Background: Despite structural reorganization during disease, conformational prion protein epitopes remain undefined.
Results: We identify specific amino acids constituting novel conformational monoclonal antibody epitopes.
Conclusion: Immunoreactivities of globular domain epitopes depend on maintenance of regional tertiary structure.
Significance: Our studies address how denatured conformational epitopes remain functional, provide insights into normal and disease-related prion protein, and expand epitope tagging options.
Keywords: Amino Acid, Antibodies, Epitope Mapping, Prions, Protein Domains
Abstract
Whereas prion replication involves structural rearrangement of cellular prion protein (PrPC), the existence of conformational epitopes remains speculative and controversial, and PrP transformation is monitored by immunoblot detection of PrP(27–30), a protease-resistant counterpart of the pathogenic scrapie form (PrPSc) of PrP. We now describe the involvement of specific amino acids in conformational determinants of novel monoclonal antibodies (mAbs) raised against randomly chimeric PrP. Epitope recognition of two mAbs depended on polymorphisms controlling disease susceptibility. Detection by one, referred to as PRC5, required alanine and asparagine at discontinuous mouse PrP residues 132 and 158, which acquire proximity when residues 126–218 form a structured globular domain. The discontinuous epitope of glycosylation-dependent mAb PRC7 also mapped within this domain at residues 154 and 185. In accordance with their conformational dependence, tertiary structure perturbations compromised recognition by PRC5, PRC7, as well as previously characterized mAbs whose epitopes also reside in the globular domain, whereas conformation-independent epitopes proximal or distal to this region were refractory to such destabilizing treatments. Our studies also address the paradox of how conformational epitopes remain functional following denaturing treatments and indicate that cellular PrP and PrP(27–30) both renature to a common structure that reconstitutes the globular domain.
Introduction
Infectious transmission in prion diseases, which include epidemics of bovine spongiform encephalopathy, scrapie of sheep and goats, chronic wasting disease (CWD)3 of cervids, and human Creutzfeldt-Jakob disease, involves conformational conversion of the prion protein (PrP), which exists in two physicochemically distinct isoforms. The cellular form, PrPC, is monomeric, sensitive to protease treatment, and soluble in detergents, whereas disease-associated PrPSc is partially protease-resistant, detergent-insoluble, and prone to aggregation. During disease, exponential accumulation of PrPSc results from template-mediated conversion of PrPC by PrPSc, leading to fatal and untreatable central nervous system neurodegeneration.
Mammalian PrP genes encode an ∼250 amino acid residue translation product, processed by removal of 22-residue amino-terminal and 23-residue carboxyl-terminal signal peptides. Cleavage of the latter facilitates addition of a glycosylphosphatidylinositol moiety by which PrP is anchored to the cell surface (1). Although the amino terminus of PrPC is largely unstructured and contains a tandem array of five copper-binding octapeptide repeats (2), the region encompassing amino acids 126–218 (mouse PrP numbering is used throughout, unless otherwise stated) consists of three α-helices and two short sections forming a β-pleated sheet (3). In both PrPC and PrPSc, asparagine-linked oligosaccharides are attached at residues 180 and 196, and a disulfide bond is formed between cysteine residues 178 and 213. In most examples of prion disease, in vitro treatment of PrPSc with proteinase K (PK) results in cleavage of ∼66 amino-terminal amino acids and persistence of a protease-resistant core referred to as PrP(27–30). Endoproteolytic cleavage of PrPSc following residue 88 results in a similar 21-kDa carboxyl-terminal fragment, referred to as C2, originally observed in the brains of patients with Creutzfeldt-Jakob disease (4), and is subsequently shown to be calpain-dependent (5). PrPC is cleaved between amino acids 110/111 to produce a 17-kDa carboxyl-terminal fragment referred to as C1 (4).
Because of the proteinaceous nature of prions, antibodies have been invaluable reagents for studying virtually all aspects of pathogenesis. The seminal observation that polyclonal antisera raised against PrP(27–30) (6) also reacted with PrP in uninfected brains (7) was instrumental in establishing the precursor-product relationship between the cellular and scrapie isoforms. Subsequent attempts to isolate anti-PrP monoclonal antibodies (mAbs) were not without significant challenges (6), in large part because the host does not mount an inflammatory response during prion infection. The availability of Prnp0/0 mice in which the PrP gene was ablated (8) circumvented the underlying immune tolerance to PrP, allowing the generation of various anti-PrP mAbs (9).
Definition of structural epitopes requires x-ray crystallography of anti-PrP mAb-antigen complexes (10, 11). Because this approach is laborious, ELISA-based peptide scanning has been the preferred method to map the approximate locations of epitopes. However, surprisingly few anti-PrP mAbs are amenable to this form of characterization, leading previous investigators to infer the existence of discontinuous PrP epitopes (12–17). To date, the involvement of specific amino acid residues in such hypothesized conformational epitopes has not been described. Moreover, this inference raises a conundrum (13), because all such antibodies bind PrP in Western blots, where PrP is defined by investigators as denatured (12, 13, 15, 18–22). We therefore set out to gain a better understanding of the involvement of specific amino acid residues in discontinuous, conformation-dependent PrP epitopes. We used a directed molecular evolution approach (23) to create shuffled genes expressing novel PrP epitopes, and by determining the reactivities of the resulting mAbs against a large panel of PrP primary structures and polymorphic variants thereof, we mapped and confirmed by mutational analysis the involvement of specific residues in epitope binding and PrPC to PrPSc conversion.
EXPERIMENTAL PROCEDURES
Transgenic Mice
Transgenic (Tg) mice expressing mouse, deer, bovine, and sheep PrP were generated using the cosSHa. Tet cosmid vector (24). Tg mice expressing deer PrP with polymorphisms at residues 95 and 96, elk, and human PrP with valine or methionine polymorphisms at residue 129 (25) were generated using the MoPrP.Xho vector (26). Tg mice expressing horse PrP were produced using a modified MoPrP.Xho vector, referred to as pJB1, in which BsiWI and FseI sites replaced the XhoI site. All mice were maintained on an inbred Prnp0/0 FVB background (FVB/Prnp0/0).
