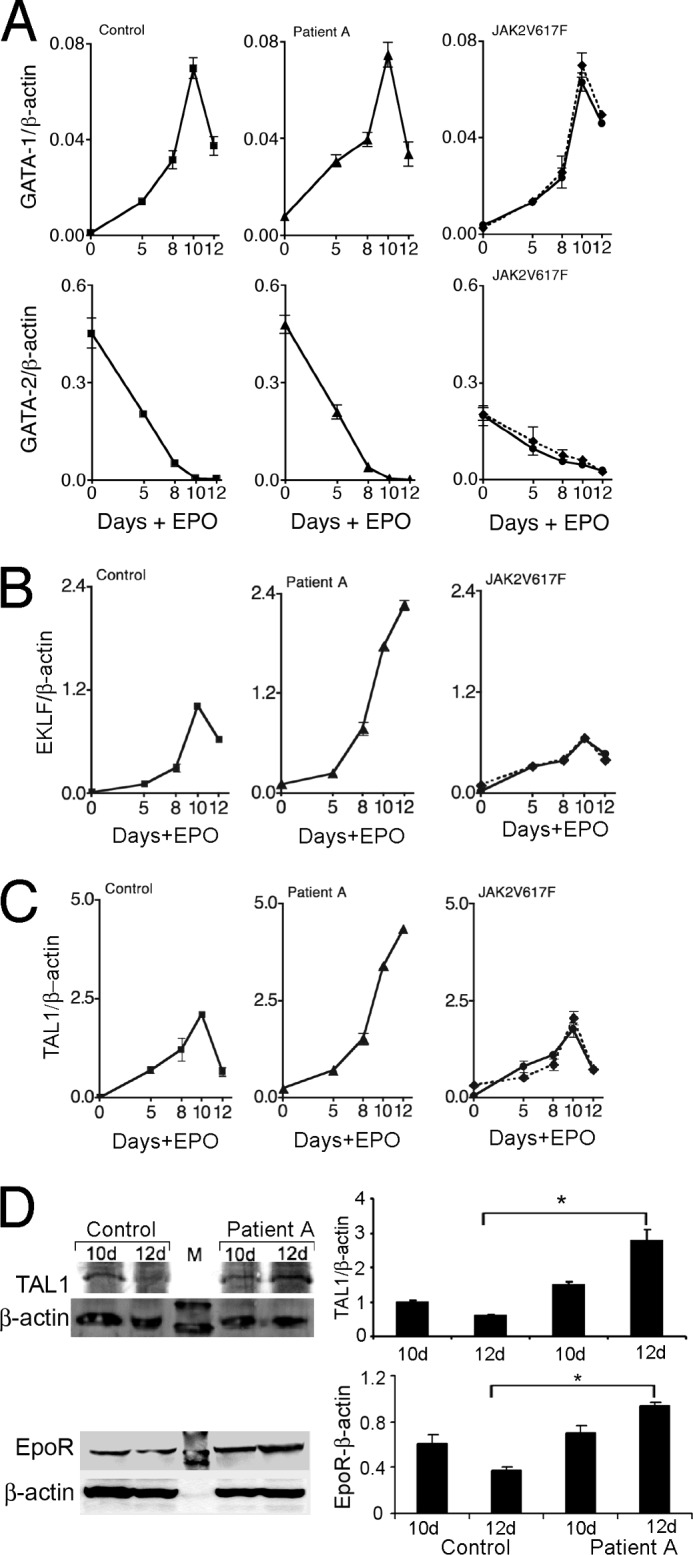
Transcription factor expression in human primary erythroid progenitor cells. A–C, expression of GATA-1 (A), GATA-2 (A), EKLF (B), and TAL1 (C) are shown following 12 days of treatment with EPO (1 unit/ml) in primary erythroid progenitor cell cultures from normal control (left), Patient A (center) and two patients with JAK2V617F associated polycythemia vera, Patient B (solid line) and Patient C (dashed line) (right). D, Western blotting is shown for TAL1, EPO-R and β-actin for cultures from normal control and Patient A from days 10 and 12 of EPO stimulation. Data represent mean ± S.D. *, p < 0.05.
Although GATA-1 expression appeared normal, we observed increased induction in TAL1 mRNA with EPO stimulation in hAEPC cultures from Patient A (Fig. 2C). In control hAEPC cultures and cultures of Jak2V617F hAEPC, EPO induction of TAL1 expression peaked between days 8 and 10 of EPO stimulation and was then down-regulated. In hAEPC cultures from Patient A, TAL1 expression continued to increase even beyond day 10 reaching expression levels 2-fold or greater than the peak level determined for control and Jak2V617F cultures. Furthermore, the TAL1 protein level at day 12 was confirmed to be increased by more than 3-fold compared with control cultures (Fig. 2D). Consistently, the EPO-R protein level was observed to be elevated at day 12 compared with day 10 in Patient A and compared with day 12 in control (Fig. 2D, bottom panel). In contrast to the normal level of GATA-1 expression, the increased EPO induction of TAL1 and EPO-R provides a potential link to the EPO hypersensitivity of erythroid progenitor cells from Patient A.
Genetic analysis was carried out to determine the presence of mutations in the major regulatory regions contributing to TAL1 gene expression or evidence of gene rearrangements that might account for elevated TAL1 expression in progenitor cells from Patient A. Southern blotting and PCR analysis of DNA did not reveal the major chromosomal rearrangements associated with T-acute lymphoblastic leukemia (33). The chromosomal mutations leading to a high level of TAL1 expression in lymphocytes such as the t(1;14)(p32;q11) translocation, the interstitial deletion that fuses the 5′ UTR of TAL1 with the 5′ UTR of the upstream SIL gene were not detected in the DNA from Patient A (supplemental Fig. 1A). No mutations were detected by sequencing 500 bp upstream and downstream in the TAL1 erythroid promoter 1a (34), the +19 enhancer necessary to drive activity of the TAL1 stem cell enhancer (35), and the +51 enhancer that drives expression in the primitive and definitive erythroid lineages (36) (data not shown). TAL1 coding region and EPO-R coding region sequencing also did not reveal any mutations (data not shown). Nevertheless, the increased induction of TAL1 by EPO treatment and resultant increase in EPO-R expression suggest aberrant regulation resulting in increased TAL1 expression as a potential explanation of excessive erythrocytosis.
