Background: Olfactomedin 1 (Olfm1) is a highly conserved secreted glycoprotein with an unknown mechanism of action.
Results: Olfm1 binds to NgR1, and its binding reduced binding of NgR1 coreceptors p75NTR and LINGO-1.
Conclusion: Olfm1 is a novel NgR1 ligand that modulates the functions of the NgR1 complex in axonal growth.
Significance: Olfm1 may be used to facilitate neuronal growth after axonal damage.
Keywords: Axon, Protein Complexes, Protein Domains, Protein Expression, Zebrafish, DiI Labeling, Dorsal Root Ganglia, Growth Cone Collapse
Abstract
Olfm1, a secreted highly conserved glycoprotein, is detected in peripheral and central nervous tissues and participates in neural progenitor maintenance, cell death in brain, and optic nerve arborization. In this study, we identified Olfm1 as a molecule promoting axon growth through interaction with the Nogo A receptor (NgR1) complex. Olfm1 is coexpressed with NgR1 in dorsal root ganglia and retinal ganglion cells in embryonic and postnatal mice. Olfm1 specifically binds to NgR1, as judged by alkaline phosphatase assay and coimmunoprecipitation. The addition of Olfm1 inhibited the growth cone collapse of dorsal root ganglia neurons induced by myelin-associated inhibitors, indicating that Olfm1 attenuates the NgR1 receptor functions. Olfm1 caused the inhibition of NgR1 signaling by interfering with interaction between NgR1 and its coreceptors p75NTR or LINGO-1. In zebrafish, inhibition of optic nerve extension by olfm1 morpholino oligonucleotides was partially rescued by dominant negative ngr1 or lingo-1. These data introduce Olfm1 as a novel NgR1 ligand that may modulate the functions of the NgR1 complex in axonal growth.
Introduction
Olfm1 belongs to the family of olfactomedin domain-containing proteins (1). This highly conserved (86% amino acid identity between human and zebrafish) secreted glycoprotein is also known as noelin in chicken and Xenopus (2, 3), pancortin in mice (4), olfactomedin-related glycoprotein in rats (5), and hOlfA in humans (6). The expression patterns of Olfm1 are similar across the species studied, although some differences exist. In general, Olfm1 is expressed preferentially in neurogenic tissues during development (2, 3, 7–9). Postnatally, Olfm1 is highly expressed in the cerebral cortex, including the olfactory bulb and hippocampus (4, 7), with different forms of Olfm1, exhibiting overlapping but not identical expression patterns (7–9). Available data suggest that Olfm1 plays a role in promoting neuronal cell death in mice (10), neural crest generation in chicken (2), maintenance of neuronal precursor cells in Xenopus (9), eye size regulation in Xenopus and zebrafish, and optic nerve arborization in the optic tectum in zebrafish (9, 11).
The Olfm1 protein contains an N-terminal signal peptide followed by a coiled-coil domain and an olfactomedin domain located in the C-terminal part of the protein molecule. Four structurally distinct mRNAs, named AMY, BMY, AMZ, and BMZ, are produced from the Olfm1 gene (2, 5, 8). These mRNAs share a common central region (M) and have two different 5′ regions (A and B) transcribed from separate promoters and two different 3′ regions (Y and Z) produced by alternative splicing of corresponding mRNAs (5). The olfactomedin domain is encoded by the last two 3′ exons found in the AMZ and BMZ forms. The AMY and BMY forms encode shorter forms of Olfm1 that lack the olfactomedin domain. Many previous studies indicate that the N-terminal part of Olfm1 contains the active domain of the functions.
The molecular mechanisms underlying Olfm1 activities remain unclear. Considering that Olfm1 is a secreted protein, one might expect its main targets to be extracellular proteins or receptor-like molecules on the cell membrane. However, several identified binding partners of Olfm1 are expressed intracellularly. In adult mouse brain, the BMY form of Olfm1 interacts with WAVE1, an actin-reorganizing protein, and Bcl-xL, a proapoptotic factor, thereby promoting the death of neurons following ischemic injury (10). β-Dystrobrevin, a component of the dystrophin-associated protein complex, was identified as a potential binding partner of the BMY form of Olfm1 (12), although the physiological implications of this interaction are unknown. The only identified extracellular binding partner of Olfm1 is secreted Wnt inhibitory factor 1. In zebrafish embryos, interaction of olfm1 with wnt inhibitory factor 1 led to a decrease of the anterior-posterior axial length of the ocular globe (11).
Axonal elongation is a primary step in neural development, required before synaptic connections can be formed between distant neurons. During axonal pathfinding, glial cells provide assistance in directing growth cones toward their targets by secreting neurotrophic factors and positive guidance molecules and by expressing inhibitory membrane proteins that obstruct aberrant growth cone migration. In the adult mammalian CNS, axonal growth is largely restricted by barriers provided by glial cells (13). These barriers are formed by membrane proteins, including myelin-associated glycoprotein (MAG),3 oligodendrocyte myelin glycoprotein (OMgp), Nogo A (14), and chondroitin sulfate proteoglycans (15) on the glial surface. Each of these membrane proteins interacts with the Nogo A receptor complex, which consists of Nogo A receptor 1 (NgR1) and putative coreceptors (p75NTR, LINGO-1, and TROY) (16). These receptors are expressed at the growth cones where ligand binding induces reorganization of the cytoskeleton, resulting in growth cone repulsion or collapse. Studies on knockdown of NgR1 or MAG suggest that the inhibition of axonal pathfinding by NgR1 is complex and may involve other unknown molecules (17, 18). Identification of new NgR1 ligands may lead to a better understanding of the inhibition of axon regrowth in the injured adult CNS and new therapeutic approaches to promote axon regeneration.
Here, we present data showing that a possible mechanism underlying axon growth stimulation by Olfm1 involves interaction with NgR1, leading to inhibition of RhoA activity. In zebrafish in vivo, olfm1 and ngr1 interact to regulate the optic nerve extension.
EXPERIMENTAL PROCEDURES
Animals and Cell Cultures
All experiments using animals were approved by the National Eye Institute Animal Care and Use Committee. Adult C57BL/6 mice that were used in this work were 2–3 months old. Mice expressing EGFP under the Olfm1 promoter {TG(Olfm1:EGFP)} were purchased from the Mutant Mouse Regional Resource Center (MMRRC, University of California). Wild-type zebrafish were maintained as described (19). COS7 cells were cultured in six-well culture plates with DMEM culture medium supplemented with 10% fetal bovine serum.
DNA Constructs and Transfection
The BMZ and AMZ Olfm1 forms with a deletion of a sequence derived from exon 4 were produced from corresponding full-length cDNAs using two-step PCR. MAG (BC053347), NgR1 (BC011787), NgR2 (BC113673), NgR3 (BC030471), EphA3 (BC063282), EphB3 (BC052968), Na(v)b1 (BC067122), Na(v)b2 (BC036793), Na(v)b3 (BC117282), SynCAM1 (BC095986), Robo2 (BC055333), LINGO-1 (BC011057), p75NTR (BC038365), and NCAM1 (BC101924) were cloned into the PCRII-TOPO vector (Invitrogen) using expressed sequence tag clones (Open Biosystems) as templates. The following oligonucleotide primers were used for cloning: AMZ-FW, 5′-aagcttgccgccaccatgcaaccggcccggaagct-3′; AMZ-RV, 5′-gaattcctacaactcatcggagcgga-3′; BMZ-FW, 5′-ataagcttgccgccaccatgcaaccggcccggaagct-3′; BMZ-RV, 5′-attctagactacaactcatcggagcggatga-3′; BMY-RV, 5′-ctagcccttgaactgcctggctagat-3′; Olfm1DelEx4-RV, 5′-cttctcgagtagctgcctcag-3′; Olfm1DelEx4-FW, 5′-ctcgagaaggcaataaaagcgaaaatgga-3′; MAG-FW, 5′-ctagctgagtatgctgaaatccgggtcaag-3′; MAG-RV, 5′-gatccttgacccggatttcagcatactcag-3′; NgR1-FW, 5′-aagcttgccgccaccatgaagagggcgtccgctgg-3′; NgR1-RV, 5′-tctagagcagggcccaagcactgtcc-3′; NgR2-FW, 5′-gcggccgccaccatgctgcccgggctcaggcg-3′; NgR2-RV, 5′-tctagagaggtggtggggcaccagga-3′; NgR3-FW, 5′-gcggccgccaccatgcttcgcaaagggtgctg-3′; NgR3-RV, 5′-tctagagcggagagtgactgccagcc-3′; EphA3-FW, 5′-gaattcgccgccaccatggattgtcagctctccat-3′; EphA3-RV, 5′-ggatcccacgggaactgggccattct-3′; EphB3-FW, 5′-aagcttgagctgccacggccatggccaga-3′; EphB3-RV, 5′-tctagagacctgcacaggcagcgtct-3′; SynCAM1-FW, 5′-gcggccgccaccatggcgagtgctgtgctgcc-3′; SynCAM1-RV, 5′-gaattcgcgatgaagtactctttctttt-3′; NCAM1-FW, 5′-aagcttgccgccaccatgctgcgaactaaggatctc-3′; NCAM1-RV, 5′-tctagatgctttgctctcattctcttt-3′; Robo2-FW, 5′-aagcttgccgccaccatgctgcgaactaaggatctc-3′; Robo2-RV, 5′-tctagatgctttgctctcattctcttt-3′; LINGO-1-FW, 5′-gcggccgccaccatgcaggtgagcaagaggat-3′; LINGO-1-RV, 5′-tctagatatcatcttcatgttgaact-3′; p75NTR-FW, 5′-gcggccgccaccatgaggagggcaggtgctgcct-3′; p75NTR-RV, 5′-tctagacacaggggacgtggcagtgga-3′; Na(v)b1, 5′-aagcttgccgccaccatggggaggctgctggcctt-3′; Na(v)b1, 5′-tctagattcggccacctggacgcccg-3′; Na(v)b2-FW, 5′-aagcttgccgccaccatgcacagagatgcctggct-3′; Na(v)b2-RV, 5′-tctagacttggcgccatcatccgggt-3′; Na(v)b3-FW, 5′-aagcttgccgccaccatgcctgccttcaatagatt-3′; and Na(v)b3-RV, 5′-tctagattcctccactggtaccgcag-3′. TrkA, TrkB, Sdk1, Sdk2, TrkC, and DsCAM were subcloned into the pcDNA3 or p3XFLAG-CMV14 vectors (Sigma). Transfection of cDNA constructs was performed using Lipofectamine 2000 (Invitrogen) or PolyJet (Signagen). Conditioned medium (CM) was replaced with fresh medium 16 h after transfection, and cells were further cultured for 48–72 h. When secretion of proteins was assessed by Western blotting, CM was replaced with serum-free medium 24 h post-transfection.