Generation of Chimeric PrP Coding Sequences by DNA Shuffling
One hundred ng of recombinant pCAGGS vectors harboring elk or mouse PrP-coding sequences were amplified with Pfu polymerase (Stratagene, La Jolla, CA) using the following conditions: predenaturation 95 °C/3 min; 95 °C/40 s; 60 °C/40 s; and 72 °C/60 s. PCR products were mixed in an equal ratio, and 3 μg of total DNA was incubated at 15 °C for 10 min, followed by addition of 0.15 units of DNase I (Roche Applied Science), and a further incubation at 15 °C for 10 min. The reaction was terminated by adding 10 μl of stop solution and incubation at 96 °C for 10 min. DNase I-digested DNA was separated on a 2% low temperature agarose gel (Invitrogen), and fragments in the range of 10–300 bp were excised and extracted in phenol/chloroform. For re-assembly of small DNA fragments by PCR, 30 μl of DNase I-digested DNA fragments, 5 μl of Pfu Ultra buffer, 1 μl of dNTP (Roche Applied Science), and 1 μl of Pfu Ultra (Stratagene, La Jolla, CA) were mixed, and the volume was adjusted to 50 μl with distilled H2O. The re-assembly PCR was performed for 45 cycles using the following conditions: 94 °C/3 min; 94 °C/30 s (27 °C + 1 °C/cycles)/1 min; 72 °C/(1 min + 4 s/cycles). The PCR product was maintained at 72 °C for 10 min and then stored at 4 °C. Reassembled DNA fragments were amplified using primers recognizing 5′ and 3′ ends of the open reading frame containing added EcoRV and BamHI sites for 35 cycles using the following conditions: predenaturation 94 °C/3 min; 94 °C/45 s; 47 °C with increases of 0.7 °C/cycle 1 min; 72 °C/1 min. PCR products were held at 72 °C for 10 min. Shuffled DNA fragments were digested with BamHI and EcoRV (New England Biolabs), purified, and then inserted into pIRESpuro3 vector (Clontech). Following transformation of bacteria to antibiotic resistance, colonies were screened for recombinant plasmids by PCR. Plasmids were extracted from clones containing the correct sized recombinants using a QIA miniprep kit (Qiagen, Valencia), and plasmids were sequenced using a CEQ8000 (Beckman Coulter, Fullerton, CA).
Preparation of Purified Recombinant PrP
Selected shuffled DNA constructs were PCR-amplified and cloned into pET100/D-TOPO® (Invitrogen). Transformed BL21 StarTM (DE3) (Invitrogen) were cultured until the absorbance value reached 1.0 at 650 nm and induced using isopropyl 1-thio-β-d-galactopyranoside (Sigma) at a final concentration of 1 mm. Bacteria were harvested after a 5-h incubation at 37 °C. Bacterial pellets were resuspended and lysed with CelLytic solution (Sigma) and centrifuged to obtain inclusion bodies, following solubilization with CelLytic IB solution (Sigma). Purification was performed using Ni-NTA column (Qiagen, Valencia) according to the manufacturer's instructions. Purified protein was refolded by dialysis in 20 mm NaAc buffer (pH 4.5). The 6-histidine tag was removed by cleavage with enterokinase (Invitrogen). Protein content was determined by the bicinchoninic acid (BCA) assay (Pierce), and purity was assessed by SDS-PAGE followed by Coomassie Blue staining.
Development and Cloning of Hybridoma Cells
FVB/Prnp0/0 mice were immunized with 50 μg of purified recombinant PrP in Freund's complete adjuvant by injection into the peritoneal cavity. Injections were repeated after 15, 30, and 45 days using 30 μg of antigen with Freund's incomplete adjuvant. Finally, 3 days prior to fusion, mice received the same amount of antigen in PBS without adjuvant. Mice with the highest antibody titer were used in cell fusions. Harvested spleen cells were fused with P3X63.AG8 myeloma cells (ATCC, TIB-9) at a ratio of 2:1 in DMEM (HyClone, Logan) with 50% (w/v) polyethylene glycol 1500 (Roche Applied Science). Hybridoma cells were selected in medium containing hypoxanthine/aminopterin/thymidine (HAT).
Characterization of Antibody Reactivity by ELISA
ELISA plates (NUNC, Rochester, NY) were coated with 100 μl of 1 μg/ml recombinant shuffled PrP solution and stored overnight at 4 °C. Plates were blocked with 200 μl of 3% bovine serum albumin in PBS, 0.05% Tween 20 for 2 h at 37 °C and washed three times with PBS, 0.05% Tween 20. Plates were incubated with 100 μl of conditioned medium from hybridoma cells for 2 h at 37 °C. After three PBS/Tween 20 washes, 100 μl of peroxidase-conjugated goat anti-mouse antibody (Sigma) at a dilution of 1:5000 was added, and the plates were incubated for 1 h at 37 °C, followed by three PBS/Tween 20 washes. Color development was accomplished by adding 100 μl of 2,2′-azinobis [3-ethylbenzothiazoline-6-sulfonic acid]-diammonium salt peroxidase solution (KPL, Gaithersburg, MD) at room temperature in the dark. The color reaction was stopped by adding 100 μl of stop solution (KPL, Gaithersburg, MD), and the absorbance of samples was measured at 405 nm in an ELISA plate reader (ELx808, Bio-Tek Instruments, Winooski, VT). Isotyping was performed by ELISA using isotype-specific anti-immunoglobulin conjugates (Pierce).
Antibody Purification
Hybridoma cells were cultured in a 50:50 solution of complete DMEM and CDM4Mab (HyClone, Logan), which is a serum-free medium for the culture of hybridoma cells. After 1 week, the medium was collected, and cell debris was removed by centrifugation at 2000 × g for 10 min. Antibodies were purified by affinity chromatography using a HiTrapTM protein G column (GE Healthcare) and the ProfiniaTM protein purification system (Bio-Rad) with preprogrammed methods for antibody purification. Following equilibration of the protein G column with 20 mm sodium phosphate (pH 7.0) binding buffer, hybridoma culture medium was applied at a rate of 1 ml min−1. After washing the column with binding buffer, antibodies were eluted with 0.1 m glycine-HCl (pH 2.7) that was neutralized by the addition of 50 μl of 1 m Tris-HCl (pH 9.0) per 1 ml of elution buffer.
Stable Transfection and Prion Infection of Cultured Cells
PrP coding sequences with or without mAb epitope mutations were synthesized (GenScript, Piscataway, NJ) with AflII and EcoRI restriction endonuclease recognition sites at the 5′ and 3′ ends, respectively. Digested amplicons were inserted into AflII- and EcoRI-cleaved pIRESpuro3 (Clontech). PrP expression cassettes containing in-frame deletions were generated by PCR-based mutagenesis using the QuikChange mutagenesis kit (Stratagene) according to the manufacturer's instructions. Mutated constructs were sequenced using a CEQ 8000 (Beckman Coulter, Fullerton, CA). Rabbit kidney epithelial cells (RK13) were plated in 6-well plates 1 day prior to transfection. Transfection mixtures were prepared by mixing 2 μg of plasmid and 10 μl of Lipofectamine 2000 (Invitrogen) in 500 μl of Opti-MEM (Invitrogen). After 5 h, the transfection solution was exchanged with complete medium containing 10% FBS, followed by passage to 10-cm plates the next day. Transfected cells were selected in complete medium containing 1 μg/ml puromycin.