Increased EPO-R Expression Promotes Erythroid Proliferation and Differentiation
To confirm that the increase of EPO-R expression contributes to increased EPO sensitivity in culture, we used a mouse Epo-R expression vector to further induce EPO-R expression in EPO-stimulated hAEPC cultures. Evidence for the regulation of EPO-R by TAL1 during EPO stimulation in vivo was suggested by Patient A with chronic, isolated erythrocytosis and is further confirmed by in vitro data. To validate some possible links and explanation for the extra erythropoiesis in the Patient A, we used hAEPC cultures derived from peripheral blood of normal individuals to determine whether increased EPO-R expression contributes to erythropoiesis and can be regulated by TAL1. EPO stimulation of hAEPC induces expression of transcription factors, GATA-1, TAL1, and EKLF that in turn stimulate expression of EPO-R, globin, and other erythroid genes (11, 15, 24, 37). Forced expression of EPO-R on day 3 of EPO stimulation increased hAEPC proliferation and erythroid differentiation as indicated by benzidine staining (Fig. 3A). Analysis of gene expression showed increased TAL1, EKLF that specifically activates β-globin, and, after day 5 of EPO stimulation, in GATA-1, and accelerated down-regulation of GATA-2 (Fig. 3B). We also observed increased expression of endogenous EPO-R at day 5 of EPO stimulation, suggesting activation of EPO-R transcription prior to the increase in GATA-1. These data illustrate that forced EPO-R expression in primary hAEPC cultures increases expression of erythroid transcription factors, and increases cell proliferation and activation of the erythroid program, which provide a possible link between increased EPO-R and extra erythropoiesis in Patient A. Increased EPO signaling via increased EPO-R expression also increased γ-globin expression, but the early increase in the γ-globin/(γ-globin + β-globin) ratio (γ/(γ + β)) did not persist to the end of the erythroid differentiation period (Fig. 3C). Increased EPO-R expression was confirmed by Western blotting (Fig. 3D) and quantitative RT-PCR showed an increased expression of “endogenous” human EPO-R mRNA and an increased exogenous mouse Epo-R expression (supplemental Fig. 1C).
FIGURE 3.
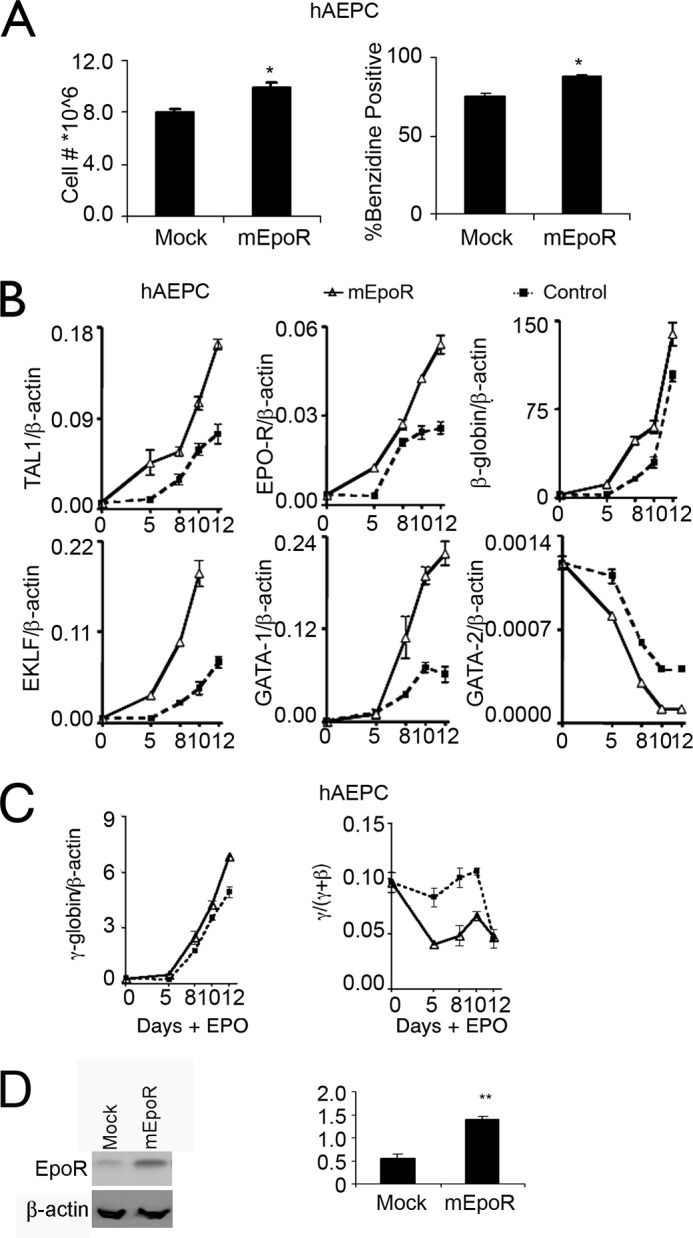
Overexpression of mEpo-R in human primary erythroid progenitor cells. A, cell proliferation (left) and % benzidine positive cells (right) following 12 days of treatment with EPO in primary adult erythroid progenitor cell (hAEPC) cultures with (mEpoR-transfected) and without (Mock-transfected) forced mEPO-R expression are shown. B, expression of endogenous TAL1, EPO-R, β-globin, EKLF, GATA-1, and GATA-2 were monitored during 12 days of EPO stimulation with (triangle, solid line) and without (square, dashed line) overexpression of mEPO-R in hAEPC. C, expression of γ-globin and the γ-globin/(γ-globin + β-globin) ratio (γ/(γ + β)) were monitored during EPO stimulation with (triangle, solid line) and without (square, dashed line) overexpression of mEpo-R in hAEPC. D, EPO-R overexpression was assessed and normalized to β-actin using Western blotting. Results are normalized to β-actin expression. Data represent mean ± S.D. *, p < 0.05.
Conserved GATA and TAL1 Binding Motifs in the EPO-R Proximal Promoter
The hAEPC cultures from Patient A exhibited normal GATA-1 levels but elevated EPO-R and TAL1. These data suggest that induction of EPO-R is not only due to transactivation by GATA-1. Unlike many erythroid genes, the EPO-R proximal promoter has high GC content and lacks TATA and CAAT sequences in the 5′ flanking region (38), features in common with housekeeping genes. The highly conserved GATA and Sp1 binding motifs in the proximal promoter are critical for EPO-R expression and GATA-1 activation of EPO-R expression (24, 26, 39). However, the conservation region between mouse and human extends downstream of the GATA and Sp1 binding sites, through the transcription start site and extends 55 to 70 bp 3′, respectively, to include three E-box sequences (CAGCTG) in the 5′ untranslated transcribed region (UTR) (supplemental Fig. 2A). These E-boxes are consensus sequences for potential binding sites for basic helix-loop-helix proteins such as TAL1.