Purification of Olfm1 Protein
PC12 cells were infected with pOlfm1 retrovirus, and a clone stably secreting the Olfm1 protein was isolated. The culture medium of Olfm1-producing cells was replaced with serum-free DMEM, which was collected 24–48 h later. Purification of Olfm1 from the CM was performed as described previously with some modifications (20). 2-Mercaptoethanol, CHAPS, and Tris-HCl (pH 7.4) were added to CM to final concentrations of 10 mm, 0.2%, and 50 mm, respectively. Lectin-agarose beads conjugated to Ricinus communis agglutinin I (20 μl/ml medium) were added to CM and incubated at 4 °C overnight. The beads were precipitated and washed five times with PBS with 0.2% CHAPS and 10 mm 2-mercaptoethanol. The bound Olfm1 was eluted with PBS-0.2% CHAPS containing 500 mm D-galactose (Sigma) at 4 °C for 1 h. After removing galactose, Olfm1 was concentrated by ultrafiltration with 10,000 kDa cutoff, filtered through a syringe filter (0.22 μm), and applied to a HiTrapTM Q XL anion exchange column (AKTA purifier, GE Healthcare) equilibrated with 20 mm Tris-HCl, 0.2% CHAPS, and 2 mm 2-mercaptoethanol. Proteins bound to the column were eluted by a gradient of 0–1 M NaCl in the same buffer over 20 ml elution volume. 100-μl fractions were collected. The ten fractions with the highest concentration of Olfm1 were combined and used in subsequent experiments.
Olfm1 Antibodies
Two different antibodies against Olfm1 were used. The monoclonal antibody was generated against the peptides SRDARTKQLRQLLEKVQN by the Custom Antibody Production Service of the University of Virginia. This monoclonal antibody detects a denatured protein on Western blot analyses and an antigen-retrieved protein on histological sections. A polyclonal antibody generated against purified Olfm1 was produced by Covance, Inc. This antibody detected intact Olfm1 and was used for immunoprecipitation and immunofluorescence experiments.
Olfm1-Alkaline Phosphatase Binding Assay
MAG, the AMZ, BMZ, BMY, and AMZdelEx4 forms of Olfm1, were subcloned into the pAPtag2 vector (GenHunter). COS7 cells were transfected with cDNAs encoding indicated alkaline phosphatase (AP) fusion proteins or vector control. The culture medium was changed to the fresh serum-free medium 24 h after the transfection, and CM was harvested 24–48 h later, filtered through a 0.22-μm filter, and stored at −80 °C until use. Absolute concentration of Olfm1-AP and other AP fusion proteins as well as the integrity of the protein were determined by Western blotting with a known amount of purified Olfm1 and AP (GenHunter). Another culture of COS7 cells was transfected with a construct encoding tested membrane protein. The expression of each protein after transfection was confirmed by Western blot analysis and, in some cases, by immunostaining. The mouse postnatal day 3 (P3) olfactory bulbs were dissected in a buffer (120 mm NaCl, 2.5 mm KCl, 2.5 mm CaCl2, 1.5 mm MgCl2, 15 mm glucose and 20 mm Hepes (pH7.4)) and sliced into 200-μm sections using a tissue chopper (Campden Instruments). The tested protein-expressing cells or tissue slices were incubated in the CM containing indicated AP fusion proteins (0–20 nm) for 90 min at 4 °C. When tissue sections were incubated with Olfm1-AP or AP, normal rabbit IgG or anti-NgR1 polyclonal IgG were added to the incubation mixtures (0.005 μg of IgG/μl). When binding of different fusion proteins was compared, equal amounts of AP were added. The samples were washed four times with PBS, fixed with 0.5% glutaraldehyde for 15 min at room temperature, and washed twice with PBS. The endogenous AP was inactivated by incubation of the samples at 65 °C for 90 min. The cell-bound AP activity was detected with AP substrate mixture (INT/BCIP, Roche) in the staining buffer (100 mm Tris-HCl (pH 9.5), 100 mm NaCl, 50 mm MgCl2) for 12–48 h. The images of stained cells were obtained with a dissection microscope (Zeiss, STEMI SV-11). For quantitative analysis of the activity of cell-bound AP, 1-StepTM PNPP (Pierce) was added to the fixed cells, and the absorbance at 405 nm in the supernatant was measured using a microplate reader (Bio-Rad, model 680).
Western Blotting and Immunoprecipitation
Cultured cells or isolated tissues were homogenized in a lysis buffer (10 mm Tris-HCl (pH 7.5), 1 mm EDTA, 150 mm NaCl, 1% Nonidet P-40, 5 mm NaF, 0.5 mm sodium orthovanadate, 10% glycerol, 1 mm PMSF, 1 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 μg/ml pepstatin) by repeated pipetting on ice. Following centrifugation, the soluble fraction was collected, and 5–15 μg of extracted proteins was separated on a 10% SDS-PAGE gel (Invitrogen) and transferred to a PVDF membrane (Invitrogen). A membrane was incubated with anti-Olfm1 (1:5,000 dilution) or anti-FLAG (M2) (1:1,000 dilution, Sigma) antibodies, followed by incubation with anti-mouse IgG antibody conjugated to HRP (1:10,000 dilution, Amersham Biosciences). The HRP signals were detected using a chemiluminescence detection kit (SuperSignal Femto Dura extended duration substrate, Pierce). For coimmunoprecipitation, protein A-agarose beads (Roche) were blocked with 0.05% (w/v) BSA (Sigma) for 1 h, incubated with anti-FLAG polyclonal antibody (0.5 μg/sample, Sigma) for 1 h, and washed with the lysis buffer. The cell lysate was incubated with unbound beads for 1 h to preclear the sample and then with the antibody-bound beads overnight at 4 °C on a rotator. The beads were precipitated and washed five times with the lysis buffer. Bound proteins were analyzed by Western blotting as described above.
Frozen Sections and Immunofluorescence
Mouse embryos and tissues were fixed with 4% paraformaldehyde in PBS. For frozen sectioning, fixed tissues were equilibrated in 20% sucrose, embedded into O.C.T. compound (Electron Microscopy Sciences), and 10-μm frozen sections were prepared. Cultured PC12 cells were seeded on coverslips (Electron Microscopy Sciences) coated with poly-D-lysine or rat tail collagen I, induced for the differentiation by nerve growth factor (100 ng/ml) for 3 days, and fixed with 4% paraformaldehyde. Slides with frozen sections or coverslips with cultured cells were blocked in a blocking buffer (PBS, 0.05% Tween 20, 1% normal horse serum, and 1% BSA) and then incubated with the same blocking buffer with primary antibodies. The primary antibodies used were anti-Olfm1 (1:100 dilution), anti-NgR1 (1:50 dilution, Alomone Laboratory), and anti-EGFP (1:2,000 dilution, Aves Labs, Inc.). Slides were washed in PBS and then incubated in a buffer (PBS, 0.05% Tween 20, 1% BSA) with Alexa Fluor 488- or Alexa Fluor 594-conjugated goat anti-mouse, anti-rabbit, or anti-chicken IgG (1:400 dilution, Invitrogen) and DAPI for nuclear staining. Images were collected using a confocal laser microscope (LSM 700, Carl Zeiss, Inc.) or an Axioplan 2 microscope (Carl Zeiss, Inc.).
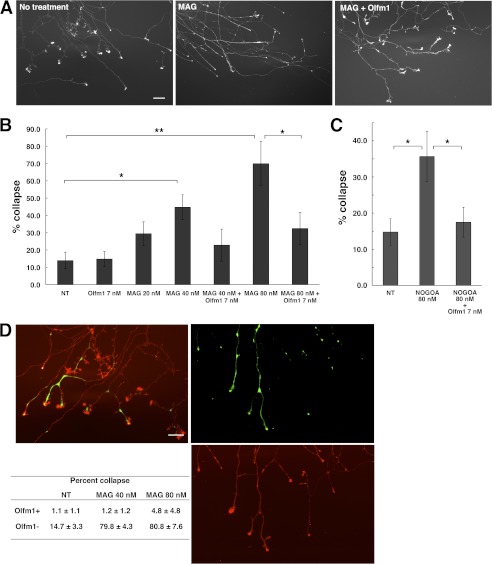
MAG-induced Growth Cone Collapse in Mouse DRG Explant Cultures
The dissected adult mouse DRG were cultured on chambered coverslips coated with laminin and poly-D-lysine (BD Biosciences) with Neurobasal A medium, 1 × ITS and 16 ng/ml nerve growth factor. DRG were grown for 2 days with daily medium change and then treated with MAG-Fc (80 nm, R&D Systems) for 30 min with or without Olfm1 (7 nm) pretreatment for 10 min. Cells were immediately fixed with 4% paraformaldehyde in PBS and stained with rhodamine-phalloidin (Invitrogen). The stained growth cones were observed using a fluorescence microscope, and images were recorded. The shape and area of all growth cones in the image were analyzed by ImageJ software. Growth cones larger than 10 μm2 with well spread filopodia were counted as healthy growth cones.