For infection, transfected cells (2 × 105 cells/well) were plated in 6-well plates, and 0.2% brain homogenates in PBS were added to cell monolayers. After 5 h, 2 ml of complete medium was added, and cells were incubated for 5 days. After three passages, lysates of confluent cell monolayers were prepared in cold lysis buffer (50 mm Tris (pH 8.0), 150 mm NaCl, 0.5% sodium deoxycholate, 0.5% Igepal CA-630) and analyzed by Western blotting.
Western Blotting
Brain homogenates were prepared in 10% (w/v) sterile PBS lacking Ca2+ and Mg2+ by repeated extrusion through 18- and then 21-gauge needles. Protein content in brain homogenates and cell lysates was determined by BCA (Pierce). Brain homogenates and cell lysates were digested with 100 or 30 μg/ml of proteinase K (PK), respectively (Roche Applied Science), in cold lysis buffer for 1 h at 37 °C. Digestion was terminated with phenylmethylsulfonyl fluoride at a final concentration of 2 μm. Deglycosylation of PrP was performed by treatment of PNGase F (New England Biolabs) for 3 h at 37 °C. Samples were prepared for SDS-PAGE either in the presence or absence of β-mercaptoethanol (βME) (Bio-Rad) and boiled for 10 min. Proteins were resolved by SDS-PAGE and transferred to polyvinylidene difluoride Immobilon (PVDF)-FL membranes (Millipore). Membranes were probed with primary mAbs followed by horseradish peroxidase-conjugated anti-mouse secondary antibody (GE Healthcare). Protein was visualized by chemiluminescence using ECL Plus (GE Healthcare) and an FLA-5000 scanner (Fujifilm Life Science, Woodbridge, CT).
Surface Plasmon Resonance (SPR) Measurements
SPR measurements were obtained with a Biacore T100 instrument (GE Healthcare). The surfaces of flow channels on a CM5 chip were activated with 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride and N-hydroxysuccinimide. Recombinant mouse PrP or elk PrP was diluted to 20 μg/ml in 10 mm sodium acetate (pH 5.5) and immobilized on a channel of the activated CM5 sensor chip. After immobilization, all surfaces were blocked with 1 m ethanolamine hydrochloride (pH 8.5). Another channel of the chip was processed through an identical coupling procedure without addition of ligand recombinant PrP and was used as reference. All kinetic SPR analyses were run at a 10 μl/min PBS flow, and antibodies were injected at concentrations ranging from 50 to 3200 nm. Association or dissociation was recorded for 180 or 360 s, respectively. After each cycle, the surface was regenerated with 50 mm NaOH. Kinetic data were calculated using Biacore T100 evaluation software according to the bivalent analyte model.
Structural and Statistical Analyses
Three-dimensional structures of mouse PrP (3) were analyzed using the 2007 iMol Molecular Visualization Program. Statistical analyses were performed using 2010 Prism 5.0d for Mac OS X, GraphPad Software Inc.
RESULTS
Generation of Chimeric PrP by DNA Shuffling
Nucleotides within the mouse Prnp and cervid PRNP gene coding sequences share ∼80% identity, and the primary structures of their translation products are ∼85% identical. We recombined the coding sequences of mouse and elk PrP in vitro, using a PCR-based technique referred to as DNA shuffling (27), to produce a library of randomly chimeric PrP. DNA sequence analysis of 96 recombinant clones identified 34 different shuffled PrP sequences. Three chimeras, referred to as Mo/CerPrP-3, Mo/CerPrP-34, and Mo/CerPrP-68, were selected for bacterial expression and recombinant (Rec) PrP purification.
Isolation of PRC Monoclonal Antibodies
High antibody titers resulted from immunization of Prnp0/0 mice with purified Rec PrP preparations. Following fusion of splenocytes from immunized mice with myeloma cells, we obtained two clones from Rec Mo/CerPrP-3 immunized mice, 25 clones from Rec Mo/CerPrP-68 immunized mice, and no clones from Rec Mo/CerPrP-34 immunized mice. Of the 25 clones from Rec Mo/CerPrP-68 immunized mice, 14 had reproducibly strong reactivity against Rec Mo/CerPrP-68 by ELISA and Western blotting. Ultimately, four mAbs, originally coded as 9E9, 5C6, 7H11, and 9H9 (28, 29), hereafter referred to as PRC (Prion Research Center) 1, 5, 7, and 9, respectively, were selected for further characterization. Hybridomas were cloned two additional times. Monoclonal antibody PRC5 is IgG2a, whereas mAbs PRC1, PRC7, and PRC9 are IgG1.
Epitope Characterization of PRC Monoclonal Antibodies
We determined the antigen binding properties of the PRC mAbs by probing Western blots of brain extracts from Tg mice expressing mouse, deer, elk, cattle, sheep, horse, or human PrP, as well as various species including Syrian, Chinese, and Armenian hamsters, mink, ferret, squirrel monkey, and cattle. Comparison of the reactive and nonreactive PrP primary structures with that of the shuffled Rec Mo/CerPrP-68 immunogen allowed us to identify specific amino acid residues that were critical for recognition by the various PRC mAbs. Of the 30 amino acid differences between mouse and elk PrP, recombinant Mo/CerPrP-68 shares 14 with mouse and 16 with elk PrP.
Monoclonal antibody PRC5 reacted with all primary structures tested except squirrel monkey PrP (Fig. 1A). However, reactivity with sheep PrP was dependent on the naturally occurring Ala/Val polymorphism at residue 136, because PRC5 also failed to react with in brain extracts from Tg mice expressing sheep PrP with Val at this position, referred to as OvPrP-V136 (Fig. 1A). Because squirrel monkey PrP contains Ala at the corresponding location (Fig. 1B), its failure to react with PRC5 (Fig. 1A) indicated that other amino acid(s), differing in this species also include the PRC5 epitope. Squirrel monkey PrP contains serine at residue 162, equivalent to residue 158 in mouse PrP, whereas Mo/CerPrP-68, and all other PrP species tested contain Asn at this position (Fig. 1B). No other amino acids differing between PRC5 reactive and nonreactive PrP primary structures correlated with antibody binding.