TAL1 Increases EPO-R Gene Expression
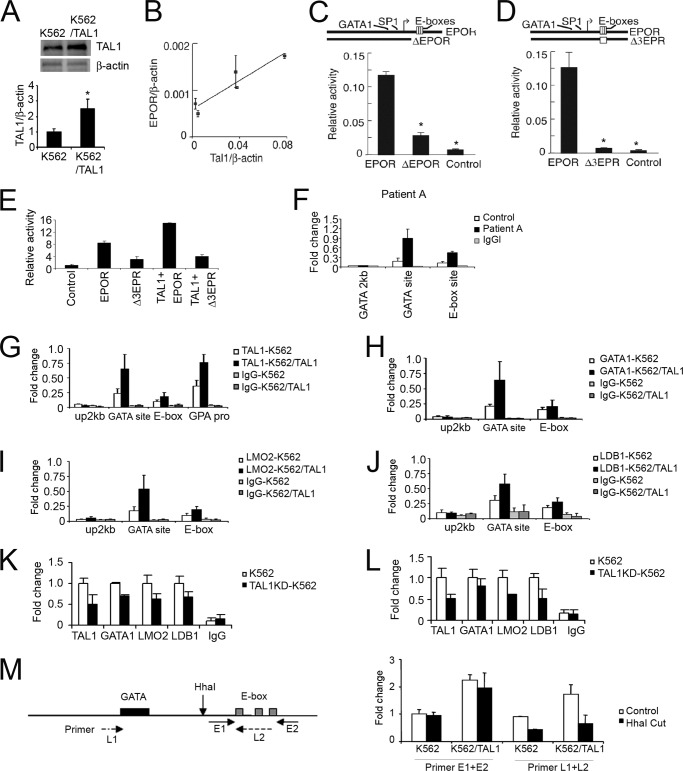
The relatively high level of TAL1 and EPO-R in hAEPC culture from Patient A raises the possibility of a link between TAL1 and EPO-R expression. We recently reported that overexpression of TAL1 in myoblasts increased EPO-R expression (40). To determine the ability of TAL1 to increase expression of endogenous EPO-R in erythroid cells, we overexpressed TAL1 in K562 cells, confirmed by Western blotting (Fig. 4A), and isolated stable clones. Quantification of mRNA expression revealed a TAL1 dose-dependent increase in EPO-R expression (Fig. 4B). To demonstrate that the 3 E-box motif region could act as a potential TAL1 binding site and contribute to regulation of EPO-R expression, a reporter gene was constructed using the EPO-R proximal promoter containing the 3 E-box motifs and extending 198 bp 5′ of the transcription start site. Deletion of the E-box region leaving the Sp1 and GATA-1 binding motifs intact resulted in down-regulated transcription activity by 4-fold (Fig. 4C). Mutating the E-box motifs decreased EPO-R promoter activity by more than 10-fold (Fig. 4D). Conversely, elevated TAL1 expression increased the transcription activity of the intact EPO-R proximal promoter, but not the E-box motif-mutated promoter (Fig. 4E). These data suggest that the E-box region contributes to high activation of EPO-R transcription and that TAL1 acts via the E-box motifs to transactivate EPO-R expression.
FIGURE 4.
Reporter gene assays and transcription factor binding for the EPO-R proximal promoter. A, overexpression of the TAL1 protein level in K562 was assessed by Western blotting using β-actin as loading control; B, endogenous EPO-R expression (EPOR) in K562 cells with stable overexpression of TAL1 is shown (each point represents a stable clone overexpressing TAL1)(r2 = 0.786). Expression is normalized to β-actin. Data represent mean ± S.D. D and E, activity for the firefly luciferase reporter gene construct containing the EPO-R proximal promoter without (EPOR) and with (ΔEPOR) truncation of the conserved E-box region (C) and for the EPO-R promoter without and with (Δ3EPR) mutation of the three conserved E-boxes (D) compared with a promoter-less construct (Control) was determined by transfection of reporter genes into K562 cells. Results were normalized to SV40 promoter activity. E, reporter gene activities were determined for Δ3EPR cotransfected with a TAL1 expression vector into K562 cells. Reporter gene activity was normalized to control activity. A Renilla luciferase construct was used for transfection control. Data represent mean ± S.D. (n = 3). F, cross-linked chromatin from EPO-treated (12 days) primary erythroid progenitor cells from Patient A and normal control was precipitated with TAL1 antibodies. The precipitated DNA fragments were amplified by using primers specific for GATA site 2 kb upstream, GATA site, linker region including the GATA site and E-box motifs, and E-box motifs. G–J, chromatin immunoprecipitation (ChIP) assay of K562 cells without and with (K562/TAL1) TAL1 overexpression was determined using antibodies for TAL1 (G), GATA-1 (H), LMO2 (I), and LDB1 (J) are shown (open bar), IgG was used as control (closed bar). The precipitated DNA fragments were amplified using primers specific for GATA site 2 kb upstream, the GATA site, the E-box motifs, and the glycophorin A promoter for TAL1 as a positive control. K and L, ChIP assay of K562 cells without and with (TALKD-K562) TAL1 knockdown determined using antibodies for TAL1, GATA-1, LMO2, and LDB1 are shown for the GATA site (K) and the E-box region (L) of the EPO-R promoter, IgG was used as negative control. M, cross-linked chromatin from K562 cells with or without overexpression was digested by the HhaI restriction enzyme and precipitated with TAL1 antibodies. The precipitated DNA fragments were amplified using primers specific for the EPO-R promoter including the HhaI site (primer pair L1–L2; dashed line) and E-box site only (primer pair E1–E2; solid line). Results are shown relative to input DNA. Data represent mean ± S.D. (n = 3), *, p < 0.05 and **, p < 0.01.
TAL1 Binds to the EPO-R Conserved E-box Region
Of note, unlike a number of erythroid genes that contain a TAL1 binding site about one helical turn away from a GATA binding motif, the EPO-R gene contains GATA and TAL1 binding E-box motifs that flank the transcription start site and are separated by 75 bp. In vivo ChIP analysis demonstrated that the EPO-R promoter GATA site and E-box motifs showed increased TAL1 binding by more than 3-fold in Patient A hAEPC cultures with EPO treatment for 12 days compared with the control hAEPC cultures and IgG negative control (Fig. 4F). Endogenous TAL1 binding to the EPO-R E-box region in K562 cells was also demonstrated by ChIP analysis. Nuclei were isolated from cross-linked K562 cells and DNA-protein complexes were isolated using anti-TAL1 antibody. Analysis of the precipitated DNA revealed direct binding of TAL1 to the EPO-R E-box region. K562 cells with forced expression of TAL1 showed an increase of TAL1 binding to the E-box region of EPO-R and the promoter region for the TAL1 target gene, glycophorin A (Fig. 4G). Amplification of the EPO-R region containing the GATA site 75 bp 5′ from the E-boxes also indicated TAL1 binding that increased with TAL1 overexpression that was not evident at a GATA site 2 kb upstream. These data suggest increased TAL1 expression and binding to the EPO-R promoter increases EPO-R expression, which provides the possibility for increased EPO response that, in turn, increases cell proliferation.
Conversely, ChIP analysis using anti-GATA-1 confirmed GATA-1 binding to the GATA site in the proximal promoter that also increased with overexpression of TAL1 without an increase in GATA-1 expression (Fig. 4H), suggesting that TAL1 can recruit GATA-1 and increase GATA-1 binding to the EPO-R promoter region to stimulate gene expression. Some GATA-1 association with the E-box region was also observed but this did not increase with overexpression of TAL1. These data suggest that TAL1 binds to the GATA binding motif and the E-box region flanking the EPO-R transcription start site, and that TAL1 can further increase GATA-1 binding to the GATA motif in the EPO-R proximal promoter region.
