Background: A sex pheromone system controls bacterial conjugation.
Results: The initial receptor PrgZ has been crystallized in complex with the sex pheromone cCF10.
Conclusion: An extensive network of hydrogen bonds explains the high peptide specificity of PrgZ.
Significance: The sex pheromone and inhibitor peptide compete for binding to PrgZ, providing new insight into the regulation of conjugation.
Keywords: ABC Transporter, Bacterial Conjugation, Bacterial Signal Transduction, Membrane Transport, Pheromone, Receptor Structure-Function
Abstract
We present the crystal structure of the pheromone receptor protein PrgZ from Enterococcus faecalis in complex with the heptapeptide cCF10 (LVTLVFV), which is used in signaling between conjugative recipient and donor cells. Comparison of PrgZ with homologous oligopeptide-binding proteins (AppA and OppA) explains the high specificity of PrgZ for hydrophobic heptapeptides versus the promiscuity of peptide binding in the homologous proteins.
Introduction
Enterococcus faecalis is a Gram-positive bacterium that inhabits the gastrointestinal tract of mammals (1). In the past decades, E. faecalis infections in humans have increased, especially in nosocomial environments (2, 3). One of the main reasons for this increase is an abundance of efficient conjugative resistance transfer systems induced by peptide sex pheromones, which allow E. faecalis to quickly become multiresistant to antimicrobial agents. The enterococcal sex pheromone systems represent a variation on quorum sensing, which allows an entire microbial population to carry out coordinated responses in a density-dependent fashion (4). In E. faecalis, oligopeptides secreted by plasmid-free recipient cells allow plasmid donors to express conjugation functions only in the presence of recipients. One of the peptides in E. faecalis is the heptapeptide cCF10 (LVTLVFV), which originates from processing of the chromosomal gene product of ccfA (5). This peptide, together with the 70-kb plasmid pCF10, which confers antibiotic resistance and carries genes for conjugation proteins, contributes to the virulence of the bacteria (6). pCF10 conjugation is activated using cCF10 as the signal.
The inhibitor peptide iCF10 (AITLIFI), encoded by the icf10 gene on pCF10, prevents self-induction of the conjugation system by endogenous cCF10. Both cCF10 and iCF10 are secreted in the process of their production, and thus, both are re-imported into the cell (6). The initial receptor for cCF10 is PrgZ, a lipid-anchored extracellular peptide-binding protein. PrgZ delivers cCF10 to the oligopeptide permease (Opp), which subsequently transports it inside the cell (7), where cCF10 binds to PrgX, a DNA repressor protein (8, 9). Inside a pCF10-harboring cell, PrgX normally has a tetrameric quaternary structure with iCF10 bound. In the tetrameric state, PrgX binds to pCF10 and represses transcription of the conjugation genes. Upon an increase in the cytoplasmic concentration of cCF10, the inhibitor peptide is displaced, and the tetrameric structure of PrgX is disrupted, leading to loss of its repressor activity (9). This initiates the transcription of the prgQ conjugation operon (10).
PrgZ is the sole extracellular receptor for cCF10 at typical concentrations of the pheromone in vivo of 10 pm (6, 11). The oligopeptide-binding protein A (OppA) of E. faecalis can also bind cCF10 and initiate pheromone import at higher (non-physiological) concentrations (12). It is clear that iCF10 and cCF10 compete for the same binding site in PrgX, but it has not been shown to date whether iCF10 also competes with cCF10 for binding to PrgZ. Furthermore, it is unknown what discriminates PrgZ from OppA in terms of structure determinants that allow the pheromone receptor to bind cCF10 with high affinity, whereas OppA, based on studies of its homologs in Salmonella typhimurium, Escherichia coli, and Lactococcus lactis, is highly promiscuous and binds a wide variety of peptides with moderate affinity (13–15). To gain detailed insight in how PrgZ binds its ligands, we present here the structure of PrgZ complexed with cCF10, as well as ligand-binding studies.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, Growth Conditions, and Protein Purification
The prgZ gene was obtained by PCR using the template vector pINY6033 (8) and the complementary primers (forward, ATGGTGAGAATTTATATTTTCAAGGTATGACAAAAGATTCACAAGATGCA; and reverse, TGGGAGGGTGGGATTTTCACCATATACCCTCCATTACTTCTCTATTCTC) to yield prgZ*, i.e. prgZ without the nucleotide sequence coding for the signal sequence. The product was subsequently ligated into the pREnLIC vector (16); the resulting plasmid (pREnLIC-PrgZ*) was transformed into E. coli MC1061. The prgZ* gene was then transferred from pREnLIC to pNZnLIC via SfiI digestion and mixing of pREnLIC-PrgZ* and pERL according to the LIC-vBEX (ligation-independent cloning-vector backbone exchange) procedure (16). The plasmid was transformed into L. lactis NZ9000.
