Background: Wnt and Notch signaling pathways belong to a highly conserved network that controls gene expression.
Results: Wnt5a enhances Notch1 signaling through down-regulation of SMRT by Ca2+/calmodulin-dependent protein kinase II (CaMKII).
Conclusion: CaMKII is crucial for the regulation between Notch and Wnt5a signaling.
Significance: The findings of this study may begin to shed some light onto what may be a signal cross-talk mechanism of Notch1 signaling and the CaMKII.
Keywords: CaMKII, Corepressor Transcription, Notch, Proteasome, Ubiquitination, Wnt Signaling
Abstract
Serine-threonine Ca2+/calmodulin-dependent protein kinase II (CaMKII) is the key component in noncanonical Wnt5a signaling and has been shown to regulate its signaling. In this study, we found that CaMKII induced by Wnt5a remarkably reduced the protein stability of the silencing mediator of retinoic acid and thyroid hormone receptor (SMRT), a co-repressor of Notch signaling, through proteasomal degradation. Wnt5a was found to enhance Notch1 intracellular domain (Notch1-IC) transcription activity, which could be inhibited by treatment with KN93, a CaMKII inhibitor. The kinase activity of CaMKII was essential for the activation of Notch signaling. We also determined that CaMKII could enhance the association between Notch1-IC and RBP-Jk. Furthermore, the physical association between RBP-Jk and SMRT was substantially suppressed by CaMKII. We demonstrated that CaMKII directly bound and phosphorylated SMRT at Ser-1407, thereby facilitating SMRT translocation from the nucleus to the cytoplasm and proteasome-dependent degradation. These results suggest that CaMKII down-regulated the protein stability of SMRT through proteasomal degradation.
Introduction
Notch proteins participate in conserved pathways that regulate cell fate determination, cell differentiation, cell proliferation, and cell death (1–4). Mammalian Notch receptors (Notch1–Notch4)3 are cell-surface type I transmembrane proteins that harbor a large extracellular domain involved in ligand binding and a cytoplasmic domain involved in signal transduction (5–7). The Notch1 receptor is processed in the trans-Golgi network by proteases of the furin (S1 cleavage) during transport to the cell surface, where it is expressed in heterodimeric form (8). At the cell surface, Notch can interact with ligands, Jagged and Delta. The transmembrane C-terminal fragment of Notch is generated via proteolytic cleavage (S2 cleavage), which subsequently cleaves the Notch fragment by γ-secretase (S3 cleavage), an enzyme complex that contains presenilin (PS1), nicastrin (NCT), anterior pharynx defective (APH-1), and presenilin enhancer-2 (PEN-2). This process then results in the release of the Notch intracellular domain (Notch-IC) (9–23). Notch1-IC translocates to the nucleus and interacts with various transcriptional activation complexes including CSL (CBF1/RBP-Jk, suppressor of hairless, Lag-1), coactivator protein MAML-1 (mastermind-like-1), and p300/CBP (CREB-binding protein), which possess histone acetyltransferase activity, leading to transcriptional activation of downstream target genes such as those encoding Hes1, Hes5, Hes7, Hey1, Hey2, HeyL, and Cyclin D (24–34). In the absence of Notch-IC, CSL recruits corepressor factors, including SKIP (Ski-interacting protein), SMRT (silencing mediator for retinoid and thyroid hormone receptors), N-CoR (nuclear receptor co-repressor), and histone deacetylases, resulting in the formation of a transcriptional repressor complex (6, 35, 36).
The SMRT corepressor participates in the repression of target gene expression through a variety of transcription factors, including the nuclear hormone receptors and promyelocytic leukemia zinc finger protein (37–39). The ability of SMRT to associate with these transcription factors and thereby to mediate repression is strongly inhibited by activation of tyrosine kinase signaling pathways, including pathways regulated by the epidermal growth factor receptor (40). Recent studies have emphasized the role of ubiquitination in regulating SMRT stability. Phosphorylation of SMRT by MEKK-1, Cdk2, and Pin1 induces conformational changes and reduces its binding affinity for transcription factors that promote its degradation (41, 42). Phosphorylation of SMRT by IKKα has an important meaning for the derepression of NF-κB and Notch target genes (43). Recent studies have emphasized the role of ubiquitination in regulating SMRT stability. The SMRT phosphorylation-dependent ubiquitination and degradation is an important negative feedback regulation that specifically terminates SMRT activation.
Wnt5a is a member of the Wnt family of secreted growth factor, which are 38–45 kDa secreted cysteine-rich proteins with hydrophobic signal peptides (44). Wnt5a is a highly conserved diffusible protein whose signal is transduced by Frizzled (Fz) receptors. In addition to Fzs, receptor tyrosine kinase-like orphan receptor 1/2 (Ror1/2), which is a single-pass transmembrane receptor with a tyrosine kinase domain, functions as a co-receptor (44, 45). However, the effect of Fzs and Ror1/2 on Wnt5a signaling has not yet been examined. Wnt can be broadly divided into three signaling pathways: the canonical Wnt/β-catenin pathway, the noncanonical Wnt/Ca2+ pathway, and the noncanonical Wnt/planar cell polarity pathway (46, 47). Wnt5a is a noncanonical pathway member of the Wnt family. Wnt5a is expressed in various adult tissues and are involved in a wide range of cellular processes. Signal transduction events through β-catenin-independent noncanonical Wnt5a elicits intracellular release of Ca2+, thereby activating the Ca2+/calmodulin-dependent protein kinase II (CaMKII), mitogen-activated protein kinase (MAPK), protein kinase C (PKC), and calpain to regulate cellular functions including cell migration (48–54). CaMKII is the key component in Wnt5a signaling and has been shown to regulate Wnt5a signaling. CaMKII is a multifunctional serine (Ser)/threonine (Thr) kinase that plays an important role in the regulation of cell growth, cytoskeletal organization, regulation of synaptic transmission, modulation of ion channel activity, learning, and memory (55–58). CaMKII is important for prostate cancer cell survival and promotes their progression to an androgen-independent state (59). Furthermore, a recent study has shown that calcium/calmodulin-dependent kinase II regulates notch-1 signaling in prostate cancer cells (60). Wnt5a is transcribed based on multiple mechanisms, such as NF-κB, Hedgehog, TGF-β, and Notch signaling cascades (61). Wnt5a expression in the human cells was induced by coculture and was blocked by γ-secretase inhibition. Likewise, stimulation of Notch signaling by immobilized Jagged-1 promoted Wnt5a expression (62). Wnt5a acts as an essential downstream mediator of Notch-CSL signaling, which is dependent on expression in the keratinocyte compartment of FoxN1, a gene that plays a key role in hair follicle regulatory function (63). Despite these observations, the precise mechanisms underlying the relationship between the Notch1 and Wnt5a-CaMKII signaling pathways are still largely unknown.
In this study, we demonstrated that Wnt-5-induced CaMKII enhances the Notch1 signaling pathway through the down-regulation of SMRT protein stability. CaMKII was shown to phosphorylate SMRT, which disrupts the SMRT-RBP-Jk transcription complex through SMRT protein degradation and polyubiquitination. In addition, the dissociation of SMRT from RBP-Jk provoked by CaMKII activates Hes1 and Hes5 transcriptional activity. The results of this study suggest that CaMKII is crucial for the regulation between Notch and Wnt signaling.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
Human embryonic kidney (HEK) 293 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin (100 units/ml), and streptomycin (100 μg/ml). Along with the HEK293 cells, PS1/2+/+ MEF cells and PS1/2−/− MEF cells were cultured in DMEM supplemented with 10% FBS, 100 units/ml of penicillin, and 100 μg/ml of streptomycin. For plasmid DNA transfection, cells were plated at a density of 2 × 106 cells/100-mm dish, grown overnight, and transfected with the appropriate expression vectors in the presence of the indicated combinations of plasmid DNAs via the calcium phosphate and liposome methods (64).
Luciferase Reporter Assay
Human embryonic kidney 293 cells were co-transfected with 4XCSL-Luc (a 4-time repeating section of the RBP-Jk target sequence, CGTGGGAA, with the luciferase gene), Hes1-Luc, Hes5-Luc, mutating the RBP-Jk binding site of Hes5-Luc, and β-galactosidase coupled with the indicated vector constructs. 48 h after transfection, the cells were lysed in chemiluminescence lysis buffer (18.3% 1 m K2HPO4, 1.7% 1 m KH2PO4, 1 mm phenylmethylsulfonyl fluoride (PMSF), and 1 mm dithiothreitol) and luciferase activity was assayed using a luciferase assay kit (Promega). The activity of the luciferase reporter protein in the transfected cells was normalized to β-galactosidase activity in the same cells (65). These results represent the means ± average deviation of three independent experiments.
Protein Accumulation Assay
For the protein accumulation assay, the cells were seeded at a density of 50–60% confluence and incubated overnight. 48 h after transfection, cells were treated with the proteasomal inhibitor MG-132 (Sigma). The MG-132 concentrations in the dosage assay were 0, 1 and 5 μm and the treatment time was 6 h. Protein levels were analyzed via immunoblotting (65).
Protein Stability Assay
For the protein stability assay, the cells were seeded at a density of 50–60% confluence and incubated overnight. 48 h after transfection, the cells were treated with 0.1 mm cycloheximide for 0, 0.5, 1, 2, 4, or 6 h to block the synthesis of new proteins. The cells were collected at each time point and the total lysates were then lysed in RIPA/Laemmli buffer. Protein levels of endogenous Notch1 were determined by immunoblotting with the indicated antibody (65).
In Vitro Binding Assay
The recombinant GST-SMRT proteins were expressed in Escherichia coli strain BL21, using the pGEX system as indicated (66). The GST-SMRT proteins were then purified using glutathione-agarose beads (Sigma), in accordance with the manufacturer's instructions. Equal amounts of GST or GST-SMRT fusion proteins were incubated with the lysates of HEK293 cells, which had been transfected for 3 h with combinations of expression vectors at 4 °C, with rotation. After incubation, the beads were washed three times with ice-cold PBS, and boiled with 20 μl of Laemmli sample buffer. The precipitates were separated via SDS-PAGE, and the pull-down proteins were detected via immunoblotting with specific antibodies.
Co-immunoprecipitation Assays
The cells were lysed in 1 ml of RIPA buffer for 30 min at 4 °C. After 20 min of centrifugation at 12,000 × g, the supernatants were subjected to immunoprecipitation with the appropriate antibodies coupled to protein A-agarose beads. The resulting immunoprecipitates were then washed three times in phosphate-buffered saline (PBS, pH 7.4). Laemmli sample buffer was then added to the immunoprecipitated pellets. The pellets were heated for 5 min at 95 °C and analyzed via SDS-PAGE. Western blotting was conducted with the indicated antibodies.
Western Blot Analysis
The cultured cells were harvested and lysed in RIPA buffer (50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mm PMSF, 1 mm DTT, and 2 μg/ml of leupeptin and aprotinin for 30 min. The cell lysates were subjected to 20 min of centrifugation at 12,000 × g and 4 °C. The resultant soluble fraction was then boiled in Laemmli buffer and subjected to SDS-PAGE. After gel electrophoresis, the separated proteins were transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore) via electroblotting. The membranes were then blocked with Tris-buffered saline solution (pH 7.4) containing 0.1% Tween 20 and 5% nonfat milk. The blotted proteins were probed with the indicated antibodies.
Antibody List
The blotted proteins were probed using anti-Myc antibody (9E10), anti-HA (12CA5) antibody, anti-Flag M2 (Sigma), anti-Notch1-IC (Cell Signaling), anti-RBP-Jk (Santa Cruz), anti-CaMKII (Santa Cruz), anti-SMRT (Santa Cruz), anti-Presenilin1 (D39D1) rabbit mAb, anti-Presenilin 2 (D30G3) rabbit mAb (Cell Signaling Technology) followed by incubation with anti-mouse horseradish peroxidase-conjugated secondary antibodies (Amersham Biosciences, Inc.). The blots were then developed using an enhanced chemiluminescence (ECL) system (Pierce).