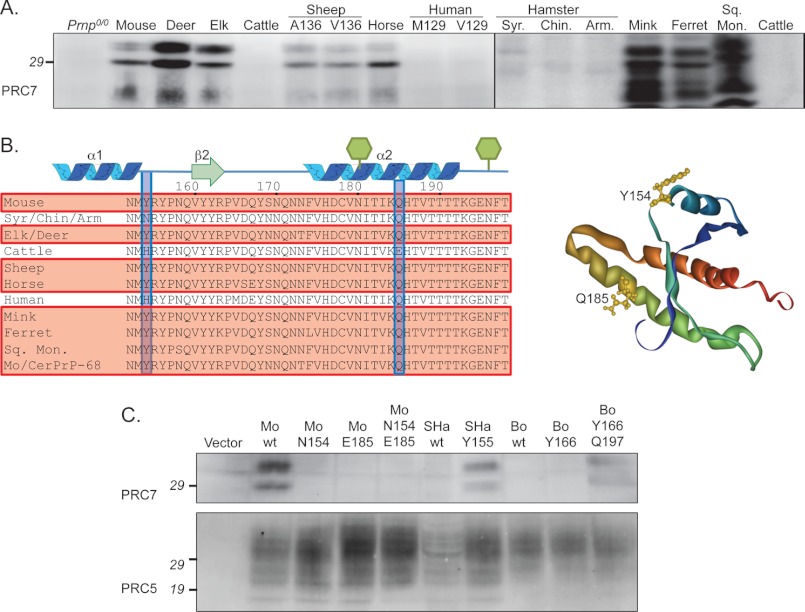
FIGURE 1.
Functional epitope of mAb PRC5. A, Western blot reactivity with various PrP primary structures. AHa, Armenian hamster; Sq. Mon., squirrel monkey. B, primary structures are aligned between mouse PrP residues 123 and 163. Reactive species are boxed and shaded red. The 6H4 and D18 epitopes are bold in cattle and mouse PrP, respectively. Residues 132 and 158 are boxed and shaded blue, and side chains are yellow in the tertiary structure. C, Western blots of RK13 cell extracts transfected with the following: Vector, pIRESPuro; Mo wt, mouse PrP; Elk wt, elk PrP; Elk V133, elk PrP with Val at residue 133; Elk S162, elk PrP with Ser at residue 162; Mo V132, mouse PrP with Val at residue 132; and Mo S158, mouse PrP with Sr at residue 158.
To confirm their role in the functional epitope of PRC5, we engineered constructs in which each of these two residues were individually mutated in mouse or elk PrP. Wild type and mutated constructs, referred to as MoPrP-V132, MoPrP-S158, ElkPrP-V133, and ElkPrP-S162, were stably expressed in RK13 cells, which do not express endogenous PrP (30). Consistent with a requirement for both residues in mAb recognition, mutation of either residue in each primary structure prevented detection by PRC5 (Fig. 1C).
Whereas mAb PRC7 also detected a wide range of PrP primary structures, it failed to react with cattle, human, or hamster PrP (Fig. 2A). In addition, PRC7 preferentially reacted with aglycosyl and monoglycosyl but not diglycosyl PrP (Figs. 2, 6A, and 7). Although all reactive primary structures contain tyrosine at the position corresponding to residue 154 in mouse PrP, hamster species contain Asn, and cattle and human PrP contain histidine (Fig. 2B). To confirm the requirement of this residue in recognition by PRC7, we changed Tyr at residue 154 in mouse PrP to Asn, and changed Asn at the corresponding residue in Syrian hamster PrP to Tyr. Mutant and wild type constructs were stably expressed in RK13 cells, and Western blotted extracts were probed with PRC7 (Fig. 2C). Consistent with a requirement for Tyr at residue 154 in mouse PrP in mAb recognition, mutation to Asn prevented detection by PRC7, whereas mutation of Asn to Tyr at position 155 in Syrian hamster PrP allowed detection by PRC7 (Fig. 2C). We also mutated His to Tyr at position 166 in cattle PrP, which corresponds to position 154 in mouse PrP, but this mutation failed to confer reactivity to PRC7 (Fig. 2C). Cattle PrP uniquely contains glutamic acid at position 197 instead of glutamine, which is conserved in all other species (Fig. 2B). To address whether this residue also partakes in PRC7 recognition, we changed the corresponding mouse PrP residue Gln-185 to Glu. This mutation, either singly or in combination with Asn-154, prevented recognition by PRC7, whereas reversal of both Asn to Tyr and Glu to Gln at the corresponding residues 166 and 197 in cattle PrP allowed detection by PRC7. We conclude that mouse PrP residues 154 and 185 participate in PRC7 recognition. No other amino acid differences between PRC7-reactive and nonreactive PrP species correlated with antibody binding.
FIGURE 2.
Functional epitope of mAb PRC7. A, Western blot reactivity with various PrP primary structures. Samples from hamsters, mink, ferret, and squirrel monkey were prion infected. Syr., Syrian; Chin., Chinese; and Arm., Armenian hamsters; Sq. Mon., squirrel monkey. B, primary structures are aligned between mouse PrP residues 152 and 198. Reactive species are boxed and shaded red. Glycans attached at residues 180 and 196 are shown as green hexagons. Residues 154 and 185 are boxed and shaded blue, and side chains are yellow in the tertiary structure. C, Western blots of RK13 cell extracts transfected with the following: Mo N154, mouse PrP with Asn at residue 154; Mo E185, mouse PrP with Glu at residue 185; Mo N154/E185, mouse PrP with Asn at residue 154 and Glu at residue 185; SHa wt, wild type Syrian hamster PrP; SHa Y155, Syrian hamster PrP with Tyr at residue 155; Bo wt, wild type cattle PrP; Bo Y166, cattle PrP with Tyr at residue 166; Bo Y166/Q197, cattle PrP with Tyr at residue 166, and Gln at residue 197.
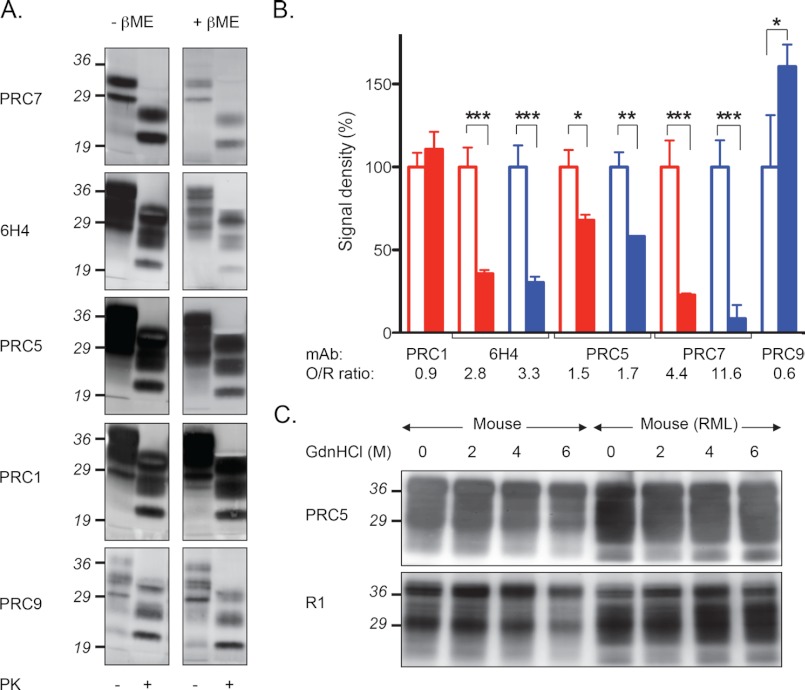
FIGURE 6.