ChIP analysis using antibodies specific to LMO2 and LDB1 showed association of both proteins in the region of the GATA motif and E-box containing EPO-R proximal promoter region particularly at the GATA binding site with overexpression of TAL1 (Fig. 4, I and J), suggesting that increased TAL1 recruits more LMO2 and LDB1 to form an active complex at the EPO-R promoter, resulting in a more open chromatin structure and promoting EPO-R expression. The association of GATA-1, LMO2, and LDB1 site in the proximal promoter appeared to be stronger relative to the GATA-binding site compared with their association to the E-box region downstream of the transcription start site (Fig. 4, H–J). Binding of TAL1 in the EPO-R promoter at the GATA site also appeared to be greater than binding at the E-box motifs. To demonstrate that binding at the E-box region is specific and not related to pull down of large DNA fragments with proteins bound to the GATA site, we first knocked down TAL1 in K562 cells and evaluated LMO2 and LDB1 binding. ChIP analysis showed a decreased binding of TAL1, LMO2, and LDB1 at the EPO-R promoter region that included the GATA site and E-box region (Fig. 4, K and L), indicating that TAL1 is essential for LMO2 and Ldb1 binding at the EPO-R promoter. Next, we used HhaI restriction enzyme digestion to cut the region between the GATA site and E-box motifs prior to immunoprecipitation of chromatin-bound DNA by anti-TAL1 antibody. Amplification of the EPO-R region spanning the GATA site and E-box motifs showed a reduction by more than 50% after HhaI digestion in contrast to amplification of the E-box region alone that showed no significant reduction, providing evidence that binding of TAL1 to the E-box region is specific and not simply due to pull down of large DNA fragments extending from GATA site binding (Fig. 4M). This observation is consistent with the view that TAL1 binding to the EPO-R promoter is necessary prior to or simultaneous with that of GATA1 (41), and suggests that increased TAL1 induction in erythroid progenitor cell cultures of Patient A can induce increased EPO-R expression without affecting expression of GATA-1.
TAL1 Increases EPO-R and Erythroid Differentiation in Erythroid Progenitor Cells
To mimic the behavior of the patient-derived hAEPC culture and to show that elevated TAL1 expression can contribute to increased EPO-R expression as a possible explanation for the link between high sensitivity of EPO stimulation and excess erythropoiesis, we overexpressed TAL1 in a hAEPC culture derived from normal individuals. On day 3, EPO-stimulated expression vectors with TAL1 in the sense or antisense orientation were transfected and gene expression was analyzed (Fig. 5A). Overexpression of TAL1 increased EPO-R expression up to 2-fold and increased the EPO-R protein level confirmed by Western blotting (Fig. 5, B and C). Transfection of expression vectors for mEpo-R and GFP were used as positive and negative controls, respectively. The hAEPC culture from Patient A also showed increased induction of EKLF in response to EPO stimulation (Fig. 2B). However, we observed that overexpression of mEklf in the control hAEPC culture did not increase EPO-R mRNA (supplemental Fig. 1B) and protein levels (Fig. 5B), but did increase erythroid differentiation, determined by benzidine staining, and decreased progenitor cell number (supplemental Fig. 1B). In contrast, down-regulation of TAL1 by the antisense construct decreased EKLF and GATA-1 expression (Fig. 5A), suggesting that TAL1 functions upstream of EKLF and GATA-1 and may be responsible for increased expression of EPO-R in Patient A. TAL1 overexpression also increased expression of β-globin and γ-globin, but without an increase in the γ-globin/(γ-globin + β-globin) ratio (γ/(γ + β)) (Fig. 5D). Only modest changes were observed for expression of GATA-1, GATA-2, and EKLF (Fig. 5A). At the end of the 12-day culture period with EPO stimulation, cell count and benzidine-positive cells were increased by 15–20% compared with control hAEPC cultures (Fig. 5E). In contrast, forced expression of TAL1 transcripts in the antisense direction significantly decreased TAL1 expression as well as expression of GATA-1, EKLF, EPO-R, and β- and γ-globin (Fig. 5, A and B), indicating repression of erythroid differentiation by expression of TAL1 antisense. Importantly, overexpression of EPO-R rescued, in part, the decreased β-globin gene expression by TAL1 knockdown (supplemental Fig. 2B), suggesting that the TAL1/EPO-R axis is important for erythrocytosis. The effect of TAL1 antisense on down-regulation of GATA-2 expression was modest although induction of GATA-1 was decreased severalfold. At the end of the EPO culture period, antisense TAL1 reduced cell count to 60% and benzidine positive cells to less than 40% compared with control cultures (Fig. 5E). These data suggest that TAL1 positively regulate EPO-R expression in erythroid progenitor cells and erythroid differentiation, which partially support that increased expression of TAL1 with EPO stimulation induces EPO-R to provide the possibility for high sensitivity of EPO leading to extra erythropoiesis.
FIGURE 5.
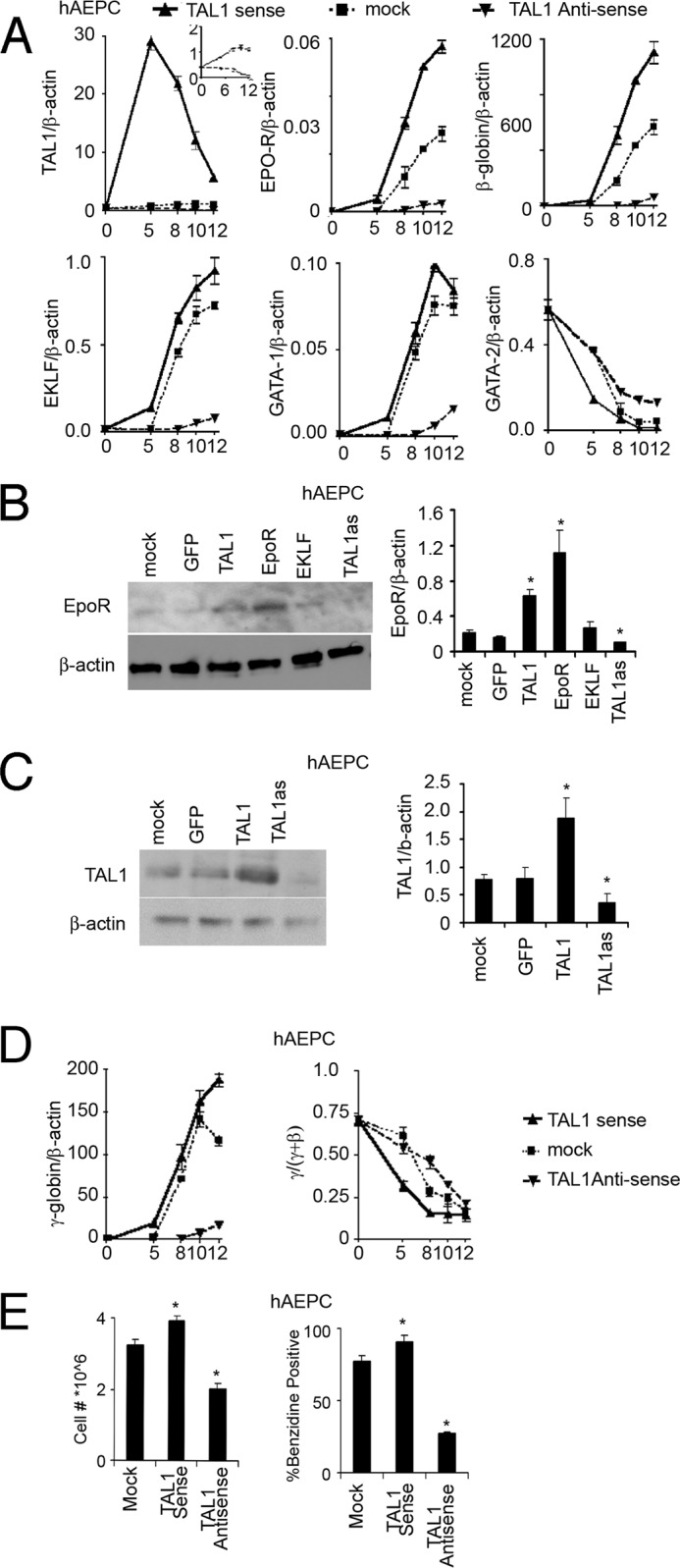
Forced expression of TAL1 in human primary erythroid progenitor cells. A, expression of endogenous TAL1, EPO-R, β-globin, EKLF, GATA-1, and GATA-2 were monitored during 12 days of EPO stimulation with TAL1 sense (triangle, solid line), antisense (inverted triangle, dashed line), and mock (square, dotted line) forced expression in hAEPC. Results are normalized to β-actin expression. TAL1 antisense and mock transfection efficiency was shown in the inset panel for clarity. B, increase in EPO-R protein expression by overexpression of GFP, TAL1, EPO-R, and EKLF and TAL1 knockdown (TAL1 as) are shown in hAEPC using Western blotting with β-actin as loading control. C, increase in TAL1 protein expression by GFP and TAL1 overexpression and TAL1 knockdown (TAL1 as) are shown in hAEPC using Western blotting with β-actin as loading control. D, expression of γ-globin and the ratio of γ/(γ + β) globin during 12 days of EPO stimulation with TAL1 sense (triangle, solid line), antisense (inverted triangle, dashed line), and mock (square, dotted line) forced expression were shown in hAEPC. E, cell proliferation (left) and % benzidine positive cells (right) following 12 days of treatment with EPO in primary erythroid progenitor cell cultures with TAL1 sense, antisense, and mock forced expression are shown in hAEPC. Data represent mean ± S.D. *, p < 0.05.