Site-directed Mutagenesis
The site-directed mutagenesis of cDNA encoding for SMRT was performed using a QuikChange kit (Stratagene), and the mutagenic primer was S1407A (5′-GGGGACCGGCCACCCgCTGTCTCCTCAGTGCAC-3′) and S1434A (5′-GAGGACAGGCCCTCGgCCGCAGGTTCCACGCC-3′) (mismatches with the SMRT cDNA template are indicated by lowercase letters). The mutations were verified by automatic DNA sequencing.
Immunocomplex Kinase Assay
The cultured cells were harvested and lysed in buffer A, containing 50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1 mm phenylmethylsulfonyl fluoride (PMSF), 2 μg/ml of leupeptin, 2 μg/ml of aprotinin, 25 mm glycerophosphate, 0.1 mm sodium orthovanadate, 1 mm sodium fluoride, 1% Nonidet P-40, 0.5% deoxycholate, and 0.1% SDS for 30 min at 4 °C. The cell lysates were then subjected to centrifugation at 12,000 × g and 4 °C for 20 min. The soluble fraction was incubated for 3 h with the appropriate antibodies against the indicated protein kinase at 4 °C. The immunocomplexes were then coupled to protein A-agarose during an additional hour of incubation at 4 °C, after which they were pelleted via centrifugation. The immunopellets were rinsed three times with lysis buffer and then twice with 20 mm HEPES (pH 7.4). The immunocomplex kinase assays were conducted by incubating the immunopellets for 30 min at 30 °C with 2 μg of substrate protein in 20 μl of reaction buffer that contained 0.2 mm sodium orthovanadate, 10 mm MgCl2, 2 μCi of [32P]ATP, 20 mm HEPES (pH 7.4). The phosphorylated substrates were separated by SDS-PAGE and quantified using a Fuji FLA7000 PhosphorImager. The GST fusion proteins that were used as substrates were expressed in E. coli using pGEX-4T (Pharmacia) and purified using glutathione-Sepharose, as described previously (66, 67). The protein concentrations were determined by the Bradford method (Shimadzu).
Immunofluorescence Staining
Assays were conducted as previously described with HEK293 cells plated at a density of 1 × 105 cells per well onto coverslips (Fisher). Cells were transfected with a total of 0.5 mg of the appropriate DNA per well using the Lipofectamine Plus reagent (Invitrogen). The transfected cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS), and then permeabilized with 0.1% Triton X-100 in PBS. Cells were blocked in 1% BSA in PBS. Anti-HA or anti-Flag M2 antibodies (Sigma) were employed as the primary antibodies at a dilution of 1:100, washed three times in PBS. Alexa Fluor 488 (Invitrogen) or Alexa Fluor 555 (Invitrogen) conjugated anti-mouse secondary antibody (1:100) was added, and then the DNA dye ToPro-3 was used for nuclear localization. The stained cells were evaluated for localization via confocal microscopy (Leica TCS SPE). Each image consisted of a single Z section at the same cellular level. The final images were obtained using a confocal microscopy and analyzed with LAS AF software (Leica) and at least 300 cells were counted for each experimental condition. Scale bars represent 25 μm as indicated (68).
Preparation of Cytosolic and Nuclear Fractions
The cells were rinsed with ice-cold PBS, and harvested via 5 min of centrifugation at 3,000 × g and 4 °C. The dispersed cells were then homogenized with buffer A (10 mm HEPES, pH 7.9, 10 mm KCl, 0.5 mm EDTA, 1 mm DTT, and 0.5 mm PMSF). After 15 min on ice, 10% Nonidet P-40 was added and vortexed vigorously for 10 s. The resultant supernatant was then used as a cytosolic fraction, via 1–2 min of centrifugation at 13,000 × g and 4 °C. The pellet was then homogenized with buffer B (20 mm HEPES, pH 7.9, 0.4 m NaCl, 1 mm EDTA, 1 mm DTT, and 1 mm PMSF). After 10 min of vigorous vortexing, the homogenates were centrifuged for 10 min at 13,000 × g at 4 °C. The resultant supernatants were then used as nuclear fractions. Two fractions were quantified using the Bradford method and 20 μg of the each fraction were analyzed by SDS-PAGE (66).
RESULTS
Wnt5a Enhances the Transcriptional Activity of Notch1 Target Genes through CaMKII
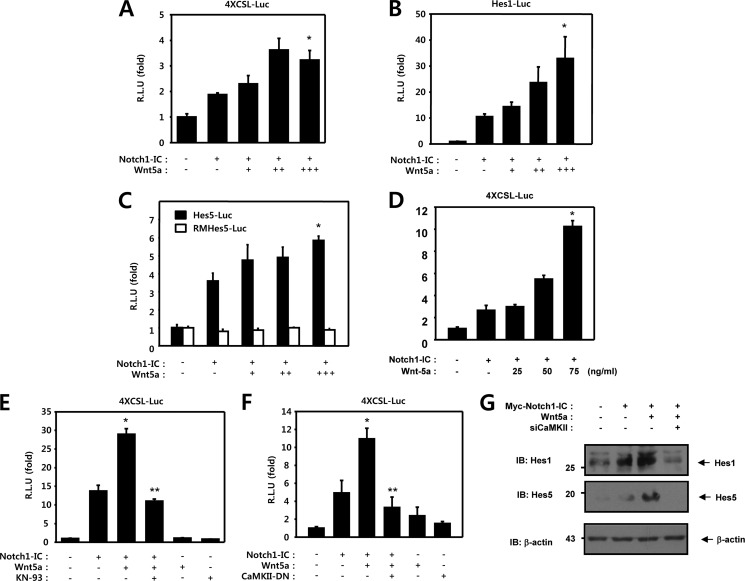
To determine whether Wnt5a is involved in regulating the transcriptional activation of Notch1 target genes, a reporter assay was performed with HEK293 cells using luciferase reporter genes. In this study, three types of luciferase reporter genes under the control of the Hes1 promoter, the Hes5 promoter, and the artificial four time repeat of RBP-Jk binding sequence were evaluated. We investigated the effect of Wnt5a on Notch1-IC transcriptional activity. HEK293 cells were transfected with 4XCSL-Luc and either the Notch1-IC or an empty vector. As expected, Notch1-IC mediated transcription activity was increased in these samples (Fig. 1A). The Notch1-IC-induced 4XCSL luciferase reporter activity was enhanced by co-transfection with Wnt5a in a dose-dependent manner (Fig. 1A). The bHLH proteins Hes1 and Hes5, both of which contain multiple CSL binding DNA sequences on their promoters, were identified as essential targets of Notch in epithelial cells. Therefore, we have confirmed the effects of Wnt5a on the Notch1 signaling pathway, using the Hes1, Hes5, and RMHes5 reporter systems, respectively. RMHes5-Luc was constructed by mutating the RBP-J binding site (−79 to −72: tgtggaa) of Hes5-Luc to tgtgctga (69). Expression of Notch1-IC was determined to significantly induce activation of the Hes1 and Hes5 reporter systems (Fig. 1, B and C). Wnt5a enhanced Notch1-IC induced natural Hes1 and Hes5 promoter transcriptional activity in a dose-dependent manner, but not RMHes5 activity (Fig. 1, B and C). Moreover, The Notch1-IC-induced 4XCSL luciferase reporter activity was enhanced by purified Wnt5a in a dose-dependent manner (Fig. 1D). Based on these results, it was assumed that Wnt5a can activate the expression of the Notch1 target genes.
FIGURE 1.
Wnt5a enhances the transcriptional activity of Notch1 target genes through CaMKII. A–C, HEK293 cells were transfected with 50 ng of 4XCSL-Luc (A), 50 ng of Hes1-Luc (B), 50 ng of Hes5-Luc and RMHes5-Luc (C), and 50 ng of β-galactosidase, along with 100 ng of Notch1-IC and 50 ng (+), 100 ng (++), and 150 ng (+++) of Wnt5a, as indicated. *, p < 0.05 versus Notch1-IC. D, HEK293 cells were transfected with 50 ng of 4XCSL-Luc and 50 ng of β-galactosidase, along with 100 ng of Notch1-IC. Before harvest, HEK293 cells were treated with 25, 50, and 100 ng/ml of Wnt5a protein for 8 h, as indicated. *, p < 0.05 (C) and p < 0.01 (D) versus Notch1-IC. E, HEK293 cells were transfected for 48 h with 50 ng of 4XCSL-Luc, 100 ng of Notch1-IC, 150 ng of Wnt5a, and 50 ng of β-galactosidase. Before harvest, HEK293 cells were treated with KN-93 (5 μm) for 6 h, the cells were lysed, and the luciferase activity was determined. *, p < 0.05 versus Notch1-IC; **, p < 0.05 versus Notch1-IC + Wnt5a. F, HEK293 cells were transfected with 50 ng of 4XCSL-Luc and β-galactosidase, along with 100 ng of Notch1-IC, 150 ng of Wnt5a, and 100 ng of CaMKII-DN, as indicated. 48 h after transfection, the cells were lysed, and the luciferase activity was determined. DN, dominant-negative. *, p < 0.05 versus Notch1-IC; **, p < 0.05 versus Notch1-IC + Wnt5a. G, HEK293 cells were transfected with 0.5 μg of Myc-Notch1-IC, 0.5 μg of Wnt5a, and 0.5 μg of siCaMKII as indicated. 48 h after transfection, the cell lysates were immunoblotted (IB) with anti-Hes1 and anti-Hes5 antibodies. Probing with an antibody to β-actin was used as a loading control. R.L.U., relative luciferase units.
To delineate the possible role of CaMKII in the regulation of Notch1 signaling by Wnt5a in intact cells, we assessed the effects of the CaMKII inhibitor and dominant-negative CaMKII on Notch signaling. We determined that the KN-93 or dominant-negative CaMKII was able to block the effects of Wnt5a on Notch1-IC-induced transcriptional activity (Fig. 1, E and F). From these results, it was assumed that Wnt5a can increase the expression of the Notch1 target gene through up-regulation of CaMKII. To demonstrate this assumption, Western blots were performed for endogenous Hes1 and Hes5. The HEK293 cells were transfected with Myc-Notch1-IC, Wnt5a, or siCaMKII. The expression levels of the Hes1 and Hes5 proteins were increased by Wnt5a, as expected, and the Wnt5a-induced expressions of Hes1 and Hes5 were decreased via coexpression of siCaMKII (Fig. 1G). Therefore, we hypothesized that CaMKII plays a role in up-regulation of the Notch1 target gene transcriptional activity.
CaMKII Activates Notch1 Transcriptional Activity
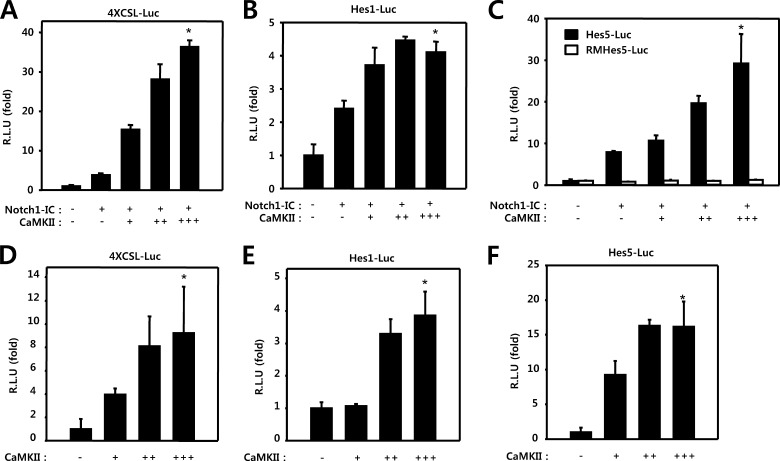
We attempted to determine whether or not CaMKII plays a role in the regulation of Notch1-IC mediated transcriptional activity. HEK293 cells were transfected with 4XCSL-Luc and either Notch1-IC or CaMKII. In these experiments, CaMKII was shown to activate Notch1-IC to stimulate transcription in a dose-dependent manner (Fig. 2A). We have also confirmed the effects of CaMKII on the Notch1 signaling pathway, using the Hes1 and Hes5 reporter systems. Expression of the active form of Notch1 was determined to induce activation of the Hes1 and Hes5 reporter systems. CaMKII expression enhanced Notch1-IC-induced natural Hes1 and Hes5 promoter transcriptional activities, but not RMH5 activity (Fig. 2, B and C).