L. lactis NZ9000 cells expressing PrgZ* were cultivated semi-anaerobically in M17 broth, 1% (w/v) glucose, and 5 μg/ml chloramphenicol at 30 °C to A600 = 0.6, at which they were induced by the addition of a 0.2% (v/v) culture supernatant of the nisin A-producing strain NZ9700 (17). The cells were harvested 2 h after the start of induction, resuspended to A600 = 200 in buffer A (50 mm potassium Pi (pH 8.0) and 200 mm NaCl) supplemented with 10% glycerol, flash-frozen in liquid nitrogen, and stored at −80 °C.
Frozen cells were thawed at room temperature and diluted to A600 = 80 with buffer A. Subsequently, 100 μg/ml deoxyribonuclease type I, 10 mm MgSO4, and 0.1 mm phenylmethanesulfonyl fluoride were added. The cells were broken by two consecutive passes in a cell disruptor (Constant Systems Ltd.) at 39,000 p.s.i. and 5 °C. Unbroken cells and cell debris were removed by ultracentrifugation at 267,000 × g for 75 min at 4 °C. The supernatant was collected (resulting in 20 ml of lysate/liter of culture), and 1.0 ml of Ni2+-Sepharose resin (Amersham Biosciences) was added per 20 ml of lysate. The mixture was incubated for 1 h at 4 °C in buffer A supplemented with 15 mm imidazole plus 10% (v/v) glycerol. Subsequently, the resin was poured into a disposable column (Bio-Rad) and washed with buffer A containing 50 mm imidazole (20 column volumes). The protein was eluted with 20 mm Na-MES (pH 6.0), 300 mm NaCl, and 500 mm imidazole (pH 6.0; 2 column volumes). PrgZ* was further purified on a Superdex 200 10/300 GL size exclusion column (Amersham Biosciences) in 20 mm Na-MES (pH 6.0) and 150 mm NaCl. Fractions containing PrgZ* were pooled and concentrated in spin concentrators with a 30-kDa cutoff (Vivaspin with polyethersulfone membrane, Sartorius) to 32 mg/ml protein. For differential scanning calorimetry (DSC)3 experiments, the Superdex 200 10/300 column was run in 20 mm Na-HEPES (pH 7.5) and 150 mm NaCl.
Crystallization and Structure Determination
To co-crystallize PrgZ* with cCF10 (LVTLVFV), 0.5 mg of solid cCF10 was added to 200 μl of 32 mg/ml PrgZ* with 0.01% n-dodecyl-β-d-maltoside added. After overnight incubation at 4 °C, this solution was used as a basis to grow crystals using the hanging drop vapor diffusion method. Crystallization conditions that yielded the open conformation consisted of 2 μl of protein (32 mg/ml PrgZ) and 1 μl of reservoir solution (0.01 m zinc sulfate, 0.1 m MES (pH 6.5), and 25% (w/v) PEG 550 monomethyl ether). Crystals were soaked in cryoprotectant (0.01 m zinc sulfate, 0.1 m Na-MES (pH 6.5), 30% (w/v) PEG 550, and 10% glycerol) for 30 s and subsequently flash-cooled in liquid nitrogen. Data were collected to 2.0 Å (wavelength of 0.874 Å) on beamline ID23-2 at the European Synchrotron Radiation Facility (Grenoble, France). Data processing and reduction were carried out using XDS (18).
The structure was solved using the MR protocol of Auto-Rickshaw, the European Molecular Biology Laboratory Hamburg automated crystal structure determination platform (19), which performs molecular replacement using BALBES (20), density modification using PIRATE (21), and model building using ARP/wARP (22). The calculated electron density was of sufficient quality to let ARP/wARP build 95% of the residues of the final model, which Auto-Rickshaw returned. A few cycles of refinement using Refmac5 (23), interspersed with manual model building using Coot (24), were necessary to complete the model. Water molecules were placed automatically in Fo − Fc Fourier difference maps at a 3σ cutoff level and validated to ensure correct coordination geometries using Coot. MolProbity (25) was used during the final refinement for model validation. Relevant statistics of the data collection and model refinement are given in Table 1.
