Background: Fatty acids activate UCP1 to catalyze mitochondrial proton leak.
Results: Palmitate changed the kinetics of two processes: mant-GDP binding to UCP1 and enzymatic proteolysis of UCP1.
Conclusion: We present the first demonstration that fatty acids induce a conformational change in UCP1.
Significance: The finding has profound implications for the mechanism of UCP1, providing crucial insights into cellular metabolic inefficiency.
Keywords: Adipose Tissue Metabolism, Bioenergetics, Mitochondria, Mitochondrial Transport, Uncoupling Proteins, Brown Adipose Tissue, mant-GDP, Proton Leak, Thermogenesis, Trypsinolysis
Abstract
UCP1 catalyzes proton leak across the mitochondrial inner membrane to disengage substrate oxidation from ATP production. It is well established that UCP1 is activated by fatty acids and inhibited by purine nucleotides, but precisely how this regulation occurs remains unsettled. Although fatty acids can competitively overcome nucleotide inhibition in functional assays, fatty acids have little effect on purine nucleotide binding. Here, we present the first demonstration that fatty acids induce a conformational change in UCP1. Palmitate dramatically changed the binding kinetics of 2′/3′-O-(N-methylanthraniloyl)-GDP, a fluorescently labeled nucleotide analog, for UCP1. Furthermore, palmitate accelerated the rate of enzymatic proteolysis of UCP1. The altered kinetics of both processes indicate that fatty acids change the conformation of UCP1, reconciling the apparent discrepancy between existing functional and ligand binding data. Our results provide a framework for how fatty acids and nucleotides compete to regulate the activity of UCP1.
Introduction
UCP1 (uncoupling protein 1) dissipates the mitochondrial protonmotive force that couples substrate oxidation to ATP synthesis (1, 2). UCP1 facilitates non-shivering thermogenesis in brown adipose tissue (BAT),4 short-circuiting the chemiosmotic process to increase the rate of heat generation (3). The protein is also functionally expressed in the rodent thymus (4, 5) and carp liver mitochondria (6), where its role is likely attenuation of reactive oxygen species production rather than heat production (7).
Concerted regulation of UCP1 is exerted at the levels of catalytic activity, transcription, and mitochondrial turnover (8). Activation of the protein by free fatty acids and inhibition by purine nucleoside di- and triphosphates have been known for decades (9). However, the explicit mechanism of this acute regulation remains contentious and unresolved.
Three competing models seek to describe catalysis of proton transport by UCP1. (i) Fatty acids are obligatory cofactors for proton transport, embedding their carboxyl side chains into the core of the protein to buffer protons (10, 11). (ii) UCP1 facilitates a protonophoric cycle by translocating fatty acid anions across the mitochondrial inner membrane (12, 13). (iii) Fatty acids and nucleotides functionally compete to regulate UCP1-catalyzed proton leak (14).
The functional competition model is likely the most physiologically relevant of the three, as it is informed by studies of the native protein in isolated mitochondria from BAT. Conversely, the first two models rely primarily on data from UCP1 reconstituted into proteoliposomes, a system that produces results that are sometimes inconsistent with those from mitochondria (15).
However, the functional competition model fits poorly with the longstanding observation that fatty acids have little effect on the binding of nucleotides to UCP1 (9, 11, 16). This uncovers a glaring question regarding the mechanism of UCP1: how do fatty acids and nucleotides functionally compete to regulate the activity of UCP1 if they bind non-competitively?
Here, we present the first demonstration that fatty acids change the conformation of UCP1. Using two independent tests, fluorescently labeled nucleotide binding and controlled trypsinolysis, we show that palmitate significantly changed the kinetics of both of these processes. This finding eliminates arguably the most substantial intellectual roadblock to acceptance of the functional competition model to describe the regulation of UCP1.
EXPERIMENTAL PROCEDURES
Animals
All animals were housed at 21 ± 2 °C with humidity of 57 ± 5% and a 12-h light/dark cycle. Food and water were available ad libitum. Female Wistar rats were euthanized between 5 and 8 weeks of age by stunning and cervical dislocation. Wild-type and sibling-paired Ucp1-null mice were derived as described previously (17, 18). All animal care and procedures were in compliance with Principles of Laboratory Animal Care (National Institutes of Health Publication 85-23, revised 1996) under Protocol A10080 approved by the Buck Institute Institutional Animal Care and Use Committee. When applicable, Home Office (United Kingdom) Guidelines for the Care and Use of Laboratory Animals were followed.
Mitochondrial Isolation
Mitochondria from rodent BAT were isolated by differential centrifugation as described (19) with all steps conducted at 4 °C. Briefly, inter- and subscapular BAT was excised and homogenized with a glass Dounce homogenizer in medium containing 250 mm sucrose, 5 mm Tris (pH 7.4 at 4 °C), 2 mm EGTA, and 1% (w/v) defatted BSA. Mitochondria were washed at 700 × g and collected at 8500 × g.
Mitochondria from rodent thymus were isolated in a similar medium containing 0.5% (w/v) defatted BSA. They were washed at 1000 × g and collected at 11,600 × g. Protein concentrations were determined using the Bradford method (20) for mitochondria from BAT or the biuret method (21) for mitochondria from thymus.
Proton Leak Kinetics
The proton leak rate was defined as the respiration needed to pump protons out of the mitochondrial matrix in non-phosphorylating mitochondria (19, 22). Therefore, the proton leak kinetics were measured as the dependence of this proton leak rate on the mitochondrial membrane potential that drives it (15).
Mitochondria from BAT (0.35 mg/ml) were incubated at 37 °C in 3.5 ml of assay medium containing 50 mm KCl, 5 mm HEPES (pH 7.2), 1 mm EGTA, 4 mm KH2PO4, and 1% (w/v) defatted BSA. Respiration was measured using a Clark-type electrode, and it was assumed that the electrode buffer contained 406 nmol of O/ml (23). Mitochondrial membrane potential was measured using an electrode responsive to the lipophilic cation triphenylmethylphosphonium (TPMP+). A TPMP+ binding correction of (0.4 μl/mg protein)−1 was applied as described previously (24).
Prior to each experiment, the TPMP+ electrode was calibrated with sequential additions of 0.5 μm TPMP+ (up to 2.5 μm). Mitochondria were energized by the addition of 10 mm sn-glycerol 3-phosphate as substrate, and after 1.5 min, membrane potential was sequentially decreased with a cyanide titration (5–255 μm). The following compounds were included in each experiment: 5 μm rotenone (added to inhibit NADH:ubiquinone oxidoreductase, Complex I), 1 μg/ml oligomycin (ATP synthase inhibitor added to ensure that all proton conductance was due to leak), and 80 ng/ml nigericin (added to decrease the pH gradient). 0.6 μm carbonyl cyanide p-trifluoromethoxyphenylhydrazone was added after each experiment to release TPMP+ from the mitochondria and to allow base-line correction.