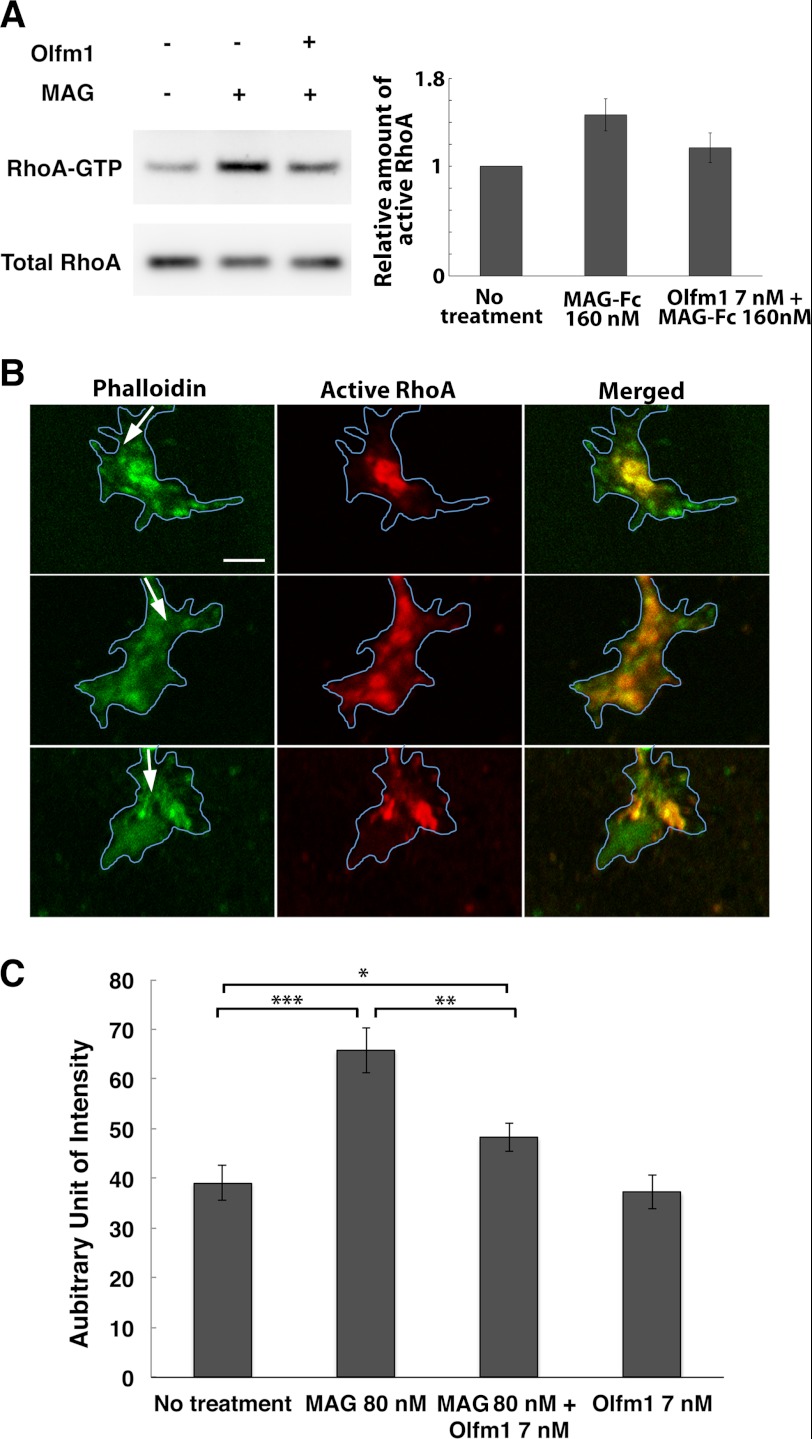
Measurement of RhoA GTP
RhoA GTP was measured using Rhotekin RDB-agarose (Millipore) and following the protocol of the company. Briefly, cells were homogenized in Mg2+ lysis buffer (25 mm HEPES (pH 7.5), 150 mm NaCl, 1% Igepal CA-630, 10 mm MgCl2, 1 mm EDTA, 5 mm NaF, 0.5 mm sodium orthovanadate, 10% glycerol, 1 mm PMSF, 1 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 μg/ml pepstatin) for 20 min on ice. After centrifugation for 15 min, the supernatants were collected, and protein concentrations were measured. Samples were adjusted to the same protein concentration, mixed with Rhotekin-RDB-conjugated agarose beads on a rotator for 45 min at 4 °C, and beads were washed three times with the Mg2+ lysis buffer. The amounts of bound active RhoA and total RhoA in the initial cell lysates were estimated by Western blot analysis with anti-RhoA antibody (Santa Cruz Biotechnology, Inc.). For active RhoA GTP measurements in the DRG growth cones, samples were stained with anti-active RhoA-GTP (New East Biosciences, 1:500 dilution), anti-mouse IgG conjugated with Alexa Fluor 647 (1:400 dilution), and rhodamine-phalloidin. Images at the growth cones before collapse (average area > 30 μm2) were captured with a confocal laser microscope, and the intensity of RhoA GTP signal at each growth cone was quantitated by ImageJ software.
Zebrafish Morpholino Oligonucleotide (MO) and RNA Injections
The antisense MOs were designed and produced by Gene Tools LLC (Philomath, OR). The sequences of MOs used in this work were as follows: Control-MO, 5′-CCTCTTACCTCAGTTACAATTTATA-3′; Olfm1-MO, 5′-AGCAAAGGCACCGACATCTCTGCTC-3′; and Missense-MO, 5′-AGGAAACGCACCCACATGTCTCCTC-3′. The MOs were dissolved in distilled water, mixed with 2 × injection buffer (0.05% phenol red, 240 mm KCl, and 40 mm HEPES (pH 7.4)) and injected into egg yolk of 1- to 4-cell-stage embryos with a pneumatic PicoPump (PV820, World Precision Instruments, Inc., Sarasota, FL). Inserts of cDNA clones CO250150 and BC076103 encoding the full-length AMZ form of zebrafish olfm1b and ngr1 obtained from Open Biosystems were subcloned into the pCS2+ vector. RNAs were synthesized using an mMESSAGE mMACHINE kit (Ambion) and different pCS2 constructs as templates. Synthesized RNAs (12.5–25 pg) were injected into one-cell stage embryos.
Optic Nerve Analysis
1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI) injection was performed using the method described previously (21). Briefly, embryos were fixed with 4% paraformaldehyde and placed in 1.2% low-melting agarose. DiI (0.5 nl, 75 mg/ml) was injected into two positions diagonal to each other (nasodorsal and temporovental) in the left eye. The embryos were incubated at 28 °C overnight, and the projection of the optic nerve stained with DiI was observed and photographed using a dissection fluorescence microscope (Stemi SV11, Zeiss). The diameter of the optic nerves was measured and calculated using National Institutes of Health Image software.
Statistical Analysis
Data were analyzed using unpaired Student's t test preceded by F test for variances.
RESULTS
Olfm1 Interacts with NgR1
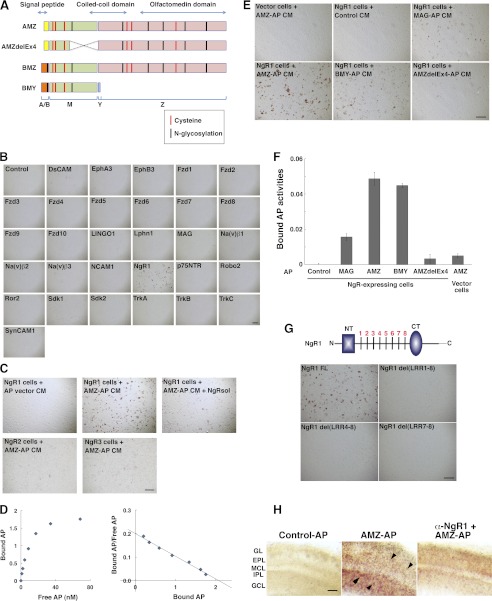
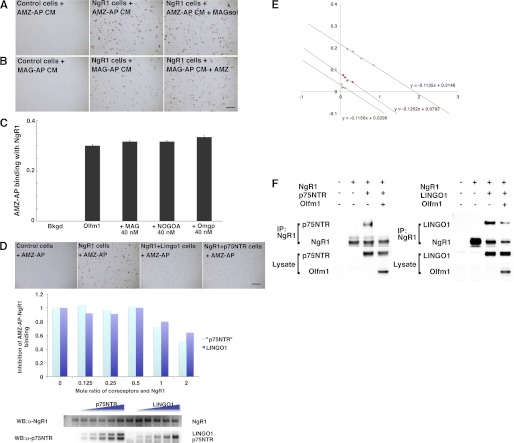
Because there are no known receptor or membrane targets for secreted Olfm1, we screened for such molecules in a series of binding experiments using different forms of Olfm1 (Fig. 1A) and a variety of neuronal membrane proteins. We first selected proteins with multiple immunoglobulin domains as well as those important for axon growth that have patterns of glycosylation sites and cysteine arrangements similar to Olfm1 because our previous results indicated that Olfm1 may be essential for axon growth (11). To test possible binding of Olfm1 to selected proteins, we used CM of COS7 cells transiently transfected with a construct encoding the Olfm1 (the AMZ form) alkaline phosphatase fusion protein (Olfm1-AP). Olfm1-AP-containing CM (10–20 nm) was added to COS7 cells that were transfected with plasmids encoding candidate proteins. COS7 cells were incubated with Olfm1-AP CM, and the activities of AP bound to cell membrane were visualized by staining using nitro-blue tetrazolium and 5-bromo-4-chloro-3′-indolyphosphate as substrates. Among 33 membrane-bound proteins tested by this assay, only NgR1 demonstrated a strong interaction with Olfm1-AP (Fig. 1, B and C). Even two receptors closely related to NgR1, NgR2 and NgR3, did not show strong Olfm1-AP binding (Fig. 1C), indicating that the interaction between Olfm1 and NgR1 is highly specific. Addition of a soluble NgR1 extracellular domain (about 5 nm) as a competitor to the Olfm1-AP CM during incubation with COS7 cells dramatically reduced binding of Olfm1-AP to cell membranes, confirming a direct interaction of Olfm1-AP and NgR1 (Fig. 1C).