TABLE 1.
Data collection and refinement statistics
| Data collection | |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 68.6, 74.1, 112.6 |
| α, β, γ | 90° |
| Wavelength (Å) | 0.872 |
| Resolution range | 35–2.0 |
| Rmeas | 0.11 (0.46) |
| I/σ(I) | 11.8 (3.7) |
| Completeness (%) | 100.0 (100.0) |
| Redundancy | 4.9 |
| Refinement statistics | |
| Resolution range (Å) | 35–2.0 |
| No. of reflections | 43,632 |
| Rwork/Rfree | 0.155/0.192 |
| No. of atoms | |
| Protein | 4053 |
| Ligand | 69 |
| Water | 380 |
| Average B-factors (Å2) | |
| Protein | 22.1 |
| Ligand | 17.5 |
| Water | 33.0 |
| r.m.s.d.a | |
| Bond lengths (Å) | 0.010 |
| Bond angles | 1.12° |
a r.m.s.d., root mean square deviation.
Differential Scanning Calorimetry
The stability of PrgZ*, with and without ligand, was measured by DSC on a VP-DSC system (MicroCal) at 25 °C and at 25 p.s.i. For measurements, PrgZ was diluted in 20 mm Na-HEPES (pH 7.5) and 150 mm NaCl to 4 μm. The samples (1 ml) were incubated in the absence or presence of 0.2 mg of lyophilized cCF10 or iCF10 at 4 °C for 14 h while rotating. After incubation, the samples were centrifuged for 10 min at 20,000 × g to remove any not dissolved peptide, and the supernatant was used for the analysis. Protein samples and buffers were degassed before injection into the cell to prevent air bubble formation. The temperature scans were performed between 25 and 80 °C at a scan rate of 60 °C/h. Data were analyzed using the MicroCal software (26).
Data Analysis
Binding pocket volumes were calculated with the program VOIDOO from the Uppsala Software Factory (27) using the probe-occupied volume for calculation. The program was run 10 times with the exact same parameters (probe radius of 1.4 Å, grid spacing of 0.5 Å, atomic fattening factor of 1.2 Å, and grid shrink factor of 0.9) but with randomly rotated copies of the molecule to average out random errors.
RESULTS
Crystallization and Structure Determination
PrgZ without a lipid anchor (termed PrgZ*) was produced in the cytoplasm of L. lactis. PrgZ* was co-crystallized together with cCF10, yielding crystals of the proteins in the closed liganded conformation in space group P212121 diffracting to 2.0 Å (Table 1). The structure was solved using molecular replacement in Auto-Rickshaw (19) with the structures of OppA from S. typhimurium (Protein Data Bank code 1JET) and YliB from E. coli (code 1UQW) as search models. The final model comprises residues 58–564. The first 57 residues and the Ser-408–Asn-410 loop are (partially) disordered and not visible in the resulting electron density.
PrgZ* Complexed with cCF10
Based on the connectivity of the β-strands, PrgZ* has the typical type II fold of substrate-binding proteins, with two α/β-domains that enclose the bound ligand (28). PrgZ has an extra domain, as has been previously described for OppA from S. typhimurium and L. lactis (14, 29), and thus has a total of three domains (domains I–III). Substrate-binding proteins are structurally related even though their sequence identity is often below 20% (30). Compared with other peptide-binding proteins for which crystal structures are available, PrgZ is most similar in sequence to OppA from E. coli (25% identity). With its 60-kDa size and extra domain (domain II), it structurally belongs to Cluster C of the substrate-binding proteins (30).