Fatty Acid Preparation
Albumin-buffered palmitate was prepared, and the free fatty acid concentration was calculated as described (25).
Nucleotide Binding
BAT mitochondria were incubated at 2.5 mg/ml in medium identical to that used to measure proton leak kinetics. Mitochondria were exposed to a range of concentrations of [3H]GDP or 2′/3′-O-(N-methylanthraniloyl)-GDP (mant-GDP) (26) for 5 min. Nonspecific binding was determined by running parallel samples in the presence of 3 mm unmodified GDP.
Following incubation with [3H]GDP, samples were pelleted, resuspended in 20% (v/v) Triton X-100, placed in scintillation vials, vigorously mixed with scintillation fluid, and counted. Following incubation with mant-GDP, samples were pelleted (16,000 × g for 5 min), washed with assay medium at 4 °C, solubilized in 1% (w/v) SDS, and briefly spun to remove insoluble material. Any mant-GDP specifically bound to UCP1 would be present in the soluble fraction and was detected by fluorescence emission at 435 nm when excited at 350 nm. As such, fluorescence was used only as a directly proportional measurement of the amount of tagged GDP specifically bound to UCP1 (analogous to the [3H]GDP measurements).
Trypsinolysis
Freeze-thawed mitochondria from BAT (0.35 mg/ml in assay medium) and thymus (0.50 mg/ml in isolation medium) were subjected to controlled enzymatic proteolysis at 37 °C. Exogenous trypsin was added (300 μg/mg of mitochondrial protein), and the reaction was quenched at defined times with excess trypsin inhibitor (1 mg/ml) at 4 °C.
Immediately upon trypsin inhibition, samples were pelleted and prepared for Western analysis. They were resuspended in SDS loading buffer (50 mm Tris (pH 6.8), 1 mm EDTA, 10% (v/v) glycerol, 2% (w/v) SDS, 1% (v/v) β-mercaptoethanol, and 0.01% (w/v) bromphenol blue) at 0.5 mg/ml and boiled for 5 min.
SDS-PAGE and Western Analysis
Samples were run on a Laemmli gel (resolving gel, 375 mm Tris (pH 8.8), 12.5% (w/v) polyacrylamide, and 0.1% (w/v) SDS; and stacking gel (<3-mm depth), 125 mm Tris (pH 6.8), 6% (w/v) polyacrylamide, and 0.1% (w/v) SDS) (27) for ∼2 h at 150 V in running buffer (25 mm Tris, 192 mm glycine, and 0.1% (w/v) SDS). Proteins were transferred to nitrocellulose membranes (0.45 μm pore; Whatman) at 20 V for 30 min in transfer buffer (25 mm Tris, 150 mm glycine, 20% (v/v) methanol, and 0.05% (w/v) SDS) using the semidry method. Membranes were blocked for 2 h in 5% (w/v) nonfat milk solution in TBS/Tween (20 mm Tris (pH 7.4), 137 mm NaCl, and 0.1% (v/v) Tween 20). They were later exposed to a primary rabbit polyclonal antibody for UCP1 (Sigma U6382) at either 0.05 μg/ml (BAT) or 0.15 μg/ml (thymus) in blocking buffer and incubated overnight at 4 °C. After washing with TBS/Tween, the membranes were incubated with peroxidase-conjugated goat anti-rabbit secondary antibody (Thermo Scientific 31463) at 0.05 μg/ml in blocking buffer for 1 h. Densitometry analysis with ImageJ was used to quantify immunoblots (28). Purified UCP1 was obtained from laboratory stocks, generated as described previously (29).
Statistics
Statistical significance was assessed by analysis of variance for repeated measures with Dunnett's post-test (95% confidence interval) using GraphPad Prism version 5.0a. Binding parameters and curve fits were also calculated with this software. Values of p < 0.05 (*) were considered statistically significant (**, p < 0.01).
RESULTS
Using mant-GDP to Study Conformational Changes in UCP1
The activation of UCP1 by fatty acids and its inhibition by nucleotides can be described by Michaelis-Menten kinetics with a competitive inhibitor (14). However, fatty acids have little effect on the affinity or maximum binding of [3H]GDP to UCP1 (9, 11, 16). This apparent discrepancy between the functional and binding properties of the protein is a persistent hurdle, preventing a thorough description of the physiological mechanism and regulation of UCP1.
To address this issue, we used a fluorescently labeled guanine nucleotide analog, mant-GDP, to determine whether fatty acids alter the binding of nucleotides to UCP1. The demonstration of such a change in nucleotide binding would imply a fatty acid-induced conformational change in UCP1 and would help explain how fatty acids can overcome nucleotide inhibition.
Tracking the mant Moiety as a Label
It is crucial to reiterate that the fluorescence of mant-GDP was used in this study simply as a proportional indicator of how much nucleotide was bound to a given sample. Initial attempts to find a UCP1-specific signal by FRET upon mant-GDP binding, as was done for purified UCP2 (30), were unsuccessful. In this study, fluorescence from mant-GDP was measured only as a straightforward label of bound nucleotide in resolubilized mitochondrial pellets; the mant substitution can be thought of as similar to the tritium substitution in [3H]GDP.
Inhibition of Proton Conductance by mant-GDP
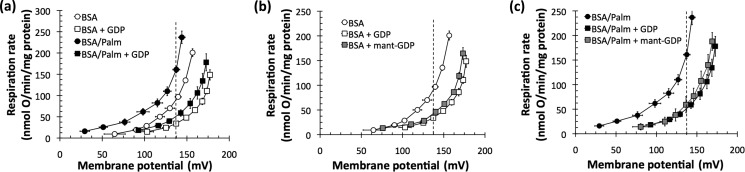
Prior to studying its binding to UCP1, it was necessary to verify that mant-GDP could inhibit proton conductance as effectively as unmodified GDP. Fig. 1a shows the classical activation and inhibition of proton conductance in mitochondria from BAT. As expected, the proton leak rate at any given mitochondrial membrane potential (Δψm) increased with the addition of 40 μm albumin-buffered palmitate. The addition of 30 μm GDP decreased the proton conductance in the absence of palmitate and almost entirely inhibited the increase in proton conductance caused by palmitate addition.