FIGURE 2.
CaMKII activates Notch1 transcriptional activity. A–C, HEK293 cells were transfected with 50 ng of 4XCSL-Luc (A), 50 ng of Hes1-Luc (B), 50 ng of Hes5-Luc and RMHes5-Luc (C), and 50 ng of β-galactosidase, along with 100 ng of Notch1-IC and 50 ng (+), 100 ng (++) and 150 ng (+++) of CaMKII, as indicated. D–F, HEK293 cells were transfected with 50 ng of 4XCSL-Luc (D), 50 ng of Hes1-Luc (E), 50 ng of Hes5-Luc (F), and 50 ng of β-galactosidase, along with 50 (+), 100 (++), and 150 ng (+++) of CaMKII, as indicated. 48 h after transfection, the cells were lysed, and the luciferase activity was determined. *, p < 0.01 (A and C) or p < 0.05 (B) versus Notch. D–F, *, p < 0.05 versus control. R.L.U., relative luciferase units.
To assess the role of CaMKII in the basal level of Notch1 signaling, we performed a transcription reporter assay. HEK293 cells were transfected with 4XCSL-Luc and CaMKII. The transcriptional activity of endogenous Notch1-IC in HEK293 cells was enhanced by CaMKII in a dose-dependent manner (Fig. 2D). Similar results were also obtained using the Hes1-Luc and Hes5-Luc reporter systems (Fig. 2, E and F). However, the absolute value of three reporter systems was higher in Notch1-IC and CaMKII cotransfected cells than in cells transfected with only CaMKII, suggesting that Notch1-IC and CaMKII produce synergic effects on reporter activity. Thus, taken together, our results indicate that CaMKII has a positive role in the regulation of Notch1 transcriptional activity.
CaMKII Activates Notch1 Transcriptional Activity through RBP-Jk-dependent and Notch1-IC-independent Manner
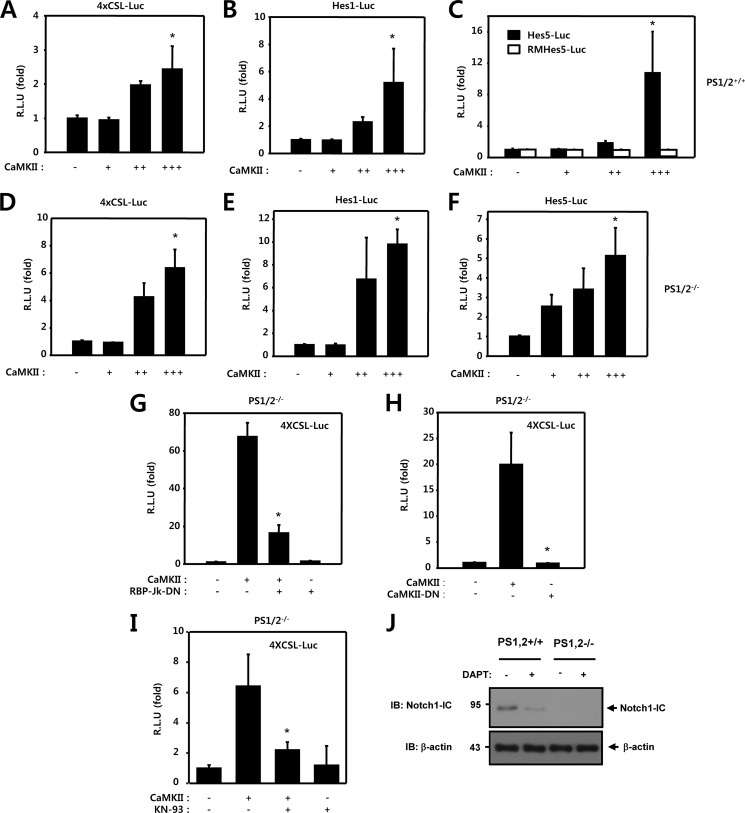
To determine whether CaMKII regulates the transcriptional activation of Notch1 target genes through a Notch1-IC dependent mechanism, we performed a transcription reporter assay using PS1 and PS2 wild-type (PS1/2+/+) and PS1 and PS2-deficient (PS1/2−/−) MEF cells. We investigated the effect of CaMKII on the basal level of Notch1 transcriptional activity. PS1/2+/+ and PS1/2−/− MEF cells were transfected with either 4XCSL-Luc or CaMKII. In these experiments, CaMKII was shown to activate the basal level of Notch1 reporter activity both in PS1/2+/+ and PS1/2−/− MEF cells (Fig. 3, A and D). Similar results were also observed when using the Hes1-Luc, Hes5-Luc, and RMHes5-Luc reporter systems in both PS1/2+/+ and PS1/2−/− MEF cells (Fig. 3, B, C, E, and F). These data demonstrate that CaMKII could activate the Notch1 reporter activity despite the absence of Notch1-IC generation, suggesting that CaMKII, at least in part, could regulate Notch1 target gene expression in a Notch1-IC independent manner. However, the absolute value of three reporter systems was higher in PS1/2+/+ MEF cells than PS1/2−/− MEF cells, suggesting that CaMKII could modulate Notch1 activity through a Notch1-IC-dependent and -independent manner.
FIGURE 3.
CaMKII activates Notch1 transcriptional activity through RBP-Jk-dependent and Notch1-IC-independent manner. A–C, PS1/2+/+ MEF cells were transfected with 50 ng of 4XCSL-Luc (A), 50 ng of Hes1-Luc (B), 50 ng of Hes5-Luc and RMHes5-Luc (C), and 50 ng of β-galactosidase, along with 50 ng (+), 100 ng (++), and 150 ng (+++) of CaMKII, as indicated. *, p < 0.05 versus control. D–F, PS1/2−/− MEF cells were transfected with 50 ng of 4XCSL-Luc (D), 50 ng of Hes1-Luc (E), 50 ng of Hes5-Luc (F), and 50 ng of β-galactosidase, along with 50 ng (+), 100 ng (++), and 150 ng (+++) of CaMKII, as indicated. *, p < 0.05 versus control. G, PS1/2−/− MEF cells were transfected with 50 ng of 4XCSL-Luc and 50 ng of β-galactosidase, along with 100 ng of CaMKII and 100 ng of RBP-Jk-DN as indicated. *, p < 0.01 versus CaMKII. H, PS1/2−/− MEF cells were transfected with 50 ng of 4XCSL-Luc and 50 ng of β-galactosidase, along with 100 ng of CaMKII and 100 ng of CaMKII-KD with 4XCSL-Luc. *, p < 0.01 versus CaMKII. I, PS1/2−/− cells were transfected with 50 ng of 4XCSL-Luc and 50 ng of β-galactosidase, along with 100 ng of CaMKII. 42 h after transfection, HEK293 cells were treated with KN-93 (5 μm) for 6 h and the luciferase activity was determined. *, p < 0.05 versus CaMKII. A–I, 48 h after transfection, the cells were lysed, and luciferase activity was determined. DN, dominant-negative. J, immunoblots of PS1/2+/+ and PS1/2−/− MEF cells treated with 1 μm N-[(3,5-difluorophenyl)acetyl]-l-alanyl-2-phenylglycine-1,2-dimethylethyl ester (DAPT) for 6 h. The cell lysates were immunoblotted (IB) with anti-Notch1-IC antibodies. Probing with an antibody to β-actin was used as a loading control. R.L.U., relative luciferase units.
To investigate whether the transcriptional activation of endogenous Notch1 target genes by CaMKII occurs through a RBP-Jk-dependent manner, we performed a transcription reporter assay using the dominant-negative form of RBP-Jk (RBP-Jk-DN). In the luciferase reporter gene assay with PS1/2−/− MEF cells, RBP-Jk-DN was transfected and the effect of this transfection on the transcriptional inhibition of Notch1-IC target genes was then assessed using 4XCSL-Luc. The transcriptional activity of endogenous Notch1-IC was activated by CaMKII but was inhibited by co-transfection with RBP-Jk-DN, suggesting that CaMKII activates Notch1 transcriptional activity through a RBP-Jk-dependent mechanism (Fig. 3G). To investigate whether the kinase activity of CaMKII is necessary for up-regulation of the transcriptional activity of Notch1 target genes, we used a dominant-negative, kinase-deficient CaMKII mutant (CaMKII-K42M, called CaMKII-DN) to block the kinase activity of CaMKII. In the luciferase reporter gene assay with PS1/2−/− MEF cells, CaMKII-DN was transfected instead of CaMKII, and the effect of this transfection on the transcriptional activation of endogenous Notch1-IC target genes was assessed using 4XCSL-Luc. The transcriptional activity of endogenous Notch1-IC was enhanced by CaMKII but was not enhanced by transfection with the dominant-negative CaMKII (Fig. 3H). We investigated the effect of CaMKII-DN on basal Notch1-IC transcriptional activity with 4CSL-Luc in PS1/2−/− MEF cells. Basal Notch1-IC transcriptional activity was decreased by CaMKII-DN in a dose-dependent manner (data not shown). Similar results were also observed using the 4XCSL-Luc reporter system with KN-93, a CaMKII chemical inhibitor. The CaMKII-induced 4XCSL luciferase reporter activity was inhibited by treating PS1/2−/− MEF cells with KN-93 (Fig. 3I). To determine the role of endogenous Notch1-IC in PS1/2+/+ and PS1/2−/− MEF cells was further evaluated by the knockdown of Notch1 using N-[(3,5-difluorophenyl)acetyl]-l-alanyl-2-phenylglycine-1,2-dimethylethyl ester, which is a γ-secretase inhibitor. These results confirmed that expression of the Notch1-IC protein level was reduced by N-[(3,5-difluorophenyl)acetyl]-l-alanyl-2-phenylglycine-1,2-dimethylethyl ester in the PS1/2+/+ MEF cells (Fig. 3J). PS1 and PS2 expression levels in PS1/2+/+ and PS1/2−/− MEF cells were tested using specific antibodies (supplemental Fig. S1). Thus, up-regulation of the Notch1-IC activity by CaMKII could be dependent on the intact kinase activity of CaMKII through RBP-Jk-dependent and Notch1-IC-independent mechanisms.
