Background: Although phosphatidylserine decarboxylase 1 (Psd1) is of central importance for the generation of cellular phosphatidylethanolamine (PE), its biogenesis is only poorly understood.
Results: Biogenesis of Psd1 involves processing by two proteases, MPP and Oct1, and an autocatalytic separation of Psd1α from the membrane-anchored Psd1β.
Conclusion: Psd1 requires integration into the inner mitochondrial membrane for full enzymatic activity.
Significance: This study presents a new model for the biogenesis and topology of Psd1.
Keywords: Decarboxylase, Lipids, Mitochondria, Phosphatidylethanolamine, Phosphatidylserine, Yeast, Processing, Saccharomyces cerevisiae
Abstract
The inner mitochondrial membrane plays a crucial role in cellular lipid homeostasis through biosynthesis of the non-bilayer-forming lipids phosphatidylethanolamine and cardiolipin. In the yeast Saccharomyces cerevisiae, the majority of cellular phosphatidylethanolamine is synthesized by the mitochondrial phosphatidylserine decarboxylase 1 (Psd1). The biogenesis of Psd1 involves several processing steps. It was speculated that the Psd1 precursor is sorted into the inner membrane and is subsequently released into the intermembrane space by proteolytic removal of a hydrophobic sorting signal. However, components involved in the maturation of the Psd1 precursor have not been identified. We show that processing of Psd1 involves the action of the mitochondrial processing peptidase and Oct1 and an autocatalytic cleavage at a highly conserved LGST motif yielding the α- and β-subunit of the enzyme. The Psd1 β-subunit (Psd1β) forms the membrane anchor, which binds the intermembrane space-localized α-subunit (Psd1α). Deletion of a transmembrane segment in the β-subunit results in mislocalization of Psd1 and reduced enzymatic activity. Surprisingly, autocatalytic cleavage does not depend on proper localization to the inner mitochondrial membrane. In summary, membrane integration of Psd1 is crucial for its functionality and for maintenance of mitochondrial lipid homeostasis.
Introduction
Mitochondrial membranes contain a high amount of non-bilayer-forming phospholipids, such as phosphatidylethanolamine (PE)3 and cardiolipin (CL) (1–4). These lipids have a characteristic conical shape due to the relatively small headgroup compared with the large hydrophobic acyl chains. Non-bilayer-forming phospholipids may locally induce inverted hexagonal (HII) phase structures, thereby increasing the tension in the lipid bilayer. This property seems to be important for vesicle formation, membrane fusion, transbilayer transport of lipids and polar solutes, protein interactions, topogenesis, and protein function (5, 6). PE and CL associate with different membrane-bound proteins and protein complexes, such as the respiratory chain complexes or the mitochondrial ATP/ADP carrier to maintain their stability and function (7–9). As a consequence, lack of CL affects the structural organization of the respiratory chain supercomplexes and protein translocases in both outer and inner mitochondrial membrane (2, 10–17). Simultaneous loss of both major non-bilayer forming phospholipids, PE and CL, is lethal for Saccharomyces cerevisiae (18). Also in mammals, biosynthesis of PE is essential (19), and alterations in content or acylation of CL lead to mitochondrial dysfunction and diseases like Barth syndrome (20, 21).
CL synthesis is restricted to mitochondria (22–27), whereas in yeast, PE biosynthesis occurs in different cellular compartments, including mitochondria, the Golgi/vacuolar compartment, and the endoplasmic reticulum with its mitochondria-associated membrane fraction (28–39). The mitochondrial phosphatidylserine decarboxylase (Psd1) synthesizes the majority of PE, which is not only supplied to mitochondrial but also to other cellular membranes (1, 40). Because the substrate of Psd1, phosphatidylserine (PS), is synthesized in the endoplasmic reticulum/mitochondria-associated membrane, transport to the site of enzymatic conversion is required. Whether this translocation process involves membrane contact sites like the endoplasmic reticulum-mitochondria tethering complex ERMES is under debate (41–43). Deletion of PSD1 leads to ethanolamine auxotrophy during growth on non-fermentable carbon sources, reduced growth on fermentable media, and morphological alterations of mitochondria (44, 45). Moreover, Psd1 is required for the regulation of pleiotropic drug resistance by inducing PDR5 gene expression (46). Although extramitochondrially synthesized PE can be imported into mitochondria, defects of psd1Δ strains cannot be completely compensated by this process, indicating that PE uptake by mitochondria has limited efficiency (44, 47).
Phosphatidylserine decarboxylases form a protein family, which is highly conserved from bacteria to humans (48). Psd1 from various cell types share a characteristic LGST motif, which is required for autocatalytic cleavage of the enzyme into α- and β-subunits (48–50). This process has been described in some detail for Psd1 homologs of various species (9, 46, 51–54). It starts with an ester bond formation between Gly-253 and Ser-254 followed by α,β-elimination and release of the mature β-subunit, leaving a dehydroalanine residue at the N terminus of the α-subunit. Hydration and subsequent elimination of ammonia lead to formation of a pyruvoyl prosthetic group at the N terminus of the α-subunit (9, 48, 50, 51). Mutations of the highly conserved LGST motif were shown to cause defects in Psd1 processing and formation of an inactive form of the enzyme (9, 46, 53). Surprisingly, such defects have no effect on the induction of PDR5 expression for regulating multidrug resistance, suggesting that this process may occur independently of PS decarboxylation (46).
Although Psd1 is of central importance for the generation of cellular PE, its biogenesis is only poorly understood. Like the vast majority of mitochondrial proteins, Psd1 is synthesized on cytosolic ribosomes as a precursor containing a cleavable signal sequence (36). Upon in vitro import into isolated yeast mitochondria, three proteolytic products are formed from the Psd1 precursor, indicating a stepwise processing of the polypeptide (55). Psd1 contains a hydrophobic stretch, which was proposed to target the polypeptide to the inner mitochondrial membrane (49, 50, 56). It was assumed that this putative inner membrane sorting sequence is removed upon import, yielding the mature Psd1 localized to the mitochondrial inner membrane/intermembrane space (50, 55). This view was supported by enzymatic analyses assigning the enzyme activity to the intermembrane space site of the inner membrane (31, 57, 58). However, experimental evidence for the functionality of the putative inner membrane sorting sequence, the import route of the Psd1 precursor, and the nature of the three processed Psd1 forms was missing. Moreover, the topology of the α- and β-subunits of Psd1 remained unclear.
In a detailed biochemical study presented here, we report that in yeast the Psd1 β-subunit (Psd1β) is integrated into the mitochondrial inner membrane and serves as an anchor for the intermembrane space-localized α-subunit (Psd1α). Analysis of the individual processing steps of Psd1 showed involvement of the matrix-localized peptidases mitochondrial processing peptidase (MPP) and Oct1, which remove the N-terminal signal peptides from Psd1. We demonstrate that lack of the hydrophobic stretch leads to mislocalization and partial inactivation of the enzyme. In contrast, self-cleavage of Psd1 into α- and β-subunits can occur upon mislocalization within mitochondria but strictly depends on the LGST motif and mitochondrial membranes. Thus, correct integration of the Psd1 β-subunit into the inner membrane is essential for proper enzymatic function.
EXPERIMENTAL PROCEDURES
Strains and Culture Conditions
Psd1 mutant strains and plasmids used in this study are listed in Table 1. Deletion strains of mitochondrial proteases, mas1 temperature-sensitive strain, TOM70His, TOM40HA, TOM22His, tom22Δ, tom20Δ, and tom70Δ strains, and their corresponding wild types have been described before (59–62). The reference wild type strain for oct1Δ is a re-expression of OCT1 in the oct1Δ background (63). Yeast cells were grown under aerobic conditions to the logarithmic growth phase at 24 or 30 °C on YPG or YPS medium (1% yeast extract (Oxoid), 2% peptone (Oxoid), 3% glycerol (Roth), or 2% sucrose adjusted to pH 5.0 with HCl). The temperature-sensitive phenotype of a mas1 temperature-sensitive mutant strain was induced by growth for 6 h at non-permissive temperature (61). In the case of yeast strains transformed with expression plasmids, cells were grown on minimal medium minus uracil containing 2% galactose (Roth), 0.67% yeast nitrogen base without amino acids (U. S. Biological), and 0.063% amino acid mix without uracil (Roth, Fluka).
TABLE 1.
Strains used in this study
| Strain | Genotype | Source/Reference |
|---|---|---|
| Wild type | BY4741 MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Euroscarf |
| psd1Δ | BY4741 MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 psd1Δ::KanMX4 | Euroscarf |
| WT+pYES2 | BY4741 MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 + pYES2 | This study |
| psd1Δ+pYES2 | BY4741 MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 psd1Δ::KanMX4 + pYES2 | This study |
| Psd1HA | BY4741 MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 psd1Δ::KanMX4 + pYES2-Psd1HA | This study |
| Psd1S463A | BY4741 MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 psd1Δ::KanMX4 + pYES2-Psd1S463A | This study |
| Psd1ΔIM | BY4741 MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 psd1Δ::KanMX4 + pYES2-Psd1ΔIM | This study |
Plasmid and Strain Constructions
For the generation of a yeast strain expressing a C-terminally HA-tagged Psd1, PSD1 was amplified using gene-specific primers from genomic DNA in a standard PCR mixture containing ExTaq DNA polymerase (Takara). The purified PCR product was inserted via BamHI and NotI (Fermentas) into the pYES2 vector (Invitrogen), leading to pYES2-PSD1HA. For the generation of Psd1S463A and Psd1ΔIM (residues from Val-81 to Ser-100 were deleted), pYES2-PSD1HA was used as a template either for site-directed mutagenesis (QuikChange XL site-directed mutagenesis kit; Stratagene) or overlap-extension PCR. A psd1Δ strain was transformed with the described constructs by lithium acetate transformation (64), generating Psd1HA, Psd1S463A, and Psd1ΔIM.