FIGURE 1.
Effects of fatty acids and nucleotides on proton leak kinetics in mitochondria from rat BAT. a, effects of 40 nm free palmitate (Palm) and 30 μm GDP. b, effects of 30 μm GDP and 30 μm mant-GDP in the absence of palmitate. c, effects of 30 μm GDP and 30 μm mant-GDP in the presence of 40 nm free palmitate. There was no significant difference in the inhibition of proton conductance between mant-GDP and [3H]GDP. All comparisons of proton leak rate were made at 137 mV, the highest shared Δψm for all biological replicates (dashed vertical lines in a–c). All data are means ± S.E. (n = 3).
Fig. 1 (b and c) shows that 30 μm mant-GDP was indeed an equally potent inhibitor of proton conductance as 30 μm unmodified GDP in either the absence (Fig. 1b) or presence (Fig. 1c) of exogenously added palmitate. We conclude that mant-GDP and unmodified GDP are functionally equivalent inhibitors of the proton conductance of UCP1.
Mitochondrial Labeling with mant-GDP
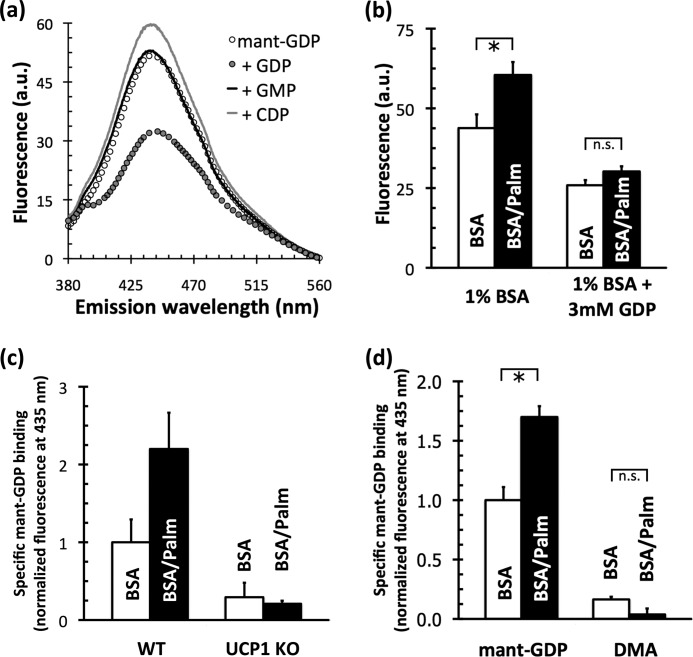
Mitochondrial labeling with mant-GDP is insensitive to nucleotides that are not ligands of UCP1. Fig. 2a shows a fluorescence emission spectrum of mant-GDP released from BAT mitochondria after binding, centrifugation, and solubilization of the pellet. The amount of mant-GDP retained in the pellet was reduced in the presence of excess unmodified GDP. The difference between these spectra represents specific mant-GDP binding.
FIGURE 2.
Binding of mant-GDP to rat and mouse BAT mitochondria. a, fluorescence emission spectrum (λex = 350 nm) of mant-GDP bound to rat BAT mitochondria. Mitochondria (2.5 mg/ml protein) were incubated for 5 min with 30 μm mant-GDP in 250 mm sucrose, 20 mm HEPES (pH 5.9), 1 mm EDTA, 3.75 μg of carboxyatractylate/mg of mitochondrial protein, and 1 mm GDP, GMP, or CDP as shown. Mitochondria were sedimented, and the pellet was resuspended in detergent before analysis. a.u., arbitrary units. b, effect of palmitate in the absence or presence of 3 mm GDP. Rat BAT mitochondria were incubated with 30 μm mant-GDP for 5 min at 23 °C in assay medium containing either 1% (w/v) BSA or 1% (w/v) BSA supplemented with 1 mm palmitate (Palm; 515 nm free palmitate). Pellets were resuspended in medium lacking palmitate, and fluorescence was measured (λex = 350 nm; λem = 435 nm). Data are means ± S.E. (n = 3). *, p < 0.05; n.s., not significant. c, effect of palmitate on specific mant-GDP binding in BAT mitochondria from wild-type and Ucp1-null mice. Binding was measured as described for b, and nonspecific binding was calculated as the signal in the presence of 3 mm GDP. Data are means ± spread from two independent mitochondrial preparations of wild-type and sibling-paired Ucp1-null animals. KO, knock-out. d, effect of palmitate on binding of mant-GDP and dimethyl anthranilate (DMA) to rat BAT mitochondria. Specific binding was measured as described for c. Data are means ± S.E. (n = 10 (mant-GDP) or n = 3 (dimethyl anthranilate)).
The nucleotide specificity for UCP1 is well documented: it binds to and is inhibited by only purine nucleoside di- and triphosphates (31). Importantly, neither excess GMP nor CDP reduced the amount of mant-GDP bound (Fig. 2a), showing that mitochondrial labeling with mant-GDP was in accordance with the nucleotide specificity of UCP1.
Fatty Acids Enhance Specific Binding of mant-GDP
Upon finding that mitochondria from BAT could specifically bind mant-GDP, a clear extension of the result was to determine whether this binding could be altered by fatty acids. Fig. 2b shows that 515 nm free palmitate (1 mm palmitate buffered with 1% (w/v) BSA) significantly increased the amount of mant-GDP bound to mitochondria from BAT after incubation for 5 min.
Critically, palmitate affected only the specific binding of mant-GDP. Nonspecific binding, calculated as the amount of mant-GDP bound in the presence of excess unmodified GDP, was unaffected (Fig. 2b, right). Although palmitate slightly increased the specific binding of mant-GTP, mant-ADP, and mant-ATP to mitochondria from BAT (data not shown), a significant, pronounced increase was seen only with mant-GDP binding.
Fatty Acid-enhanced Binding Is Mediated by UCP1
To ensure that the palmitate-stimulated increase in mant-GDP binding was a genuine interaction between UCP1 and the nucleotide, two controls were required. First, Fig. 2c shows that UCP1 was almost wholly responsible for this effect. In mitochondria from BAT of wild-type mice, specific mant-GDP binding was more than doubled in the presence of palmitate (Fig. 2c, left). This increase was almost entirely absent in mitochondria from BAT of Ucp1-null mice (Fig. 2c, right), where trace binding was likely attributable to the adenine nucleotide translocator (32).