FIGURE 1.
Secreted Olfm1-AP binds to NgR1 on the surface of COS7 cells. A, schematic diagram showing three different isoforms (AMZ, BMZ, and BMY), and a deletion mutant (AMZdelEx4) of the Olfm1 protein, their domain structures, and possible modification sites. The alternative N-terminal sequences, A and B, that include signal peptides are shown in yellow and orange, the central M part of Olfm1 is common for all isoforms and is shown in green. The olfactomedin domain is marked in pink. B, Olfm1-AP binds to NgR1 but not to other tested receptor and membrane proteins on the surface of COS7 cells. COS7 cells transfected with receptor-expressing constructs are indicated in each panel. Two days later, cells were incubated with 10 nm Olfm1 (AMZ)-AP and stained for bound AP. Scale bar = 500 μm. C, Olfm1 (AMZ)-AP (10 nm) binds to NgR1 more strongly than to NgR2 or NgR3, and this binding was inhibited by soluble NgR1 protein (NgRsol) (5 nm). Scale bar = 500 μm. D, Scatchard plot of Olfm1 (AMZ)-AP binding to NgR1. These experiments were repeated four times, and results of typical experiment are shown. E, binding of different isoforms of Olfm1-AP, a deletion mutant of Olfm1-AP and MAG-AP, to COS7 cells transfected with NgR1. The AP activities in all CM were adjusted to the same concentration before adding to the NgR1 cells. Note that mutated Olfm1 with a deletion in the central region (delEx4) exhibits reduced activity. Scale bar = 500 μm. F, quantification of the results shown in E. G, binding of Olfm1-AP to COS7 cells transfected with full-length and deletion mutants of NgR1. The schematic diagram shows the main structural features of the NgR1 protein. The N- and C-terminal cysteine-rich cupping domains are shown as a blue rectangle and oval, respectively. Eight leucine-rich repeats are marked by transverse black lines. NT, N-terminal capping domain; CT, C-terminal capping domain. Scale bar = 500 μm. H, binding of Olfm1-AP to the sections from mouse P3 olfactory bulbs. Fresh sections (200 μm) were incubated with AP (left panel) or Olfm1-AP (center and right panels) together with normal rabbit IgG (left and center panels) or anti-NgR1 polyclonal IgG (right panel) at the concentration of 5 ng IgG/μl. The arrowheads show positive cells in different cell layers. EPL, external plexiform layer; GCL, granule cell layer; GL, glomerular layer; IPL, internal plexiform layer; MCL, mitral cell layer. Scale bar = 100 μm.
The affinity of Olfm1 for NgR1 was estimated on the basis of binding of increased amounts of the Olfm1-AP protein to NgR1-expressing COS7 cells. The calculated Kd was 9.5 ± 0.7 (Fig. 1D). The interaction of Olfm1 with NgR1 did not require the presence of the olfactomedin domain because the BMY form of Olfm1 (10–20 nm) also bound to NgR1 in this test (see Fig. 1, E and F). Deletion of the central portion of the N-terminal part of Olfm1 (M domain, AMZdelEx4, see Fig. 1A) dramatically reduced the binding ability of the modified protein compared with wild-type Olfm1, confirming the importance of the intact N-terminal half of Olfm1 for the interaction with NgR1 (Fig. 1, E and F). NgR1 contains eight leucine-rich repeats in the extracellular domain (22). Removal of all these repeats or several of these repeats disrupted binding of Olfm1 to NgR1 (Fig. 1G). Olfm1-AP binding to NgR1 was also tested in native tissues using P3 mouse olfactory bulb sections. Both Olfm1 and NgR1 are expressed intensely in the olfactory bulbs at this stage. Olfm1-AP showed the strongest binding to cells located in the granule cell and internal plexiform layers and weaker binding to cells located in other layers (Fig. 1H). This binding was markedly reduced by addition of antibodies against NgR1 but not by unrelated control antibodies (Fig. 1H).
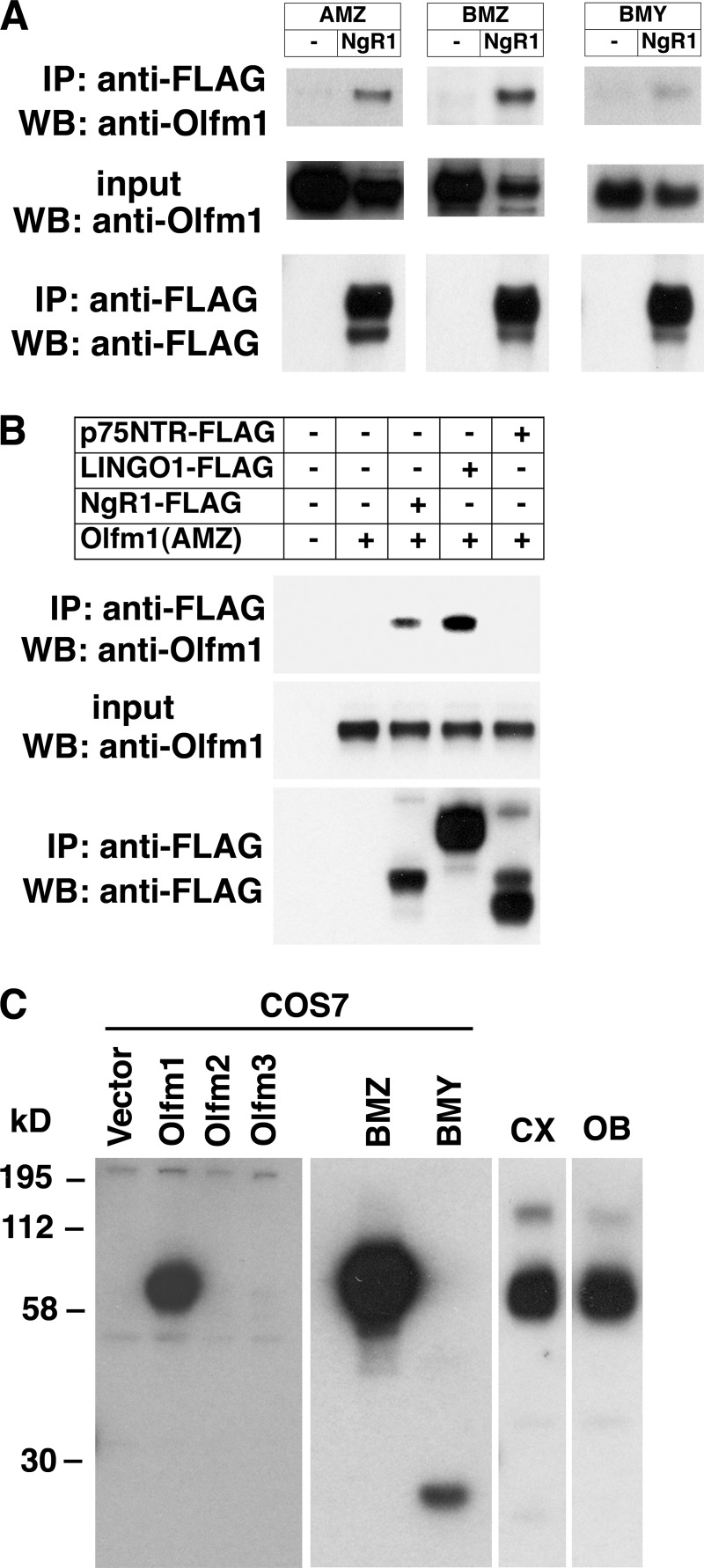
The interactions between Olfm1 and NgR1 were further confirmed in coimmunoprecipitation experiments. All tested forms of Olfm1 coprecipitated with NgR1 (Fig. 2A). Unlike in the Olfm1-AP binding test, Olfm1 was coimmunoprecipitated not only with NgR1 but also with a protein in the same NgR1 complex, LINGO-1 (Fig. 2B). Another protein interacting with NgR1, p75NTR, did not show a coprecipitation with Olfm1. The difference between AP-binding and coimmunoprecipitation tests may be due to the fact that interactions in the former test occurred extracellularly with heavily glycosylated Olfm1, whereas in the latter test it may have occurred intracellularly with a less modified form of Olfm1, exhibiting a different three-dimensional organization. We concluded that Olfm1 may function as a novel ligand of NgR1 and proceeded to study the physiological consequences of their interaction in more detail.