Isolation of Mitochondria and Submitochondrial Localization of Psd1
Mitochondria were isolated by differential centrifugation according to standard procedures (4, 65). Mitochondrial fractions were adjusted to a protein concentration of 10 mg/ml in SEM buffer (250 mm sucrose, 1 mm EDTA, and 10 mm MOPS-KOH (pH 7.2)), and aliquots were shock-frozen in liquid nitrogen and stored at −80 °C. Enrichment of marker proteins and cross-contamination of subcellular fractions were assessed as described (4).
The submitochondrial localization of Psd1 was determined by accessibility of Psd1 to externally added proteinase K in intact, hypotonically swollen or lysed mitochondria. For hypo-osmotic swelling of mitochondria, mitoplasts were generated by treating mitochondria with a 9:1 ratio of EM buffer (10 mm MOPS-KOH (pH 7.2) and 1 mm EDTA) and SEM buffer for 10 min on ice. Subsequently, samples were treated with 20–50 μg/ml proteinase K for 15 min on ice. For lysis, mitochondria were treated with Triton X-100 at a final concentration of 0.5% (v/v) prior to the addition of proteinase K. In general, proteinase K activity was stopped by the addition of 2 mm PMSF and incubation for 10 min on ice. Subsequently, mitochondria were re-isolated by centrifugation (16,000 × g, 10 min, 4 °C) and washed with SEM buffer. Samples were subjected to SDS-PAGE and Western blot analysis. To determine membrane association of proteins, carbonate extraction was used as described previously (65–68). In brief, isolated mitochondria were resuspended in freshly prepared 0.1 m sodium carbonate buffer at pH 10.8 or pH 11.5 and incubated on ice for 30 min. Mitochondrial membranes were re-isolated by ultracentrifugation (100,000 × g, 40 min, 4 °C). The pellet was solubilized in SDS-PAGE loading dye, whereas proteins remaining in the supernatant were precipitated by trichloroacetic acid. Samples were subjected to SDS-PAGE and Western blot analysis.
Import of Precursor Proteins into Isolated Mitochondria and Microsomes
For in vitro transcription, a PCR-generated template containing the SP6 promoter was used. RNA was purified (MEGAclear kit; Invitrogen) and used for in vitro translation (TNT kit, Promega) in the presence of 35S-labeled methionine. Import of 35S-labeled precursor proteins into isolated yeast mitochondria (corresponding to 50 μg of protein content) was performed at 25 °C in the presence of 2 mm NADH, 2 mm ATP, and an ATP-regenerating system (5 mm creatine phosphate and 0.1 mg/ml creatine kinase) in import buffer (3% BSA (w/v), 250 mm sucrose, 80 mm KCl, 5 mm MgCl2, 2 mm KH2PO4, 5 mm methionine, 10 mm MOPS-KOH (pH 7.2)) (65). Import reactions were stopped on ice and by dissipation of the membrane potential with 8 μm antimycin A, 1 μm valinomycin, and 20 μm oligomycin. For protease treatment, samples were incubated with 50 μg/ml proteinase K for 15 min on ice. Mitochondria were re-isolated by centrifugation (16,000 × g, 10 min, 4 °C) and washed with SEM buffer. For pulse-chase experiments, 35S-labeled Psd1 was imported into mitochondria for 5 min under standard import conditions (pulse). Subsequently, mitochondria were re-isolated, washed, and incubated a second time under import conditions but without the addition of new 35S-labeled Psd1 precursor (chase). Samples were subjected to SDS-PAGE, and 35S-labeled proteins were detected by autoradiography. For blue native electrophoresis, mitochondria were solubilized with digitonin buffer (20 mm Tris-HCl (pH 7.4), 50 mm NaCl, 0.1 mm EDTA, 10% glycerol) containing 1% (w/v) digitonin. After a clarifying spin, mitochondrial protein complexes were separated by blue native electrophoresis as described (69, 70).
The import of 35S-labeled precursor proteins into dog pancreatic microsomes (Promega) was carried out following the standard import protocol for isolated mitochondria. Membranes were re-isolated by centrifugation (125,000 × g, 30 min, 4 °C) and washed with SEM buffer. Membranes were lysed under denaturing conditions and subjected to SDS-PAGE analysis.
Affinity Purification of Tagged Proteins
Mitochondria were solubilized in digitonin buffer containing 1% (w/v) digitonin at a protein concentration of 1 mg/ml for 20 min on ice. After a clarifying spin, the supernatant was incubated with an anti-HA matrix (Roche Applied Science) under constant rotation for 1 h at 4 °C. Following several washing steps with digitonin buffer containing 0.3% (w/v) digitonin, proteins were eluted and subjected to SDS-PAGE, Western blot analysis, and immunodetection with the indicated antisera. For affinity purification of arrested precursor, 35S-labeled Psd1 was imported in the absence of a membrane potential into mitochondria containing Tom22His, Tom40HA, or Tom70His. Subsequently, pull-down experiments were performed as described (59–62).
Phospholipid Analysis
Lipids were extracted from total cell free homogenate and mitochondria by the procedure of Folch et al. (1957) using chloroform/methanol (2:1, v/v) (71). Remaining contaminants (e.g. proteins and salts) were removed by additional washing steps of the organic phase with 0.034% MgCl2 solution (w/v), 2 n KCl, methanol (4:1, v/v), and methanol/water/chloroform (48:47:3, v/v/v), respectively. Individual phospholipids were separated by two-dimensional thin layer chromatography on Silica gel 60 plates (Merck) by using chloroform, methanol, 25% NH3 (68:35:5, v/v/v) as the first and chloroform/acetone/methanol/acetic acid/water (53:20:10:10:5, v/v/v/v) as the second developing solvent. Phospholipids were visualized on thin-layer chromatography plates by staining with iodine vapor, scraped off, and quantified as described (72).
Psd1 Activity Assay
The enzymatic activity of Psd1 was determined in vitro by measuring the conversion of 3H-labeled PS to 3H-labeled PE in isolated mitochondria from yeast cells grown to the logarithmic growth phase as reported by Kuchler et al. (31). Radioactively labeled [3H]PS was synthesized in vitro by incubating isolated yeast microsomes (8 mg of protein) with 0.02/0.2 mm CDP-DAG, 5 mm hydroxylamine, 0.6 mm MnCl2, 0.2% Triton X-100, 0.5 mm l-serine, 0.01 μCi of l-[3H]serine (specific activity 21.99 Ci/mmol), and 0.1 m Tris-HCl (pH 8.0) in a total volume of 2 ml for 2 h at 30 °C. The reaction was stopped by the addition of 3 ml of chloroform/methanol (2:1, v/v), and lipids were extracted as described previously (71). The Psd1 activity assay was performed in a final volume of 1.5 ml containing 100 nmol of [3H]PS (specific activity, 0.64 nCi/nmol), 0.1 m Tris-HCl (pH 7.2), 10 mm EDTA, and 1 mg of protein from isolated mitochondria (31, 47). Incubations were carried out for 7 min at 30 °C and terminated by the addition of 3 ml of chloroform/methanol (2:1, v/v). Lipids were extracted, and individual phospholipids were separated as described above. After visualization with iodine vapor, PE was scraped off, and radioactivity was measured by liquid scintillation counting using LSC Safety (Baker) with 5% water as scintillation mixture.
RESULTS
The β-Subunit of Psd1 Anchors the α-Subunit to the Inner Mitochondrial Membrane
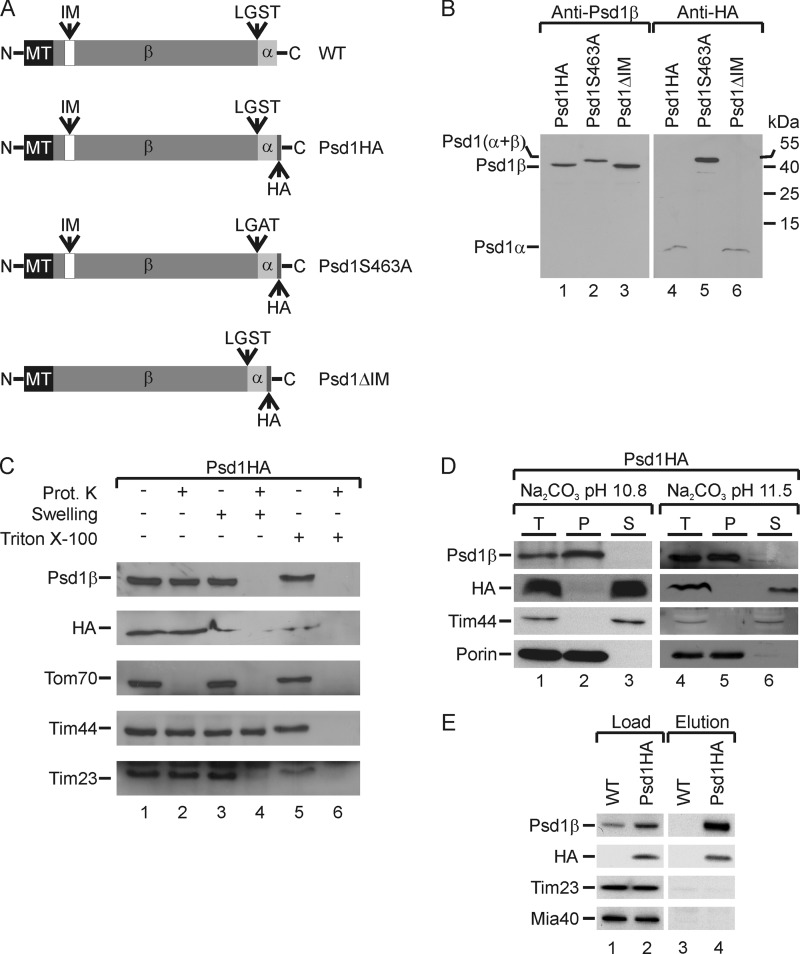
A special feature of Psd1 maturation is its structural arrangement in the form of an α- and a β-subunit (Fig. 1, A and B, lanes 1 and 4). Interestingly, the precise topology of Psd1, especially of the two non-identical subunits within mitochondrial compartments, has not yet been determined. To address this issue in detail, we used a specific antibody recognizing the β-subunit of the protein (Fig. 1B, lane 1) and generated a yeast strain expressing Psd1 with an HA tag C-terminally fused to the α-subunit (Fig. 1A). HA tagging did not affect maturation of Psd1 because the mature β-subunit of Psd1-HA behaved like wild type Psd1 (data not shown). Use of an HA-specific antibody allowed us to detect a 4-kDa α-subunit of Psd1 in isolated mitochondria (Fig. 1B, lane 4). The appearance of the α-subunit was not affected by deletion of the predicted inner membrane sorting signal of Psd1 (Psd1ΔIM) (Fig. 1B, lane 6). To further test the specificity of both antibodies, we constructed a yeast strain expressing a Psd1S463A variant with a mutation at the LGST cleavage site, which cannot undergo autocatalytic processing (Fig. 1A) (9, 46, 53). As expected, both antibodies recognized unprocessed Psd1 (Fig. 1B, lanes 2 and 5) and could therefore be used to determine the submitochondrial localization of both Psd1 subunits.