Second, it was necessary to prove that there was no specific interaction between UCP1 and the mant moiety attached to the nucleotide. Fig. 2d shows that GDP is the functional group responsible for mant-GDP binding. Substitution of GDP with a methyl group, resulting in dimethyl anthranilate, which fluoresces similarly to mant-GDP, all but eliminated both the specific binding of mant-GDP and the increase in binding stimulated by palmitate. The results from these two experiments led to the following conclusion: the palmitate-stimulated increase in mant-GDP binding is due exclusively to UCP1 interacting with the nucleotide.
Competition between mant-GDP and [3H]GDP
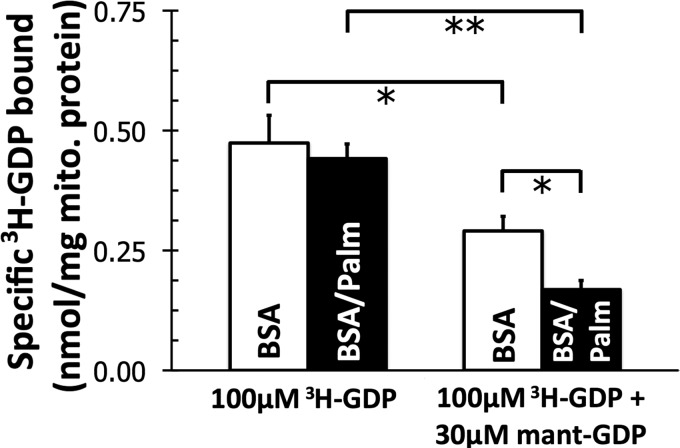
The discovery that palmitate changed the amount of mant-GDP bound to UCP1 is of particular importance because of the multiple independent confirmations that fatty acids do not substantially change the ability of UCP1 to bind [3H]GDP (9, 11, 16). It was therefore necessary to verify that mant-GDP and [3H]GDP both bound to the established nucleotide-binding site in UCP1.
As expected, palmitate did not change the amount of [3H]GDP bound to mitochondria from BAT (Fig. 3, left). Fig. 3 (right) shows two significant effects when mitochondria were incubated with both mant-GDP and [3H]GDP. First, a half-maximum concentration of mant-GDP (30 μm) (Table 1) decreased the specific binding of [3H]GDP when mitochondria were incubated with a near maximum concentration of [3H]GDP (100 μm) (9, 33) in the presence of BSA. Second, the displacement of [3H]GDP by mant-GDP was enhanced when palmitate was present in the medium. Therefore, not only does mant-GDP bind to the classical nucleotide-binding site of UCP1, but also the palmitate-stimulated increase in binding occurs at this site.
FIGURE 3.
Effect of 30 μm mant-GDP, a concentration approximating its K0.5 (Table 1), on specific binding of 100 μm [3H]GDP to mitochondria from rat BAT. Data are means ± S.E. (n = 4). mito., mitochondrial. *, p < 0.05; **, p < 0.01.
TABLE 1.
Half-maximal concentrations (K0.5) and apparent maximum binding capacities (Bmax) of UCP1 in rat BAT mitochondria for mant-GDP and [3H]GDP in the absence or presence of 515 nm free palmitate
Data are means ± S.E. (n = 5).
|
K0.5 |
Bmax |
|||
|---|---|---|---|---|
| BSA | BSA/palmitate | BSA | BSA/palmitate | |
| μm | nmol/mg protein | |||
| mant-GDP | 12.6 ± 3.0 | 28.2 ± 6.3 | 0.29 ± 0.02a | 0.63 ± 0.04a |
| [3H]GDP | 8.2 ± 1.5 | 10.9 ± 3.2 | 0.57 ± 0.02 | 0.63 ± 0.04 |
a Statistically significant pairing (p < 0.05).
Affinities and Maximum Binding of mant-GDP
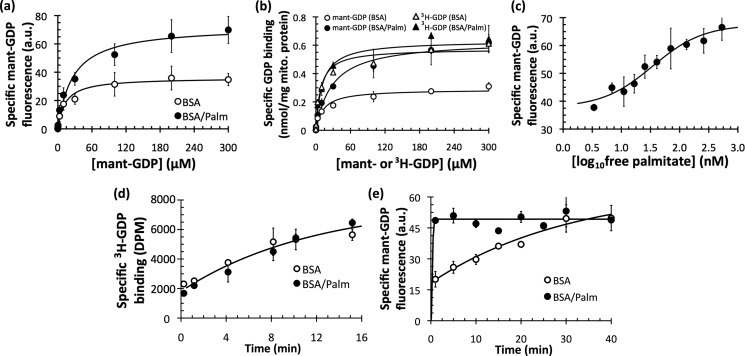
Upon finding that fatty acids increased mant-GDP binding to UCP1, it was important to determine the mechanism by which this occurred. In principle, fatty acids could have increased the amount of nucleotide bound to UCP1 by changing either the apparent affinity of UCP1 for mant-GDP (K0.5) or the maximum capacity of UCP1 to bind mant-GDP (Bmax).
Fig. 4a shows that palmitate substantially changed the apparent Bmax for mant-GDP. At any given concentration, palmitate increased the amount of mant-GDP specifically bound to mitochondria from BAT after a 5-min incubation. There was no decrease in K0.5 (Table 1).
FIGURE 4.
Kinetics of mant-GDP and [3H]GDP binding to rat BAT mitochondria. a, mitochondria from BAT were incubated with a range of mant-GDP concentrations for 5 min in the absence or presence of 515 nm free palmitate (Palm), and specific binding to the pellet was measured in arbitrary fluorescence units (a.u.). Data are means ± S.E. (n = 5). Aggregate data were modeled as ligand binding to a single site of an enzyme (Michaelis-Menten equation). b, data from a were calibrated with a standard curve of known mant-GDP concentrations and compared with a similar experiment to measure the specific binding of [3H]GDP. Data are means ± S.E. (n = 4). mito., mitochondrial. c, specific binding of mant-GDP as a function of free palmitate concentration. BAT mitochondria (1.25 mg/ml) were incubated with 30 μm mant-GDP for 5 min at 23 °C with a range of free palmitate concentrations. Aggregate data were modeled with a sigmoidal dose response. K0.5 = 35 nm (calculated as the inflection point of the curve, midway between the upper and lower bounds). Data are means ± S.E. (n = 3). d, time course of specific binding of 500 nm [3H]GDP to BAT mitochondria in the absence and presence of 515 nm free palmitate at 4 °C. Data are means ± S.E. (n = 3). e, time course of specific binding of 100 μm mant-GDP to BAT mitochondria in the absence and presence of 515 nm free palmitate at 21 °C. Data are means ± spread (n = 2).