TAL1 Binds to the EPO-R Promoter during EPO-R Nucleosome Positioning
Considering possible mechanisms by which increased TAL1 expression up-regulates EPO-R expression, we note that forced expression of TAL1 promotes hematopoiesis but, unlike erythroid differentiation, TAL1 activity in hematopoietic stem cell development does not require TAL1 binding to DNA (15, 34, 42). In contrast, during erythropoiesis, TAL1 binding directly to DNA, including TAL1 binding to E-box DNA motifs in proximity to GATA-1 binding sites, are important for activation of erythroid-specific genes such as glycophorin A, β-globin, and erythroid transcription factors, such as GATA-1 and EKLF (21, 22, 43). As observed with housekeeping genes, the EPO-R promoter is TATA-less. Nucleosome shifting can facilitate TAL1 binding that is linked to regulation of target gene transcription (44). Furthermore, the nucleosome linker position has been shown to be related to active gene expression (31).
Therefore, we performed nucleosome positioning analysis in primary human CD34+ hematopoietic stem cells and CD36+ differentiated erythrocyte precursor cells. CD34+ and CD36+ cells show low and high EPO-R expression, respectively (Fig. 6A). The GATA-binding motif and E-boxes flank the transcription start site in the EPO-R gene and in CD34+ cells with a low level of EPO-R transcription, we found that nucleosomes also flanked the EPO-R transcription start site and were about 220 bp apart. In contrast, when EPO-R was highly expressed in CD36+ cells, the nucleosomes that flank the EPO-R transcription start site were shifted further apart and the linker region was expanded by about 50 bp (Fig. 6B). The reposition of nucleosomes in the EPO-R promoter region results in increased exposure of the E-box motifs and the GATA site in the linker region between the two nucleosomes. This raises the possibility for increasing interaction with transcription factors such as TAL1 with the EPO-R promoter and increasing EPO-R gene expression (Fig. 6B). Importantly, shifting the nucleosome positioning can specifically facilitate binding of TAL1 and is linked to subsequent transcriptional regulation of target genes (44), providing further support for the suggestion that the expanded linker region of EPO-R in CD36+ cells facilitates TAL1 binding to the EPO-R promoter and increases EPO-R transcription. Although the cause of the elevated EPO induction of TAL1 in erythroid progenitor cells remains uncertain in Patient A, these data are consistent with regulation of EPO-R by TAL1 resulting in increased EPO-R expression and provide a possible link to hypersensitivity of EPO with erythroid differentiation.
FIGURE 6.
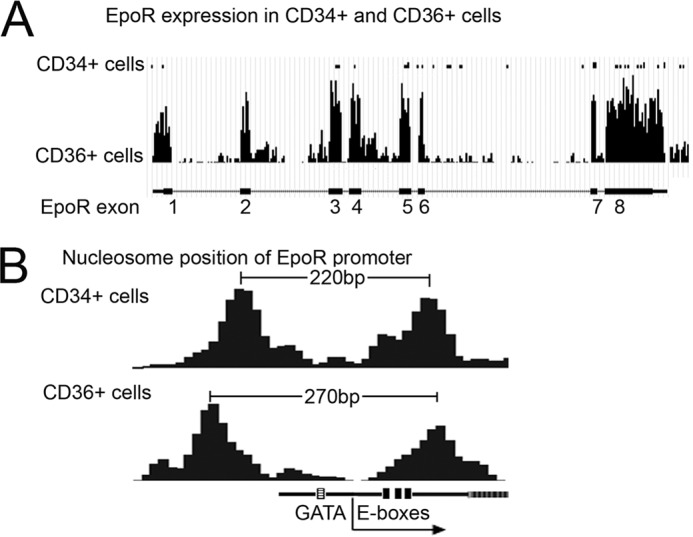
Comparison of TAL1 chromatin occupancy at EPO-R promoter by nucleosome position of EPO-R promoter. A, comparison of EPO-R expression in CD34+ cells and CD36+ cells. B, comparison of nucleosome position of EPO-R promoter in CD34+ and CD36+ cells.
DISCUSSION
TAL1 is required for hematopoietic stem cell formation in the yolk sac during early embryonic development and is essential for normal erythropoiesis in adult (45). In differentiating ES cells in vitro, forced expression of TAL1 promotes hematopoiesis (42). TAL1 activity in hematopoietic stem cell development is proposed to be mediated via TAL1 participation in a complex with other binding partners, GATA-1/2, LMO2, LDB1, E2A, and possibly Sp1, but does not require direct TAL1 binding to DNA (15, 47). The importance of this complex is suggested by the Lmo2 knock-out mouse that exhibits a phenotype resembling the Tal1 knock-out (43).
The sequence conservation in the EPO-R proximal promoter includes the GATA-1 and Sp1 sites required for high level EPO-R gene activation and extends 3′ beyond the transcription start site to include a 37-bp region with 3 E-box motifs that are separated from the GATA-1 site by 60 and 75 bp, respectively, in the mouse and human genomes. We determined that increasing TAL1 increases its binding to the EPO-R E-boxes and increases binding of the GATA-1·TAL1 complex to the EPO-R promoter region. These TAL1 associated activities also contribute to the high level of EPO-R expression in erythroid cells. A 300-bp fragment containing the GATA-1 and Sp1 binding motifs and the E-box region was sufficient to drive luciferase reporter gene activity, whereas mutation of the E-box region decreased reporter gene expression by more than 10-fold. TAL1 binds directly to the EPO-R E-box region and forced TAL1 expression increases EPO-R expression and provides the possibility to increase EPO-stimulated erythroid differentiation in primary erythroid progenitor cells. These data identify EPO-R as a direct target gene of TAL1 requiring direct binding. The decrease in EPO-R expression and resultant decrease in EPO stimulated differentiation observed here as a consequence of Tal1 knockdown in cultures of CD34+ erythroid progenitor cells is consistent with the resultant decrease in mouse erythroid differentiation in vivo with loss of TAL1, indicating a link between regulation of EPO-R expression by TAL1 and erythropoiesis.