FIGURE 1.
The β-subunit of Psd1 tethers the Psd1 α-subunit to the inner membrane. A, the Psd1 constructs used are shown schematically. MT, predicted mitochondrial targeting sequence; IM, putative inner membrane sorting signal. B, mitochondrial proteins from yeast strains expressing Psd1HA, Psd1S463A, or Psd1ΔIM were separated by SDS-PAGE and visualized by immunodetection with the indicated antibodies. C, intact, swollen, or lysed (with Triton X-100) mitochondria were incubated with or without proteinase K. Subsequently, samples were subjected to SDS-PAGE, and the indicated components were visualized by immunodetection with the respective antisera. D, mitochondria were subjected to carbonate extraction. Total (T), membrane-bound (P), and soluble proteins (S) were separated by SDS-PAGE, and components were visualized by immunodetection with the indicated antisera. E, Psd1HA was precipitated with an anti-HA affinity matrix. Load (5%) and eluted proteins (100%) were separated by SDS-PAGE, and proteins were visualized by immunodetection with the indicated antisera.
For this purpose, we treated intact, hypotonically swollen or lysed mitochondria with proteinase K (Fig. 1C). Both α- and β-subunits were protected in intact mitochondria but accessible to the protease after rupture of the outer membrane by osmotic swelling (Fig. 1C, lanes 2 and 4). Similarly, the inner membrane protein Tim23 with a domain exposed to the intermembrane space was degraded after rupture of the outer membrane (Fig. 1C, lanes 2 and 4). The outer membrane protein Tom70 was proteolytically cleaved on intact mitochondria, whereas the matrix-localized Tim44 was digested only after lysis of mitochondria with detergent (Fig. 1C, lane 6). These results indicate that both subunits of Psd1 face the intermembrane space, which is in line with the reported detection of Psd1 activity (31, 57, 58).
Previously, it was speculated that Psd1 might be released into the intermembrane space by cleavage of a proposed inner membrane sorting signal (50, 55), although experimental evidence for this hypothesis was missing. To test a possible membrane association of Psd1, we used a well established alkaline extraction method (66, 67). This approach revealed that the Psd1 β-subunit (Psd1β) remained membrane-associated after carbonate extraction at pH 10.8 and pH 11.5 similar to the outer membrane protein porin, whereas the peripheral membrane protein Tim44 was efficiently removed (Fig. 1D). We concluded from this result that the β-subunit was integrated into the inner mitochondrial membrane. In contrast, the α-subunit was extracted under all tested conditions and therefore localized to the intermembrane space (Fig. 1D). Furthermore, we tested attachment of both subunits to each other after autocatalytic processing in the PSD1-HA strain by pull-down experiments utilizing an anti-HA affinity matrix (Fig. 1E). The Psd1 β-subunit was efficiently co-purified with the HA-tagged α-subunit (Fig. 1E, lane 4), indicating stable association of both Psd1 subunits. Thus, the β-subunit of Psd1 contains a membrane anchor tethering the soluble α-subunit to the inner mitochondrial membrane.
Tom70 and Tom22 Are the Main Mitochondrial Receptors for the Psd1 Precursor
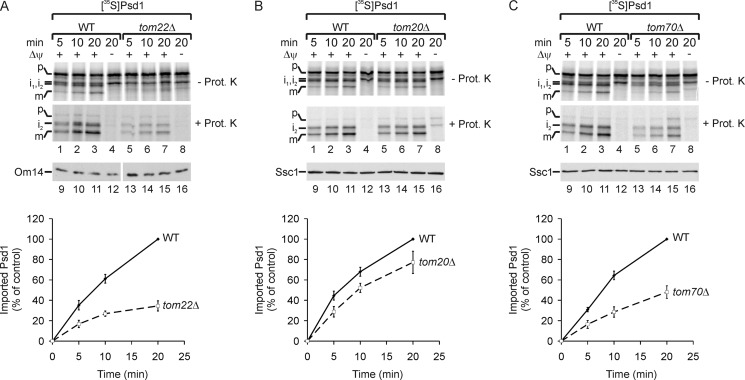
Localization of Psd1 to the inner membrane raises the question of the import mechanism of this protein into mitochondria. In general, the majority of mitochondrial proteins are translated on cytosolic ribosomes as precursors with an N-terminal signal sequence, which is sufficient for targeting and import into mitochondria. The translocase of the outer mitochondrial membrane (TOM complex) is crucial for the translocation of precursor proteins across the outer membrane (73–77). First, we tested whether Psd1 was imported via the TOM complex at all by importing 35S-labeled Psd1 precursor into mitochondria isolated from a yeast strain lacking Tom22 (Fig. 2A). Deletion of Tom22 results in destabilization of the TOM complex and blocks the import of the majority of TOM-dependent substrates into mitochondria (78). Upon import into isolated mitochondria, three forms of Psd1 were detected (Fig. 2A, lanes 1–3), which were absent in the reticulocyte lysate, showing that processing does not occur spontaneously (data not shown). The three fragments represent intermediates 1 and 2 (i1 and i2) formed by unknown processing steps and presumably the mature Psd1 β-subunit (m) (Fig. 2A, lanes 1–3) (55). Intermediate 1 was not completely taken up by mitochondria because it was accessible to externally added proteinase K and predominantly formed in the absence of a membrane potential (Fig. 2A, lanes 1–4). In contrast, intermediate 2 and the mature Psd1β were only formed in the presence of the membrane potential and showed protease resistance (Fig. 2A, lanes 1–3). Thus, we conclude that intermediate 2 and the mature Psd1β were fully imported into mitochondria. In the absence of Tom22, import of the precursor into isolated mitochondria was strongly reduced, indicating the general requirement of the TOM complex for biogenesis of Psd1 (Fig. 2A). The TOM complex has two peripheral receptors, Tom20 and Tom70. Tom20 was reported to recognize precursor proteins with cleavable signal information, whereas Tom70 mainly interacts with hydrophobic precursors harboring internal non-cleavable targeting information (3, 73–79). To test which peripheral receptor was involved in the import of Psd1 precursor, we performed import experiments with mitochondria from yeast strains lacking either Tom20 or Tom70. Surprisingly, import of Psd1 into mitochondria lacking Tom20 was only mildly affected (Fig. 2B) but significantly reduced in the absence of Tom70 (Fig. 2C). Thus, although Psd1 harbors a cleavable signal sequence, Tom70 and Tom22 are the main receptors for the import of Psd1 precursor into mitochondria.
FIGURE 2.
Tom70 and Tom22 are the main receptors for the import of Psd1 precursor into mitochondria. A–C, 35S-labeled Psd1 precursor was imported in the presence or absence of a membrane potential (Δψ) into mitochondria from wild type, tom22Δ (A), tom20Δ (B), or tom70Δ (C). Subsequently, mitochondria were incubated with or without proteinase K (Prot. K). 35S-Labeled proteins were detected by autoradiography. The time-dependent formation of the mature form of Psd1β (m) in mitochondria was quantified by using ImageJ. The longest import time point of Psd1β (m) into wild type mitochondria was set to 100%. Mean values of four measurements with S.E. (error bars) are shown. Immunodetection with the indicated antisera was used as a loading control. p, precursor; i1, i2, intermediates 1 and 2; m, mature Psd1β.
MPP and Oct1 Cleave the Signal Sequence of Psd1
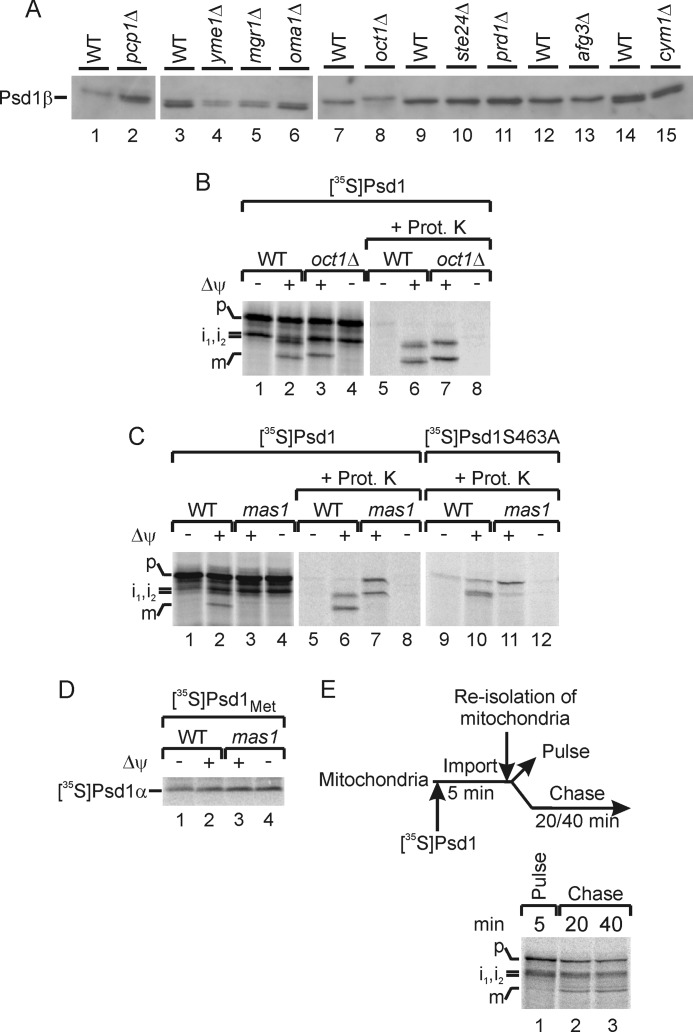
The import of 35S-labeled Psd1 precursor into mitochondria leads to formation of three fragments (see Fig. 2A) (53, 55). However, the molecular mechanism of proteolytic cleavage and the nature of these fragments remained unclear. Previously, it was speculated that the inner membrane sorting sequence might be removed by a protease like Yme1 (50, 55). However, the observed membrane integration of the β-subunit points to a different processing mode. To identify the processing proteases required for Psd1 maturation, we analyzed the protein pattern of Psd1 in mitochondria from strains lacking described or predicted mitochondrial proteases (61) using the antibody specific for Psd1 β-subunit (Fig. 3A). Surprisingly, deletion of Yme1 did not affect the size of mature Psd1 in mitochondria at all (Fig. 3A, lane 4). Also processing of freshly imported Psd1 was not affected in the absence of Yme1 (data not shown). Similarly, the inner membrane proteases Imp1 and Imp2, both required for maturation of mitochondrial proteins destined for the intermembrane space (74, 76, 80, 81), had no effect on Psd1 maturation (data not shown).