The observations that palmitate changed the Bmax of mant-GDP binding (Fig. 4a) and that the palmitate-stimulated increase in binding occurred at the classical nucleotide-binding site of UCP1 (Fig. 3) led to the following hypothesis: mant-GDP binding to UCP1 is occluded in the absence of palmitate. Comparisons of the binding of mant-GDP with [3H]GDP supported this hypothesis (Fig. 4b). As expected, palmitate again changed neither the K0.5 nor Bmax of [3H]GDP binding to UCP1 (Fig. 4b, triangles). In stark contrast, palmitate had a dramatic effect on the ability of mant-GDP to bind to UCP1. When the medium lacked palmitate, the apparent maximum capacity of UCP1 to bind mant-GDP was severely restricted (Fig. 4b, open circles). With palmitate present, however, the Bmax was restored, and the binding of mant-GDP was in line with that of [3H]GDP (Fig. 4b, closed circles; and Table 1).
Physiological Relevance of Palmitate Concentration
A significant change in nucleotide binding due to fatty acids has never before been reported for UCP1. Therefore, it was important to confirm that this effect could occur at physiologically relevant palmitate concentrations because the experiments described above used saturating concentrations (515 nm) of free palmitate.
Fig. 4c shows that physiologically relevant concentrations of palmitate stimulated an increase in mant-GDP binding. 35 nm palmitate was required for a half-maximum increase in mant-GDP binding, which falls between the reported values of 5 nm (14) and 80 nm (34) for the K0.5 of fatty acid activation of UCP1.
Time Course of mant-GDP Binding
How might palmitate restore the apparent maximum capacity of UCP1 to bind mant-GDP? A time course of nucleotide binding revealed that palmitate changed the rate constant of mant-GDP binding to UCP1. Fig. 4d shows the longstanding result with [3H]GDP binding: the time course of binding of this nucleotide to UCP1 (conducted on ice to slow the rate) was independent of the presence or absence of palmitate. However, Fig. 4e shows this was not the case for mant-GDP, as palmitate drastically increased the rate at which mant-GDP bound to UCP1 (conducted at room temperature to give higher rates). Without palmitate, mant-GDP eventually reached its maximum binding after 30 min. It was not, however, anywhere near its maximum at 5 min, the duration of the binding incubation used in the experiments described above.
We therefore conclude that palmitate induced a change (presumably conformational) in UCP1 that resulted in an increased rate constant for mant-GDP binding. Our results are consistent with the inability of previous studies to diagnose this conformational change using [3H]GDP, as there was no change in any binding parameters for [3H]GDP upon palmitate addition.
This finding has profound implications for the regulation and catalytic mechanism of UCP1. Therefore, it was important to use another method to independently confirm that palmitate induces a conformational change in UCP1.
Trypsinolysis of UCP1
Controlled enzymatic proteolysis has been used to correctly identify the topology of mitochondrial carriers (35). This has been done for UCP1, and the technique can link the functional inhibition of UCP1 by GDP with a conformational change that protects Lys-292 from tryptic cleavage (36). Therefore, a natural extension of the finding discussed above was to determine whether trypsinolysis could detect a palmitate-induced conformational change in UCP1.
Selectivity and Calibration of Western Analysis
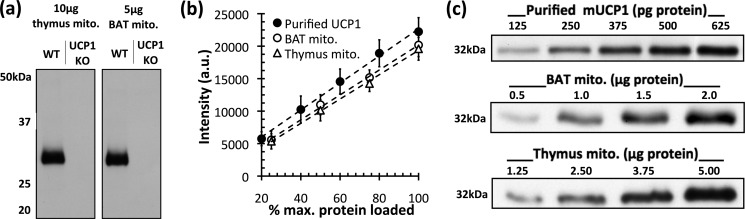
Prior to conducting trypsinolysis of UCP1, it was necessary to confirm that the analysis was both specific and appropriately calibrated. Fig. 5a shows that immunodetection of UCP1 was specific, as there was no signal in mitochondria isolated from BAT or thymus of Ucp1−/− mice. Furthermore, Fig. 5 (b and c) shows that a 4–5-fold pseudo-linear range for Western analysis existed using mitochondria and purified UCP1. No measurements were made subsequently outside of the range of protein concentrations shown in Fig. 5 (37). To ensure that the transfer events were reliable, two samples of mitochondria were collected prior to exogenous trypsin addition (time = 0) and run on opposite ends of the SDS-polyacrylamide gel (data not shown). Each experiment was conducted with five biological replicates.
FIGURE 5.
Selectivity and linearity of Western analysis of UCP1. a, antibody specificity in mitochondria (mito.) from mouse thymus (left) and mouse BAT (right). The 32-kDa band present in mouse mitochondria from wild-type thymus and BAT was not present in mitochondria from sibling-paired knock-out (KO) mice with Ucp1 ablated. b, the working range at which the immunoblot signal was pseudo-linearly dependent on protein concentration was determined for all samples used in this study. Three independent mitochondrial preparations and Western analyses revealed a 4–5-fold working linear range. Data are means ± S.E. (n = 3). a.u., arbitrary units. c, representative UCP1 immunoblots of purified UCP1 (125–625 pg of protein) and mitochondrial protein from BAT (0.5–2.0 μg of protein) and thymus (1.25–5.00 μg of protein). mUCP1, recombinant mouse UCP1.