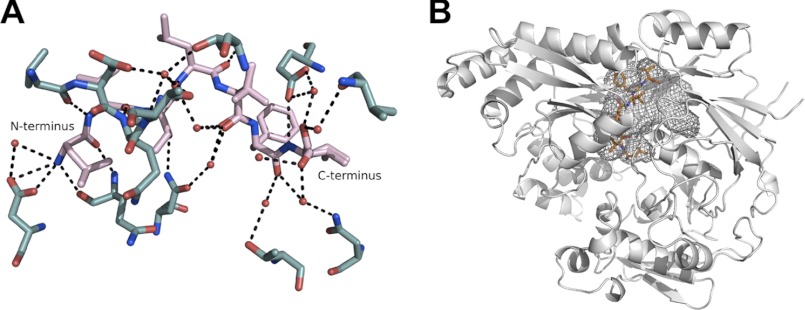
During refinement, electron density became apparent between the two domains and accounted for the bound peptide cCF10 (LVTLVFV). The peptide was buried inside PrgZ*, with no access to the bulk solvent. The electron density of the ligand was of excellent quality, allowing for straightforward model building of the heptapeptide. An extensive network of hydrogen bonds is present between the protein and the ligand, with 10 direct hydrogen bonds between cCF10 and PrgZ* and an additional eight via bridging water molecules within the binding cavity (Fig. 1A). The N terminus of cCF10 forms a salt bridge with Asp-472, whereas the C terminus is not fixed.
FIGURE 1.
A, hydrogen-bonding network forming the binding site for cCF10 (pink) within PrgZ* (teal). B, schematic representation of PrgZ*, with the bound cCF10 ligand in orange and the binding cavity outlined in gray mesh.
The volume of the binding cavity of PrgZ* is 1600 Å3, which is smaller than that of AppA from Bacillus subtilis (2500 Å3) or OppA from L. lactis (4900 Å3) but larger than that of OppA from S. typhimurium (1000 Å3) (14). The side chains of cCF10 are located in defined, mostly hydrophobic, side chain pockets, and with the exception of a hydrogen bond to the Thr-3 side chain, the side chains do not form specific interactions with the protein. Instead, hydrophobic and van der Waals bonds shape the interaction between the peptide side chains and the protein. The C terminus of cCF10 is located in a spacious area, with the binding cavity extending farther within the protein (Fig. 1B).
Ligand Binding to PrgZ*
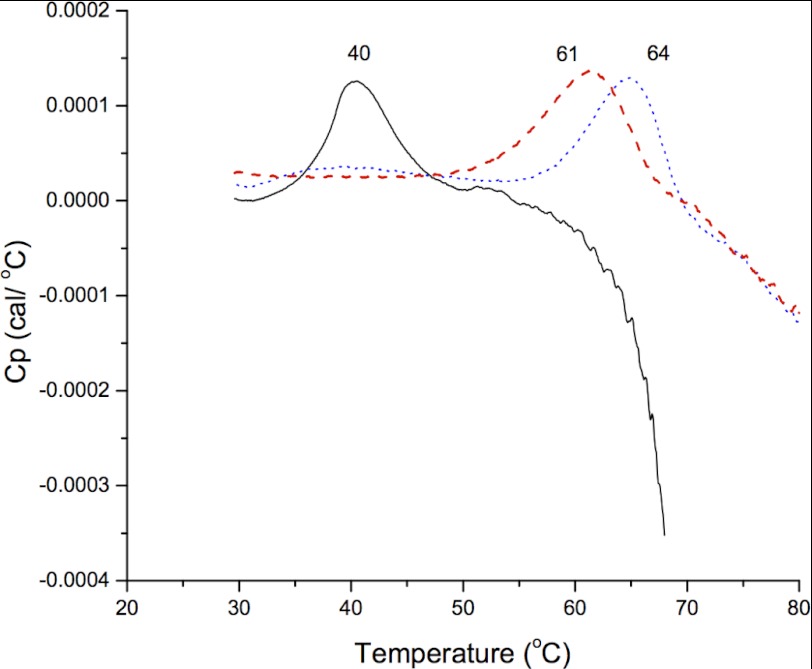
cCF10 and iCF10 are poorly soluble in aqueous buffers. Solubility increases to 1 and 0.2 mm, respectively, when a minimum of 10% (v/v) Me2SO is present. Unfortunately, PrgZ was not stable in the presence of 10% Me2SO, which prohibited the use of intrinsic protein fluorescence titration or isothermal titration calorimetry to determine binding affinities. Instead, the binding of cCF10 and iCF10 to PrgZ* was characterized by DSC. PrgZ* was incubated for 16 h with 0.2 mg of solid cCF10 or iCF10 in the presence of 0.01% of the detergent n-dodecyl-β-d-maltoside, which was added to dissolve the peptides but lowered the melting temperature (Tm) of PrgZ from 42.5 ± 0.5 to 40.5 ± 0.5 °C (n = 3). Upon peptide binding, the Tm of PrgZ* increased from 40.5 °C (apoprotein) to 64 °C (cCF10-bound) or 61 °C (iCF10-bound) (Fig. 2). The variation in Tm in two independent experiments was <1 °C. The exact Tm of PrgZ* complexed to either peptide could not be determined because the protein began to aggregate within this temperature interval. The endotherms are relatively broad, which may be attributed to the tendency of PrgZ to aggregate in both the absence and presence of ligand (pheromone or inhibitor peptide). Moreover, neither PrgZ nor any other substrate-binding protein is a standard globular protein but instead has two similarly sized domains. It is likely that the two domains unfold at slightly different temperatures that are not resolved in the DSC scans, which leads to a broadening of the endotherms.