FIGURE 2.
Coimmunoprecipitation of Olfm1 with NgR1 and LINGO-1. A and B, different forms of Olfm1 were cotransfected with FLAG-tagged NgR1, p75NTR, or LINGO-1. Cell lysates were immunoprecipitated (IP) with anti-FLAG antibody, and coprecipitated Olfm1 was detected in Western blot analysis (WB) with anti-Olfm1 antibody. All tested forms (AMZ, BMZ, and BMY) were coprecipitated with NgR1 (A). Olfm1 (AMZ) was also coprecipitated with LINGO-1 but not with p75NTR (B). All immunoprecipitation experiments were repeated at least three times. Input lines contained about 10% of lysates that were used for immunoprecipitation. C, validation of Olfm1 polyclonal antibody used for immunofluorescence study. Western blot analysis of lysates of COS7 cells transfected with the vector or the expression constructs encoding Olfm1, Olfm2, Olfm3, or different forms of Olfm1. The polyclonal Olfm1 antibody (1:2000) recognized only Olfm1 protein. The two right lanes show a Western blot analysis of mouse brain cortex (CX) or OB lysates. The antibody recognized one major band of Olfm1 protein in mouse CX and OB. The minor band represents Olfm1 dimers.
Olfm1 and NgR1 Have Overlapping Intracellular and Tissue Distribution
To investigate the intracellular and tissue distribution of Olfm1 and compare it with the distribution of NgR1, we produced a novel polyclonal antibody against full-length Olfm1 protein expressed in mammalian cells. This was necessary because available Olfm1 monoclonal antibodies function poorly in tissue sections. Our polyclonal Olfm1 antibody recognized different isoforms of mammalian Olfm1 but not closely related Olfactomedin 2 (Olfm2) or Olfactomedin 3 (Olfm3) proteins expressed in COS7 cells transiently transfected with corresponding expression constructs. Olfm1 antibody gave one major band in Western blotting experiments using adult mouse cerebral cortex or olfactory bulb extracts (Fig. 2C).
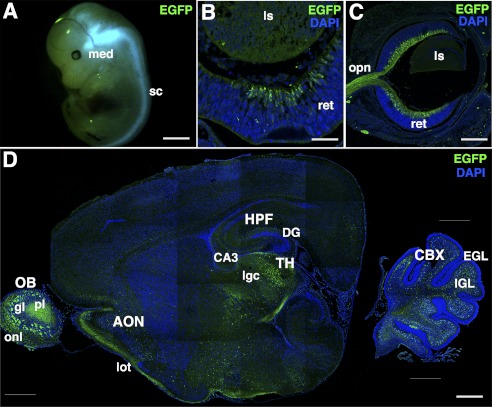
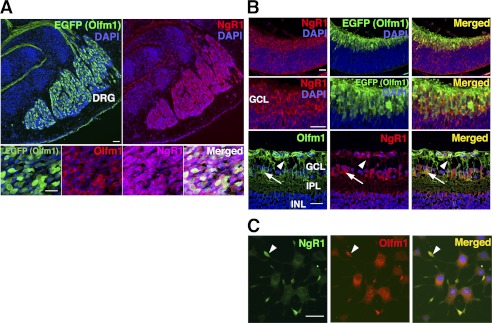
The specificity of the antibody and Olfm1 expression was also evaluated using transgenic mice containing bacterial artificial chromosome with the EGFP sequence inserted downstream of the Olfm1 AMZ promoter (23). In these transgenic mice, EGFP expression in the developing and adult brain coincides with known sites of Olfm1 expression, as determined by in situ hybridization (4, 24). Expression of Olfm1 reaches the maximum level between the late embryonic and the early postnatal stages, as judged by in situ hybridization and quantitative PCR (25). At the embryonic stage E13, EGFP expression was detected mainly in the neuronal tissues, with the highest levels of expression in the medulla oblongata, outer area of the midbrain, DRG, and spinal cord. In the eye, expression was detected mainly in the central retina in differentiating retinal ganglion cells (Fig. 3, A and B). At E15, EGFP expression in the eye was observed in differentiating retinal ganglion cells that migrated from the central region to the peripheral region as well as in the retinal nerve fiber layer and in retinal ganglion cells axons (Fig. 3C). At P3, EGFP continued to be expressed in the retinal ganglion cells and their axons. In the brain, the main sites of EGFP expression were the olfactory bulbs, anterior olfactory nucleus, thalamus, hippocampus, and cerebellum. Axons from the OB to the lateral olfactory tract were also positive (Fig. 3D). The polyclonal Olfm1 antibody recognized all EGFP-positive cells in the mouse brain and retina, confirming the specificity of the antibody. Staining of the brain and eye sections with antibodies against NgR1 showed colocalization of NgR1 and Olfm1 that was particularly pronounced in DRG and retina (Fig. 4, A and B). In DRG, NgR1 was detected in a larger population of neurons compared with Olfm1-expressing neurons that stained about 30% of neuronal population. Costaining of the retina of E13, E15, and P3 mice with NgR1 antibodies demonstrated that NgR1 and Olfm1 were colocalized in most of Olfm1-expressing neurons and their processes (Fig. 4B). The colocalization was detected not only in the cell bodies but also in nerve fibers, suggesting that the interaction of Olfm1 and NgR1 may play a role in neurite development or activity.
FIGURE 3.
EGFP expression in TG(Olfm1:EGFP) mice. A, E13 mouse embryo. med, medulla oblongata; sc, spinal cord. Scale bar = 2 mm. B, frozen section cut through the eye of E13 embryo. ret, retina; ls, lens. Scale bar = 50 μm. C, as in B but at E15. Opn, optic nerve. Scale bar = 200 μm. D, frozen section of the brain of 3-day-old TG(Olfm1:EGFP) mice. Nuclei were stained with DAPI (blue) in B–D. The major sites of expression are observed in olfactory bulb (OB), anterior olfactory nucleus (AON), thalamus (TH), CA3 region, and dentate gyrus (DG) in hippocampus (HPF) and internal granular layer (IGL) in cerebellum (CBX). Axons from OB to lateral olfactory tract (lot) are also positive. EGL, external granular layer; gl, glomerular layer; lgc, lateral geniculate complex; onl, olfactory nerve layer; pl, plexiform layer. Scale bar = 500 μm.
FIGURE 4.
Colocalization of Olfm1 and NgR1 in mouse tissues and PC12 cells. A, DRG from E13.5 TG(Olfm1:EGFP) mice was stained with indicated antibodies. Upper panels, green, EGFP; red, NgR1. Lower panels, green, EGFP; red, Olfm1; purple, NgR1. Nuclei were stained with DAPI (blue). Note that only about 30% of NgR1-expressing cells were EGFP- and Olfm1-positive. Scale bar = 25 μm. B, retina from E13.5 (upper and center panels) and P3 (lower panels) TG(Olfm1:EGFP) mice stained for Olfm1 and NgR1. Colocalization of Olfm1 and NgR1 is observed in cell soma (arrows) and neurites (arrowheads). GCL, ganglion cell layer; INL, inner nuclear layer; IPL, inner plexiform layer. Scale bar = 25 μm. C, PC12 cells stained for Olfm1 (red) and NgR1 (green). Both Olfm1 and NgR1 accumulate in growth cones (arrowheads). Scale bars = 25 μm.
Finally, staining of differentiating PC12 cells demonstrated that endogenously expressed Olfm1 and NgR1 showed overlapping intracellular localization. Olfm1 staining was equally strong in the endoplasmic reticulum and the growth cones, whereas NgR1 staining was much stronger in the growth cone compared with the endoplasmic reticulum (Fig. 4C). We concluded that intracellular and tissue distribution of endogenously expressed Olfm1 and NgR1 supports their possible functional interaction during development. Localization of Olfm1 in the growth cones of neuronal cells suggests that Olfm1 may play a role in axon growth and/or guidance.
Olfm1 Inhibits Growth Cone Collapse Induced by Myelin Inhibitors
Myelin-associated inhibitors (Nogo A, MAG, and OMgp) interact with NgR1 and limit axonal outgrowth (16). To assess the effects of Olfm1 on NgR1 activity stimulated by myelin-associated inhibitors, we first used mouse DRG explant culture. DRG explant in culture started extending axons to all directions. The tip of each growth cone demonstrated a healthy shape as shown (Fig. 5A, left panel). The addition of 20–80 nm of MAG-Fc into the culture medium induced a dose-dependent growth cone collapse after 30 min (Fig. 5A, center panel). This effect was also observed with another myelin-associated inhibitor tested, Nogo A-Fc, although the collapsing effect of Nogo A-Fc were less pronounced than collapsing effects of MAG at the same concentration (Fig. 5C). The addition of Olfm1 (7 nm) to DRG cultures 10 min prior to myelin-associated inhibitor treatment significantly reduced their growth cone-collapsing activity (Fig. 5, A–C).
FIGURE 5.