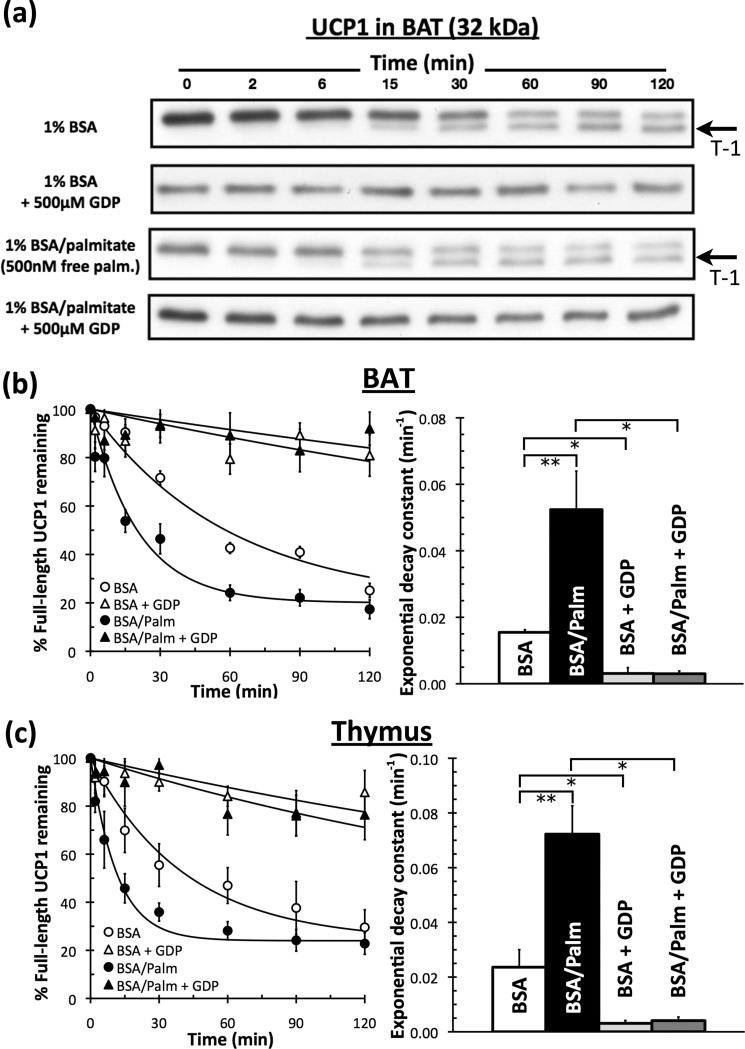
Trypsinolysis of UCP1 in Mitochondria from BAT
Fig. 6 (a and b) confirms previous work showing that purine nucleotides protect UCP1 from tryptic cleavage at the C terminus (36, 38). However, palmitate significantly accelerated the rate of degradation of UCP1 in mitochondria from BAT. The exponential decay constant associated with UCP1 trypsinolysis was 3-fold greater upon the addition of 500 nm free palmitate (Fig. 6b), indicating an acute molecular interaction with palmitate that induced a conformational change in UCP1.
FIGURE 6.
Trypsinolysis of UCP1 in the absence and presence of palmitate and GDP. Isolated rat BAT or rat thymus mitochondria were treated with exogenous trypsin. The reaction was stopped at defined times and probed for UCP1. a, representative immunoblot of matched treatments for UCP1 in BAT mitochondria. 1.5 μg of mitochondrial protein was loaded in each lane. The T-1 degradation fragment is indicated with an arrow. palm., palmitate. b, trypsinolysis in mitochondria from BAT was modeled with simple exponential decay kinetics to a non-zero value on each day to reflect incomplete degradation (perhaps due to incomplete rupturing of the outer membrane). Left, aggregate data are from five independent mitochondrial preparations. Right, individual rate constants from each experiment. Data are means ± S.E. (n = 5). c, aggregate data and rate constants for trypsinolysis of UCP1 in mitochondria from thymus as described for b. Data are means ± S.E. (n = 5). *, p < 0.05; **, p < 0.01.
Trypsin initially cleaves UCP1 at Lys-292, and the resultant UCP1 molecule lacking its C terminus is historically called “T-1” (36, 38). Preincubation of mitochondria from BAT with 500 μm GDP completely inhibited the formation of this species (Fig. 6a).
Importantly, the accelerated degradation profile in response to palmitate also included the appearance of the T-1 product (Fig. 6a). It can therefore be concluded that the conformation induced by palmitate, in direct contrast to GDP, made Lys-292 more accessible to trypsinolysis. This conformational change was entirely inhibited when mitochondria were incubated with 500 μm GDP prior to the addition of 500 nm palmitate.
Measurements of cleavage of N-benzoyl-l-arginine ethyl ester showed unperturbed signals in the presence of palmitate or GDP (data not shown), indicating that the specific activity of trypsin was not compromised by these effectors. Moreover, no endogenous degradation was seen in the absence of trypsin over 2 h (data not shown), suggesting that palmitate was directly affecting the conformation of UCP1 and not stimulating endogenous proteolysis.
Trypsinolysis of UCP1 in Mitochondria from Thymus
Trypsinolysis of UCP1 expressed in thymus resulted in effects similar to those obtained by trypsinolysis of the protein expressed in BAT. (i) The protein was almost entirely protected by GDP. (ii) Palmitate induced a conformational change that accelerated the rate of degradation. (iii) The hallmark T-1 band was present upon trypsin addition. (iv) Induction of this conformation was completely inhibited upon preincubation with GDP (Fig. 6c).
DISCUSSION
Although fatty acids can overcome nucleotide inhibition in BAT mitochondria to activate UCP1, this functional competition has never been explained at the molecular level, as ligand binding experiments using [3H]GDP have revealed little or no effect of fatty acids on nucleotide binding. Our results can resolve this apparent discrepancy.
We provide the first evidence that fatty acids change the conformation of UCP1: fatty acids increase the rate constant of mant-GDP binding and accelerate the trypsinolysis of UCP1. This conformational change provides the means by which fatty acids can overcome inhibition by nucleotides to activate UCP1.
At first, the observation that palmitate induces a conformation that promotes mant-GDP binding may seem counterintuitive, as fatty acids are activators of UCP1 activity, and nucleotides are potent inhibitors. Rather, the observation strongly supports a model in which UCP1 broadly exists in two conformations: a “loose” proton-conducting conformation and a “tight” inhibited conformation (36, 39).
The data show that [3H]GDP can bind equally well to each conformation but that mant-GDP strongly favors the loose conformation. Presumably, fatty acids facilitate mant-GDP binding by inducing an open, proton-translocating conformation that can better accommodate the bulky, substituted nucleotide. Such a model not only supports the existence of two conformations of UCP1 but can also accommodate a two-stage model for nucleotide binding (39, 40).
Differential binding of modified nucleotides to UCP1 is not without precedent. For both mant and dansyl nucleotides, bulky substitutions attached to 2′-O or 3′-O of the ribose cause preference for different conformations of UCP1 depending on the nucleotide involved (40).
Fatty acids did not significantly change the capacity of UCP1 to bind mant-ADP, mant-ATP, or mant-GTP. This reinforces two principal observations. First, the altered binding of mant-GDP is not due to a promiscuous reaction between fatty acids and the “mant” moiety, and second, the conformational change promoted by fatty acids is rather subtle at the level of ligand binding. It accelerates mant-GDP binding but is not so pronounced that it alters the binding of [3H]GDP or other mant-substituted nucleotide analogs.