A genome-wide analysis of transcription factor occupancy and mRNA expression indicated that TAL1 occupancy is associated with selected GATA-1-activated genes but is not usually associated with selected GATA-1 repressed genes (21, 22, 46). In addition, 83% of GATA-1 induced genes were reported to exhibit co-occupancy by TAL1 at all GATA-1 occupancy sites, although this did not indicate the amount of TAL1 binding (46). ChIP-Seq analysis of TAL1 binding in hematopoietic cells also revealed overrepresentation of GATA motifs in 212 of 228 TAL1-bound regions (23). TAL1 binding directly to DNA contributes importantly to erythropoiesis. In particular, TAL1 binding to the E-box DNA sequence motifs in proximity to GATA-1 binding sites separated by 9 to 12 nucleotides associates with the GATA-1/TAL1 multiprotein complex and activation of erythroid specific genes such as glycophorin A, β-globin, and transcription factors, TAL1, GATA-1, and EKLF. Binding and activation by TAL1 via two E-box-GATA elements in the proximal promoter (beginning at −318 and −25) of the protein 4.2 gene provided evidence for LDB1 as a member of the GATA-1·TAL1 complex (47). The E-box·GATA binding complex of GATA-1·TAL1·LMO2·LDB1 also contributes to long-range gene activation in the β-globin gene locus between the strong upstream erythroid enhancer, the locus control region, and the distal downstream β-globin gene (48). TAL1 interaction with these E-box·GATA motifs are examples of the relatively close interactions involved in the GATA-1·TAL1 multiprotein complex as well as its ability to participate in long-range interaction that bridges these complexes via LDB1 such as that described for the interaction of the locus control region and the downstream β-globin promoter.
The EPO-R gene shares a common feature with constitutive genes that have a TATA-less promoter. The region containing the GATA binding motif in the EPO-R proximal promoter and the conserved E-boxes 75 bp downstream appears to be located in a linker region between two nucleosomes (31). Nucleosome shifting specifically facilitates binding of TAL1 but not GATA1 and is linked to subsequent transcriptional regulation of target genes (44). Our data show the interaction of TAL1 with the GATA binding motif and the conserved E-box region flanking the EPO-R transcription start site and the shift in nucleosome positioning in the EPO-R promoter region in erythroid progenitor cells stimulated with EPO. Together, these data suggest that nucleosome phasing shifts to an opened state after EPO-R activation. Furthermore, increased TAL1 expression by overexpression in cultured cells or by endogenous mechanisms in erythroid progenitor cells from Patient A increases TAL1 binding to the EPO-R promoter region and, therefore, increases TAL1-associated EPO-R expression activation and EPO response.
Increased EPO signaling via genetic mutations that delete the EPO-R carboxyl-terminal negative regulatory domain have been identified in patients with increased erythropoiesis. Other genetic mutations resulting in increased signal transduction during EPO-stimulated erythropoiesis include the Jak2V617F mutation associated with polycythemia vera patients who exhibit an increase in BFU-E and erythroid progenitor cell proliferation even in the absence of EPO. TAL1 induction of EPO-R provides an alternate mechanism for increased EPO sensitivity. We show here that the direct increase of EPO-R expression in primary hAEPC cultures also promotes EPO-dependent erythropoiesis. Induction of EPO-R by TAL1 is relevant to the increased EPO sensitivity we observe in primary hAEPC cultures from Patient A with increased erythrocytosis. The increase in erythroid progenitor cell proliferation is only observed in the presence of EPO and no mutations in the EPO-R or JAK2 genes could be identified. EPO induces an unusually elevated and prolonged level of EPO-R in hAEPC culture from Patient A concomitant with high induction of TAL1, but not of GATA-1, suggesting that the induced level of EPO-R expression following EPO stimulation is a result of the increased expression of TAL1. This is in contrast to the forced expression of EPO-R in control hAEPC culture that results in increased induction of both TAL1 and GATA-1. High induction of either TAL1 or GATA-1 is not observed in hAEPC cultures derived from polycythemia patients with the Jak2V617F mutation. The increased EPO induction of EKLF as well as TAL1 in hAEPC culture from Patient A explains the increased expression of β-globin. However, overexpression of EKLF in the control hAEPC culture did not increase EPO-R expression. Given that the E-box motif in the EKLF promoter is required for EKLF expression (49) and the failure of induction of EKLF in TAL1-null mouse embryonic stem cells (50), the induction of EKLF may be in part a consequence of the increasing expression of TAL1 or relate to a shared mechanism of transcription activation with TAL1.
Overexpression of EPO-R resulting from gene rearrangements could be important in the leukemogenic process. A novel translocation involving EPO-R and the immunoglobulin heavy chain locus, t(14;19)(q32;p13) resulted in a greater than 230-fold increase in EPO-R identified in two patients with B-cell precursor acute lymphoblastic leukemia (51). High EPO-R expression has also been associated with the TEL-AML1 subtype of B-cell precursor acute lymphoblastic leukemia that gives rise to a ETV6/RUNX1 fusion gene product (52). In contrast to these examples of high EPO-R expression associated with acute lymphoblastic leukemia, the increased EPO-R expression in progenitor cell cultures from Patient A was detected only after EPO stimulation, consistent with hematopoietic changes only in the erythroid lineage with normal white blood cell count, differential white blood cell count, and platelet count. In addition, rearrangements associated with TAL1 translocations and leukemia are absent in Patient A. No mutations have been revealed upon sequencing known regulatory elements in the TAL1 gene such as the promoter region and the +19 enhancer; the regulatory event giving rise to high EPO induction of TAL1 remains to be identified. Nevertheless, the elevated EPO-R concomitant with the increased expression of TAL1 but not other erythroid-specific factors during erythropoiesis provides a naturally occurring example of elevated TAL1 and EPO-R and suggests a potential link between mechanisms driving TAL1 expression and increased EPO sensitivity in erythroid progenitor cells resulting in erythrocytosis.
This work was supported, in whole or in part, by the National Institutes of Health Intramural Research Program of the NIDDK.

This article contains supplemental Figs. S1 and S2.