FIGURE 2.
DSC scans of PrgZ* without ligand (solid black line), with cCF10 added (dotted blue line), and with iCF10 added (dashed red line). The Tm of each measurement is indicated above each peak.
DISCUSSION
Previous in vivo activity assays indicated that PrgZ selectively binds cCF10 and does not tolerate large deviations in its peptide sequence (12). However, the assay could not discriminate between peptide binding to PrgZ or to the DNA regulatory protein PrgX. The measured response of the in vivo assays was the induction of cell clumping in the culture, for which binding to both proteins is required. Based on the DSC experiments and consistent with the crystal structure, it is clear that PrgZ binds both cCF10 and iCF10, two related but different peptides.
We conclude that iCF10 will thus act as a competitive inhibitor for cCF10 binding to PrgZ. The apparent Tm values, derived from the DSC measurements, indicate that PrgZ may have a slightly higher affinity for cCF10 than for iCF10. The affinities have to be high, as the typical extracellular concentration of cCF10 in vivo is only 10 pm (6, 11). High-affinity pheromone binding is achieved because PrgZ forms an extensive network of hydrogen bonds with cCF10 (Fig. 1A), mainly to the backbone of the peptide. The peptide sequence selectivity of PrgZ must originate from the physical shape, charge, and polarity of the side chain pockets, which determine how well a side chain fits. Side chain pockets 2, 4, 6, and 7 form a confined space, which is lined mostly by hydrophobic residues. Side chain pockets 1, 3, and 5 have a larger volume, and pockets 3 and 5 are lined mostly by hydrophilic residues (Table 2). We note that the most hydrophilic residue of the pheromone, i.e. Thr, is found in pocket 3. Interestingly, the results of a previous genetic screen suggested the importance of this residue in a specific interaction with PrgZ (12). The properties of the side chain pockets suggest that not just a unique side chain is tolerated at any given position. However, for high-affinity binding of a given peptide, the physicochemical properties of the side chains (size and hydrophobicity) should match those of the PrgZ pockets. Further support for this notion comes from the observation that water molecules reside in all side chain pockets, and these could form bridging hydrogen bonds to bound peptides, as seen previously in OppA (14, 15, 29). The more hydrophilic pockets harbor more defined water molecules inside compared with the more hydrophobic ones (Table 2). That the side chain pockets can harbor different residues is also evident from the fact that PrgZ binds both cCF10 and iCF10 (LVTLVFV and AITLIFI, respectively). Both pheromones are hydrophobic, and although related in sequence, they differ at positions 1, 2, 5, and 7. The largest difference is the N-terminal residue, which is changed from leucine to alanine in iCF10. Even though the hydrophobic side chain pockets form a confined space for the peptide side chains, there are no obvious specific interactions between the peptide pheromone and the protein. Although both iCF10 and cCF10 bind PrgZ and compete with one another for import into the donor cell, the striking differential effects of these two peptides when bound to PrgX (9, 31) account for a major difference in their biological activities.
TABLE 2.