The conformational change induced by fatty acids can also be diagnosed by controlled enzymatic proteolysis. The protection from trypsinolysis afforded by GDP has previously been linked to a conformation of UCP1 that does not translocate protons. It is therefore likely that the conformation induced by palmitate, which accelerates tryptic cleavage, is protonophoric.
When the medium was supplemented with both 500 nm palmitate and 500 μm GDP, UCP1 was entirely protected regardless of the order of effector addition (data not shown). Initially, this may seem more in line with UCP1 reconstituted into liposomes, where fatty acids cannot overcome nucleotide inhibition of proton translocation (16), rather than the functional competition seen in mitochondria (14). No such determination can be made, however, without a proper titration of both effectors. The established K0.5 values for nucleotides (9, 33) and fatty acids (14, 34) indicate that the concentrations used routinely in this study were saturating for both.
The results obtained from trypsinolysis in mitochondria from thymus do more than simply corroborate the results from BAT. Foremost, they provide independent evidence to support the functional expression of UCP1 in the rodent thymus, which heretofore has been shown only by one research group (4, 5).
Additionally, showing that fatty acids changed the conformation of UCP1 expressed in this tissue could be relevant to the evolutionary history of uncoupling proteins. It remains controversial as to whether UCP1 and its fatty acid activation evolved part and parcel with BAT and non-shivering thermogenesis (41, 42). At the very least, our demonstration that fatty acids can acutely activate UCP1 in mitochondria from thymus shows that fatty acid-activated uncoupling does not strictly imply a thermogenic function. Emerging biophysical methods to study conformational changes in UCP1 could prove powerful, provided expression in these systems exhibits regulatory properties similar to BAT (43).
Although all three competing models of UCP1 activation can accommodate the demonstration that palmitate induces a conformational change in the protein, only the functional competition model explicitly suggests an allosteric change induced by fatty acids. A conformational change is also compatible with evidence that fatty acids and nucleotides functionally compete to regulate UCP1 activity without fatty acids changing the binding of unmodified nucleotides. In principle, palmitate and GDP can compete to induce conformational changes in the protein to modulate activity with neither molecule preventing the other from binding to the protein.
This work was supported, in whole or in part, by National Institutes of Health Grants P01 AG025901 and PL1 AG032118 (to M. D. B.). This work was also supported by the Medical Research Council (United Kingdom) and a British Marshall scholarship and a National Science Foundation graduate research fellowship (to A. S. D.).
- BAT
- brown adipose tissue
- TPMP
- triphenylmethylphosphonium
- mant-GDP
- 2′/3′-O-(N-methylanthraniloyl)-GDP.
REFERENCES
- 1. Heaton G. M., Wagenvoord R. J., Kemp A., Jr., Nicholls D. G. (1978) Brown adipose tissue mitochondria: photoaffinity labeling of the regulatory site of energy dissipation. Eur. J. Biochem. 82, 515–521 [DOI] [PubMed] [Google Scholar]
- 2. Lin C. S., Klingenberg M. (1980) Isolation of the uncoupling protein from brown adipose tissue mitochondria. FEBS Lett. 113, 299–303 [DOI] [PubMed] [Google Scholar]
- 3. Cannon B., Nedergaard J. (2004) Brown adipose tissue: function and physiological significance. Physiol. Rev. 84, 277–359 [DOI] [PubMed] [Google Scholar]
- 4. Adams A. E., Hanrahan O., Nolan D. N., Voorheis H. P., Fallon P., Porter R. K. (2008) Images of mitochondrial UCP1 in mouse thymocytes using confocal microscopy. Biochim. Biophys. Acta 1777, 115–117 [DOI] [PubMed] [Google Scholar]
- 5. Carroll A. M., Haines L. R., Pearson T. W., Fallon P. G., Walsh C. M., Brennan C. M., Breen E. P., Porter R. K. (2005) Identification of a functioning mitochondrial uncoupling protein 1 in thymus. J. Biol. Chem. 280, 15534–15543 [DOI] [PubMed] [Google Scholar]
- 6. Jastroch M., Buckingham J. A., Helwig M., Klingenspor M., Brand M. D. (2007) Functional characterization of UCP1 in the common carp: uncoupling activity in liver mitochondria and cold-induced expression in the brain. J. Comp. Physiol. B 177, 743–752 [DOI] [PubMed] [Google Scholar]
- 7. Brand M. D. (2000) Uncoupling to survive? The role of mitochondrial inefficiency in aging. Exp. Gerontol. 35, 811–820 [DOI] [PubMed] [Google Scholar]
- 8. Azzu V., Brand M. D. (2010) The on-off switches of the mitochondrial uncoupling proteins. Trends Biochem. Sci. 35, 298–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rial E., Poustie A., Nicholls D. G. (1983) Brown adipose tissue mitochondria: the regulation of the 32,000 Mr uncoupling protein by fatty acids and purine nucleotides. Eur. J. Biochem. 137, 197–203 [DOI] [PubMed] [Google Scholar]
- 10. Klingenberg M., Winkler E. (1985) The reconstituted isolated uncoupling protein is a membrane potential-driven H+ translocator. EMBO J. 4, 3087–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Winkler E., Klingenberg M. (1994) Effect of fatty acids on H+ transport activity of the reconstituted uncoupling protein. J. Biol. Chem. 269, 2508–2515 [PubMed] [Google Scholar]
- 12. Garlid K. D., Jabůrek M., Jezek P. (1998) The mechanism of proton transport mediated by mitochondrial uncoupling proteins. FEBS Lett. 438, 10–14 [DOI] [PubMed] [Google Scholar]
- 13. Breen E. P., Gouin S. G., Murphy A. F., Haines L. R., Jackson A. M., Pearson T. W., Murphy P. V., Porter R. K. (2006) On the mechanism of mitochondrial uncoupling protein 1 function. J. Biol. Chem. 281, 2114–2119 [DOI] [PubMed] [Google Scholar]
- 14. Shabalina I. G., Jacobsson A., Cannon B., Nedergaard J. (2004) Native UCP1 displays simple competitive kinetics between the regulators purine nucleotides and fatty acids. J. Biol. Chem. 279, 38236–38248 [DOI] [PubMed] [Google Scholar]