Inhibition of MAG-induced growth cone collapse in mouse DRG explant cultures by Olfm1. A, DRG explants cultured for 2 days were treated with MAG-Fc (80 nm) for 30 min with or without Olfm1 (7 nm) pretreatment for 10 min. Growth cones were stained with rhodamine-phalloidin to count healthy or collapsed growth cones. Scale bar = 50 μm. B, quantification of growth cone collapse in DRG cultures treated with increasing concentrations of MAG with or without Olfm1 pretreatment. Five explant cultures were used in these experiments. *, p < 0.05; **, p < 0.01. C, quantification of growth cone collapse in DRG cultures treated with Nogo A-Fc (80 nm) with and without Olfm1 pretreatment. These experiments were performed with at least nine explant cultures per condition. *, p < 0.05. D, growth cones of Olfm1(EGFP)-positive and negative DRG axons after MAG treatment (right panels). The left panel shows untreated growth cones. n = 8 (NT), 7 (MAG 40 nm), and 6 (MAG 80 nm). **, p < 0.01.
About 30% of DRG neurons produced high levels of Olfm1 (see Figs. 4A and 5D). We tested whether DRG neurons expressing Olfm1 behave differently in the MAG-induced growth cone collapse assay than neurons not expressing Olfm1. MAG-Fc (40–80 nm) was added to the DRG explants from TG(Olfm1:EGFP) mice and growth cone collapse was counted separately for EGFP-positive and -negative axons. Staining of DRG cultures with antibodies against Olfm1 confirmed that EGFP-positive axons express Olfm1. The results of these experiments showed that axons expressing Olfm1 were more resistant to the MAG-induced collapse than axons expressing low levels of Olfm1 (Fig. 5D). This observation suggested that endogenous Olfm1 transported to the growth cones and secreted protects them from endogenous inhibitory molecules, such as MAG, and may facilitate axon growth in development.
Olfm1 Inhibits the Interaction of NgR1 and p75NTR or LINGO-1
Inhibition of the NgR1 activity by Olfm1 may occur through the competition for binding with one or some of its ligands. Three myelin ligands of NgR1, MAG, Nogo A, and OMgp, were tested for inhibition of Olfm1-AP binding to NgR1. Soluble MAG did not inhibit the binding of Olfm1-AP to NgR1 and Olfm1 did not inhibit binding of soluble MAG-AP to NgR1, indicating that MAG and Olfm1 may interact with distinct domains of NgR1 (Fig. 6, A and B). Similarly, none of tested NgR1 ligands at the concentration of 40 nm inhibited the Olfm1-NgR1 binding (Fig. 6C). Even 400 nm of MAG and Nogo A failed to inhibit the Olfm1 binding to NgR1. Because tested concentrations of ligands are much higher than their Kd values (5–10 nm), we concluded that these ligands do not compete with Olfm1 for the NgR1 binding.
FIGURE 6.
Inhibition of NgR1 complex formation by Olfm1. A, B, and C, Olfm1 and NgR1 ligands do not compete for binding with NgR1. A, Olfm1-AP (10 nm) binding to NgR1 with and without pretreatment with soluble MAG (40 nm) for 30 min. B, MAG-AP (10 nm) binding to NgR1 with or without pretreatment with Olfm1 (30 nm) for 30 min. C, quantification of Olfm1-AP (10 nm) binding to NgR1 after pretreatment with different NgR1 ligands. D and E, inhibition of Olfm1-AP binding to NgR1 by coexpression of p75NTR or LINGO-1. COS7 cells were transfected with NgR1 together with increasing amounts of p75NTR or LINGO-1. The expression levels of NgR1, p75NTR, and LINGO-1 in transfected cells were confirmed by Western blotting (lower panel in D). Olfm1-AP binding was quantified as described under “Experimental Procedures.” E, Scatchard plots of Olfm1 (AMZ)-AP binding to NgR1 in the presence of p75NTR or LINGO-1. These experiments were repeated three times, and results of typical experiment are shown. Blue circle, NgR1 alone; red square, NgR1 plus p75NTR; green triangle, NgR1 plus LINGO-1. F, inhibition of coimmunoprecipitation of NgR1 and p75NTR or LINGO-1 by Olfm1. COS7 cells were transfected with NgR1 together with indicated combinations of coreceptors. NgR1 was immunoprecipitated with anti-NgR1 antibody, and coprecipitated p75NTR or LINGO-1 were detected by Western blot analysis using corresponding antibodies.
Next, we tested effects of p75NTR and LINGO-1 on Olfm1-AP binding with NgR1. Coexpression of p75NTR or LINGO-1 with NgR1 inhibited Olfm1-AP binding, and these inhibitions were dependent upon the amount of the expressed p75NTR protein or LINGO-1 (Fig. 6D). The Kd value of Olfm1 for NgR1 was not changed by the addition of p75NTR or LINGO-1. Instead, these coreceptors were competing with Olfm1 for binding with NgR1 (Fig. 6E). We also tested whether Olfm1 inhibits interactions of NgR1 with p75NTR or LINGO-1. Binding of NgR1 and p75NTR was detected by coimmunoprecipitation from cell culture lysates. The p75NTR protein was coimmunoprecipitated together with NgR1 by anti-NgR1 antibody (Fig. 6F). Coexpression of Olfm1 with NgR1 and p75NTR strongly inhibited their binding. The binding of NgR1 and LINGO-1 was similarly inhibited by Olfm1 coexpression.
Finally, we tested whether the inhibition of NgR1-coreceptor binding by Olfm1 leads to the inhibition of downstream signaling that may lead the axonal growth inhibition. Addition of the soluble MAG-Fc protein to COS7 cells expressing NgR1, p75NTR, and LINGO-1 activated RhoA, as demonstrated by up-regulation of RhoA-GTP in 15 min. Olfm1 pretreatment for 10 min prior to MAG-Fc addition reduced the RhoA-GTP to the control level, suggesting that binding of Olfm1 to NgR1 inhibited coreceptor interactions with NgR1 and attenuated intracellular signal transductions (Fig. 7A). Changes in the RhoA activity induced by MAG and Olfm1 were also measured in DRG explants. Active RhoA-GTP was increased at the tip of growth cones after MAG-Fc treatment. This increase in RhoA-GTP was inhibited by a pretreatment with Olfm1 (Fig. 7, B and C).
FIGURE 7.
Inhibition of NgR1-mediated and MAG-induced RhoA activation by Olfm1. A, COS7 cells were transfected with cDNAs encoding NgR1, p75 NTR and/or LINGO-1. 48 h after the transfection, purified Olfm1 or buffer control was added for 10 min, and then cells were stimulated with MAG-Fc (160 nm). Cells were harvested 15 min later, and the active form of RhoA (RhoA-GTP) was pulled down and detected by Western blotting. These experiments were repeated seven times. B, DRG growth cones were treated with MAG (80 nm) for 10 min with (lower panels) or without (center panels) Olfm1 (7 nm) pretreatment. Upper panels, untreated control. The explants were stained with anti-active RhoA antibody (red) together with phalloidin (green). Arrows show the direction of axon growth. Blue lines represent the outlines of growth cones. Scale bar = 5 μm. C, quantification of the fluorescence intensities for active RhoA in the immunostained growth cones (>30 growth cones from three to six explants) measured using ImageJ. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Functional Interaction of Olfm1 and NgR1 in Zebrafish in Vivo
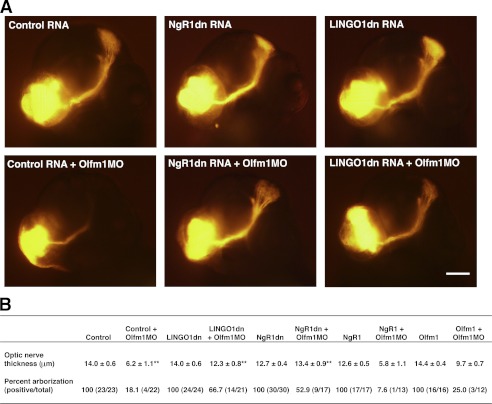
The expression of olfm1a and olfm1b (8) and ngr1 (26) have been analyzed previously in the course of zebrafish development. Expression of these genes overlaps in the neuronal tissues, including the cranial ganglia, telencephalon, optic tectum, olfactory bulb, and retina at 30–48 h post-fertilization. We have also shown that overexpression of the zebrafish or mammalian Olfm1 proteins in zebrafish facilitated optic nerve extension and the arborization in the optic tectum (11). Oppositely, knockdown of the olfm1 protein by MOs inhibited the optic nerve extension and arborization (11). However, the mechanisms underlining these effects of Olfm1 were not understood. We hypothesized that the effect of olfm1 knockdown by morpholino may result from an up-regulation of ngr1 activity. In this case, suppression of ngr1 activity could reduce the effect of olfm1 MO. To test this hypothesis, we first injected RNA encoding dominant negative forms of ngr1 (ngr1dn) or lingo-1 (lingo-1dn) to suppress ngr1 activity (27) and then olfm1 MO. Optic nerve extension was measured at 3 days post-fertilization. In accordance with previous results (11), embryos injected with olfm1 MO alone demonstrated a reduced extension of the optic nerve and a dramatic reduction in the arborization in the optic tectum (Fig. 8A). The thickness of the optic nerve bundle was reduced more than 2-fold in the injected samples as compared with control embryos injected with RNA encoding EGFP. Injection of RNA encoding ngr1dn or lingo-1dn (12.5 or 25 pg/embryo) alone did not significantly affect the optic nerve extension. Expression of dominant negative constructs did not produce additional effects because endogenous olfm1 already blocked the ngr1 activity. However, injections of RNA encoding ngr1dn or lingo-1dn together with olfm1 MO dramatically reduced optic nerve defects observed after injection of olfm1 MO alone. In particular, optic nerve thickness was restored to the level close to the control embryos (Fig. 8B). The arborization in the optic tectum was also restored in the half of embryos injected with RNA encoding ngr1dn or lingo-1dn together with olfm1 MO. Injection of ngr1 RNA (20 pg/embryo) together with olfm1MO, as predicted, reduced optic nerve thickness and reduced the arborization in the optic tectum more dramatically than injection of olfm1 MO alone (Fig. 8B). These in vivo results supported the idea that Olfm1 is a NgR1 ligand participating in the regulation of axon growth.