FIGURE 3.
MPP and Oct1 process the Psd1 precursor. A, mitochondrial proteins from several deletion strains were analyzed on SDS-PAGE. The β-subunit of Psd1 was visualized by immunodetection with a Psd1-specific antibody. B, 35S-labeled Psd1 precursor was imported in the presence or absence of a membrane potential (Δψ) into mitochondria from wild type or oct1Δ. Subsequently, mitochondria were incubated with or without proteinase K (Prot. K). 35S-Labeled proteins were detected by autoradiography. P, precursor; i1, i2, intermediates 1 and 2; m, mature Psd1β. C, 35S-labeled Psd1 and Psd1S463A precursor were imported in the presence or absence of a membrane potential (Δψ) into mitochondria from wild type or mas1ts mutant strain. Subsequently, mitochondria were incubated with or without proteinase K. 35S-Labeled proteins were detected by autoradiography. D, 35S-labeled Psd1Met containing six methionines at the C terminus of Psd1 was imported in the presence or absence of a membrane potential (Δψ) into mitochondria from wild type or mas1ts mutant strain. 35S-Labeled Psd1α was detected by autoradiography. E, experimental scheme and pulse-chase experiment of 35S-labeled Psd1 into wild type mitochondria. 35S-Labeled Psd1 was imported into wild type mitochondria for 5 min at 25 °C under standard import conditions (lane 1; Pulse). After re-isolation and washing steps, mitochondria were further incubated under import conditions but without the addition of new 35S-labeled precursor (lanes 2 and 3, Chase). 35S-Labeled proteins were detected by autoradiography.
Whereas deletion of several other mitochondrial proteases did not affect the formation of Psd1β, a small size shift of Psd1 was observed in mitochondria lacking Oct1 (octapeptidyl aminopeptidase) (Fig. 3A, lane 8). It is known that the matrix-localized Oct1 typically removes an octapeptide from a precursor intermediate after cleavage by MPP to stabilize the client protein (63). To confirm processing of the Psd1 precursor by Oct1, we imported 35S-labeled Psd1 into mitochondria lacking this protease (Fig. 3B). We observed a size shift of intermediate 2 (i2) and of the mature Psd1 β-subunit (m), indicating that Oct1 indeed removed a peptide from the Psd1 precursor (Fig. 3B, lanes 3 and 7).
The amino acid sequence of the Psd1 precursor suggests the presence of a classical N-terminal mitochondrial signal sequence (36). Such signal sequences are typically removed by MPP of the mitochondrial matrix, which consists of the two essential subunits Mas1 and Mas2 (74, 80, 81). To address the role of MPP in processing of Psd1, we imported the Psd1 precursor into mitochondria isolated from a temperature-sensitive Mas1 mutant (61, 62). Mas1ts was inactivated in vivo by shifting the culture to non-permissive growth conditions. In isolated mitochondria of the in vivo heat-shocked mas1ts cells, removal of the signal peptide from intermediate 2 and the mature Psd1β was blocked (Fig. 3C, lanes 3 and 7). Thus, the N-terminal presequence of the Psd1 precursor is cleaved by the matrix-located proteases MPP and Oct1, yielding Psd1β (m).
So far, we only speculated that the mature Psd1β is formed by self-processing, which leads to the formation of two protease-protected bands upon import of Psd1 in mas1ts (Fig. 3C, lane 7). To test this notion experimentally, we imported a Psd1 variant into wild type and mas1ts mitochondria, which contains a point mutation in the conserved LGST motif to prevent autocatalytic cleavage (9, 46, 53). This Psd1S463A construct was imported with the same efficiency as wild type, but cleavage into α- and β-subunit was blocked (Fig. 3C, lane 10). In mas1ts mitochondria, only the upper band was formed, indicating that the mature Psd1β emerged upon self-cleavage in an MPP-independent manner (Fig. 3C, lane 11). To confirm this conclusion, we analyzed formation of the α-subunit of Psd1 in mas1ts mitochondria directly. The C terminus of Psd1 was tagged with six methionines to allow detection of 35S-labeled Psd1α. This Psd1Met construct was imported like wild type Psd1 (data not shown), and the formation of Psd1α with a size of 4 kDa was not affected in mas1ts mitochondria (Fig. 3D, lanes 3 and 4). To determine at which import step autocatalysis occurred, we performed a pulse-chase experiment with 35S-labeled Psd1 (Fig. 3E). 35S-Labeled Psd1 was imported for a short time period (Fig. 3E, lane 1, Pulse). Subsequently, mitochondria were re-isolated and further incubated under import conditions but without addition of new 35S-labeled precursor (Fig. 3E, lanes 2 and 3, Chase). Interestingly, the mature Psd1β was formed more efficiently after the chase but only to a minor extent during the short pulse period (Fig. 3E). This observation indicated that self-cleavage of Psd1 occurs at late import time points and follows processing by MPP and Oct1 under normal import conditions.
TOM Complex-arrested Psd1 Precursor Is Processed into α- and β-Subunit
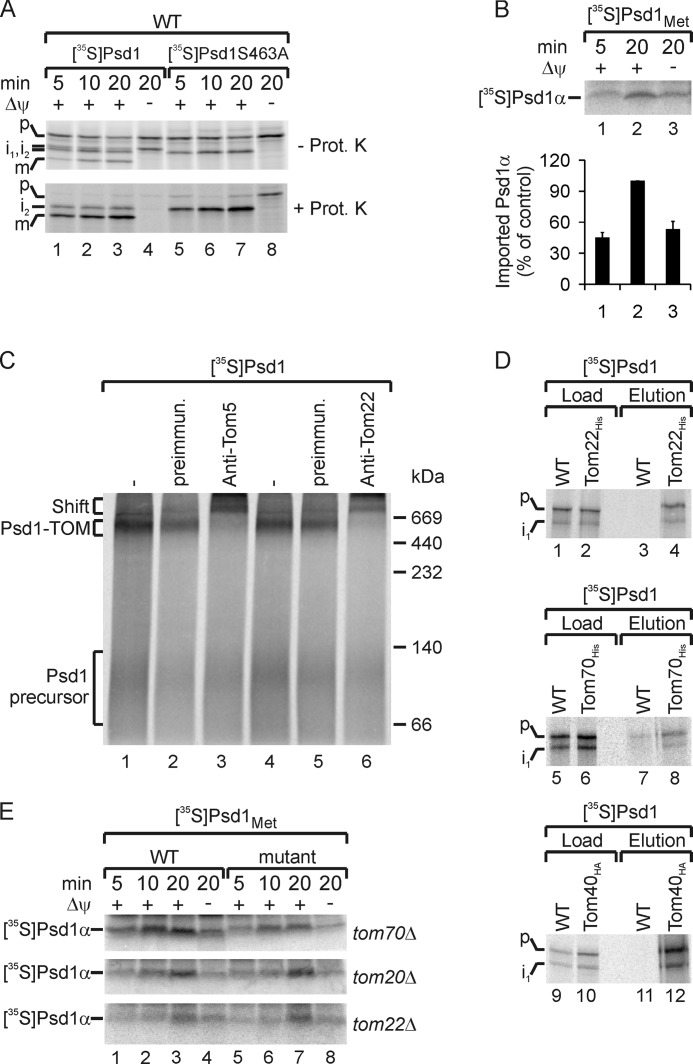
Strikingly, Psd1α is also formed in the absence of a membrane potential, which indicates that self-cleavage of Psd1 can occur independently of its import into mitochondria (Fig. 3D, lane 1). Because intermediate 1 is also formed in the absence of a membrane potential (Fig. 3B, lane 1), we wondered whether intermediate 1 represented a self-processed Psd1 precursor. To address this issue, we performed import experiments with the Psd1S463A precursor in wild type mitochondria (Fig. 4A, lanes 5–8). Interestingly, intermediate 1 was not generated during import of the Psd1S463A precursor, demonstrating that intermediate 1 is formed by self-processing of Psd1 into α- and β-subunit. To assess the efficiency of the self-cleavage by arrested Psd1, we quantified the formation of Psd1α in the presence and absence of a membrane potential (Fig. 4B). We found that albeit import into mitochondria favors self-processing, a marked amount of arrested Psd1 precursor undergoes autocatalysis as well (Fig. 4B, lane 3).
FIGURE 4.