- 15. Divakaruni A. S., Brand M. D. (2011) The regulation and physiology of mitochondrial proton leak. Physiology 26, 192–205 [DOI] [PubMed] [Google Scholar]
- 16. Jezek P., Orosz D. E., Modriansky M., Garlid K. D. (1994) Transport of anions and protons by the mitochondrial uncoupling protein and its regulation by nucleotides and fatty acids. A new look at old hypotheses. J. Biol. Chem. 269, 26184–26190 [PubMed] [Google Scholar]
- 17. Enerback S., Jacobsson A., Simpson E. M., Guerra C., Yamashita H., Harper M. E., Kozak L. P. (1997) Mice lacking mitochondrial uncoupling protein are cold-sensitive but not obese. Nature 387, 90–94 [DOI] [PubMed] [Google Scholar]
- 18. Parker N., Crichton P. G., Vidal-Puig A. J., Brand M. D. (2009) Uncoupling protein 1 (UCP1) contributes to the basal proton conductance of brown adipose tissue mitochondria. J. Bioenerg. Biomembr. 41, 335–342 [DOI] [PubMed] [Google Scholar]
- 19. Esteves T. C., Parker N., Brand M. D. (2006) Synergy of fatty acid and reactive alkenal activation of proton conductance through uncoupling protein 1 in mitochondria. Biochem. J. 395, 619–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 21. Gornall A. G., Bardawill C. J., David M. M. (1949) Determination of serum proteins by means of the biuret reaction. J. Biol. Chem. 177, 751–766 [PubMed] [Google Scholar]
- 22. Parker N., Affourtit C., Vidal-Puig A., Brand M. D. (2008) Energization-dependent endogenous activation of proton conductance in skeletal muscle mitochondria. Biochem. J. 412, 131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reynafarje B., Costa L. E., Lehninger A. L. (1985) O2 solubility in aqueous media determined by a kinetic method. Anal. Biochem. 145, 406–418 [DOI] [PubMed] [Google Scholar]
- 24. Brand M. D. (1995) in Bioenergetics: A Practical Approach (Brown G. C., Cooper C. E., eds) pp, 39–62, IRL Press, Oxford [Google Scholar]
- 25. Richieri G. V., Anel A., Kleinfeld A. M. (1993) Interactions of long-chain fatty acids and albumin: determination of free fatty acid levels using the fluorescent probe ADIFAB. Biochemistry 32, 7574–7580 [DOI] [PubMed] [Google Scholar]
- 26. Hiratsuka T. (1983) New ribose-modified fluorescent analogs of adenine and guanine nucleotides available as substrates for various enzymes. Biochim. Biophys. Acta 742, 496–508 [DOI] [PubMed] [Google Scholar]
- 27. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 28. Abramoff M. D., Magelhaes P. J., Ram S. J. (2004) Image processing with ImageJ. Biophotonics Int. 11, 36–42 [Google Scholar]
- 29. Stuart J. A., Harper J. A., Brindle K. M., Jekabsons M. B., Brand M. D. (2001) A mitochondrial uncoupling artifact can be caused by expression of uncoupling protein 1 in yeast. Biochem. J. 356, 779–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jekabsons M. B., Echtay K. S., Brand M. D. (2002) Nucleotide binding to human uncoupling protein 2 refolded from bacterial inclusion bodies. Biochem. J. 366, 565–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lin C. S., Klingenberg M. (1982) Characteristics of the isolated purine nucleotide-binding protein from brown fat mitochondria. Biochemistry 21, 2950–2956 [DOI] [PubMed] [Google Scholar]
- 32. Khailova L. S., Prikhodko E. A., Dedukhova V. I., Mokhova E. N., Popov V. N., Skulachev V. P. (2006) Participation of ATP/ADP antiporter in oleate- and oleate hydroperoxide-induced uncoupling suppressed by GDP and carboxyatractylate. Biochim. Biophys. Acta 1757, 1324–1329 [DOI] [PubMed] [Google Scholar]
- 33. Klingenberg M. (1988) Nucleotide binding to uncoupling protein. Mechanism of control by protonation. Biochemistry 27, 781–791 [DOI] [PubMed] [Google Scholar]
- 34. González-Barroso M. M., Fleury C., Bouillaud F., Nicholls D. G., Rial E. (1998) The uncoupling protein UCP1 does not increase the proton conductance of the inner mitochondrial membrane by functioning as a fatty acid anion transporter. J. Biol. Chem. 273, 15528–15532 [DOI] [PubMed] [Google Scholar]
- 35. Marty I., Brandolin G., Gagnon J., Brasseur R., Vignais P. V. (1992) Topography of the membrane-bound ADP/ATP carrier assessed by enzymatic proteolysis. Biochemistry 31, 4058–4065 [DOI] [PubMed] [Google Scholar]
- 36. Eckerskorn C., Klingenberg M. (1987) In the uncoupling protein from brown adipose tissue, the C terminus protrudes to the c-side of the membrane as shown by tryptic cleavage. FEBS Lett. 226, 166–170 [DOI] [PubMed] [Google Scholar]
- 37. Aksamitiene E., Hoek J. B., Kholodenko B., Kiyatkin A. (2007) Multistrip Western blotting to increase quantitative data output. Electrophoresis 28, 3163–3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huang S. G. (2003) Limited proteolysis reveals conformational changes in uncoupling protein 1 from brown adipose tissue mitochondria. Arch. Biochem. Biophys. 420, 40–45 [DOI] [PubMed] [Google Scholar]
- 39. Huang S. G., Klingenberg M. (1996) Two-stage nucleotide binding mechanism and its implications to H+ transport inhibition of the uncoupling protein from brown adipose tissue mitochondria. Biochemistry 35, 7846–7854 [DOI] [PubMed] [Google Scholar]
- 40. Huang S. G., Klingenberg M. (1995) Fluorescent nucleotide derivatives as specific probes for the uncoupling protein: thermodynamics and kinetics of binding and the control by pH. Biochemistry 34, 349–360 [DOI] [PubMed] [Google Scholar]
- 41. Hughes D. A., Jastroch M., Stoneking M., Klingenspor M. (2009) Molecular evolution of UCP1 and the evolutionary history of mammalian non-shivering thermogenesis. BMC Evol. Biol. 9, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Saito S., Saito C. T., Shingai R. (2008) Adaptive evolution of the uncoupling protein 1 gene contributed to the acquisition of novel non-shivering thermogenesis in ancestral eutherian mammals. Gene 408, 37–44 [DOI] [PubMed] [Google Scholar]
- 43. Blesneac I., Ravaud S., Juillan-Binard C., Barret L. A., Zoonens M., Polidori A., Miroux B., Pucci B., Pebay-Peyroula E. (2012) Production of UCP1, a membrane protein, from the inner mitochondrial membrane using the cell-free expression system in the presence of a fluorinated surfactant. Biochim. Biophys. Acta 1818, 798–805 [DOI] [PubMed] [Google Scholar]