CaMKII Modulates the Binding of RBP-Jk to Notch1-IC or SMRT
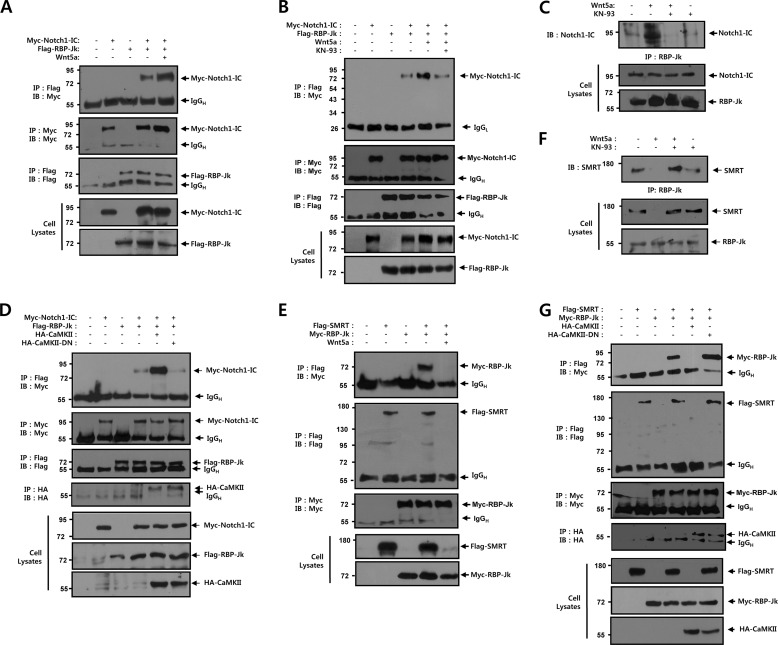
To observe the effects of Wnt5a on the molecular interactions between Notch1-IC and RBP-Jk, co-immunoprecipitation was performed in HEK293 cells that had been co-transfected with Myc-tagged Notch1-IC, Flag-tagged RBP-Jk, and Wnt5a. Immunoprecipitation was performed on cell lysates using an anti-Flag antibody, and immunoblotting was performed using the anti-Myc antibody. Notch1-IC and RBP-Jk were co-immunoprecipitated, but when they were co-transfected with Wnt5a, the band intensity corresponding to the interaction between Notch1-IC and RBP-Jk was enhanced (Fig. 4A). We evaluated the physical association between Notch1-IC and RBP-Jk by Wnt5a in immunocomplexes that were collected using anti-Myc antibody. These results confirmed that the interaction between Notch1-IC and RBP-Jk was enhanced in the presence of Wnt5a. In the presence of KN-93, the association between Notch1-IC and RBP-Jk by Wnt5a was reduced, suggesting that CaMKII kinase activity might play an important role in the Notch1-IC-RBP-Jk interaction (Fig. 4B). We next evaluated the effects of KN-93 on the association of endogenous Notch1-IC and RBP-Jk by Wnt5a in intact cells. In these experiments, HEK293 cells were transiently transfected with Wnt5a. 42 h after transfection, cells were treated with exogenous KN-93 for 6 h, and then the affinity between these two proteins was measured. Wnt5a facilitated the physical association between Notch1-IC and RBP-Jk and this binding was substantially suppressed by KN-93 treatment, suggesting that Wnt5a induced CaMKII activation plays a critical role in the endogenous Notch1-IC-RBP-Jk binding (Fig. 4C). To further assess whether CaMKII plays a positive role in Notch1-IC-RBP-Jk binding, co-immunoprecipitation was performed in HEK293 cells with co-transfection of Myc-tagged Notch1-IC, Flag-tagged RBP-Jk, HA-tagged CaMKII, and HA-tagged CaMKII-DN. In the presence of CaMKII, the interaction between Notch1-IC and RBP-Jk was higher than in the absence of CaMKII transfection and had no effect on CaMKII-DN (Fig. 4D).
FIGURE 4.
CaMKII modulates the binding of RBP-Jk to Notch1-IC or SMRT. A, HEK293 cells were transfected for 48 h with the indicated combinations of 0.5 μg of Myc-Notch1-IC, 0.5 μg of Flag-RBP-Jk, and 0.5 μg of Wnt5a. Cell lysates were subjected to immunoprecipitation (IP) with anti-Flag antibody and the immunoprecipitates were immunoblotted with anti-Myc antibody. The cell lysates were also subjected to immunoblot analysis with anti-Myc and anti-Flag antibodies, respectively. B, HEK293 cells were transfected for 42 h with the indicated combinations of 0.5 μg of Myc-Notch1-IC, 0.5 μg of Flag-RBP-Jk, and 0.5 μg of Wnt5a. Before harvest, cells were treated with KN-93 (4 μm) for 6 h. The cell lysates were subjected to immunoprecipitation with anti-Myc antibody and the immunoprecipitates were immunoblotted with anti-Flag antibody. The cell lysates were also subjected to immunoblot analysis with anti-Flag and anti-Myc antibodies, respectively. C, HEK293 cells were transfected for 42 h with 0.5 μg of Wnt5a. Before harvest, HEK293 cells were treated with KN-93 (5 μm) for 6 h. Cell lysates were subjected to immunoprecipitation with anti-RBP-Jk antibody and the immunoprecipitates were immunoblotted with anti-Notch1-IC antibody. The cell lysates were also subjected to immunoblot analysis with anti-Notch1-IC and anti-RBP-Jk antibodies, respectively. D, HEK293 cells were transfected for 48 h with the indicated combinations of 0.5 μg of Myc-Notch1-IC, 0.5 μg of Flag-RBP-Jk, 0.5 μg of HA-CaMKII, and 0.5 μg of HA-CaMKII-DN. Cell lysates were subjected to immunoprecipitation with anti-Flag antibody and the immunoprecipitates were immunoblotted with anti-Myc antibody. The cell lysates were also subjected to immunoblot analysis with anti-Myc anti-HA and anti-Flag antibodies, respectively. E, HEK293 cells were transfected for 48 h with the indicated combinations of 0.5 μg of Flag-SMRT, 0.5 μg of Myc-RBP-Jk, and 0.5 μg of Wnt5a. Cell lysates were subjected to immunoprecipitation with anti-Flag antibody and the immunoprecipitates were immunoblotted with anti-Myc antibody. The cell lysates were also subjected to immunoblot analysis with anti-Myc and anti-Flag antibodies, respectively. F, HEK293 cells were transfected for 42 h with 0.5 μg of Wnt5a. Before harvest, HEK293 cells were treated with KN-93 (5 μm) for 6 h. Cell lysates were subjected to immunoprecipitation with anti-RBP-Jk antibody and the immunoprecipitates were immunoblotted with anti-SMRT antibody. The cell lysates were also subjected to immunoblot analysis with anti-SMRT and anti-RBP-Jk antibodies, respectively. G, HEK293 cells were transfected for 48 h with the indicated combinations of 0.5 μg of Flag-SMRT, 0.5 μg of Myc-RBP-Jk, 0.5 μg of HA-CaMKII, and 0.5 μg of HA-CaMKII-DN. Cell lysates were subjected to immunoprecipitation with anti-Flag antibody and the immunoprecipitates were immunoblotted with anti-Myc antibody. The cell lysates were also subjected to immunoblot analysis with anti-Flag anti-HA and anti-Myc antibodies, respectively. A, B, D, E, and G, cell lysates were subjected to immunoprecipitation with anti-Myc, anti-Flag, and anti-HA antibody and immunoprecipitates were immunoblotted with anti-Myc, anti-Flag, and anti-HA antibody, respectively. DN, dominant-negative; IgGH, immunoglobulin heavy chain; IgGL, immunoglobulin light chain; IB, immunoblot.
To evaluate the effects of Wnt5a on the molecular interactions between SMRT and RBP-Jk, co-immunoprecipitation was performed in HEK293 cells that had been co-transfected with Flag-tagged SMRT, Myc-tagged RBP-Jk, and Wnt5a. SMRT and RBP-Jk were co-immunoprecipitated, but when they were co-transfected with Wnt5a, the band corresponding to the SMRT-RBP-Jk complex disappeared (Fig. 4E). We also examined the effects of KN-93 on the association of endogenous RBP-Jk and SMRT by Wnt5a in intact cells. In these experiments, HEK293 cells were transiently transfected with Wnt5a. 42 h after transfection, the cells were treated with exogenous KN-93 for 6 h, and then affinity between these two proteins was measured. Wnt5a inhibited the physical association between RBP-Jk and SMRT and this binding was substantially rescued by KN-93 treatment, suggesting that Wnt5a induced CaMKII activation plays a critical role in the endogenous RBP-Jk-SMRT binding (Fig. 4F). To confirm the role of CaMKII in the regulation of RBP-Jk-SMRT transcription repressor complex, co-immunoprecipitation was performed in HEK293 cells that had been co-transfected with Flag-tagged SMRT, Myc-tagged RBP-Jk, HA-tagged CaMKII, and HA-tagged CaMKII-DN. SMRT and RBP-Jk were co-immunoprecipitated, but when they were co-transfected with CaMKII, the band corresponding to the SMRT-RBP-Jk complex disappeared (Fig. 4G). Furthermore, the dominant-negative mutant of CaMKII (CaMKII-DN) had no effect on the binding between RBP-Jk and SMRT, suggesting that the kinase activity of CaMKII is essential for the physical binding between RBP-Jk and SMRT (Fig. 4G). Interestingly, in the cell lysate immunoblot, the level of endogenous and ectopically expressed SMRT proteins, but not Notch1-IC and RBP-Jk, was significantly down-regulated upon coexpression with Wnt5a or CaMKII (Fig. 4, E–G), which shows that CaMKII may regulate the steady state level of the SMRT protein. These results indicate that Wnt5a-mediated CaMKII activation facilitates not only the formation of the Notch1-IC-RBP-Jk transcription activator complex but also the dissociation of the RBP-Jk-SMRT transcription repressor complex due to down-regulation of the SMRT protein level.
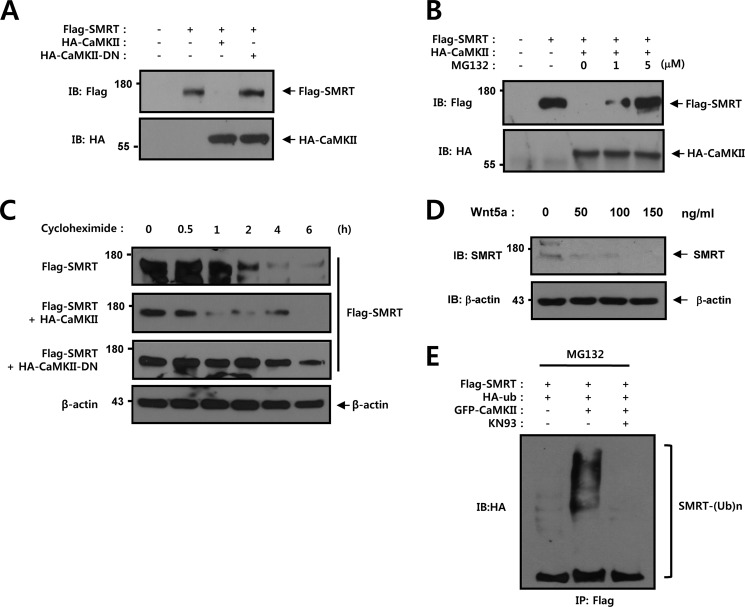
CaMKII Down-regulates the Steady State Level of SMRT Protein
Next, HEK293 cells were subjected to Western blot analysis to determine whether CaMKII plays a role in regulation of the SMRT protein level. Cells were co-transfected with Flag-tagged SMRT, HA-tagged CaMKII, and HA-tagged CaMKII-DN. We found that the SMRT protein level was reduced upon co-transfection of CaMKII but was not reduced upon co-transfection of CaMKII-DN (Fig. 5A). This result showed that the kinase activity of CaMKII is essential for regulation of the SMRT protein level. In an effort to determine whether the degradation of SMRT proteins by CaMKII is mediated by the proteasome pathway, the proteasome inhibitor MG132 was administered to SMRT and CaMKII-expressing cells. The cells were then subjected to 6 h of proteasome inhibitor treatment, and the SMRT protein level was measured via an immunoblot assay. Our studies demonstrated that the SMRT protein level was reduced in the presence of CaMKII, but was significantly restored by treatment with MG132 (Fig. 5B). In an effort to evaluate the possible role of CaMKII in the regulation of SMRT protein stability, we transfected HEK293 cells with Flag-tagged SMRT, HA-tagged CaMKII, and HA-tagged CaMKII-DN, and the quantity of the remaining SMRT was analyzed after various periods of cycloheximide treatment. We determined the protein stability of SMRT in HEK293 cells by cycloheximide treatment with or without CaMKII. After cycloheximide treatment, the SMRT protein level gradually decreased, where approximately half of the protein was degraded after 1.5 h without CaMKII (Fig. 5C). The SMRT protein was also rapidly decreased in the presence of CaMKII, where approximately half of the protein was degraded after 0.8 h (Fig. 5C). However, the SMRT protein level gradually increased in the presence of dominant-negative CaMKII, where approximately half of the protein was degraded after 5 h (Fig. 5C). No reduction in the level of β-actin, which was used as a control, was observed (Fig. 5C). HEK293 cells were subjected to Western blot analysis to determine whether purified Wnt5a plays a role in regulation of the endogenous SMRT protein level. The expression level of SMRT protein was reduced by purified Wnt5a in a dose-dependent manner (Fig. 5D). We then attempted to characterize the involvement of CaMKII activity in the polyubiquitination of SMRT. Cells were co-transfected with Flag-tagged SMRT, GFP-tagged CaMKII, and HA-tagged ubiquitin, and then subjected to ubiquitination analysis. Immunoblot analysis of the Flag immunoprecipitates from the transfected cells with an anti-HA antibody showed that CaMKII facilitated the ubiquitination of SMRT and KN93 prevented the ubiquitination of SMRT (Fig. 5E). These results showed that the kinase activity of CaMKII is essential for regulating the SMRT protein level.