Effect of mAb recognition following PrP tertiary structure disruption by disulfide bond reduction. A, Western blots showing relative immunoreactivities of mAbs with brain extracts of RML-infected wild type mice in the presence (+βME) or absence (−βME) of reducing agent. Samples treated with proteinase K (PK) as indicated. B, densitometric analysis of mouse (blue) and elk (red) PrP levels in the presence (shaded bars) and absence (unshaded bars) of βME. *, p < 0.05; **, p < 0.005; ***, p < 0.001. O/R, ratio of oxidized to reduced PrP. C, Western blots demonstrating the effects of treatment with various concentrations of guanidinium hydrochloride on PrP detection in the brains of uninfected or RML-infected mice.
FIGURE 7.
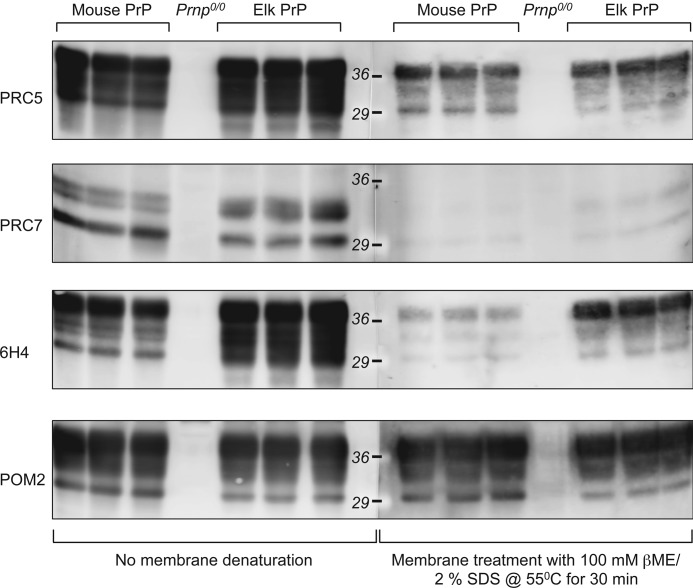
Immunoreactivity of conformation-dependent but not conformation-independent mAbs is reduced following post-transfer denaturation of PrP. Brain extracts were separated by SDS-PAGE and transferred to PVDF-FL membranes. Membranes were then either not treated or treated with 100 mm βME, 2% SDS at 55 °C for 30 min prior to immunoprobing with the indicated antibodies.
PRC1 reacted with deer, elk, cattle, horse, and sheep PrP and failed to react with mouse (Fig. 3, A and D) or human PrP (Fig. 3A). PRC1 also failed to react with deer PrP expressing His at residue 95, referred to as deer PrP-H95 (deer PrP residue numbering). Immunodetection with mAb 6H4 (31) confirmed deer PrP-H95 transgene expression (Fig. 3A). The primary structures of deer PrP-H95 and wild type deer PrP expressing glutamine at residue 95 differ at only this single residue.
FIGURE 3.
Functional epitope of mAb PRC1. A, Western blot reactivity with various PrP primary structures. Deer H95 and S96, deer PrP with His and Ser at residues 95 and 96, respectively. B, primary structures are aligned between mouse PrP residues 81 and 115. Reactive species are boxed and shaded red. The HumP and POM2 epitopes are bold in cattle and mouse PrP, respectively. Amino acids controlling PRC1 reactivity are boxed and shaded blue. Arrows depict the locations of protease cleavage to generate C2 and C1. C, Western blots of RK13 cell extracts transfected with the following: Vector, pIRESPuro; Mo wt, mouse PrP; Elk wt, elk PrP; Mo 2G, mouse PrP with two Gly residues after residue 90; Elk 3G, elk PrP with three Gly residues. D, Western blots showing the reactivity of mAb PRC1 with PrP in the brains of uninfected and RML prion infected wild type mice and uninfected and CWD-infected Tg mice expressing elk PrP. Samples were treated with proteinase K (PK) or PNGase F as indicated. Positions of unglycosylated full-length (F), C1, and C2 fragments are shown.
Conservation of Gln at this position between reactive and nonreactive species (Fig. 3B) demonstrated that additional primary structural elements controlled PRC1 binding. Although species that failed to react with PRC1 contained three consecutive Gly residues immediately following this Gln, reactive species contained only two (Fig. 3B), suggesting that the number of Gly residues at this location controlled recognition by PRC1. To address this, we created mutated mouse and elk PrP coding sequences, referred to as MoPrP-2G and ElkPrP-3G, in which the number of Gly residues was changed from three to two in the mouse PrP sequence and from two to three in elk PrP. Mutant and wild type constructs were stably expressed in RK13 cells, and Western blotted extracts were probed with PRC1. Each construct had the expected reactivity with PRC1 as follows: deletion of one of the three Gly residues between 91 and 93 in mouse PrP conferred susceptibility to MoPrP-2G, whereas insertion of an additional Gly residue at the corresponding location in elk PrP abolished detection in ElkPrP-3G (Fig. 3C). Immunoblot probing with mAb 6H4 confirmed transgene expression in all cases. Consistent with this epitope location, PRC1 detected full-length elk PrP, C2, and PrP(27–30) but failed to react with C1 following PNGase F-mediated removal of N-linked glycans; in contrast, mAb 6H4 detected full-length mouse and elk PrP, and both endogenously cleaved fragments (Fig. 3D).
Of the four PRC mAbs, PRC9 had the most restricted reactivity, responding only to mouse and hamster PrP (Fig. 4A). By comparing the PrP primary structures of Rec Mo/CerPrP-68 with those of reactive and nonreactive species, we located the PRC9 epitope within the C terminus of PrP (Fig. 4B). Mouse and hamster PrP primary structures contain aspartic acid and Gly residues at positions 226 and 227, whereas nonreactive species lacked these two adjacent residues. Expression of additional mouse PrP mutants in RK13 cells showed that PRC9 binding was inhibited by mutation of Tyr to Ala at residue 224, which is conserved in all tested species except horse, but not mutation of Tyr to Ala at residue 225 (Fig. 4B). Our results demonstrate that the PRC9 epitope overlaps the binding locations of recombinant R1 and R2 antibodies, which have been mapped to the region containing residues 224–230 (16).
FIGURE 4.