TOM complex-arrested Psd1 precursor undergoes autocatalytic processing. A, 35S-labeled Psd1 and Psd1S463A precursor were imported in the presence or absence of a membrane potential (Δψ) into wild type mitochondria. Subsequently, mitochondria were incubated with or without proteinase K (Prot. K). 35S-Labeled proteins were detected by autoradiography. p, precursor; i1, i2, intermediates 1 and 2; m, mature Psd1β. B, 35S-labeled Psd1Met containing six methionines at the C terminus of Psd1 was imported in the presence or absence of a membrane potential (Δψ) into mitochondria from wild type. 35S-Labeled Psd1α was detected by autoradiography. The time-dependent formation of Psd1α in mitochondria was quantified by using ImageJ. The longest import time point of Psd1α into wild type mitochondria in the presence of a membrane potential was set to 100%. Mean values of three measurements with S.E. (error bars) are shown. C, 35S-labeled Psd1 precursor was imported in the absence of a membrane potential into wild type mitochondria. Mitochondria were incubated with or without antibodies against Tom5 or Tom22 or preimmune sera as a control. Subsequently, protein complexes of lysed mitochondria were separated by blue native gel electrophoresis. 35S-Labeled proteins were detected by autoradiography. D, 35S-labeled Psd1 precursor was imported in the absence of a membrane potential into wild type, Tom22His, Tom70His, and Tom40HA mitochondria. Subsequently, mitochondria were lysed and subjected to affinity purification utilizing the His and HA tag. Load (4%) and eluted proteins (100%) were separated by SDS-PAGE and analyzed by autoradiography. i1, intermediate 1. E, 35S-labeled Psd1Met precursor was imported in the presence or absence of a membrane potential (Δψ) into mitochondria from wild type, tom70Δ, tom20Δ, and tom22Δ. 35S-Labeled Psd1α was detected by autoradiography.
It has been shown before that upon depletion of the membrane potential, some hydrophobic precursors accumulate at the TOM complex (12, 82). To test whether or not the arrested Psd1 precursor binds to the TOM complex as well, we incubated 35S-labeled Psd1 precursor with isolated mitochondria in the absence of a membrane potential (Fig. 4C). After the import, mitochondria were lysed using the mild detergent digitonin, and samples were analyzed by blue native electrophoresis to detect the TOM-bound intermediate. In the absence of a membrane potential, the Psd1 precursor was stably bound to the TOM complex, as shown by a size shift of the intermediate after adding antibodies against Tom5 or Tom22 to mitochondria (Fig. 4C, lanes 3 and 6). To analyze whether both precursor and intermediate 1 bind to the TOM complex, we arrested 35S-labeled Psd1 at the translocases in mitochondria containing His-tagged Tom22, Tom70, or HA-tagged Tom40 and performed affinity purification (Fig. 4D). These experiments revealed that intermediate 1 and unprocessed precursors were co-purified with tagged TOM subunits (Fig. 4D, lanes 4, 8, and 12). To determine if Tom receptors play a role in self-cleavage of Psd1, we analyzed the formation of Psd1α in mutants lacking one Tom receptor (Fig. 4E). Only the absence of Tom70 slightly affected Psd1α formation (Fig. 4E, top). This observation indicated that Tom70 is involved in the initial recognition of the Psd1 precursor, whereas Tom22 is involved in later import steps. Altogether, in the absence of a membrane potential, autocatalytic cleavage of the Psd1 precursor into α- and β-subunits occurred when the precursor was exposed on the outer membrane and bound to the TOM complex.
Psd1 Requires Integration into the Inner Mitochondrial Membrane for Full Enzymatic Activity
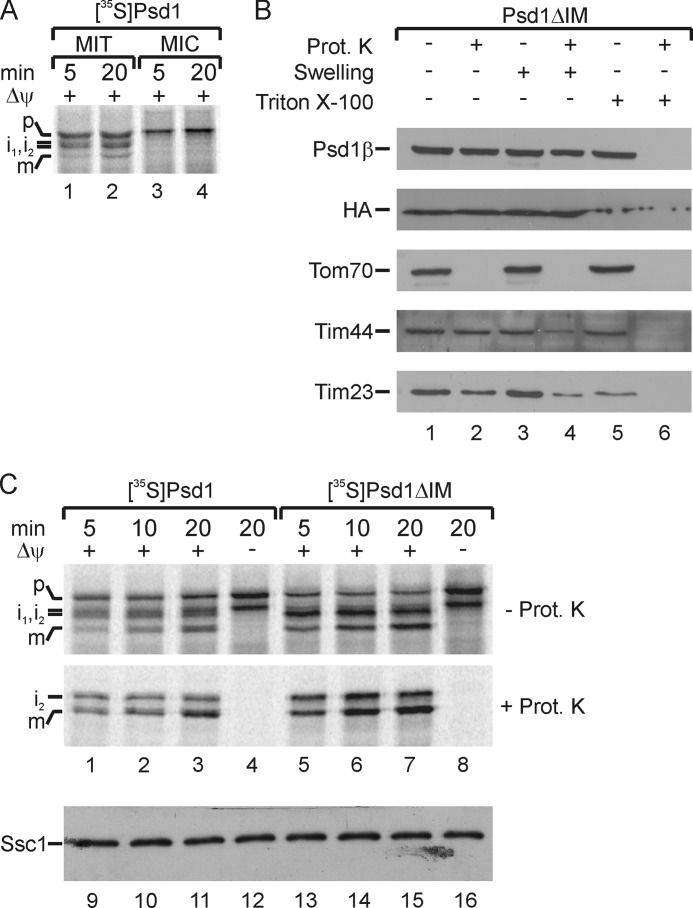
The surprising finding that an arrested Psd1 precursor probably undergoes self-processing at the outer membrane tempted us to speculate that separation of α- and β-subunit may also occur in other mitochondrial compartments or even on other membranes. To test this hypothesis, we followed the processing of 35S-labeled Psd1 precursor after incubation with mitochondria and microsomes under the same import conditions. We could not detect any cleavage of the Psd1 precursor when canine pancreatic microsomes were added (Fig. 5A, lanes 3 and 4). This result indicates that Psd1 maturation depends on mitochondrial membranes, which is consistent with the results obtained with Plasmodium knowlesi (PkPSD) (54). We also tested whether sorting of Psd1 to the inner membrane is required for self-processing and function. To address this issue, we generated a yeast strain expressing a Psd1 variant that lacks the predicted inner membrane anchor (Val-81 to Ser-100; Psd1ΔIM) (Fig. 1A) (46). Detection of both α- and β-subunits using the subunit-specific antibodies described above demonstrated that Psd1ΔIM undergoes autocatalytic processing (Figs. 1B, lanes 3 and 6, and 5B). In contrast to Psd1HA, the Psd1ΔIM α- and β-subunit were not accessible to externally added proteases after rupturing the outer membrane by osmotic swelling (Fig. 5B, lane 4). Like the matrix-localized Tim44, Psd1ΔIM was only degraded by proteinase K when membranes were lysed with detergent (Fig. 5B, lane 6). This result indicated that Psd1ΔIM was fully transported to the matrix site of the inner membrane and that the predicted inner membrane anchor was crucial for correct topology. To compare the processing efficiency of both constructs, import experiments with 35S-labeled Psd1ΔIM and wild type Psd1 into isolated mitochondria were performed (Fig. 5C). Deletion of the putative inner membrane anchor in Psd1ΔIM even stimulated import and processing steps of the precursor by MPP and Oct1 (Fig. 5C, lanes 5–8).
FIGURE 5.
Autocatalytic processing of Psd1 is restricted to mitochondria but does not depend on the proper sorting of Psd1. A, 35S-labeled Psd1 precursor was imported in the presence of a membrane potential (Δψ) into wild type mitochondria (MIT) and canine pancreatic microsomes (MIC). P, precursor; i1, i2, intermediates 1 and 2; m, mature Psd1β. B, intact, swollen, or lysed (with Triton X-100) mitochondria from a yeast strain expressing Psd1ΔIM were incubated with or without proteinase K (Prot. K). Subsequently, samples were subjected to SDS-PAGE, and proteins were visualized by immunodetection with the indicated antisera. C, 35S-labeled Psd1 or Psd1ΔIM was imported in the presence or absence of a membrane potential (Δψ) into wild type mitochondria. Subsequently, mitochondria were incubated with or without proteinase K. 35S-Labeled proteins were detected by autoradiography. For loading controls, the mitochondrial Hsp70 (Ssc1 (stress-seventy subfamily C/mitochondrial Hsp70)) was immunodetected with specific antibodies. p, precursor; i1 and i2, intermediates 1 and 2; m, mature Psd1.