FIGURE 8.
Inhibition of optic nerve growth on zebrafish embryos by olfm1 MO and rescues by ngr1 inhibition. A and B, zebrafish 1- to 4-cell-stage embryos were injected with or without indicated RNAs and olfm1 MO. As control RNA, EGFP RNA was injected. At 75 h post-fertilization, embryos were fixed, and DiI was injected to the retina in the unilateral eye. A, representative image of embryo head (frontal view) injected with DiI. The lower left round orange signals are the dye-injected position. Optic nerves are diagonally extended to the optic tectum (upper right panel) and make arborizations. B, optic nerve thickness was measured in the middle of the optic nerves and the percent arborizations were counted.
DISCUSSION
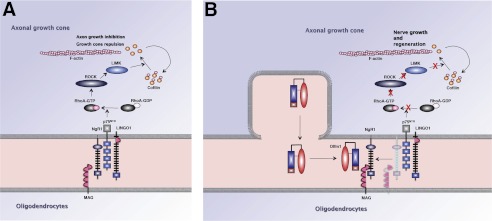
During nervous system development, each neuron extends an axon that navigates through a changing environment to reach its final destination. Neuronal growth cones, specialized structures at the tips of extending axons, play a critical role in axonal growth, pathfinding, and synaptogenesis. In the adult mammalian CNS, axons have very limited capacity for regrowth following injury compared with the embryonic CNS and the peripheral nervous system. Although their role in nerve regeneration in vivo is still controversial, myelin-associated inhibitory factors are thought to contribute to the inhibition of regrowth of injured axons in the adult CNS. Four classes of major myelin-associated proteins, MAG, Nogo A, OMgp, and chondroitin sulfate proteoglycans are potent inhibitors of neurite outgrowth in vitro that interact with the NgR1 complex. The downstream effectors of this signaling are Rho and Rho-associated kinase (28). The existing data suggest that the NgR1-ligand signaling system may contain additional components (29). In this study, we demonstrate that Olfm1 is a component of neuronal growth cones and is a novel soluble ligand of NgR1, which regulates axonal outgrowth and axon bundle formation during brain development. If binding of MAG or other known ligands to the NgR1 complex leads to RhoA GTPase activation with subsequent growth cone inhibition through the cytoskeletal reorganization (30), binding of Olfm1 to NgR1 reduces the level of RhoA-GTP, suggesting that Olfm1 attenuates receptor complex activity (Fig. 9).
FIGURE 9.
Schematic diagrams illustrating the involvement of Olfm1 in the regulation of axon growth through the NgR1 complex and RhoA signaling. A, ligands such as MAG on the glial surface (bottom compartment) or unknown soluble molecules in the extracellular space (center compartment) bind to NgR1-p75NTR-LINGO-1 complex on the surface of the axonal growth cone (upper compartment). The signal is transduced to the intracellular space of the growth cone, converts RhoA to the active GTP-bound form, activates a cascade of proteins including ROCK and LIMK, and removes G-actin from cofilin. The released cofilin depolymerizes actin filaments, leading to growth cone collapse or retraction. B, when Olfm1 is secreted from the tips of the growth cone or neighboring cells, it binds to NgR1, dissociates NgR1-coreceptor interactions, and inhibits the activation of RhoA. This facilitates axon growth and may stimulate regeneration of damaged nerves.
The well known ligands of NgR1, such as MAG, are membrane-bound proteins, whereas Olfm1 is a secreted protein. We used the AP assay to look for physiological interactions between different membrane receptors and Olfm1. We found that Olfm1 could bind only NgR1 in this assay, and that all other receptors tested in this assay failed to bind, even though some proteins showed potent interactions with Olfm1 in other methods, such as coimmunoprecipitation and immunofluorescence colocalization. We believe that secreted Olfm1, being more heavily glycosylated as compared with intracellular Olfm1, may have a different three-dimensional organization and altered binding affinity for certain proteins as compared with the less glycosylated intracellular Olfm1. Although the binding of Olfm1 to NgR1 seems to be potent but slightly weaker compared with other known ligands such as MAG on the basis of the Kd value, we showed that the binding of Olfm1 is disturbing the interaction of NgR1 and coreceptors such as p75NTR and LINGO-1 and inhibiting further signal transductions (Fig. 6). In many respects, effects of Olfm1 on NgR1 are similar to those observed for a recently identified ligand of NgR1, LGI1 (29). The latter secreted leucine-rich repeat protein binds to NgR1 but not to NgR2 or NgR3 and antagonizes myelin-induced growth cone collapse (29). It will be interesting to determine whether Olfm1 and LGI1 compete for NgR1 binding and produce additive effects.
Previous studies have established that the N-terminal half of the Olfm1 protein is essential for its physiological activity and is capable of binding with target proteins (9–11). Consistent with these studies, an Olfm1 shorter form has a binding ability to NgR1, and a deletion mutant has a reduced ability of binding to NgR1 in the AP assay. Olfm1 being an additional NgR1 ligand may help in understanding the complicated but strictly organized patterning of axonal connections in the brain.
The interaction of Olfm1 with NgR1 appears to be especially important in the development of the sensory and motor nervous systems. In our previous study using a zebrafish developmental model, olfm1 was highly expressing in the retina, sensory ganglia, olfactory bulb, and spinal cord. Knockdown of the olfm1 gene by MOs caused strong defects in nerve formation along the spinal cord and in optic nerve arborization. In this study, this defect was reduced by coinjection of dominant negative ngr1 or lingo-1, indicating that the phenotype of olfm1 morphants may involve up-regulation of ngr1 activity. Considering that olfm1 is expressed in other sensory and motor systems, including DRG and retina, it will be important to investigate potential structural and functional effects of Olfm1 in these tissues using the same experimental models.
Functions of olfactomedin domain-containing proteins and mechanisms of their action are largely unknown. Olfm1 is closely related to two other family members, Olfm2 (31) and Olfm3 (32), and these proteins may act through similar mechanisms. Knockdown of olfm2 gene expression by MO in zebrafish perturbs the formation of axonal projections from branchiomotor neurons and disrupts anterior head and CNS development, which manifests as severe defects in olfactory pit, eye, and optic tectum development. On the other hand, overexpression of Olfm3 in PC12 cells facilitates cell attachment and inhibits neuronal differentiation. It would be interesting to determine whether Olfm2 and Olfm3 interact with the same target proteins as Olfm1.
Recent data suggest that myelin-associated inhibitors and their receptors are associated with different neural diseases including schizophrenia, amyotrophic lateral sclerosis, multiple sclerosis, and Alzheimer's disease (see Ref. 16 for references). Mutations in LGI1 may lead to autosomal dominant lateral temporal lobe epilepsy (33, 34). A growing number of cases indicate that defects in different proteins that contribute to a common functional protein complex may lead to similar pathologies. It is reasonable to suggest that mutations of Olfm1 may lead to a spectrum of neurological disorders. In conclusion, our data imply that Olfm1 is an endogenous antagonist of myelin-associated axon growth inhibitors. Because other NgR1 antagonists may facilitate axon growth in vivo (35, 36), it is reasonable to propose that application of Olfm1 or stimulation of its expression may be used to facilitate neuronal growth in vivo after axonal damage or injury.
Acknowledgments
We thank Drs. F. F. Lee, J. McNamara, B. Nelkin, K. L. Guan, L. Kaufman, J. Nathans, and Y. Minami for providing different constructs; Dr. L. Li and Barbara Norman for technical help; and Drs. S. Ikeda and F. Ono for advice.
This work was supported, in whole or in part, by the Intramural Research Program of the National Eye Institute, National Institutes of Health.
- MAG
- myelin-associated glycoprotein
- EGFP
- enhanced green fluorescent protein
- CM
- conditioned media
- AP
- alkaline phosphatase
- DRG
- dorsal root ganglia
- MO
- morpholino oligonucleotide
- DiI
- 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate
- E13
- embryonic day 13
- P3
- postnatal day 3
- OB
- olfactory bulb.