- EPO
- erythropoietin
- EPO-R
- erythropoietin receptor
- TAL1
- T-cell acute leukemia 1
- hAEPC
- human erythroid progenitor cell
- BFU-E
- burst forming unit erythroid
- CFU-E
- colony forming unit erythroid
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Wu H., Liu X., Jaenisch R., Lodish H. F. (1995) Generation of committed erythroid BFU-E and CFU-E progenitors does not require erythropoietin or the erythropoietin receptor. Cell 83, 59–67 [DOI] [PubMed] [Google Scholar]
- 2. Livnah O., Stura E. A., Middleton S. A., Johnson D. L., Jolliffe L. K., Wilson I. A. (1999) Crystallographic evidence for preformed dimers of erythropoietin receptor before ligand activation. Science 283, 987–990 [DOI] [PubMed] [Google Scholar]
- 3. Percy M. J., Furlow P. W., Lucas G. S., Li X., Lappin T. R., McMullin M. F., Lee F. S. (2008) A gain-of-function mutation in the HIF2A gene in familial erythrocytosis. N. Engl. J. Med. 358, 162–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ang S. O., Chen H., Hirota K., Gordeuk V. R., Jelinek J., Guan Y., Liu E., Sergueeva A. I., Miasnikova G. Y., Mole D., Maxwell P. H., Stockton D. W., Semenza G. L., Prchal J. T. (2002) Disruption of oxygen homeostasis underlies congenital Chuvash polycythemia. Nat. Genet. 32, 614–621 [DOI] [PubMed] [Google Scholar]
- 5. Percy M. J., Zhao Q., Flores A., Harrison C., Lappin T. R., Maxwell P. H., McMullin M. F., Lee F. S. (2006) A family with erythrocytosis establishes a role for prolyl hydroxylase domain protein 2 in oxygen homeostasis. Proc. Natl. Acad. Sci. U.S.A. 103, 654–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baxter E. J., Scott L. M., Campbell P. J., East C., Fourouclas N., Swanton S., Vassiliou G. S., Bench A. J., Boyd E. M., Curtin N., Scott M. A., Erber W. N., Green A. R. (2005) Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet 365, 1054–1061 [DOI] [PubMed] [Google Scholar]
- 7. Scott L. M., Tong W., Levine R. L., Scott M. A., Beer P. A., Stratton M. R., Futreal P. A., Erber W. N., McMullin M. F., Harrison C. N., Warren A. J., Gilliland D. G., Lodish H. F., Green A. R. (2007) JAK2 exon 12 mutations in polycythemia vera and idiopathic erythrocytosis. N. Engl. J. Med. 356, 459–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Meyer L., Deau B., Forejtníková H., Duménil D., Margottin-Goguet F., Lacombe C., Mayeux P., Verdier F. (2007) β-Trcp mediates ubiquitination and degradation of the erythropoietin receptor and controls cell proliferation. Blood 109, 5215–5222 [DOI] [PubMed] [Google Scholar]
- 9. Agarwal N., Gordeuk R. V., Prchal J. T. (2007) Genetic mechanisms underlying regulation of hemoglobin mass. Adv. Exp. Med. Biol. 618, 195–210 [DOI] [PubMed] [Google Scholar]
- 10. Arcasoy M. O., Harris K. W., Forget B. G. (1999) A human erythropoietin receptor gene mutant causing familial erythrocytosis is associated with deregulation of the rates of Jak2 and Stat5 inactivation. Exp. Hematol. 27, 63–74 [DOI] [PubMed] [Google Scholar]
- 11. Rogers H. M., Yu X., Wen J., Smith R., Fibach E., Noguchi C. T. (2008) Hypoxia alters progression of the erythroid program. Exp. Hematol. 36, 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pope N. J., Bresnick E. H. (2010) Differential coregulator requirements for function of the hematopoietic transcription factor GATA-1 at endogenous loci. Nucleic Acids Res. 38, 2190–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hu X., Ybarra R., Qiu Y., Bungert J., Huang S. (2009) Transcriptional regulation by TAL1. A link between epigenetic modifications and erythropoiesis. Epigenetics 4, 357–361 [DOI] [PubMed] [Google Scholar]
- 14. Ravet E., Reynaud D., Titeux M., Izac B., Fichelson S., Roméo P. H., Dubart-Kupperschmitt A., Pflumio F. (2004) Characterization of DNA-binding-dependent and -independent functions of SCL/TAL1 during human erythropoiesis. Blood 103, 3326–3335 [DOI] [PubMed] [Google Scholar]
- 15. Kassouf M. T., Chagraoui H., Vyas P., Porcher C. (2008) Differential use of SCL/TAL-1 DNA-binding domain in developmental hematopoiesis. Blood 112, 1056–1067 [DOI] [PubMed] [Google Scholar]
- 16. Schuh A. H., Tipping A. J., Clark A. J., Hamlett I., Guyot B., Iborra F. J., Rodriguez P., Strouboulis J., Enver T., Vyas P., Porcher C. (2005) ETO-2 associates with SCL in erythroid cells and megakaryocytes and provides repressor functions in erythropoiesis. Mol. Cell. Biol. 25, 10235–10250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meier N., Krpic S., Rodriguez P., Strouboulis J., Monti M., Krijgsveld J., Gering M., Patient R., Hostert A., Grosveld F. (2006) Novel binding partners of Ldb1 are required for haematopoietic development. Development 133, 4913–4923 [DOI] [PubMed] [Google Scholar]
- 18. Huang S., Qiu Y., Shi Y., Xu Z., Brandt S. J. (2000) P/CAF-mediated acetylation regulates the function of the basic helix-loop-helix transcription factor TAL1/SCL. EMBO J. 19, 6792–6803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hu X., Li X., Valverde K., Fu X., Noguchi C., Qiu Y., Huang S. (2009) LSD1-mediated epigenetic modification is required for TAL1 function and hematopoiesis. Proc. Natl. Acad. Sci. U.S.A. 106, 10141–10146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang S., Brandt S. J. (2000) mSin3A regulates murine erythroleukemia cell differentiation through association with the TAL1 (or SCL) transcription factor. Mol. Cell. Biol. 20, 2248–2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tripic T., Deng W., Cheng Y., Zhang Y., Vakoc C. R., Gregory G. D., Hardison R. C., Blobel G. A. (2009) SCL and associated proteins distinguish active from repressive GATA transcription factor complexes. Blood 113, 2191–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu M., Riva L., Xie H., Schindler Y., Moran T. B., Cheng Y., Yu D., Hardison R., Weiss M. J., Orkin S. H., Bernstein B. E., Fraenkel E., Cantor A. B. (2009) Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol. Cell 36, 682–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wilson N. K., Miranda-Saavedra D., Kinston S., Bonadies N., Foster S. D., Calero-Nieto F., Dawson M. A., Donaldson I. J., Dumon S., Frampton J., Janky R., Sun X. H., Teichmann S. A., Bannister A. J., Göttgens B. (2009) The transcriptional program controlled by the stem cell leukemia gene Scl/Tal1 during early embryonic hematopoietic development. Blood 113, 5456–5465 [DOI] [PubMed] [Google Scholar]
- 24. Fujiwara T., O'Geen H., Keles S., Blahnik K., Linnemann A. K., Kang Y. A., Choi K., Farnham P. J., Bresnick E. H. (2009) Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol. Cell 36, 667–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vyas P., McDevitt M. A., Cantor A. B., Katz S. G., Fujiwara Y., Orkin S. H. (1999) Different sequence requirements for expression in erythroid and megakaryocytic cells within a regulatory element upstream of the GATA-1 gene. Development 126, 2799–2811 [DOI] [PubMed] [Google Scholar]