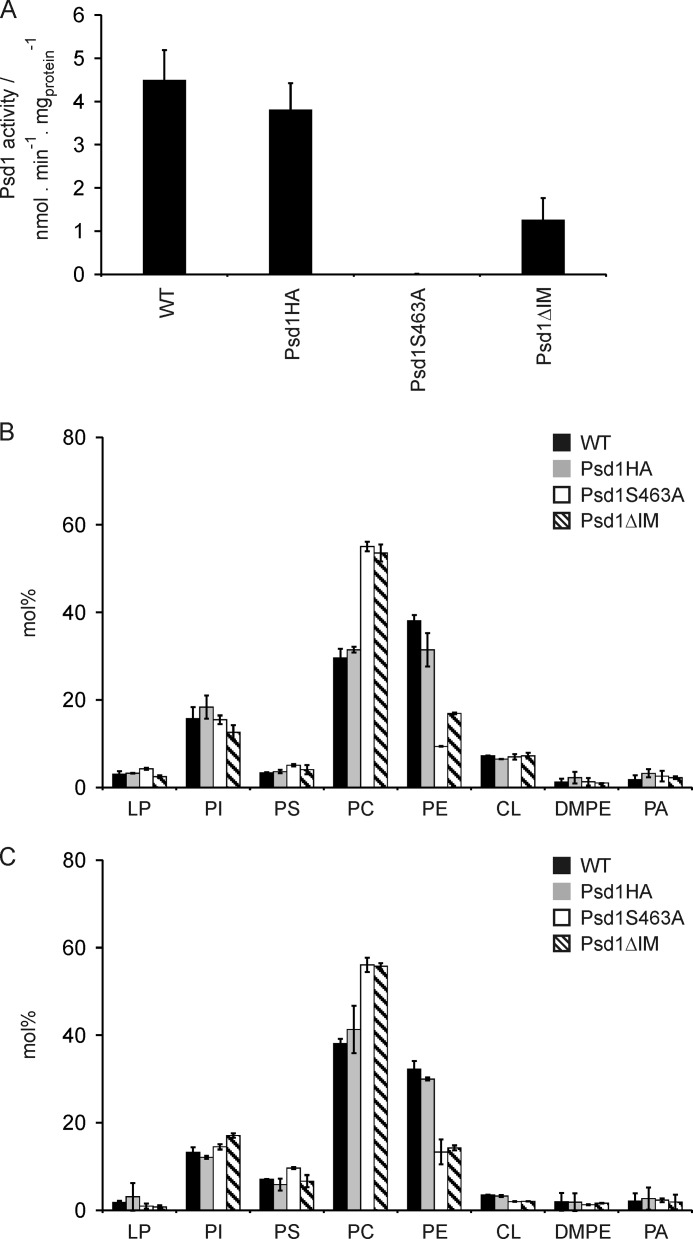
Finally, we determined the effect of Psd1ΔIM mislocalization on the activity of the enzyme (Fig. 6A) and on the mitochondrial phospholipid pattern (Fig. 6B). Mitochondria from a strain expressing Psd1S463A did not display detectable decarboxylase activity due to the inability to generate the active site of the enzyme. In contrast, mislocalization of Psd1 by removing the membrane anchor of the β-subunit (Psd1ΔIM) led to a reduced enzymatic activity compared with wild type and HA-tagged Psd1 (Fig. 6A). The loss of Psd1 activity in Psd1S463A led to a typical psd1Δ phospholipid pattern, which is characterized by an enrichment of phosphatidylcholine at the expense of PE in mitochondria and homogenate (Fig. 6, B and C) (18, 47). The level of PS within mitochondria was not increased because it is used as a substrate for Psd2, which is localized to the Golgi/vacuolar compartment and responsible for extramitochondrially synthesized PE (37, 38).
FIGURE 6.
Inner membrane localization of Psd1 is required for optimal enzymatic activity. A, phosphatidylserine decarboxylase activity was measured using isolated mitochondria from wild type and the indicated Psd1 mutants. Strains were grown on minimal galactose medium at 30 °C. Mean values of three independent measurements and S.D. values are shown. B and C, phospholipids were extracted from mitochondria (B) and homogenate (C) isolated from wild type and the indicated Psd1 mutants. Strains were grown on minimal galactose medium at 30 °C. LP, lysophospholipids; PI, phosphatidylinositol; PC, phosphatidylcholine; DMPE, dimethylphosphatidylethanolamine; PA, phosphatidic acid. Mean values of two independent measurements and S.D. values (error bars) are shown.
Interestingly, the phospholipid pattern of Psd1ΔIM in homogenate resembled that of Psd1S463A (Fig. 6C). These findings indicated that mislocalization of Psd1 to the mitochondrial matrix leads to a reduced activity of the enzyme (Fig. 6A) and consequently to insufficient supply of mitochondrially synthesized PE to the total cellular PE pool (Fig. 6C).
DISCUSSION
In yeast, Psd1 catalyzes formation of the majority of cellular PE by decarboxylation of PS (1, 28, 31, 37–39, 83). Deletion of PSD1 leads to severe cell damage, including alterations of mitochondrial morphology and reduced growth under respiratory conditions caused by dramatically reduced PE levels in mitochondrial and other cellular membranes (44, 45). For this reason, functional Psd1 is highly important to maintain cell integrity and function.
Here we show that correct import and sorting is required for the enzymatic activity of Psd1. In previous experiments, it was shown that GFP-tagged Psd1 localizes to the inner mitochondrial membrane (47). However, the question remained open whether the protein was integrated into the membrane or sorted into the intermembrane space, especially due to the occurrence of two non-identical subunits (46, 50, 53, 55). In the present study, we demonstrate that the β-subunit of Psd1 is tightly anchored into the inner membrane, most likely via a single transmembrane segment. This conclusion is supported by the prediction of at least one transmembrane region, as reported previously (53, 55). Deletion of this hydrophobic segment (amino acids Val-81 to Ser-100) (46, 54) leads to mislocalization of Psd1 to the matrix side of the inner membrane, partial loss of enzymatic activity, and, consequently, reduced PE levels (Figs. 5B and 6). In such a construct, however, cleavage of the proenzyme to its non-identical subunits was not inhibited (Fig. 1B). This result suggests that not only correct processing of Psd1 but also the membrane environment plays a crucial role for the formation of an active enzyme. Because the soluble α-subunit harbors the active site of the enzyme or at least an essential part of this domain, the recruitment to the inner membrane surface with orientation to the intermembrane space appears to be crucial for the ability of Psd1 to decarboxylate the polar headgroup of PS. The soluble α-subunit present in the intermembrane space interacts with the membrane-bound β-subunit (Fig. 1, D and E).
To support the topological model of Psd1 presented in this study, we investigated the mitochondrial import route of this protein and identified components required for its processing. Here, we show that the two matrix-localized processing peptidases MPP and Oct1 remove the targeting signals from the Psd1 precursor (Fig. 3C). Finally, the processed Psd1 undergoes self-cleavage into α- and β-subunits (Fig. 3E). MPP usually cleaves the mitochondrial signal sequence, and subsequently Oct1 removes a small peptide, typically an octapeptide (80, 81). The predicted MPP cleavage site is after leucine 48 of Psd1. Moreover, the Psd1 precursor contains a sequence typical for an Oct1 cleavage motif, namely arginine at position −10 and phenylalanine at position −8 of the putative Oct1 cleavage site after serine at position 56. In general, the second processing step catalyzed by Oct1 is considered to stabilize the N terminus (63). Sequential cleavage by MPP and Oct1 presumably leads to a stabilizing glycine at the N terminus of the mature Psd1 (61, 63). Proteases, which typically remove inner membrane sorting sequences, like Imp1 and Imp2 (80, 81), had no effect on Psd1 processing. This finding is in line with the observation that the hydrophobic inner membrane sorting sequence is not proteolytically removed and can therefore serve as a membrane anchor for Psd1. Thus, the yeast mitochondrial Psd1 β-subunit is embedded into the inner membrane like its bacterial counterpart (48).
Surprisingly, we discovered that cleavage of the Psd1 precursor into α- and β-subunits, which is a crucial step in the formation of the active center of the enzyme, can also take place when the precursor is arrested at the TOM complex of the outer mitochondrial membrane by depletion of the membrane potential (Fig. 4, B and D). Even mislocalization of the protein to the matrix site of the inner membrane did not affect its self-cleavage (Fig. 5, B and C). Thus, separation of the two subunits does not depend on proper maturation and targeting of Psd1 within mitochondria. Interestingly, self-processing of the Psd1 precursor into Psd1α at the outer membrane is only mildly affected by deletion of Tom70, whereas deletion of other Tom receptors had no effect on this processing step (Fig. 4E). Thus, Tom70 provides an initial docking site for the incoming Psd1 precursor but is not strictly required for autocatalysis of Psd1. Other components of the mitochondrial surface or specific membrane properties may be important for the autocatalytic processing of Psd1 as well. Choi et al. (54) reported that processing of Psd from P. knowlesi (PkPSD) into α- and β-subunits was influenced by its lipid surrounding. In vitro experiments using liposomes demonstrated that processing of PkPSD to the mature enzyme form and Psd activity were enhanced by increasing concentrations of dioleoyl phosphatidylserine, whereas other anionic phospholipids, such as dioleoyl phosphatidic acid or dioleoyl phosphatidylglycerol, displayed an inhibitory effect (54). Future studies will be necessary to reveal whether a mitochondria-specific lipid composition contributes to induce the self-processing of Psd1 because this step does not occur when the precursor is incubated with microsomal membranes (Fig. 5A).
In summary, our studies demonstrate clearly that insertion of Psd1 into the inner membrane/intermembrane space is paramount for the formation of an enzymatically active protein. The specific membrane environment may contribute to this process. Correct topology of Psd1 at the inner membrane-intermembrane space interface appears to be important for proper access of the amphipathic substrate PS with its polar headgroup and hydrophobic tails to the active center of the enzyme.
Acknowledgments
We thank Drs. Nikolaus Pfanner, Martin van der Laan, Michael P. Yaffe, and Trevor Lithgow for materials and discussions. We thank Nicole Zufall for expert technical assistance.
This work was supported by the Austrian Science Fund (project 21429 and DK Molecular Enzymology W901-B05 (to G. D.)), the Deutsche Forschungsgemeinschaft, Sonderforschungs-bereich 746, and the Excellence Initiative of the German Federal and State Governments (EXC 294 BIOSS).
- PE
- phosphatidylethanolamine
- CL
- cardiolipin
- Psd1MET
- phosphatidylserine decarboxylase 1 containing six C-terminal methionines
- PS
- phosphatidylserine
- TOM
- translocase of the outer mitochondrial membrane
- MPP
- mitochondrial processing peptidase.