FIGURE 5.
CaMKII down-regulates the steady state level of SMRT protein. A, HEK293 cells were transfected with 0.5 μg of Flag-SMRT, 0.5 μg of HA-CaMKII, and 0.5 μg of HA-CaMKII-DN as indicated. 48 h after transfection, the cell lysates were immunoblotted with anti-Flag and anti-HA antibodies. B, HEK293 cells were transfected with 0.5 μg of Flag-SMRT and 0.5 μg of HA-CaMKII as indicated. 42 h after transfection, the cells were treated with various concentrations (0, 1, and 5 μm) of MG132 for 6 h. 48 h after transfection, the cell lysates were immunoblotted with anti-Flag and anti-HA antibodies. C, HEK293 cells were transfected with 0.5 μg of Flag-SMRT, 0.5 μg of HA-CaMKII, and 0.5 μg of HA-CaMKII-DN as indicated. 42 h after transfection, the cells were then treated with 0.1 mm cycloheximide for the indicated times. D, HEK293 cell were treated with the indicated amounts of purified Wnt5a protein for 8 h. The cell lysates were also subjected to immunoblotting analysis with anti-SMRT. Probing with an antibody to β-actin was used as a loading control. E, HEK293 cells were transfected with 0.5 μg of Flag-SMRT, 0.5 μg of HA-ubiquitin, and 0.5 μg of GFP-CaMKII as indicated. 42 h after transfection, HEK293 cells were treated with KN-93 (5 μm) for 6 h. Cell lysates were subjected to immunoprecipitation with anti-Flag antibody and the immunoprecipitates were immunoblotted with anti-HA antibody. The cell lysates were also subjected to immunoblot analysis with anti-HA, anti-Flag, and anti-GFP antibodies, respectively. Probing with an antibody to β-actin was used as a loading control. DN, dominant-negative; IB, immunoblot.
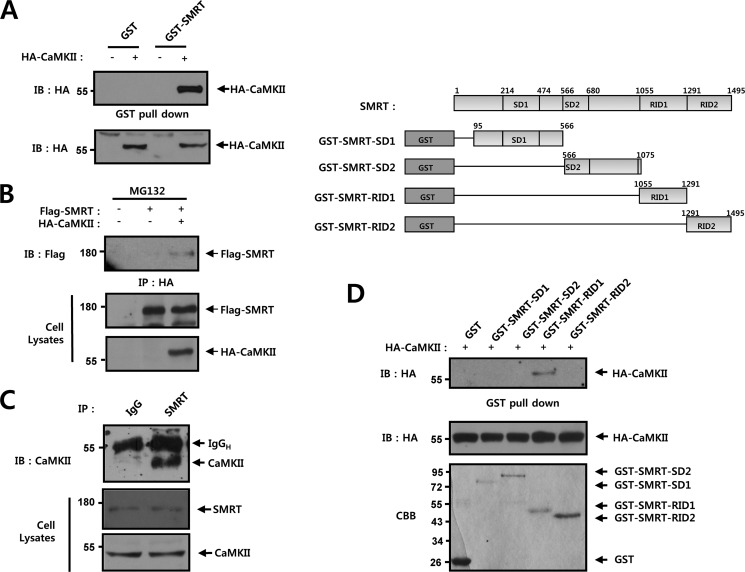
Physical Interaction of CaMKII with SMRT in Intact Cells
To delineate more precisely the manner in which CaMKII facilitates Notch1-IC transcriptional activity, we conducted a series of in vitro binding and coimmunoprecipitation experiments. In the in vitro binding experiments, purified GST or GST-SMRT proteins were immobilized onto GSH-agarose. HA-CaMKII expressing cell lysates were incubated with either GST or GST-SMRT, both of which were immobilized onto GSH-agarose. The interaction between GST-SMRT and CaMKII was detected on bead complexes (Fig. 6A). However, no interaction was observed between GST and CaMKII (Fig. 6A). We next attempted to determine whether CaMKII could physically associate with SMRT. Here, HEK293 cells were cotransfected with vectors encoding Flag-tagged wild-type SMRT and HA-tagged CaMKII and then subjected to coimmunoprecipitation analysis after treatment with MG132 (5 μm) for 6 h. Immunoblot analysis of HA immunoprecipitates from cells transfected with an anti-Flag antibody revealed that Flag-SMRT physically associated with HA-CaMKII in the cells (Fig. 6B). We also examined whether endogenous CaMKII and SMRT could interact in intact cells. Using HEK293 cells, immunoblot analysis of the SMRT immunoprecipitates using an anti-SMRT antibody indicated that endogenous CaMKII and SMRT were physically associated (Fig. 6C). SMRT has the positions of two silencing domains (SD1 and SD2) and two nuclear receptor interaction domains (RID1 and RID2) in its structure. We investigated which of these domains might be involved in the interaction between SMRT and CaMKII. We used purified GST and GST-SMRT deletion mutant proteins: SMRT-SD1, SMRT-SD2, SMRT-RID1, and SMRT-RID2. In the in vitro binding studies, purified GST and GST-SMRT deletion mutant proteins were immobilized on GSH-agarose. Cell lysates expressing HA-CaMKII were incubated with either GST or GST-SMRT deletion mutants immobilized on GSH-agarose. The interaction between GST-SMRT-RID1 and CaMKII was detected on bead complexes (Fig. 6D). In these experiments, we demonstrated that SMRT physically interacts with CaMKII via the RID1 domain.
FIGURE 6.
Physical interaction of CaMKII with SMRT in intact cells. A, recombinant GST or GST-SMRT was immobilized onto GSH-agarose. HEK293 cells were transfected with 0.5 μg of HA-CaMKII. 48 h after transfection, the cell lysates were subjected to GST pull-down experiments with immobilized GST or GST-SMRT. Proteins bound to GST or GST-Notch1-IC were analyzed via immunoblotting with an anti-HA antibody. B, HEK293 cells were transfected with 0.5 μg of Flag-SMRT and 0.5 μg of HA-CaMKII as indicated. 42 h after transfection, the cells were treated with MG132 (5 μm) for 6 h. The cell lysates were subjected to immunoprecipitation (IP) with an anti-HA antibody. The immunoprecipitates were then immunoblotted (IB) with an anti-Flag antibody. Cell lysates were also immunoblotted with anti-Flag and anti-HA antibodies. C, HEK293 cells were lysed and subjected to immunoprecipitation with immunoglobulin G (IgG) and anti-SMRT antibodies as indicated. The immunoprecipitates were immunoblotted with an anti-CaMKII antibody. Cell lysates were immunoblotted with anti-SMRT and anti-CaMKII. D, recombinant GST or GST-SMRT deletion proteins (GST-SMRT-SD1, GST-SMRT-SD2, GST-SMRT-RID1, and GST-SMRT-RID2) were immobilized onto GSH-agarose. HEK293 cells were transfected with 0.5 μg of HA-CaMKII. 48 h after transfection, the cell lysates were subjected to GST pull-down experiments with immobilized GST or GST-SMRT. Proteins bound to GST or GST-SMRT were analyzed via immunoblotting with an anti-HA antibody. CBB, Coomassie Brilliant Blue staining; IgGH, immunoglobulin heavy chain; IB, immunoblot.
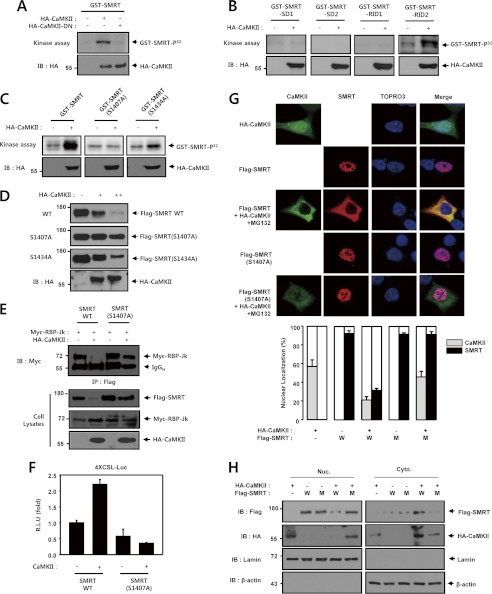
The Phosphorylation of SMRT by CaMKII Is Critical for Degradation of SMRT Protein
We next conducted an in vitro kinase assay using CaMKII and purified GST-SMRT. The CaMKII immunocomplexes prepared from HEK293 cells catalyzed the phosphorylation of purified recombinant GST-SMRT. Dominant-negative CaMKII did not phosphorylate the SMRT protein (Fig. 7A). Next, we conducted an in vitro kinase assay with HA-CaMKII and purified GST-SMRT deletion mutants (GST-SMRT-SD1, GST-SMRT-SD2, GST-SMRT-RID1, and GST-SMRT-RID2). Serial deletion mutants of SMRT were employed in the CaMKII phosphorylation reaction to determine the phosphorylation site of SMRT. CaMKII phosphorylated GST-SMRT-RID2 but did not phosphorylate the other three deletion mutants (Fig. 7B). These results demonstrated that the phosphorylation sites were located between residues 1291 and 1495 of SMRT. This region harbors two conserved sequences for CaMKII phosphorylation (RXX(S/T)): Ser-1407 and Ser-1434. Furthermore, using site-directed mutagenesis, we determined that replacement of Ser-1407 of SMRT with alanine reduced in vitro phosphorylation of the recombinant protein by the CaMKII immunoprecipitates (Fig. 7C). Moreover, we found that SMRT (S1407A) was resistant to CaMKII-induced degradation, which implies that the CaMKII-induced phosphorylation of SMRT is crucial for the degradation of the SMRT protein (Fig. 7D). We then used coimmunoprecipitation to examine the role of phosphorylation in the physical association between RBP-Jk and SMRT. In these experiments, HEK293 cells were cotransfected with vectors coding for Myc-tagged RBP-Jk, HA-tagged CaMKII, Flag-tagged SMRT, and Flag-tagged SMRT (S1407A) and then subjected to coimmunoprecipitation analysis. Immunoblot analysis of the Flag immunoprecipitates from the cells transfected showed that CaMKII does not prevent the physical association between RBP-Jk and SMRT (S1407A) in cells (Fig. 7E). Next, we examined the effect of CaMKII on the transcriptional repressive activity of SMRT and SMRT (S1407A) mutant in HEK293 cells. CaMKII was shown to activate the basal level of Notch1-induced transcriptional activity in the presence of wild-type SMRT; however, CaMKII did not activate the basal level of Notch1-induced transcriptional activity in the presence of wild-type SMRT(S1407A) (Fig. 7F). Moreover, we attempted to determine whether CaMKII could modulate SMRT localization within cells. To determine the effect of CaMKII on SMRT localization, HEK293 cells were cotransfected with vectors coding for HA-tagged CaMKII, Flag-tagged SMRT, and Flag-tagged SMRT (S1407A) and then subjected immunofluorescence staining after treatment with MG132 (5 μm) for 6 h. As expected, SMRT was localized predominantly in the nucleus and CaMKII was distributed throughout the nucleus and cytoplasm (Fig. 7, G and H). As shown in Fig. 7, G and H, coexpression of CaMKII facilitated the translocation of SMRT from the nucleus to cytoplasm. We also found that CaMKII was localized predominantly in the cytoplasm in the presence of SMRT. Furthermore, CaMKII did not affect the subcellular localization of SMRT (S1407A). We found that SMRT (S1407A) was localized predominantly in the nucleus, suggesting that CaMKII-mediated phosphorylation of SMRT was critical to the regulation of cellular localization of the SMRT protein. These data suggest that CaMKII-mediated phosphorylation of SMRT is an important step in CaMKII-mediated activation of Notch1 signaling in intact cells.