Functional epitope of mAb PRC9. A, Western blot reactivity with various PrP primary structures. B, primary structures are aligned between mouse PrP residues 218 and 241. Reactive species are boxed and shaded red. The R1 epitope is bold in hamster PrP. Amino acids controlling PRC9 reactivity are boxed and shaded blue. Arrow depicts the glycosylphosphatidylinositol anchor signal peptidase cleavage site. C, Western blots of RK13 cell extracts transfected with the following: Vector, pIRESPuro; Mo wt, mouse PrP; Mo 224A and Mo 225A, mouse PrP harboring a mutation of Tyr to Ala at residue 224 and 225, respectively.
Unexpected mAb Reactivities with Redacted PrP Constructs
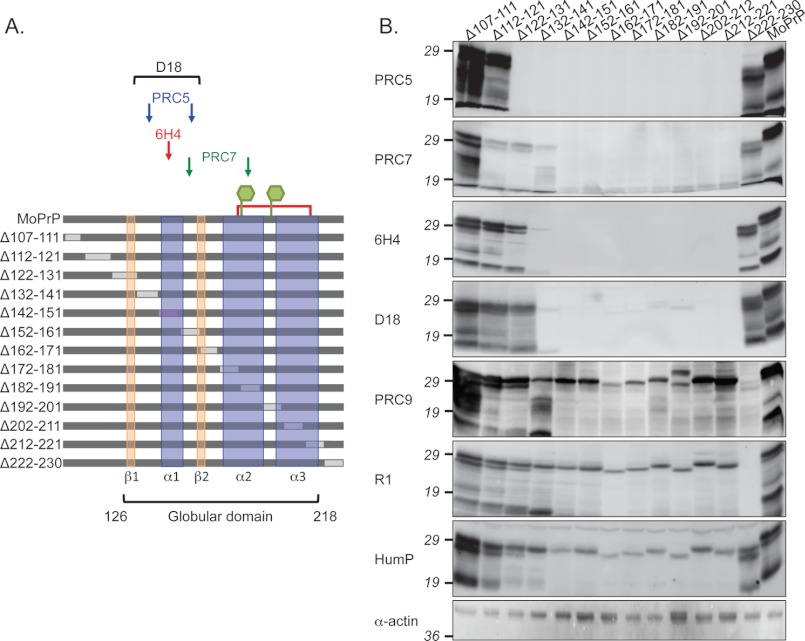
As an additional approach to map the locations of PRC mAb-binding epitopes, we constructed a series of in-frame, nonoverlapping deletions across the primary structure of mouse PrP between residues 107 and 230 (Fig. 5A). We expressed these redacted constructs in RK13 cells, and following PNGase F removal of N-linked sugars from PrP constructs, we probed cell extracts with various antibodies on Western blots. We expected that mAbs would fail to react with recombinant PrP deletions containing sequences comprising their epitopes. Based on our demonstration that PRC5 recognition involved mouse PrP residues 132 and 158, and PRC7 recognition involved mouse PrP residues 154 and 185, we therefore anticipated that PRC5 would fail to recognize PrP containing deletions between residues 132–141 and 152–161 and that PRC7 would fail to recognize PrP containing deletions between residues 152–161 and 182–191 (Fig. 5A). However, PRC5 and PRC7 unexpectedly either failed to detect or had reduced reactivity with all constructs harboring deletions between residues 122 and 222, including those that were not predicted to harbor epitope components (Fig. 5B). This region corresponds to the globular carboxyl-terminal domain that contains regions of secondary structure, defined by NMR analyses as being composed of residues 126–218 (3) (Fig. 5A). PRC5 and PRC7 recognized all constructs harboring deletions outside this domain between 107–121 and 222–230 (Fig. 5B). In contrast to the behavior of PRC5 and PRC7, PRC9 satisfied our initial expectations because it reacted with all deletion constructs except MoPrPΔ222–230, which contains its epitope (Fig. 5B). The reactivity of PRC9 also confirmed that compromised PRC5 and PRC7 binding to deletions between residues 122 and 222 was not the result of unstable mutant PrP expression in mammalian cells and bacteria. We obtained similar antibody reactivities when we expressed redacted constructs in bacteria and probed cell extracts with antibodies on Western blots (supplemental Fig. 1).
FIGURE 5.
Effect of mAb recognition following PrP tertiary structure disruption by deletion mutagenesis. A, scheme showing the various deletion constructs and locations of mAb epitopes. Arrows or brackets indicate which deletions harbor epitope components for the various conformation-dependent mAbs. N-Linked glycan attachment sites at residues 180 and 196 are indicated by green hexagons, and the disulfide bond between cysteine residues 178 and 213 as a red line. B, Western blot reactivity of conformation-dependent and -independent mAbs with PNGase F-treated extracts of RK13 cells expressing PrP deletion constructs.
We next expanded our analyses to include previously characterized mAbs. The epitopes of 6H4 and D18 have previously been mapped to the region encompassing residues 132–156 of mouse PrP (Fig. 1B) (16, 31). Similar to the behavior of PRC5 and PRC7, deletions affecting regions of secondary structure between residues 126 and 218 also prevented or reduced detection by mAbs 6H4 and D18, whereas deletions within the unstructured regions had no effect on immunodetection (Fig. 5B). These findings supported the notion that failure of PRC5 and PRC7 to bind deletions within the structured domain was not the result of undiscovered epitope-binding elements but rather that deletion mutations within the PrP globular domain perturb the correct tertiary structure of that region, and thus affect the topology of conformation-dependent epitopes of mAbs PRC5, PRC7, 6H4, and D18. In contrast to mAbs binding epitopes in the structured region, HumP, the epitope of which is adjacent to the PRC1-binding site (Fig. 3B) (32), and POM2, which binds mouse PrP at an adjacent location in the octapeptide repeat region (13), recognized all deletions and wild type PrP (Fig. 5B and supplemental Fig. 1), whereas R1 bound all constructs except MoPrPΔ222–230, which harbors its epitope (Fig. 5B).