Description of the side chain pockets, including the amino acids lining the pockets, the number of ordered water molecules per pocket, and the overall hydrophobicity of pockets in PrgZ, OppA, AppA, and DppA
The numbering of the pockets is based on the cCF10 ligand in PrgZ; the numbering of the pockets of OppA and AppA is shown in parentheses. The hydrophobicity description is based on inspection of the side chain pockets in each of the proteins. “Mixed” means that both hydrophilic and hydrophobic residues line the pocket. A dash indicates that the side chain pocket does not exist in that protein. We also note that in OppA from L. lactis, most of the pockets are not well defined, and instead, a large cavity is present that even allows peptides to bind in different register (14, 32). The data were generated using the following Protein Data Bank codes: OppA, 3DRG; AppA, 1XOC; and DppA, 1DPP.
| Side chain pocket | Residues lining the pocket (PrgZ) | Ordered water molecules (PrgZ) | Hydrophobicity |
|||
|---|---|---|---|---|---|---|
| PrgZ | OppA | AppA | DppA | |||
| – | – | Hydrophilic (1) | Hydrophilic (1) | – | ||
| – | – | – | Hydrophilic (2) | Hydrophilic (2) | – | |
| – | – | – | Mixed (3) | Mixed (3) | – | |
| 1 | Gln-81, Ile-84, Gln-470 | 1 | Mixed | Mixed (4) | Hydrophilic (4) | Mixed |
| 2 | Val-79, Met-454, Met-489, His-491 | 3 | Hydrophobic | Hydrophobic (5) | Hydrophobic (5) | Hydrophobic |
| 3 | Asn-318, Met-320, Gly-468, Gln-470 | 3 | Hydrophilic | Mixed (6) | Mixed (6) | – |
| 4 | Phe-453, Met-454, Leu-457, Ser-467 | 1 | Hydrophobic | Mixed (7) | Mixed (7) | – |
| 5 | Gly-67, Thr-68, Ser-418, Glu-423 | 4 | Hydrophilic | Hydrophilic (8) | Mixed (8) | – |
| 6 | Met-320, Leu-356, Phe-422, Glu-423, Ala-426, Phe-531 | 2 | Hydrophobic | Hydrophobic (9) | Mixed (9) | – |
| 7 | Ile-280, Pro-296, Ala-298, Phe-422 | 3 | Hydrophobic | – | – | – |
OppA from L. lactis is the most promiscuous peptide-binding protein known to date. It binds peptides in the range of 4–35 residues and tolerates small, neutral, charged, and large bulky hydrophobic amino acids at any position in the peptide. There is a voluminous “nonspecific” binding cavity, and the peptides are bound via hydrogen bonds with up to 6 backbone residues. There is only one pocket in OppA from L. lactis that favors binding of a hydrophobic peptide side chain. This pocket mimics side chain pockets 2, 4, 6, and 7 of PrgZ; pocket 5 of OppA corresponds structurally to side chain pocket 2 of PrgZ (Table 2). The four hydrophobic pockets in PrgZ do not provide much extra space for larger side chains, and they will strongly favor the binding by PrgZ of hydrophobic peptides such as cCF10 and iCF10. Most side chain pockets of AppA from B. subtilis and the nonspecific binding cavity of OppA from L. lactis can more easily adapt to a much greater diversity of amino acids (13, 14, 32, 33). These proteins also have far more hydrophilic side chain pockets than PrgZ (Table 2). Thus, the structural comparison of PrgZ with the homologous chemosensors explains the high selectivity of PrgZ for hydrophobic peptides (cCF10 and iCF10) compared with the promiscuous binding of peptides by the OppA and AppA proteins (14, 29, 33). It is likely that in E. faecalis, PrgZ and OppA have diverged to obtain different functions to discriminate hydrophobic pheromones from nutritional peptides.
Many peptide-binding proteins for which structures are available fixate their peptide termini with salt bridges. DppA and MppA from E. coli, OppA from S. typhimurium, and AppA from B. subtilis all form salt bridges to both the N and C termini of the bound peptides (29, 33–35). OppA from L. lactis is an important exception. This protein leaves both peptide termini free, allowing binding of small (4–5 residues) and very large (up to 30 residues) peptides in the most optimal position (14). PrgZ fixes the N terminus of cCF10, but not the C terminus. When inspecting the binding cavity of PrgZ, it is evident that there is space for an additional amino acid at the C-terminal end of the peptide. We hypothesize that an octapeptide could also bind to PrgZ. Finally, given the similarity of the overall structures of PrgZ, AppA, and OppA, it is most likely that PrgZ operates according to a similar Venus flytrap mechanism (30), providing a direct path for the entry of pheromone into the binding site.