FIGURE 7.
The phosphorylation of SMRT by CaMKII is critical for degradation of SMRT protein. A, HEK293 cells were transfected with 0.5 μg of HA-CaMKII or 0.5 μg of HA-CaMKII-DN as indicated. 48 h after transfection, the cell lysates were subjected to immunoprecipitation with an anti-HA antibody, and the resultant precipitates were evaluated for phosphorylation of SMRT by an immune complex kinase assay using GST-SMRT. B, HEK293 cells were transfected with 0.5 μg of HA-CaMKII or an empty vector as indicated. 48 h after transfection, the cell lysates were subjected to immunoprecipitation with an anti-HA antibody and the resulting precipitates were examined for phosphorylation of SMRT using an immune complex kinase assay with purified recombinant GST-SMRT-SD1, GST-SMRT-SD2, GST-SMRT-RID1, and GST-SMRT-RID2 proteins. C, HEK293 cells were transfected with 0.5 μg of HA-CaMKII. 48 h after transfection, the cell lysates were subjected to immunoprecipitation with an anti-HA antibody, and the resultant precipitates were evaluated for phosphorylation of SMRT using an immune complex kinase assay with GST-SMRT, GST-SMRT (S1407A), and GST-SMRT (S1434A). D, HEK293 cells were transfected with 0.5 μg of Flag-SMRT, 0.5 μg of Flag-SMRT (S1407A), 0.5 μg of Flag-SMRT (S1434A), and 0.5 μg of HA-CaMKII as indicated. 48 h after transfection, the cell lysates were immunoblotted with an anti-Flag antibody. Cell lysates were also immunoblotted with anti-Flag and anti-HA antibodies. E, HEK293 cells were transfected with 0.5 μg of Flag-SMRT, 0.5 μg of Flag-SMRT (S1407A), 0.5 μg of Myc-RBP-Jk, and 0.5 μg of HA-CaMKII as indicated. 48 h after transfection, the cell lysates were subjected to immunoprecipitation (IP) using an anti-Flag antibody. The immunoprecipitates were then immunoblotted with an anti-Myc antibody. Cell lysates were also immunoblotted with anti-Flag, anti-Myc, and anti-HA antibodies. F, HEK293 cells were transfected with 50 ng of 4XCSL-Luc, 50 ng of SMRT, 50 ng of SMRT (S1407A), and 50 ng of β-galactosidase, along with 100 ng of CaMKII, as indicated. 48 h after transfection, the cells were lysed, and luciferase activity was determined. The data were normalized with β-galactosidase. These results represent the means ± average deviation of three independent experiments. R.L.U., relative luciferase units. G, HEK293 cells were transfected with empty vector, 0.5 μg of HA-CaMKII, 0.5 μg of Flag-SMRT, and 0.5 μg of Flag-SMRT (S1407A) as indicated. 42 h after transfection, the cells were treated with MG132 (5 μm) for 6 h. 48 h after transfection, the cells were fixed and CaMKII and SMRT were stained with Alexa Fluor 488 (green) and Alexa Fluor 555 (red) and examined by confocal microscopy. The DNA dye ToPro3 was used to visualize nuclei. For each experiment, at least 300 cells were examined, and the images represent the typical staining pattern for a majority of the cells. The mean nuclear localization intensity per cell was analyzed by imaging cytometric analysis. White scale bar, 25 μm. H, HEK293 cells were transfected with empty vector, 0.5 μg of HA-CaMKII, 0.5 μg of Flag-SMRT, and 0.5 μg of Flag-SMRT (S1407A) as indicated. 48 h after transfection, the cells were fractionated into cytosolic (Cyto) and nuclear (Nuc) fractions. Cell lysates were also immunoblotted with anti-Flag and anti-HA antibodies. LaminB and β-actin were used as nuclear and cytoplasmic fraction marker. IgGH, immunoglobulin heavy chain; IB, immunoblot.
DISCUSSION
Wnt and Notch signaling pathways belong to a highly conserved network that controls gene expression during development, adult tissue maintenance, and disease (70). Recent studies have implied that these pathways cross-talk in mammals (71, 72). A recent study has suggested that CaMKII, a component of the noncanonical Wnt signaling pathway, regulates the Notch1 signaling pathway (60). However, the mechanism of this regulation has not been clearly established. In this study, we demonstrated that Wnt5a-induced CaMKII enhances Notch transcriptional activity. CaMKII physically interacts with SMRT through the RID1 domain and disrupts the SMRT-RBP-Jk transcription complex through degradation of SMRT. Dissociation of SMRT from RBP-Jk provoked by CaMKII activates Hes1 and Hes5 transcriptional activity. CaMKII can phosphorylate SMRT and activate the Notch1 signaling pathway. The kinase activity of CaMKII was required for up-regulation of the transcription activity of Notch1 target genes. To the best of our knowledge, this is the first demonstration of CaMKII-mediated positive regulation of Notch signaling through proteasomal degradation of SMRT.
Several recent studies have shown that ubiquitination-mediated protein degradation plays a role in the regulation of Notch1 signaling through degradation of Notch1 ligands, Notch1 itself, and RBP-Jk (65, 66, 73–75). This study provides the first evidence that the CaMKII is an endogenous positive regulator of the Notch signaling pathway through degradation of the SMRT corepressor. When CaMKII activity was enhanced by Wnt5a, the Notch1 transcriptional activity was significantly increased in HEK293 cells. This was examined in two different experiments: treating the cells with a CaMKII inhibitor, and expressing the dominant-negative mutant CaMKII. In both experiments, the basal Notch1-IC transcriptional activity was substantially suppressed. On the other hand, when CAMKII was overexpressed, the Notch1 transcriptional activity was markedly increased in a dose-dependent manner. Our findings provide unequivocal evidence that CAMKII is an endogenous signaling agent that up-regulates Notch1-IC transcriptional activity. Previous studies have suggested that CaMKII regulates Notch1 signaling in C4-2B cells. Inhibition of CaMKII activity by KN93 leads to a decrease in the steady state level of Notch1-IC protein and down-regulates the expression of the Notch1 target gene Hes1 and its promoter activity (60). In contrast, the steady state level of Notch1-IC was not changed under these experimental conditions.
The transcriptional activation of downstream target genes by Notch1-IC depends on the association of Notch1-IC and RBP-Jk within the nucleus (6). Wnt5a and CaMKII facilitate the interaction between Notch1-IC and RBP-Jk and this interaction was inhibited by a CaMKII inhibitor or dominant-negative CaMKII in intact cells. After binding to DNA, RBP-Jk recruits a co-repressor complex that includes SMRT and histone deacetylases 1 to exert its inhibitory effect on transcription (6). We found that Wnt5a and CaMKII inhibit the interaction between Notch1-IC and RBP-Jk and this interaction was rescued using a CaMKII inhibitor or dominant-negative CaMKII in intact cells. Based on these results, we propose that endogenous CaMKII, which was induced by Wnt5a, may facilitate Notch1 transcriptional activity due to the stable association between Notch1-IC and RBP-Jk and dissociation between RBP-Jk and SMRT corepressor. Surprisingly, in the cell lysate immunoblot, the SMRT protein level was down-regulated upon co-transfection of Wnt5a or CaMKII and the SMRT protein level was restored using a CaMKII inhibitor or dominant-negative CaMKII. These results show that Wnt5a may regulate the level of the SMRT protein.
We also demonstrated that the RID1 domain of SMRT physically interacts with CaMKII and the kinase activity of CaMKII was shown to be essential for regulation of the SMRT protein level. Recent studies have suggested that SMRT, a member of the Notch co-repressor complex, is phosphorylated and degraded in a proteasome-dependent manner (41–43, 76). Degradation of SMRT is associated with regulation of the Notch1 signaling pathway. In this study, we found that CaMKII stimulated the proteasomal degradation of ectopically expressed SMRT, and the reduction of SMRT levels by CaMKII was also observed through a proteasome-dependent mechanism. Our findings show that the kinase activity of CaMKII plays a crucial role in the proteasomal degradation of SMRT. Furthermore, the kinase activity of CaMKII was critical to the binding and inhibition of the Notch1 signaling pathway. The amino acid Arg-X-X-Ser/Thr motif was preferentially phosphorylated by CaMKII and SMRT contains several putative serine and threonine phosphorylation sites (77, 78). We found that SMRT contains a conserved sequence for phosphorylation by CaMKII. CaMKII phosphorylated the SMRT protein and the phosphorylation sites were shown to be located between residues 1291 and 1495 of SMRT, a region that harbors two conserved serine residues: Ser-1407 and Ser-1434. Furthermore, we demonstrated that CaMKII-mediated SMRT phosphorylation on Ser-1407 results in a decrease in the degradation of SMRT protein. CaMKII was also shown to positively regulate transcriptional activation of the Notch1-IC target genes and the stability of the SMRT protein by phosphorylation of SMRT at the Ser-1407 residue. Thus, enhanced SMRT phosphorylation may be a possible mechanism for CaMKII-mediated ubiquitination and proteasomal degradation of SMRT (Fig. 8). Post-translational modifications are important to SMRT-mediated transcriptional repression. Depression is associated with phosphorylation and nuclear export of the SMRT corepressors protein. Previous reports have shown that MEKK1, Cdk2 and Pin1, CK2, and IKKα induced phosphorylation mediated SMRT translocation from the nucleus to the cytoplasm and proteasome-dependent degradation (41–43, 76). In this report, we also found that CaMKII mediates a similar phosphorylation event on SMRT corepressors at Ser-1407 and thereby facilitates translocation from the nucleus to the cytoplasm. These results explain why CaMKII-induced phosphorylation of SMRT was required for proteasomal degradation and translocation from the nucleus to the cytoplasm.
FIGURE 8.
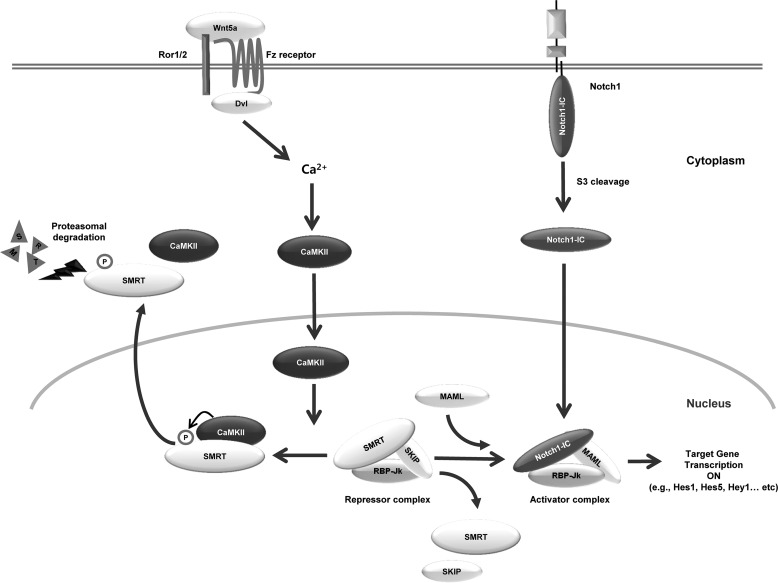
Schematic diagram of CaMKII regulates Notch1 signaling through proteasomal degradation of SMRT. Frizzled (Fz), and Ror1/2 that mediates Wnt5a/Ca2+ signaling has not yet been elucidated, however, the formation of a trimeric complex with Fz and Ror1/2 and the PDZ domain-mediated binding with dishevelled (Dvl) could be important. In this study, we evaluated that the mechanism by which Wnt up-regulates Notch1 target genes. We demonstrated that Wnt5a induced CaMKII enhances Notch transcriptional activity. CaMKII physically interacts with SMRT and disrupts the SMRT-RBP-Jk transcription complex through degradation of SMRT. Dissociation of SMRT from RBP-Jk induced by CaMKII activates Hes1 and Hes5 transcriptional activity. Furthermore, CaMKII can phosphorylate SMRT and activate the Notch1 signaling pathway. The kinase activity of CaMKII is necessary for up-regulation of the transcription activity of Notch1 target genes. The results of this study suggest that CaMKII is a crucial regulator for signal cross-talk between Wnt5a and Notch signaling via down-regulation of the SMRT protein level.