Conformation-dependent Antibody Reactivities
The almost 30-residue separation between residues 132 and 158 demonstrates that the epitope recognized by PRC5 is discontinuous. The adjacent proximity of residues 132 and 158 in the tertiary structure of PrP (Fig. 1B) (3) therefore indicates that PRC5 reactivity is dependent on PrP conformation. In previous studies, the inability to map the locations of epitopes by peptide scanning and reduced reactivity of the corresponding mAbs after disulfide bond reduction were cited as evidence that those epitopes were discontinuous and therefore that recognition depended on intact PrP tertiary structure (15). In keeping with these previous observations, reactivity of mAb PRC5 with mouse and cervid PrP on Western blots was increased 1.5-fold (p < 0.05) and 1.7-fold (p < 0.005) in the absence of βME (Fig. 6, A and B). Consistent with conformation-dependent reactivity of PRC7 involving discontinuous residues 154 and 185, immunodetection of mouse PrP by PRC7 was increased ∼4-fold and with cervid PrP ∼12-fold (p < 0.01) under nonreducing conditions compared with reducing Western blotting conditions (Fig. 6, A and B). Surprisingly, because its binding has been mapped to a single sequential length of the polypeptide chain (31), mAb 6H4 reactivity with both cervid and mouse PrP was also increased ∼3-fold (p < 0.01) under nonreducing conditions (Fig. 6, A and B), indicating that its epitope also shares the property of conformational dependence with the discontinuous epitopes of PRC5 and PRC7. In accordance with the notion that PRC1 recognizes a conformation-independent epitope, reactivity of cervid PrP with PRC1 was unaffected by treatment with reducing agents, whereas PRC9 immunoreactivity was enhanced 1.5-fold (p < 0.05) following treatment of mouse PrP with βME (Fig. 6, A and B).
The conformational dependence of PRC5, PRC7, 6H4, and D18 recognition appeared incompatible with the ability of these mAbs to recognize PrP on Western blots, because PrP is generally regarded as denatured under these conditions (12, 13, 15, 18, 20–22). Furthermore, treatment of samples with guanidinium hydrochloride concentrations as high as 6 m prior to SDS-PAGE and immunoblotting failed to prevent PrP detection by PRC5 by Western blotting (Fig. 7C). To reconcile these discrepant observations, we hypothesized that, subsequent to denaturing treatments, PrP refolds during Western blotting to a structure amenable to detection by conformation-dependent antibodies. To address this, we denatured PrP bound to PVDF membrane following SDS-PAGE and Western blot transfer by treatment with 100 mm βME, 2% SDS at 55 °C for 30 min prior to immunoprobing with conformation-dependent or conformation-independent mAbs. In accordance with our expectations, recognition of mouse and elk PrP by the conformation-independent mAb POM2 (supplemental Fig. 1) was unaffected by post-Western denaturation, whereas the reactivities of conformation-dependent antibodies PRC5, PRC7, and 6H4 with mouse or elk PrP were consistently reduced compared with membranes not receiving post-transfer denaturing treatments (Fig. 7).
Reactivities of PRC mAbs with Mouse and Cervid PrP under Native Conditions
Antibody binding of PrP expressed on the surface of RK13 cells was monitored by FACS (supplemental Fig. 2, A and B), and the ability of mAbs to react with PrP in solution was monitored by immunoprecipitation (supplemental Fig. 2C). Measurements of the KD values for mAb binding to immobilized mouse and elk PrP were determined by SPR (Table 1). Consistent with its reactivity in Western blotting conditions, PRC5 efficiently recognized native mouse and elk PrPC expressed on the surface of RK13 cells by FACS and immunoprecipitated mouse and elk PrP from brain homogenates of wild type and Tg(ElkPrP) mice, respectively. Measurements of the KD values for PRC5 binding to immobilized mouse and elk PrP by SPR gave values of 2.1 × 10−6 and 9.1 × 10−7, respectively. PRC1 efficiently recognized elk but not mouse PrP by FACS and immunoprecipitation, and the KD value for binding to recombinant elk PrP was 7.0 × 10−7. In contrast, PRC9 immunoprecipitated mouse but not elk PrP and recognized mouse PrP expressed on the surface of RK13 cells, but less strongly than PRC5 or 6H4. Accordingly, the KD value for binding to mouse PrP was 1.2 × 10−5. PRC7 also recognized cell surface expressed mouse and elk PrP but less efficiently than PRC5. Reflecting this, the KD value for PRC7 binding to elk PrP was 1.7 × 10−4. PRC7 failed to immunoprecipitate either species of PrP.
TABLE 1.
Properties of novel epitopes raised against randomly shuffled chimeric PrP
ND means not determined.
| mAb |
KD (m) |
Epitope characteristics |
Effect of mutation on conversion of PrPC to PrPSc |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Elk PrP | Mouse PrP | Constitutive amino acids | Linear/discontinuous | Conformation-dependent | Globular domain | Mutation | Mouse PrP | Elk PrP | |
| PRC mAbs | |||||||||
| PRC1 | 7.0 × 10−7 | Gln-90 | Linear | No | No | PrP3G | Converts | Inhibits | |
| PrP2G | Converts | Converts | |||||||
| PRC5 | 9.1 × 10−7 | 2.1 × 10−6 | Ala-132 | Discontinuous | Yes | Yes | A132V | Inhibits | Inhibits |
| Asn-158 | N158S | Converts | Inhibits | ||||||
| PRC7 | 1.7 × 10−4 | ND | Tyr-154 | Discontinuous | Yes | Yes | Y154N | Converts | ND |
| Gln-185 | Q185E | Inhibits | ND | ||||||
| PRC9 | 1.2 × 10−5 | Tyr-224, Asp-226, Gly-227 | Linear | No | No | ||||
| Previously reported mAbs | |||||||||
| 6H4 | Unknown, within 143–151 | Linear | Yes | Yes | |||||
| D18 | Unknown, within 132–156 | Discontinuous | Yes | Yes | |||||
| R1 | Unknown, within 224–230 | Linear | No | No | |||||
| HumP | Unknown, within 95–107 | Linear | No | No | |||||
| POM2 | Unknown, within 81–89 | Linear | No | No | |||||
Involvement of Residues Required for mAb Binding on PrPSc Formation
Having confirmed the roles of specific amino acid residues in the epitopes of different PRC mAbs, we investigated the effects of mutations at these positions on the ability of either elk or mouse PrPC to be converted into PrPSc. To address this, we infected RK13 cells stably expressing wild type and mutated versions of mouse or elk PrP with the respective species of prions, and we monitored the ability of mutated PrP to be converted to mouse or elk PrP(27–30). The effects of mutations of various epitope components depended on whether they were expressed in the context of mouse or elk PrP. For PRC5, mutation of either A133V or N162S inhibited conversion of elk PrPC to PrPSc (Fig. 8A). In contrast, whereas mutation of A132V also inhibited conversion of mouse PrPC to PrPSc, mutation of N158S did not inhibit conversion of mouse PrPC to PrPSc, but instead it augmented PrPSc levels (Fig. 8B). In the case of PRC7, although mutation of Gln to Glu at mouse PrP residue 185 prevented conversion to PrP(27–30), mutation of Tyr at residue 154 to Asn had no impact on the ability of PrPC to form protease-resistant PrP (Fig. 8C). Variable numbers of Gly residues immediately distal to mouse PrP Gln-90 controlled PrP detection by PRC1 (Fig. 3). Insertion of an additional Gly residue at this location in elk PrP, which prevented detection by PRC1 (Fig. 3C), abolished conversion to PrPSc (Fig. 8D). In contrast, deletion of a Gly residue in mouse PrP, which facilitated detection by PRC1 (Fig. 3C), did not prevent conversion to PrPSc (Fig. 8E), and the resulting MoPrPSc-2G could be detected by PRC1 (Fig. 8F). The effects of various epitope mutations on the ability of mouse and elk PrP to be converted to PrP(27–30) are summarized in Table 1.