- 26. Chin K., Oda N., Shen K., Noguchi C. T. (1995) Regulation of transcription of the human erythropoietin receptor gene by proteins binding to GATA-1 and Sp1 motifs. Nucleic Acids Res. 23, 3041–3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fibach E. (1998) Techniques for studying stimulation of fetal hemoglobin production in human erythroid cultures. Hemoglobin 22, 445–458 [DOI] [PubMed] [Google Scholar]
- 28. Fernández L. A., Winkler M., Grosschedl R. (2001) Matrix attachment region-dependent function of the immunoglobulin mu enhancer involves histone acetylation at a distance without changes in enhancer occupancy. Mol. Cell. Biol. 21, 196–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aplan P. D., Lombardi D. P., Reaman G. H., Sather H. N., Hammond G. D., Kirsch I. R. (1992) Involvement of the putative hematopoietic transcription factor SCL in T-cell acute lymphoblastic leukemia. Blood 79, 1327–1333 [PubMed] [Google Scholar]
- 30. Chepelev I., Wei G., Tang Q., Zhao K. (2009) Detection of single nucleotide variations in expressed exons of the human genome using RNA-Seq. Nucleic Acids Res. 37, e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schones D. E., Cui K., Cuddapah S., Roh T. Y., Barski A., Wang Z., Wei G., Zhao K. (2008) Dynamic regulation of nucleosome positioning in the human genome. Cell 132, 887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nussenzveig R. H., Swierczek S. I., Jelinek J., Gaikwad A., Liu E., Verstovsek S., Prchal J. F., Prchal J. T. (2007) Polycythemia vera is not initiated by JAK2V617F mutation. Exp. Hematol. 35, 32–38 [DOI] [PubMed] [Google Scholar]
- 33. Aplan P. D. (2006) Causes of oncogenic chromosomal translocation. Trends Genet. 22, 46–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bockamp E. O., McLaughlin F., Göttgens B., Murrell A. M., Elefanty A. G., Green A. R. (1997) Distinct mechanisms direct SCL/tal-1 expression in erythroid cells and CD34 positive primitive myeloid cells. J. Biol. Chem. 272, 8781–8790 [DOI] [PubMed] [Google Scholar]
- 35. Göttgens B., Nastos A., Kinston S., Piltz S., Delabesse E. C., Stanley M., Sanchez M. J., Ciau-Uitz A., Patient R., Green A. R. (2002) Establishing the transcriptional programme for blood. The SCL stem cell enhancer is regulated by a multiprotein complex containing Ets and GATA factors. EMBO J. 21, 3039–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Delabesse E., Ogilvy S., Chapman M. A., Piltz S. G., Gottgens B., Green A. R. (2005) Transcriptional regulation of the SCL locus. Identification of an enhancer that targets the primitive erythroid lineage in vivo. Mol. Cell. Biol. 25, 5215–5225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sengupta T., Cohet N., Morlé F., Bieker J. J. (2009) Distinct modes of gene regulation by a cell-specific transcriptional activator. Proc. Natl. Acad. Sci. U.S.A. 106, 4213–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Noguchi C. T., Bae K. S., Chin K., Wada Y., Schechter A. N., Hankins W. D. (1991) Cloning of the human erythropoietin receptor gene. Blood 78, 2548–2556 [PubMed] [Google Scholar]
- 39. Zon L. I., Youssoufian H., Mather C., Lodish H. F., Orkin S. H. (1991) Activation of the erythropoietin receptor promoter by transcription factor GATA-1. Proc. Natl. Acad. Sci. U.S.A. 88, 10638–10641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang L., Jia Y., Rogers H., Wu Y. P., Huang S., Noguchi C. T. (2012) GATA-binding protein 4 (GATA-4) and T-cell acute leukemia 1 (TAL1) regulate myogenic differentiation and erythropoietin response via cross-talk with Sirtuin1 (Sirt1). J. Biol. Chem. 287, 30157–30169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kassouf M. T., Hughes J. R., Taylor S., McGowan S. J., Soneji S., Green A. L., Vyas P., Porcher C. (2010) Genome-wide identification of TAL1's functional targets. Insights into its mechanisms of action in primary erythroid cells. Genome Res. 20, 1064–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ismailoglu I., Yeamans G., Daley G. Q., Perlingeiro R. C., Kyba M. (2008) Mesodermal patterning activity of SCL. Exp. Hematol. 36, 1593–1603 [DOI] [PubMed] [Google Scholar]
- 43. Yamada Y., Warren A. J., Dobson C., Forster A., Pannell R., Rabbitts T. H. (1998) The T cell leukemia LIM protein Lmo2 is necessary for adult mouse hematopoiesis. Proc. Natl. Acad. Sci. U.S.A. 95, 3890–3895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hu G., Schones D. E., Cui K., Ybarra R., Northrup D., Tang Q., Gattinoni L., Restifo N. P., Huang S., Zhao K. (2011) Regulation of nucleosome landscape and transcription factor targeting at tissue-specific enhancers by BRG1. Genome Res. 21, 1650–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hall M. A., Slater N. J., Begley C. G., Salmon J. M., Van Stekelenburg L. J., McCormack M. P., Jane S. M., Curtis D. J. (2005) Functional but abnormal adult erythropoiesis in the absence of the stem cell leukemia gene. Mol. Cell. Biol. 25, 6355–6362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cheng Y., Wu W., Kumar S. A., Yu D., Deng W., Tripic T., King D. C., Chen K. B., Zhang Y., Drautz D., Giardine B., Schuster S. C., Miller W., Chiaromonte F., Zhang Y., Blobel G. A., Weiss M. J., Hardison R. C. (2009) Erythroid GATA1 function revealed by genome-wide analysis of transcription factor occupancy, histone modifications, and mRNA expression. Genome Res. 19, 2172–2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xu Z., Huang S., Chang L. S., Agulnick A. D., Brandt S. J. (2003) Identification of a TAL1 target gene reveals a positive role for the LIM domain-binding protein Ldb1 in erythroid gene expression and differentiation. Mol. Cell. Biol. 23, 7585–7599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Song S. H., Hou C., Dean A. (2007) A positive role for NLI/Ldb1 in long-range β-globin locus control region function. Mol. Cell 28, 810–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Anderson K. P., Crable S. C., Lingrel J. B. (2000) The GATA-E box-GATA motif in the EKLF promoter is required for in vivo expression. Blood 95, 1652–1655 [PubMed] [Google Scholar]
- 50. Elefanty A. G., Robb L., Birner R., Begley C. G. (1997) Hematopoietic-specific genes are not induced during in vitro differentiation of scl-null embryonic stem cells. Blood 90, 1435–1447 [PubMed] [Google Scholar]
- 51. Russell L. J., De Castro D. G., Griffiths M., Telford N., Bernard O., Panzer-Grümayer R., Heidenreich O., Moorman A. V., Harrison C. J. (2009) A novel translocation, t(14;19)(q32;p13), involving IGHα and the cytokine receptor for erythropoietin. Leukemia 23, 614–617 [DOI] [PubMed] [Google Scholar]
- 52. Fine B. M., Stanulla M., Schrappe M., Ho M., Viehmann S., Harbott J., Boxer L. M. (2004) Gene expression patterns associated with recurrent chromosomal translocations in acute lymphoblastic leukemia. Blood 103, 1043–1049 [DOI] [PubMed] [Google Scholar]