In summary, Wnt5a induced CaMKII enhances Notch transcriptional activity. CaMKII physically interacts with SMRT and disrupts the SMRT-RBP-Jk transcription complex through degradation of SMRT protein. Dissociation of SMRT from RBP-Jk induced by CaMKII activates Hes1 and Hes5 transcriptional activity. Furthermore, CaMKII can phosphorylate SMRT and activate the Notch1 signaling pathway. The kinase activity of CaMKII was required for up-regulation of the transcription activity of Notch1 target genes. The results of this study suggest that CaMKII plays a pivotal role in the signal cross-talk between Wnt5a and Notch signaling through down-regulation of the SMRT corepressor protein.
Acknowledgments
We thank Raphael Kopan (Washington University Medical School, St. Louis, MO) for providing Notch-related constructs, Martin L. Privalsky (University of California, Davis, CA) for SMRT-related constructs, S. Diane Hayward (Johns Hopkins University School of Medicine, Baltimore, MD) for SMRT, Kunihiro Matsumoto (Nagoya University, Japan) for CaMKII and Wnt5a, and Tetsuya Taga (Tokyo Medical and Dental University, Japan) for Hes5-Luc and RMHes-5-Luc constructs.
This study was supported by Grant A100455 of the Korea Healthcare Technology R&D Project, Ministry of Health & Welfare, Republic of Korea.

This article contains supplemental Fig. S1.
- Notch1-IC
- Notch1 intracellular domain
- CaMKII
- Ca2+/calmodulin-dependent protein kinase II
- SMRT
- silencing mediator of retinoic acid and thyroid hormone receptor
- RBP-Jk
- recombining binding protein suppressor of hairless
- Ror1/2
- receptor tyrosine kinase-like orphan receptor 1/2
- PEN-2
- presenilin enhancer-2
- HEK
- human embryonic kidney
- PS
- presenilin
- MEF
- mouse embryonic fibroblasts
- DN
- dominant-negative.
REFERENCES
- 1. Artavanis-Tsakonas S., Matsuno K., Fortini M. E. (1995) Notch signaling. Science 268, 225–232 [DOI] [PubMed] [Google Scholar]
- 2. Egan S. E., St-Pierre B., Leow C. C. (1998) Notch receptors, partners and regulators. From conserved domains to powerful functions. Curr. Top. Microbiol. Immunol. 228, 273–324 [DOI] [PubMed] [Google Scholar]
- 3. Lai E. C. (2004) Notch signaling. Control of cell communication and cell fate. Development 131, 965–973 [DOI] [PubMed] [Google Scholar]
- 4. Weinmaster G. (1998) Notch signaling. Direct or what? Curr. Opin. Genet. Dev. 8, 436–442 [DOI] [PubMed] [Google Scholar]
- 5. Kopan R., Cagan R. (1997) Notch on the cutting edge. Trends Genet. 13, 465–467 [DOI] [PubMed] [Google Scholar]
- 6. Mumm J. S., Kopan R. (2000) Notch signaling. From the outside in. Dev. Biol. 228, 151–165 [DOI] [PubMed] [Google Scholar]
- 7. Weinmaster G. (1997) The ins and outs of Notch signaling. Mol. Cell Neurosci. 9, 91–102 [DOI] [PubMed] [Google Scholar]
- 8. Logeat F., Bessia C., Brou C., LeBail O., Jarriault S., Seidah N. G., Israël A. (1998) The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc. Natl. Acad. Sci. U.S.A. 95, 8108–8112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lindsell C. E., Boulter J., diSibio G., Gossler A., Weinmaster G. (1996) Mol. Cell Neurosci. 8, 14–27 [DOI] [PubMed] [Google Scholar]
- 10. Lindsell C. E., Shawber C. J., Boulter J., Weinmaster G. (1995) Jagged, a mammalian ligand that activates Notch1. Cell 80, 909–917 [DOI] [PubMed] [Google Scholar]
- 11. De Strooper B. (2003) Aph-1, Pen-2, and Nicastrin with Presenilin generate an active γ-secretase complex. Neuron 38, 9–12 [DOI] [PubMed] [Google Scholar]
- 12. Kopan R., Goate A. (2002) Aph-2/Nicastrin, an essential component of γ-secretase and regulator of Notch signaling and Presenilin localization. Neuron 33, 321–324 [DOI] [PubMed] [Google Scholar]
- 13. Wolfe M. S., Haass C. (2001) The role of presenilins in γ-secretase activity. J. Biol. Chem. 276, 5413–5416 [DOI] [PubMed] [Google Scholar]
- 14. Brou C., Logeat F., Gupta N., Bessia C., LeBail O., Doedens J. R., Cumano A., Roux P., Black R. A., Israël A. (2000) A novel proteolytic cleavage involved in Notch signaling. The role of the disintegrin-metalloprotease TACE. Mol. Cell 5, 207–216 [DOI] [PubMed] [Google Scholar]
- 15. De Strooper B., Annaert W., Cupers P., Saftig P., Craessaerts K., Mumm J. S., Schroeter E. H., Schrijvers V., Wolfe M. S., Ray W. J., Goate A., Kopan R. (1999) A presenilin-1-dependent γ-secretase-like protease mediates release of Notch intracellular domain. Nature 398, 518–522 [DOI] [PubMed] [Google Scholar]
- 16. Struhl G., Greenwald I. (1999) Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature 398, 522–525 [DOI] [PubMed] [Google Scholar]
- 17. Ye Y., Lukinova N., Fortini M. E. (1999) Neurogenic phenotypes and altered Notch processing in Drosophila Presenilin mutants. Nature 398, 525–529 [DOI] [PubMed] [Google Scholar]
- 18. Yu G., Nishimura M., Arawaka S., Levitan D., Zhang L., Tandon A., Song Y. Q., Rogaeva E., Chen F., Kawarai T., Supala A., Levesque L., Yu H., Yang D. S., Holmes E., Milman P., Liang Y., Zhang D. M., Xu D. H., Sato C., Rogaev E., Smith M., Janus C., Zhang Y., Aebersold R., Farrer L. S., Sorbi S., Bruni A., Fraser P., St George-Hyslop P. (2000) Nicastrin modulates presenilin-mediated notch/glp-1 signal transduction and βAPP processing. Nature 407, 48–54 [DOI] [PubMed] [Google Scholar]
- 19. Francis R., McGrath G., Zhang J., Ruddy D. A., Sym M., Apfeld J., Nicoll M., Maxwell M., Hai B., Ellis M. C., Parks A. L., Xu W., Li J., Gurney M., Myers R. L., Himes C. S., Hiebsch R., Ruble C., Nye J. S., Curtis D. (2002) aph-1 and pen-2 are required for Notch pathway signaling, γ-secretase cleavage of βAPP, and presenilin protein accumulation. Dev. Cell 3, 85–97 [DOI] [PubMed] [Google Scholar]
- 20. Goutte C., Tsunozaki M., Hale V. A., Priess J. R. (2002) APH-1 is a multipass membrane protein essential for the Notch signaling pathway in Caenorhabditis elegans embryos. Proc. Natl. Acad. Sci. U.S.A. 99, 775–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kimberly W. T., LaVoie M. J., Ostaszewski B. L., Ye W., Wolfe M. S., Selkoe D. J. (2003) γ-Secretase is a membrane protein complex comprised of presenilin, nicastrin, Aph-1, and Pen-2. Proc. Natl. Acad. Sci. U.S.A. 100, 6382–6387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee S. F., Shah S., Li H., Yu C., Han W., Yu G. (2002) Mammalian APH-1 interacts with presenilin and nicastrin and is required for intramembrane proteolysis of amyloid-β precursor protein and Notch. J. Biol. Chem. 277, 45013–45019 [DOI] [PubMed] [Google Scholar]
- 23. Luo W. J., Wang H., Li H., Kim B. S., Shah S., Lee H. J., Thinakaran G., Kim T. W., Yu G., Xu H. (2003) PEN-2 and APH-1 coordinately regulate proteolytic processing of presenilin 1. J. Biol. Chem. 278, 7850–7854 [DOI] [PubMed] [Google Scholar]
- 24. Abu-Issa R., Cavicchi S. (1996) Genetic interactions among vestigial, hairy, and Notch suggest a role of vestigial in the differentiation of epidermal and neural cells of the wing and halter of Drosophila melanogaster. J. Neurogenet. 10, 239–246 [DOI] [PubMed] [Google Scholar]
- 25. de Celis J. F., de Celis J., Ligoxygakis P., Preiss A., Delidakis C., Bray S. (1996) Functional relationships between Notch, Su(H) and the bHLH genes of the E(spl) complex. The E(spl) genes mediate only a subset of Notch activities during imaginal development. Development 122, 2719–2728 [DOI] [PubMed] [Google Scholar]
- 26. Jennings B., Preiss A., Delidakis C., Bray S. (1994) The Notch signalling pathway is required for Enhancer of split bHLH protein expression during neurogenesis in the Drosophila embryo. Development 120, 3537–3548 [DOI] [PubMed] [Google Scholar]
- 27. Jouve C., Palmeirim I., Henrique D., Beckers J., Gossler A., Ish-Horowicz D., Pourquié O. (2000) Notch signalling is required for cyclic expression of the hairy-like gene HES1 in the presomitic mesoderm. Development 127, 1421–1429 [DOI] [PubMed] [Google Scholar]
- 28. Ligoxygakis P., Yu S. Y., Delidakis C., Baker N. E. (1998) A subset of notch functions during Drosophila eye development require Su(H) and the E(spl) gene complex. Development 125, 2893–2900 [DOI] [PubMed] [Google Scholar]
- 29. Ohtsuka T., Ishibashi M., Gradwohl G., Nakanishi S., Guillemot F., Kageyama R. (1999) Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 18, 2196–2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bessho Y., Miyoshi G., Sakata R., Kageyama R. (2001) Hes7, a bHLH-type repressor gene regulated by Notch and expressed in the presomitic mesoderm. Genes Cells 6, 175–185 [DOI] [PubMed] [Google Scholar]
- 31. Fischer A., Schumacher N., Maier M., Sendtner M., Gessler M. (2004) The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev. 18, 901–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Maier M. M., Gessler M. (2000) Comparative analysis of the human and mouse Hey1 promoter. Hey genes are new Notch target genes. Biochem. Biophys. Res. Commun. 275, 652–660 [DOI] [PubMed] [Google Scholar]
- 33. Leimeister C., Schumacher N., Steidl C., Gessler M. (2000) Analysis of HeyL expression in wild-type and Notch pathway mutant mouse embryos. Mech Dev. 98, 175–178 [DOI] [PubMed] [Google Scholar]
- 34. Ronchini C., Capobianco A. J. (2001) Induction of cyclin D1 transcription and CDK2 activity by Notch(IC). Implication for cell cycle disruption in transformation by Notch(IC). Mol. Cell Biol. 21, 5925–5934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhou S., Fujimuro M., Hsieh J. J., Chen L., Miyamoto A., Weinmaster G., Hayward S. D. (2000) SKIP, a CBF1-associated protein, interacts with the ankyrin repeat domain of NotchIC to facilitate NotchIC function. Mol. Cell. Biol. 20, 2400–2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kao H. Y., Ordentlich P., Koyano-Nakagawa N., Tang Z., Downes M., Kintner C. R., Evans R. M., Kadesch T. (1998) A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 12, 2269–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nagy L., Kao H. Y., Chakravarti D., Lin R. J., Hassig C. A., Ayer D. E., Schreiber S. L., Evans R. M. (1997) Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 89, 373–380 [DOI] [PubMed] [Google Scholar]