REFERENCES
- 1. Bürgermeister M., Birner-Grünberger R., Nebauer R., Daum G. (2004) Contribution of different pathways to the supply of phosphatidylethanolamine and phosphatidylcholine to mitochondrial membranes of the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1686, 161–168 [DOI] [PubMed] [Google Scholar]
- 2. Osman C., Voelker D. R., Langer T. (2011) Making heads or tails of phospholipids in mitochondria. J. Cell Biol. 192, 7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van Meer G., Voelker D. R., Feigenson G. W. (2008) Membrane lipids. Where they are and how they behave. Nat. Rev. Mol. Cell. Biol. 9, 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zinser E., Daum G. (1995) Isolation and biochemical characterization of organelles from the yeast, Saccharomyces cerevisiae. Yeast 11, 493–536 [DOI] [PubMed] [Google Scholar]
- 5. Dowhan W., Bogdanov M. (2009) Lipid-dependent membrane protein topogenesis. Annu. Rev. Biochem. 78, 515–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. van den Brink-van der Laan E., Killian J. A., de Kruijff B. (2004) Nonbilayer lipids affect peripheral and integral membrane proteins via changes in the lateral pressure profile. Biochim. Biophys. Acta 1666, 275–288 [DOI] [PubMed] [Google Scholar]
- 7. Klingenberg M. (2009) Cardiolipin and mitochondrial carriers. Biochim. Biophys. Acta 1788, 2048–2058 [DOI] [PubMed] [Google Scholar]
- 8. Lange C., Nett J. H., Trumpower B. L., Hunte C. (2001) Specific roles of protein-phospholipid interactions in the yeast cytochrome bc1 complex structure. EMBO J. 20, 6591–6600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li Q. X., Dowhan W. (1990) Studies on the mechanism of formation of the pyruvate prosthetic group of phosphatidylserine decarboxylase from Escherichia coli. J. Biol. Chem. 265, 4111–4115 [PubMed] [Google Scholar]
- 10. Claypool S. M., Oktay Y., Boontheung P., Loo J. A., Koehler C. M. (2008) Cardiolipin defines the interactome of the major ADP/ATP carrier protein of the mitochondrial inner membrane. J. Cell Biol. 182, 937–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gallas M. R., Dienhart M. K., Stuart R. A., Long R. M. (2006) Characterization of Mmp37p, a Saccharomyces cerevisiae mitochondrial matrix protein with a role in mitochondrial protein import. Mol. Biol. Cell 17, 4051–4062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gebert N., Joshi A. S., Kutik S., Becker T., McKenzie M., Guan X. L., Mooga V. P., Stroud D. A., Kulkarni G., Wenk M. R., Rehling P., Meisinger C., Ryan M. T., Wiedemann N., Greenberg M. L., Pfanner N. (2009) Mitochondrial cardiolipin involved in outer membrane protein biogenesis. Implications for Barth syndrome. Curr. Biol. 19, 2133–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gohil V. M., Greenberg M. L. (2009) Mitochondrial membrane biogenesis. Phospholipids and proteins go hand in hand. J. Cell Biol. 184, 469–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kutik S., Rissler M., Guan X. L., Guiard B., Shui G., Gebert N., Heacock P. N., Rehling P., Dowhan W., Wenk M. R., Pfanner N., Wiedemann N. (2008) The translocator maintenance protein Tam41 is required for mitochondrial cardiolipin biosynthesis. J. Cell Biol. 183, 1213–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mileykovskaya E., Dowhan W. (2009) Cardiolipin membrane domains in prokaryotes and eukaryotes. Biochim. Biophys. Acta 1788, 2084–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pfeiffer K., Gohil V., Stuart R. A., Hunte C., Brandt U., Greenberg M. L., Schägger H. (2003) Cardiolipin stabilizes respiratory chain supercomplexes. J. Biol. Chem. 278, 52873–52880 [DOI] [PubMed] [Google Scholar]
- 17. Tamura Y., Harada Y., Yamano K., Watanabe K., Ishikawa D., Ohshima C., Nishikawa S., Yamamoto H., Endo T. (2006) Identification of Tam41 maintaining integrity of the TIM23 protein translocator complex in mitochondria. J. Cell Biol. 174, 631–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gohil V. M., Thompson M. N., Greenberg M. L. (2005) Synthetic lethal interaction of the mitochondrial phosphatidylethanolamine and cardiolipin biosynthetic pathways in Saccharomyces cerevisiae. J. Biol. Chem. 280, 35410–35416 [DOI] [PubMed] [Google Scholar]
- 19. Steenbergen R., Nanowski T. S., Beigneux A., Kulinski A., Young S. G., Vance J. E. (2005) Disruption of the phosphatidylserine decarboxylase gene in mice causes embryonic lethality and mitochondrial defects. J. Biol. Chem. 280, 40032–40040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Barth P. G., Valianpour F., Bowen V. M., Lam J., Duran M., Vaz F. M., Wanders R. J. (2004) X-linked cardioskeletal myopathy and neutropenia (Barth syndrome). An update. Am. J. Med. Genet. A 126A, 349–354 [DOI] [PubMed] [Google Scholar]
- 21. Chicco A. J., Sparagna G. C. (2007) Role of cardiolipin alterations in mitochondrial dysfunction and disease. Am. J. Physiol. Cell Physiol. 292, C33–C44 [DOI] [PubMed] [Google Scholar]
- 22. Chang S. C., Heacock P. N., Clancey C. J., Dowhan W. (1998) The PEL1 gene (renamed PGS1) encodes the phosphatidylglycerophosphate synthase of Saccharomyces cerevisiae. J. Biol. Chem. 273, 9829–9836 [DOI] [PubMed] [Google Scholar]
- 23. Jiang F., Rizavi H. S., Greenberg M. L. (1997) Cardiolipin is not essential for the growth of Saccharomyces cerevisiae on fermentable or non-fermentable carbon sources. Mol. Microbiol. 26, 481–491 [DOI] [PubMed] [Google Scholar]
- 24. Kelly B. L., Greenberg M. L. (1990) Characterization and regulation of phosphatidylglycerolphosphate phosphatase in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1046, 144–150 [DOI] [PubMed] [Google Scholar]
- 25. Osman C., Haag M., Wieland F. T., Brügger B., Langer T. (2010) A mitochondrial phosphatase required for cardiolipin biosynthesis. The PGP phosphatase Gep4. EMBO J. 29, 1976–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tamai K. T., Greenberg M. L. (1990) Biochemical characterization and regulation of cardiolipin synthase in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1046, 214–222 [DOI] [PubMed] [Google Scholar]
- 27. Tuller G., Hrastnik C., Achleitner G., Schiefthaler U., Klein F., Daum G. (1998) YDL142c encodes cardiolipin synthase (Cls1p) and is non-essential for aerobic growth of Saccharomyces cerevisiae. FEBS Lett. 421, 15–18 [DOI] [PubMed] [Google Scholar]
- 28. Daum G., Lees N. D., Bard M., Dickson R. (1998) Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast 14, 1471–1510 [DOI] [PubMed] [Google Scholar]
- 29. Kennedy E. P., Weiss S. B. (1956) The function of cytidine coenzymes in the biosynthesis of phospholipides. J. Biol. Chem. 222, 193–214 [PubMed] [Google Scholar]
- 30. Kim K., Kim K. H., Storey M. K., Voelker D. R., Carman G. M. (1999) Isolation and characterization of the Saccharomyces cerevisiae EKI1 gene encoding ethanolamine kinase. J. Biol. Chem. 274, 14857–14866 [DOI] [PubMed] [Google Scholar]
- 31. Kuchler K., Daum G., Paltauf F. (1986) Subcellular and submitochondrial localization of phospholipid-synthesizing enzymes in Saccharomyces cerevisiae. J. Bacteriol. 165, 901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rajakumari S., Daum G. (2010) Janus-faced enzymes yeast Tgl3p and Tgl5p catalyze lipase and acyltransferase reactions. Mol. Biol. Cell 21, 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Riekhof W. R., Voelker D. R. (2006) Uptake and utilization of lyso-phosphatidylethanolamine by Saccharomyces cerevisiae. J. Biol. Chem. 281, 36588–36596 [DOI] [PubMed] [Google Scholar]
- 34. Riekhof W. R., Wu J., Jones J. L., Voelker D. R. (2007) Identification and characterization of the major lysophosphatidylethanolamine acyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 282, 28344–28352 [DOI] [PubMed] [Google Scholar]
- 35. Saba J. D., Nara F., Bielawska A., Garrett S., Hannun Y. A. (1997) The BST1 gene of Saccharomyces cerevisiae is the sphingosine-1-phosphate lyase. J. Biol. Chem. 272, 26087–26090 [DOI] [PubMed] [Google Scholar]
- 36. Trotter P. J., Pedretti J., Voelker D. R. (1993) Phosphatidylserine decarboxylase from Saccharomyces cerevisiae. Isolation of mutants, cloning of the gene, and creation of a null allele. J. Biol. Chem. 268, 21416–21424 [PubMed] [Google Scholar]
- 37. Trotter P. J., Pedretti J., Yates R., Voelker D. R. (1995) Phosphatidylserine decarboxylase 2 of Saccharomyces cerevisiae. Cloning and mapping of the gene, heterologous expression, and creation of the null allele. J. Biol. Chem. 270, 6071–6080 [DOI] [PubMed] [Google Scholar]
- 38. Trotter P. J., Voelker D. R. (1995) Identification of a non-mitochondrial phosphatidylserine decarboxylase activity (PSD2) in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 270, 6062–6070 [DOI] [PubMed] [Google Scholar]
- 39. Zinser E., Sperka-Gottlieb C. D., Fasch E. V., Kohlwein S. D., Paltauf F., Daum G. (1991) Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J. Bacteriol. 173, 2026–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Voelker D. R. (1984) Phosphatidylserine functions as the major precursor of phosphatidylethanolamine in cultured BHK-21 cells. Proc. Natl. Acad. Sci. U.S.A. 81, 2669–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kornmann B., Currie E., Collins S. R., Schuldiner M., Nunnari J., Weissman J. S., Walter P. (2009) An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325, 477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nguyen T. T., Lewandowska A., Choi J. Y., Markgraf D. F., Junker M., Bilgin M., Ejsing C. S., Voelker D. R., Rapoport T. A., Shaw J. M. (2012) Gem1 and ERMES do not directly affect phosphatidylserine transport from ER to mitochondria or mitochondrial inheritance. Traffic 13, 880–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vance J. E. (1990) Phospholipid synthesis in a membrane fraction associated with mitochondria. J. Biol. Chem. 265, 7248–7256 [PubMed] [Google Scholar]