In conclusion, we propose that cytosolic accumulation of both cCF10 and iCF10 is PrgZ-dependent. PrgZ selects these peptides at the very low extracellular concentrations among high concentrations of nutritional peptides. iCF10, which inhibits expression of the conjugation system, acts as a competitive inhibitor of cCF10 at the level of both PrgZ and PrgX.
This work was supported, in whole or in part, by National Institutes of Health Grant GM49530 (to G. D.). This work was also supported by a Vici grant (to D.-J. S.) and TOP Subsidy Grant 700.56.302 (to B. P.) from The Netherlands Organisation for Scientific Research (NWO) and by the European Union European Drug Initiative on Channels and Transporters (EDICT) Program.
The atomic coordinates and structure factors (code 4FAJ) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- DSC
- differential scanning calorimetry.
REFERENCES
- 1. Jett B. D., Huycke M. M., Gilmore M. S. (1994) Virulence of enterococci. Clin. Microbiol. Rev. 7, 462–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hunt C. P. (1998) The emergence of enterococci as a cause of nosocomial infection. Br. J. Biomed. Sci. 55, 149–156 [PubMed] [Google Scholar]
- 3. Courvalin P. (2006) Vancomycin resistance in Gram-positive cocci. Clin. Infect. Dis. 42, S25–S34 [DOI] [PubMed] [Google Scholar]
- 4. Miller M. B., Bassler B. L. (2001) Quorum sensing in bacteria. Annu. Rev. Microbiol. 55, 165–199 [DOI] [PubMed] [Google Scholar]
- 5. Antiporta M. H., Dunny G. M. (2002) ccfA, the genetic determinant for the cCF10 peptide pheromone in Enterococcus faecalis OG1RF. J. Bacteriol. 184, 1155–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dunny G. M. (2007) The peptide pheromone-inducible conjugation system of Enterococcus faecalis plasmid pCF10: cell-cell signaling, gene transfer, complexity, and evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 362, 1185–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Leonard B. A., Podbielski A., Hedberg P. J., Dunny G. M. (1996) Enterococcus faecalis pheromone-binding protein PrgZ recruits a chromosomal oligopeptide permease system to import sex pheromone cCF10 for induction of conjugation. Proc. Natl. Acad. Sci. U.S.A. 93, 260–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bae T., Clerc-Bardin S., Dunny G. M. (2000) Analysis of expression of prgX, a key negative regulator of the transfer of the Enterococcus faecalis pheromone-inducible plasmid pCF10. J. Mol. Biol. 297, 861–875 [DOI] [PubMed] [Google Scholar]
- 9. Shi K., Brown C. K., Gu Z. Y., Kozlowicz B. K., Dunny G. M., Ohlendorf D. H., Earhart C. A. (2005) Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis. Proc. Natl. Acad. Sci. U.S.A. 102, 18596–18601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Johnson C. M., Manias D. A., Haemig H. A., Shokeen S., Weaver K. E., Henkin T. M., Dunny G. M. (2010) Direct evidence for control of the pheromone-inducible prgQ operon of Enterococcus faecalis plasmid pCF10 by a countertranscript-driven attenuation mechanism. J. Bacteriol. 192, 1634–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mori M., Sakagami Y., Narita M., Isogai A., Fujino M., Kitada C., Craig R. A., Clewell D. B., Suzuki A. (1984) Isolation and structure of the bacterial sex pheromone, cAD1, that induces plasmid transfer in Streptococcus faecalis. FEBS Lett. 178, 97–100 [DOI] [PubMed] [Google Scholar]
- 12. Fixen K. R., Chandler J. R., Le T., Kozlowicz B. K., Manias D. A., Dunny G. M. (2007) Analysis of the amino acid sequence specificity determinants of the enterococcal cCF10 sex pheromone in interactions with the pheromone-sensing machinery. J. Bacteriol. 189, 1399–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tame J. R., Dodson E. J., Murshudov G., Higgins C. F., Wilkinson A. J. (1995) The crystal structures of the oligopeptide-binding protein OppA complexed with tripeptide and tetrapeptide ligands. Structure 3, 1395–1406 [DOI] [PubMed] [Google Scholar]