- 38. Chen J. D., Umesono K., Evans R. M. (1996) SMRT isoforms mediate repression and anti-repression of nuclear receptor heterodimers. Proc. Natl. Acad. Sci. U.S.A. 93, 7567–7571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hong S. H., David G., Wong C. W., Dejean A., Privalsky M. L. (1997) SMRT corepressor interacts with PLZF and with the PML-retinoic acid receptor α (RARα) and PLZF-RARα oncoproteins associated with acute promyelocytic leukemia. Proc. Natl. Acad. Sci. U.S.A. 94, 9028–9033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hong S. H., Wong C. W., Privalsky M. L. (1998) Signaling by tyrosine kinases negatively regulates the interaction between transcription factors and SMRT (silencing mediator of retinoic acid and thyroid hormone receptor) corepressor. Mol. Endocrinol 12, 1161–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jonas B. A., Privalsky M. L. (2004) SMRT and N-CoR corepressors are regulated by distinct kinase signaling pathways. J. Biol. Chem. 279, 54676–54686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stanya K. J., Liu Y., Means A. R., Kao H. Y. (2008) Cdk2 and Pin1 negatively regulate the transcriptional corepressor SMRT. J. Cell Biol. 183, 49–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hoberg J. E., Yeung F., Mayo M. W. (2004) SMRT derepression by the IκB kinase α. A prerequisite to NF-κB transcription and survival. Mol. Cell 16, 245–255 [DOI] [PubMed] [Google Scholar]
- 44. Nishita M., Enomoto M., Yamagata K., Minami Y. (2010) Cell/tissue-tropic functions of Wnt5a signaling in normal and cancer cells. Trends Cell Biol. 20, 346–354 [DOI] [PubMed] [Google Scholar]
- 45. Green J. L., Kuntz S. G., Sternberg P. W. (2008) Ror receptor tyrosine kinases. Orphans no more. Trends Cell Biol. 18, 536–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Veeman M. T., Axelrod J. D., Moon R. T. (2003) A second canon. Functions and mechanisms of β-catenin-independent Wnt signaling. Dev. Cell 5, 367–377 [DOI] [PubMed] [Google Scholar]
- 47. Korswagen H. C. (2002) Canonical and noncanonical Wnt signaling pathways in Caenorhabditis elegans. Variations on a common signaling theme. Bioessays 24, 801–810 [DOI] [PubMed] [Google Scholar]
- 48. Küuhl M., Sheldahl L. C., Park M., Miller J. R., Moon R. T. (2000) The Wnt/Ca2+ pathway. A new vertebrate Wnt signaling pathway takes shape. Trends Genet. 16, 279–283 [DOI] [PubMed] [Google Scholar]
- 49. Kühl M., Sheldahl L. C., Malbon C. C., Moon R. T. (2000) Ca2+/calmodulin-dependent protein kinase II is stimulated by Wnt and Frizzled homologs and promotes ventral cell fates in Xenopus. J. Biol. Chem. 275, 12701–12711 [DOI] [PubMed] [Google Scholar]
- 50. Slusarski D. C., Corces V. G., Moon R. T. (1997) Interaction of Wnt and a Frizzled homologue triggers G-protein-linked phosphatidylinositol signalling. Nature 390, 410–413 [DOI] [PubMed] [Google Scholar]
- 51. Slusarski D. C., Yang-Snyder J., Busa W. B., Moon R. T. (1997) Modulation of embryonic intracellular Ca2+ signaling by Wnt-5A. Dev. Biol. 182, 114–120 [DOI] [PubMed] [Google Scholar]
- 52. Ishitani T., Kishida S., Hyodo-Miura J., Ueno N., Yasuda J., Waterman M., Shibuya H., Moon R. T., Ninomiya-Tsuji J., Matsumoto K. (2003) The TAK1-NLK mitogen-activated protein kinase cascade functions in the Wnt-5a/Ca2+ pathway to antagonize Wnt/β-catenin signaling. Mol. Cell Biol. 23, 131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Weeraratna A. T., Jiang Y., Hostetter G., Rosenblatt K., Duray P., Bittner M., Trent J. M. (2002) Wnt5a signaling directly affects cell motility and invasion of metastatic melanoma. Cancer Cell 1, 279–288 [DOI] [PubMed] [Google Scholar]
- 54. Struewing I. T., Barnett C. D., Zhang W., Yadav S., Mao C. D. (2007) Frizzled-7 turnover at the plasma membrane is regulated by cell density and the Ca2+-dependent protease calpain-1. Exp. Cell Res. 313, 3526–3541 [DOI] [PubMed] [Google Scholar]
- 55. Fink C. C., Meyer T. (2002) Molecular mechanisms of CaMKII activation in neuronal plasticity. Curr. Opin. Neurobiol. 12, 293–299 [DOI] [PubMed] [Google Scholar]
- 56. Lisman J., Schulman H., Cline H. (2002) The molecular basis of CaMKII function in synaptic and behavioral memory. Nat. Rev. Neurosci. 3, 175–190 [DOI] [PubMed] [Google Scholar]
- 57. Bayer K. U., Schulman H. (2001) Regulation of signal transduction by protein targeting. The case for CaMKII. Biochem. Biophys. Res. Commun. 289, 917–923 [DOI] [PubMed] [Google Scholar]
- 58. Lisman J. E., Zhabotinsky A. M. (2001) A model of synaptic memory. A CaMKII/PP1 switch that potentiates transmission by organizing an AMPA receptor anchoring assembly. Neuron 31, 191–201 [DOI] [PubMed] [Google Scholar]
- 59. Rokhlin O. W., Taghiyev A. F., Bayer K. U., Bumcrot D., Koteliansk V. E., Glover R. A., Cohen M. B. (2007) Calcium/calmodulin-dependent kinase II plays an important role in prostate cancer cell survival. Cancer Biol. Ther 6, 732–742 [DOI] [PubMed] [Google Scholar]
- 60. Mamaeva O. A., Kim J., Feng G., McDonald J. M. (2009) Calcium/calmodulin-dependent kinase II regulates notch-1 signaling in prostate cancer cells. J. Cell Biochem. 106, 25–32 [DOI] [PubMed] [Google Scholar]
- 61. Katoh M. (2009) Transcriptional mechanisms of WNT5A based on NF-κB, Hedgehog, TGFβ, and Notch signaling cascades. Int. J. Mol. Med. 23, 763–769 [DOI] [PubMed] [Google Scholar]
- 62. Koyanagi M., Bushoven P., Iwasaki M., Urbich C., Zeiher A. M., Dimmeler S. (2007) Notch signaling contributes to the expression of cardiac markers in human circulating progenitor cells. Circ. Res. 101, 1139–1145 [DOI] [PubMed] [Google Scholar]
- 63. Hu B., Lefort K., Qiu W., Nguyen B. C., Rajaram R. D., Castillo E., He F., Chen Y., Angel P., Brisken C., Dotto G. P. (2010) Control of hair follicle cell fate by underlying mesenchyme through a CSL-Wnt5a-FoxN1 regulatory axis. Genes Dev. 24, 1519–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kim M. Y., Ann E. J., Kim J. Y., Mo J. S., Park J. H., Kim S. Y., Seo M. S., Park H. S. (2007) Tip60 histone acetyltransferase acts as a negative regulator of Notch1 signaling by means of acetylation. Mol. Cell Biol. 27, 6506–6519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mo J. S., Ann E. J., Yoon J. H., Jung J., Choi Y. H., Kim H. Y., Ahn J. S., Kim S. M., Kim M. Y., Hong J. A., Seo M. S., Lang F., Choi E. J., Park H. S. (2011) Serum- and glucocorticoid-inducible kinase 1 (SGK1) controls Notch1 signaling by down-regulation of protein stability through Fbw7 ubiquitin ligase. J. Cell Sci. 124, 100–112 [DOI] [PubMed] [Google Scholar]
- 66. Mo J. S., Kim M. Y., Han S. O., Kim I. S., Ann E. J., Lee K. S., Seo M. S., Kim J. Y., Lee S. C., Park J. W., Choi E. J., Seong J. Y., Joe C. O., Faessler R., Park H. S. (2007) Integrin-linked kinase controls Notch1 signaling by down-regulation of protein stability through Fbw7 ubiquitin ligase. Mol. Cell. Biol. 27, 5565–5574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kim M. Y., Ann E. J., Mo J. S., Dajas-Bailador F., Seo M. S., Hong J. A., Jung J., Choi Y. H., Yoon J. H., Kim S. M., Choi E. J., Hoe H. S., Whitmarsh A. J., Park H. S. (2010) JIP1 binding to RBP-Jk mediates cross-talk between the Notch1 and JIP1-JNK signaling pathway. Cell Death Differ. 17, 1728–1738 [DOI] [PubMed] [Google Scholar]
- 68. Mo J. S., Kim M. Y., Ann E. J., Hong J. A., Park H. S. (2008) DJ-1 modulates UV-induced oxidative stress signaling through the suppression of MEKK1 and cell death. Cell Death Differ. 15, 1030–1041 [DOI] [PubMed] [Google Scholar]
- 69. Takizawa T., Ochiai W., Nakashima K., Taga T. (2003) Enhanced gene activation by Notch and BMP signaling cross-talk. Nucleic Acids Res. 31, 5723–5731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hayward P., Kalmar T., Arias A. M. (2008) Wnt/Notch signaling and information processing during development. Development 135, 411–424 [DOI] [PubMed] [Google Scholar]
- 71. Couso J. P., Martinez Arias A. (1994) Notch is required for wingless signaling in the epidermis of Drosophila. Cell 79, 259–272 [DOI] [PubMed] [Google Scholar]
- 72. Hing H. K., Sun X., Artavanis-Tsakonas S. (1994) Modulation of wingless signaling by Notch in Drosophila. Mech Dev. 47, 261–268 [DOI] [PubMed] [Google Scholar]
- 73. Lai E. C. (2002) Protein degradation. Four E3s for the notch pathway. Curr. Biol. 12, R74–78 [DOI] [PubMed] [Google Scholar]
- 74. Kim M. Y., Jung J., Mo J. S., Ann E. J., Ahn J. S., Yoon J. H., Park H. S. (2011) The intracellular domain of Jagged-1 interacts with Notch1 intracellular domain and promotes its degradation through Fbw7 E3 ligase. Exp. Cell Res. 317, 2438–2446 [DOI] [PubMed] [Google Scholar]
- 75. Kim M. Y., Mo J. S., Ann E. J., Yoon J. H., Jung J., Choi Y. H., Kim S. M., Kim H. Y., Ahn J. S., Kim H., Kim K., Hoe H. S., Park H. S. (2011) Regulation of Notch1 signaling by the APP intracellular domain facilitates degradation of the Notch1 intracellular domain and RBP-Jk. J. Cell Sci. 124, 1831–1843 [DOI] [PubMed] [Google Scholar]
- 76. Zhou Y., Gross W., Hong S. H., Privalsky M. L. (2001) The SMRT corepressor is a target of phosphorylation by protein kinase CK2 (casein kinase II). Mol. Cell Biochem. 220, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kennelly P. J., Krebs E. G. (1991) Consensus sequences as substrate specificity determinants for protein kinases and protein phosphatases. J. Biol. Chem. 266, 15555–15558 [PubMed] [Google Scholar]
- 78. Colbran R. J., Schworer C. M., Hashimoto Y., Fong Y. L., Rich D. P., Smith M. K., Soderling T. R. (1989) Calcium/calmodulin-dependent protein kinase II. Biochem. J. 258, 313–325 [DOI] [PMC free article] [PubMed] [Google Scholar]