REFERENCES
- 1. Tomarev S. I., Nakaya N. (2009) Olfactomedin domain-containing proteins. Possible mechanisms of action and functions in normal development and pathology. Mol. Neurobiol. 40, 122–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barembaum M., Moreno T. A., LaBonne C., Sechrist J., Bronner-Fraser M. (2000) Noelin-1 is a secreted glycoprotein involved in generation of the neural crest. Nat. Cell Biol. 2, 219–225 [DOI] [PubMed] [Google Scholar]
- 3. Moreno T. A., Bronner-Fraser M. (2001) The secreted glycoprotein Noelin-1 promotes neurogenesis in Xenopus. Dev. Biol. 240, 340–360 [DOI] [PubMed] [Google Scholar]
- 4. Nagano T., Nakamura A., Mori Y., Maeda M., Takami T., Shiosaka S., Takagi H., Sato M. (1998) Differentially expressed olfactomedin-related glycoproteins(pancortins) in the brain. Brain Res. 53, 13–23 [DOI] [PubMed] [Google Scholar]
- 5. Danielson P. E., Forss-Petter S., Battenberg E. L., deLecea L., Bloom F. E., Sutcliffe J. G. (1994) Four structurally distinct neuron-specific olfactomedin-related glycoproteins produced by differential promoter utilization and alternative mRNA splicing from a single gene. J. Neurosci. Res. 38, 468–478 [DOI] [PubMed] [Google Scholar]
- 6. Kulkarni N. H., Karavanich C. A., Atchley W. R., Anholt R. R. (2000) Characterization and differential expression of a human gene family of olfactomedin-related proteins. Genet. Res. 76, 41–50 [DOI] [PubMed] [Google Scholar]
- 7. Ando K., Nagano T., Nakamura A., Konno D., Yagi H., Sato M. (2005) Expression and characterization of disulfide bond use of oligomerized A2-Pancortins. Extracellular matrix constituents in the developing brain. Neurosci. 133, 947–957 [DOI] [PubMed] [Google Scholar]
- 8. Nakaya N., Tomarev S. (2007) Expression patterns of alternative transcripts of the zebrafish olfactomedin 1 genes. Gene. Expr. Patterns 7, 723–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Moreno T. A., Bronner-Fraser M. (2005) Noelins modulate the timing of neuronal differentiation during development. Dev. Biol. 288, 434–447 [DOI] [PubMed] [Google Scholar]
- 10. Cheng A., Arumugam T. V., Liu D., Khatri R. G., Mustafa K., Kwak S., Ling H. P., Gonzales C., Xin O., Jo D. G., Guo Z., Mark R. J., Mattson M. P. (2007) Pancortin-2 interacts with WAVE1 and Bcl-xL in a mitochondria-associated protein complex that mediates ischemic neuronal death. J. Neurosci. 27, 1519–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nakaya N., Lee H. S., Takada Y., Tzchori I., Tomarev S. I. (2008) Zebrafish olfactomedin 1 regulates retinal axon elongation in vivo and is a modulator of Wnt signaling pathway. J. Neurosci. 28, 7900–7910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Veroni C., Grasso M., Macchia G., Ramoni C., Ceccarini M., Petrucci T. C., Macioce P. (2007) β-Dystrobrevin, a kinesin-binding receptor, interacts with the extracellular matrix components pancortins. J. Neurosci. Res. 85, 2631–2639 [DOI] [PubMed] [Google Scholar]
- 13. Cafferty W. B., McGee A. W., Strittmatter S. M. (2008) Axonal growth therapeutics. Regeneration or sprouting or plasticity? Trends Neurosci. 31, 215–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McGee A. W., Strittmatter S. M. (2003) The Nogo-66 receptor. Focusing myelin inhibition of axon regeneration. Trends Neurosci. 26, 193–198 [DOI] [PubMed] [Google Scholar]
- 15. Dickendesher T. L., Baldwin K. T., Mironova Y. A., Koriyama Y., Raiker S. J., Askew K. L., Wood A., Geoffroy C. G., Zheng B., Liepmann C. D., Katagiri Y., Benowitz L. I., Geller H. M., Giger R. J. (2012) NgR1 and NgR3 are receptors for chondroitin sulfate proteoglycans. Nat. Neurosci. 15, 703–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Llorens F., Gil V., del Río J. A. (2011) Emerging functions of myelin-associated proteins during development, neuronal plasticity, and neurodegeneration. FASEB J. 25, 463–475 [DOI] [PubMed] [Google Scholar]
- 17. Kim J. E., Liu B. P., Park J. H., Strittmatter S. M. (2004) Nogo-66 receptor prevents raphespinal and rubrospinal axon regeneration and limits functional recovery from spinal cord injury. Neuron 44, 439–451 [DOI] [PubMed] [Google Scholar]
- 18. Bartsch U., Bandtlow C. E., Schnell L., Bartsch S., Spillmann A. A., Rubin B. P., Hillenbrand R., Montag D., Schwab M. E., Schachner M. (1995) Lack of evidence that myelin-associated glycoprotein is a major inhibitor of axonal regeneration in the CNS. Neuron 15, 1375–1381 [DOI] [PubMed] [Google Scholar]
- 19. Westerfield M. (2000) in The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio), University of Oregon Press, Eugene, OR [Google Scholar]
- 20. Snyder D. A., Rivers A. M., Yokoe H., Menco B. P., Anholt R. R. (1991) Olfactomedin. Purification, characterization, and localization of a novel olfactory glycoprotein. Biochem. 30, 9143–9153 [DOI] [PubMed] [Google Scholar]
- 21. Yoda H., Hirose Y., Yasuoka A., Sasado T., Morinaga C., Deguchi T., Henrich T., Iwanami N., Watanabe T., Osakada M., Kunimatsu S., Wittbrodt J., Suwa H., Niwa K., Okamoto Y., Yamanaka T., Kondoh H., Furutani-Seiki M. (2004) Mutations affecting retinotectal axonal pathfinding in Medaka (Oryzias latipes). Mech. Dev. 121, 715–728 [DOI] [PubMed] [Google Scholar]
- 22. Fournier A. E., GrandPre T., Strittmatter S. M. (2001) Identification of a receptor mediating Nogo-66 inhibition of axonal regeneration. Nature 409, 341–346 [DOI] [PubMed] [Google Scholar]
- 23. Gong S., Zheng C., Doughty M. L., Losos K., Didkovsky N., Schambra U. B., Nowak N. J., Joyner A., Leblanc G., Hatten M. E., Heintz N. (2003) A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425, 917–925 [DOI] [PubMed] [Google Scholar]
- 24. Nagano T., Nakamura A., Konno D., Kurata M., Yagi H., Sato M. (2000) A2-Pancortins (Pancortin-3 and -4) are the dominant pancortins during neocortical development. J. Neurochem. 75, 1–8 [DOI] [PubMed] [Google Scholar]
- 25. Sultana A., Nakaya N., Senatorov V. V., Tomarev S. I. (2011) Olfactomedin 2. Expression in the eye and interaction with other olfactomedin domain-containing proteins. Invest. Ophthalmol. Vis. Sci. 52, 2584–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brösamle C., Halpern M. E. (2009) Nogo-Nogo receptor signalling in PNS axon outgrowth and pathfinding. Mol. Cell. Neurosci. 40, 401–409 [DOI] [PubMed] [Google Scholar]
- 27. O'Brien G. S., Martin S. M., Söllner C., Wright G. J., Becker C. G., Portera-Cailliau C., Sagasti A. (2009) Developmentally regulated impediments to skin reinnervation by injured peripheral sensory axon terminals. Curr. Biol. 19, 2086–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schwab M. E. (2010) Functions of Nogo proteins and their receptors in the nervous system. Nat. Rev. Neurosci. 11, 799–811 [DOI] [PubMed] [Google Scholar]
- 29. Thomas R., Favell K., Morante-Redolat J., Pool M., Kent C., Wright M., Daignault K., Ferraro G. B., Montcalm S., Durocher Y., Fournier A., Perez-Tur J., Barker P. A. (2010) LGI1 is a Nogo receptor 1 ligand that antagonizes myelin-based growth inhibition. J. Neurosci. 30, 6607–6612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yamashita T., Higuchi H., Tohyama M. (2002) The p75 receptor transduces the signal from myelin-associated glycoprotein to Rho. J. Cell Biol. 157, 565–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tomarev S. I., Wistow G., Raymond V., Dubois S., Malyukova I. (2003) Gene expression profile of the human trabecular meshwork. NEIBank sequence tag analysis. Invest. Ophthalmol. Vis. Sci. 44, 2588–2596 [DOI] [PubMed] [Google Scholar]
- 32. Torrado M., Trivedi R., Zinovieva R., Karavanova I., Tomarev S. I. (2002) Optimedin. A novel olfactomedin-related protein that interacts with myocilin. Hum. Molec. Genet. 11, 1291–1301 [DOI] [PubMed] [Google Scholar]
- 33. Kalachikov S., Evgrafov O., Ross B., Winawer M., Barker-Cummings C., Martinelli Boneschi F., Choi C., Morozov P., Das K., Teplitskaya E., Yu A., Cayanis E., Penchaszadeh G., Kottmann A. H., Pedley T. A., Hauser W. A., Ottman R., Gilliam T. C. (2002) Mutations in LGI1 cause autosomal-dominant partial epilepsy with auditory features. Nat. Genet. 30, 335–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Morante-Redolat J. M., Gorostidi-Pagola A., Piquer-Sirerol S., Saenz A., Poza J. J., Galan J., Gesk S., Sarafidou T., Mautner V. F., Binelli S., Staub E., Hinzmann B., French L., Prud'homme J. F., Passarelli D., Scannapieco P., Tassinari C. A., Avanzini G., Marti-Masso J. F., Kluwe L., Deloukas P., Moschonas N. K., Michelucci R., Siebert R., Nobile C., Perez-Tur J., Lopez de Munain A. (2002) Mutations in the LGI1/Epitempin gene on 10q24 cause autosomal dominant lateral temporal epilepsy. Hum. Mol. Genet. 11, 1119–1128 [DOI] [PubMed] [Google Scholar]
- 35. GrandPré T., Li S., Strittmatter S. M. (2002) Nogo-66 receptor antagonist peptide promotes axonal regeneration. Nature 417, 547–551 [DOI] [PubMed] [Google Scholar]
- 36. Harvey P. A., Lee D. H., Qian F., Weinreb P. H., Frank E. (2009) Blockade of Nogo receptor ligands promotes functional regeneration of sensory axons after dorsal root crush. J. Neurosci. 29, 6285–6295 [DOI] [PMC free article] [PubMed] [Google Scholar]