- 14. Berntsson R. P., Doeven M. K., Fusetti F., Duurkens R. H., Sengupta D., Marrink S. J., Thunnissen A. M., Poolman B., Slotboom D. J. (2009) The structural basis for peptide selection by the transport receptor OppA. EMBO J. 28, 1332–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Klepsch M. M., Kovermann M., Löw C., Balbach J., Permentier H. P., Fusetti F., de Gier J. W., Slotboom D. J., Berntsson R. P. (2011) Escherichia coli peptide-binding protein OppA has a preference for positively charged peptides. J. Mol. Biol. 414, 75–85 [DOI] [PubMed] [Google Scholar]
- 16. Geertsma E. R., Poolman B. (2007) High-throughput cloning and expression in recalcitrant bacteria. Nat. Methods 4, 705–707 [DOI] [PubMed] [Google Scholar]
- 17. de Ruyter P. G., Kuipers O. P., de Vos W. M. (1996) Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62, 3662–3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kabsch W. (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 19. Panjikar S., Parthasarathy V., Lamzin V. S., Weiss M. S., Tucker P. A. (2005) Auto-Rickshaw: an automated crystal structure determination platform as an efficient tool for the validation of an x-ray diffraction experiment. Acta Crystallogr. D Biol. Crystallogr. 61, 449–457 [DOI] [PubMed] [Google Scholar]
- 20. Long F., Vagin A. A., Young P., Murshudov G. N. (2008) BALBES: a molecular replacement pipeline. Acta Crystallogr. D Biol. Crystallogr. 64, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cowtan K. (2000) General quadratic functions in real and reciprocal space and their application to likelihood phasing. Acta Crystallogr. D Biol. Crystallogr. 56, 1612–1621 [DOI] [PubMed] [Google Scholar]
- 22. Langer G., Cohen S. X., Lamzin V. S., Perrakis A. (2008) Automated macromolecular model building for x-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Murshudov G. N., Vagin A. A., Dodson E. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 24. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 25. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wiseman T., Williston S., Brandts J. F., Lin L. N. (1989) Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 179, 131–137 [DOI] [PubMed] [Google Scholar]
- 27. Kleywegt G. J., Jones T. A. (1994) Detection, delineation, measurement, and display of cavities in macromolecular structures. Acta Crystallogr. D Biol. Crystallogr. 50, 178–185 [DOI] [PubMed] [Google Scholar]
- 28. Fukami-Kobayashi K., Tateno Y., Nishikawa K. (1999) Domain dislocation: a change of core structure in periplasmic binding proteins in their evolutionary history. J. Mol. Biol. 286, 279–290 [DOI] [PubMed] [Google Scholar]
- 29. Tame J. R., Murshudov G. N., Dodson E. J., Neil T. K., Dodson G. G., Higgins C. F., Wilkinson A. J. (1994) The structural basis of sequence-independent peptide binding by OppA protein. Science 264, 1578–1581 [DOI] [PubMed] [Google Scholar]
- 30. Berntsson R. P., Smits S. H., Schmitt L., Slotboom D. J., Poolman B. (2010) A structural classification of substrate-binding proteins. FEBS Lett. 584, 2606–2617 [DOI] [PubMed] [Google Scholar]
- 31. Kozlowicz B. K., Shi K., Gu Z. Y., Ohlendorf D. H., Earhart C. A., Dunny G. M. (2006) Molecular basis for control of conjugation by bacterial pheromone and inhibitor peptides. Mol. Microbiol. 62, 958–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Berntsson R. P., Thunnissen A. M., Poolman B., Slotboom D. J. (2011) Importance of a hydrophobic pocket for peptide binding in lactococcal OppA. J. Bacteriol. 193, 4254–4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Levdikov V. M., Blagova E. V., Brannigan J. A., Wright L., Vagin A. A., Wilkinson A. J. (2005) The structure of the oligopeptide-binding protein AppA from Bacillus subtilis in complex with a nonapeptide. J. Mol. Biol. 345, 879–892 [DOI] [PubMed] [Google Scholar]
- 34. Nickitenko A. V., Trakhanov S., Quiocho F. A. (1995) 2 Å resolution structure of DppA, a periplasmic dipeptide transport/chemosensory receptor. Biochemistry 34, 16585–16595 [DOI] [PubMed] [Google Scholar]
- 35. Maqbool A., Levdikov V. M., Blagova E. V., Hervé M., Horler R. S., Wilkinson A. J., Thomas G. H. (2011) Compensating stereochemical changes allow murein tripeptide to be accommodated in a conventional peptide-binding protein. J. Biol. Chem. 286, 31512–31521 [DOI] [PMC free article] [PubMed] [Google Scholar]