- 44. Birner R., Bürgermeister M., Schneiter R., Daum G. (2001) Roles of phosphatidylethanolamine and of its several biosynthetic pathways in Saccharomyces cerevisiae. Mol. Biol. Cell 12, 997–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kuroda T., Tani M., Moriguchi A., Tokunaga S., Higuchi T., Kitada S., Kuge O. (2011) FMP30 is required for the maintenance of a normal cardiolipin level and mitochondrial morphology in the absence of mitochondrial phosphatidylethanolamine synthesis. Mol. Microbiol. 80, 248–265 [DOI] [PubMed] [Google Scholar]
- 46. Gulshan K., Schmidt J. A., Shahi P., Moye-Rowley W. S. (2008) Evidence for the bifunctional nature of mitochondrial phosphatidylserine decarboxylase. Role in Pdr3-dependent retrograde regulation of PDR5 expression. Mol. Cell. Biol. 28, 5851–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Birner R., Nebauer R., Schneiter R., Daum G. (2003) Synthetic lethal interaction of the mitochondrial phosphatidylethanolamine biosynthetic machinery with the prohibitin complex of Saccharomyces cerevisiae. Mol. Biol. Cell 14, 370–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dowhan W. (1997) Phosphatidylserine decarboxylases. Pyruvoyl-dependent enzymes from bacteria to mammals. Methods Enzymol. 280, 81–88 [DOI] [PubMed] [Google Scholar]
- 49. Schuiki I., Daum G. (2009) Phosphatidylserine decarboxylases, key enzymes of lipid metabolism. IUBMB Life 61, 151–162 [DOI] [PubMed] [Google Scholar]
- 50. Voelker D. R. (1997) Phosphatidylserine decarboxylase. Biochim. Biophys. Acta 1348, 236–244 [DOI] [PubMed] [Google Scholar]
- 51. Dowhan W., Li Q. X. (1992) Phosphatidylserine decarboxylase from Escherichia coli. Methods Enzymol. 209, 348–359 [DOI] [PubMed] [Google Scholar]
- 52. Igarashi K., Kaneda M., Yamaji A., Saido T. C., Kikkawa U., Ono Y., Inoue K., Umeda M. (1995) A novel phosphatidylserine-binding peptide motif defined by an anti-idiotypic monoclonal antibody. Localization of phosphatidylserine-specific binding sites on protein kinase C and phosphatidylserine decarboxylase. J. Biol. Chem. 270, 29075–29078 [DOI] [PubMed] [Google Scholar]
- 53. Kuge O., Saito K., Kojima M., Akamatsu Y., Nishijima M. (1996) Post-translational processing of the phosphatidylserine decarboxylase gene product in Chinese hamster ovary cells. Biochem. J. 319, 33–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Choi J. Y., Augagneur Y., Ben Mamoun C., Voelker D. R. (2012) Identification of gene encoding Plasmodium knowlesi phosphatidylserine decarboxylase by genetic complementation in yeast and characterization of in vitro maturation of encoded enzyme. J. Biol. Chem. 287, 222–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nebauer R., Schuiki I., Kulterer B., Trajanoski Z., Daum G. (2007) The phosphatidylethanolamine level of yeast mitochondria is affected by the mitochondrial components Oxa1p and Yme1p. FEBS J. 274, 6180–6190 [DOI] [PubMed] [Google Scholar]
- 56. Rontein D., Wu W. I., Voelker D. R., Hanson A. D. (2003) Mitochondrial phosphatidylserine decarboxylase from higher plants. Functional complementation in yeast, localization in plants, and overexpression in Arabidopsis. Plant Physiol. 132, 1678–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Simbeni R., Tangemann K., Schmidt M., Ceolotto C., Paltauf F., Daum G. (1993) Import of phosphatidylserine into isolated yeast mitochondria. Biochim. Biophys. Acta 1145, 1–7 [DOI] [PubMed] [Google Scholar]
- 58. Zborowski J., Dygas A., Wojtczak L. (1983) Phosphatidylserine decarboxylase is located on the external side of the inner mitochondrial membrane. FEBS Lett. 157, 179–182 [DOI] [PubMed] [Google Scholar]
- 59. Becker T., Wenz L. S., Krüger V., Lehmann W., Müller J. M., Goroncy L., Zufall N., Lithgow T., Guiard B., Chacinska A., Wagner R., Meisinger C., Pfanner N. (2011) The mitochondrial import protein Mim1 promotes biogenesis of multispanning outer membrane proteins. J. Cell Biol. 194, 387–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Meisinger C., Ryan M. T., Hill K., Model K., Lim J. H., Sickmann A., Müller H., Meyer H. E., Wagner R., Pfanner N. (2001) Protein import channel of the outer mitochondrial membrane. A highly stable Tom40-Tom22 core structure differentially interacts with preproteins, small tom proteins, and import receptors. Mol. Cell. Biol. 21, 2337–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vögtle F. N., Wortelkamp S., Zahedi R. P., Becker D., Leidhold C., Gevaert K., Kellermann J., Voos W., Sickmann A., Pfanner N., Meisinger C. (2009) Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell 139, 428–439 [DOI] [PubMed] [Google Scholar]
- 62. Witte C., Jensen R. E., Yaffe M. P., Schatz G. (1988) MAS1, a gene essential for yeast mitochondrial assembly, encodes a subunit of the mitochondrial processing protease. EMBO J. 7, 1439–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vögtle F. N., Prinz C., Kellermann J., Lottspeich F., Pfanner N., Meisinger C. (2011) Mitochondrial protein turnover. Role of the precursor intermediate peptidase Oct1 in protein stabilization. Mol. Biol. Cell 22, 2135–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gietz D., St Jean A., Woods R. A., Schiestl R. H. (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Stojanovski D., Pfanner N., Wiedemann N. (2007) Import of proteins into mitochondria. Methods Cell Biol. 80, 783–806 [DOI] [PubMed] [Google Scholar]
- 66. Fujiki Y., Hubbard A. L., Fowler S., Lazarow P. B. (1982) Isolation of intracellular membranes by means of sodium carbonate treatment. Application to endoplasmic reticulum. J. Cell Biol. 93, 97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Thornton N., Stroud D. A., Milenkovic D., Guiard B., Pfanner N., Becker T. (2010) Two modular forms of the mitochondrial sorting and assembly machinery are involved in biogenesis of α-helical outer membrane proteins. J. Mol. Biol. 396, 540–549 [DOI] [PubMed] [Google Scholar]
- 68. Truscott K. N., Voos W., Frazier A. E., Lind M., Li Y., Geissler A., Dudek J., Müller H., Sickmann A., Meyer H. E., Meisinger C., Guiard B., Rehling P., Pfanner N. (2003) A J-protein is an essential subunit of the presequence translocase-associated protein import motor of mitochondria. J. Cell Biol. 163, 707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dekker P. J., Ryan M. T., Brix J., Müller H., Hönlinger A., Pfanner N. (1998) Preprotein translocase of the outer mitochondrial membrane. Molecular dissection and assembly of the general import pore complex. Mol. Cell. Biol. 18, 6515–6524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Schägger H., von Jagow G. (1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199, 223–231 [DOI] [PubMed] [Google Scholar]
- 71. Folch J., Lees M., Sloane Stanley G. H. (1957) A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226, 497–509 [PubMed] [Google Scholar]
- 72. Broekhuyse R. M. (1968) Phospholipids in tissues of the eye. I. Isolation, characterization, and quantitative analysis by two-dimensional thin-layer chromatography of diacyl and vinyl-ether phospholipids. Biochim. Biophys. Acta 152, 307–315 [DOI] [PubMed] [Google Scholar]
- 73. Baker M. J., Frazier A. E., Gulbis J. M., Ryan M. T. (2007) Mitochondrial protein-import machinery. Correlating structure with function. Trends Cell Biol. 17, 456–464 [DOI] [PubMed] [Google Scholar]
- 74. Chacinska A., Koehler C. M., Milenkovic D., Lithgow T., Pfanner N. (2009) Importing mitochondrial proteins. Machineries and mechanisms. Cell 138, 628–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Dolezal P., Likic V., Tachezy J., Lithgow T. (2006) Evolution of the molecular machines for protein import into mitochondria. Science 313, 314–318 [DOI] [PubMed] [Google Scholar]
- 76. Neupert W., Herrmann J. M. (2007) Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76, 723–749 [DOI] [PubMed] [Google Scholar]
- 77. Schleiff E., Becker T. (2011) Common ground for protein translocation. Access control for mitochondria and chloroplasts. Nat. Rev. Mol. Cell Biol. 12, 48–59 [DOI] [PubMed] [Google Scholar]
- 78. van Wilpe S., Ryan M. T., Hill K., Maarse A. C., Meisinger C., Brix J., Dekker P. J., Moczko M., Wagner R., Meijer M., Guiard B., Hönlinger A., Pfanner N. (1999) Tom22 is a multifunctional organizer of the mitochondrial preprotein translocase. Nature 401, 485–489 [DOI] [PubMed] [Google Scholar]
- 79. Yamamoto H., Fukui K., Takahashi H., Kitamura S., Shiota T., Terao K., Uchida M., Esaki M., Nishikawa S., Yoshihisa T., Yamano K., Endo T. (2009) Roles of Tom70 in import of presequence-containing mitochondrial proteins. J. Biol. Chem. 284, 31635–31646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gakh O., Cavadini P., Isaya G. (2002) Mitochondrial processing peptidases. Biochim. Biophys. Acta 1592, 63–77 [DOI] [PubMed] [Google Scholar]
- 81. Mossmann D., Meisinger C., Vögtle F. N. (2012) Processing of mitochondrial presequences. Biochim. Biophys. Acta 1819, 1098–1106 [DOI] [PubMed] [Google Scholar]
- 82. Frazier A.E., Chacinska A., Truscott K. N., Guiard B., Pfanner N., Rehling P. (2003) Mitochondria use different mechanisms for transport of multispanning membrane proteins through the intermembrane space. Mol. Cell. Biol. 23, 7818–7828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Schuiki I., Schnabl M., Czabany T., Hrastnik C., Daum G. (2010) Phosphatidylethanolamine synthesized by four different pathways is supplied to the plasma membrane of the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1801, 480–486 [DOI] [PubMed] [Google Scholar]