FIGURE 8.
Effects of mutating PRC mAb epitopes on PrPSc formation. Western blots of extracts from RK13 cells expressing wild type and mutant PrP infected with CWD or RML prions. A and B, residues involved in the PRC5 epitope. C, residues involved in the PRC7 epitope. Mo N154, mouse PrP harboring mutation of Tyr to Asn at 154; Mo E185, mouse PrP harboring a mutation of Gln to Glu at 185; Mo N154/E185, mouse PrP harboring mutations of Tyr to Asn at 154 and Gln to Glu at 185. D–F, residues involved in the PRC1 epitope. Elk3G, elk PrP with three instead of two Gly residues; MoPrP2G, mouse PrP harboring two instead of three glycine residues; MoPrP2G/RML, infected with RML prions. Extracts were either treated (+) or not treated (−) with proteinase K (PK).
DISCUSSION
DNA Shuffling to Create Chimeric PrP
We used a directed molecular evolutionary approach to create shuffled genes expressing novel PrP epitopes as immunogens. Our approach was based on previous studies that used DNA shuffling to produce chimeric envelope proteins capable of inducing neutralizing antibodies against all four dengue virus serotypes (33, 34) and to optimize immunogenicity and protective efficacy of Venezuelan equine encephalitis and type 1 human immunodeficiency virus envelope proteins (35, 36). Our strategy resulted in the isolation of mAbs that recognized cervid but not mouse PrP (PRC1) and mouse but not cervid PrP (PRC9), as well as antibodies that recognized both mouse and cervid PrP (PRC5 and PRC7). Not unexpectedly, all PRC mAbs had variable cross-reactivities with PrP from different species, although in the case of the PRC9, cross-reactivity was restricted to hamster PrP. The properties of epitopes recognized by these newly developed mAbs, as well as the known properties of previously characterized mAbs used in these studies, are detailed in Table 1.
Characterization of Discontinuous Conformation-dependent PrP Epitopes
In previous studies the lack of mAb binding to peptide fragments in ELISA led investigators to infer the existence of discontinuous PrP epitopes (15, 16). In the case of the IgM mAb 15B3, which has been used to selectively immunoprecipitate PrP aggregates in infectious prion disease (31), as well as a Tg mouse model of a genetically programmed prion disease (37), three distinct peptide sequences were found to react with 15B3 (human PrP amino acids 142–148, 162–170, and 214–226) (31). However, the direct involvement of specific amino acid residues in this, or any other conformation-dependent PrP epitope, has not been previously reported. Moreover, 15B3 and possibly other purported PrPSc-specific mAbs (38) have been shown to interact with noninfectious as well as infectious PrP aggregates (39), and additional studies provide evidence that 15B3 interactions with such aggregates are nonspecific and paratope-independent (40).
Our epitope mapping studies show that the highly conserved Ala residue at mouse PrP position 132, as well as the Asn at residue 158, contribute to recognition by mAb PRC5, whereas residues 154 and 185 are components of the PRC7 epitope. Because antibody paratopes are occupied at most by 15–22 amino acid antigen residues (41), the ∼30-residue separation between amino acid residues in each case unequivocally defines the epitopes of PRC5 and PRC7 as discontinuous.
Consistent with the conformational dependence of these discontinuous epitopes, we show that PRC5 and PRC7 immunoreactivity is reliant on the integrity of the disulfide bond linking α-helices 2 and 3, which stabilizes the tertiary structure of PrP. Our mutational analyses showing that recognition by PRC5, PRC7, 6H4, and D18 is inhibited exclusively by deletions disrupting the structured globular region are also consistent with our characterization of these epitopes as conformation-dependent. Our findings indicate that deletions invading any aspect of the structured globular domain, even sequence elements far removed from defined epitope components, destabilize the overall tertiary structure of the region, preventing correct epitope presentation for antibody recognition. In the case of mAb PRC5, Ala-132 and Asn-158 are adjacent in PrP tertiary structure at the base of the loop containing α-helix 1, which is formed during antiparallel juxtaposition of β-sheet regions 1 and 2 (Fig. 1B) (3). Disulfide bond disruption or destabilizing deletion mutations abolish the overall tertiary structure of the globular region, which in turn precludes residues 132 and 158 from acquiring the required proximity for PRC5 epitope formation. A similar explanation accounts for the behavior of the discontinuous conformation-dependent PRC7 epitope spanning residues 154 and 185.
Because its epitope has been mapped to the region containing sequential amino acids DYEDRYYRE corresponding to mouse PrP residues 143–151 (31), the behavior of mAb 6H4 indicates that linear as well as discontinuous epitopes within the structured globular domain exhibit conformational dependence, which is consistent with the previously observed behaviors of this mAb (20, 42). Previous studies showing loss of 6H4 immunoreactivity following PrP reduction and alkylation (22) were interpreted to mean that its epitope in α-helix 1 became masked and therefore unavailable for detection. However, our results favor the alternative hypothesis that immunoreactivity of 6H4, and other mAbs with epitope components in this region, depends on correct local folding of α-helix 1 and, moreover, that the sustained integrity of this secondary structural element is co-dependent on the overall tertiary structure of the globular domain.
In contrast to these conformation-dependent epitopes with the globular domain, the epitopes of PRC9, R1, HumP, and POM2, located in relatively unstructured regions proximal and distal to the globular region, are independent of PrP conformation for their recognition, because their ability to recognize PrP is unaffected by structure-disrupting deletions or by treatment with disulfide bond-reducing agents. Coincidentally, although deletion analysis has suggested a discontinuous epitope for anti-PrP mAb T2 (43), our results also indicate that mapping approaches relying exclusively on a lack of mAb binding to deletions within the structured domain of PrP (43) may lead to spurious interpretations of epitope locations.
Renaturation of Conformational PrP Epitopes
Our results shed light on previously reported difficulties surrounding anti-PrP mAb epitope mapping, which resulted in speculation that PrP may be idiosyncratically processed by antigen-presenting cells (12), or to inferences that the epitopes of such mAbs are discontinuous, and/or that peptide fragments in this format are unable to adopt specific conformations required for antibody binding (12–17). This latter hypothesis was rejected as highly unlikely (13), because all such antibodies bind PrP in Western blots, where PrP is defined by investigators as denatured (12, 13, 15, 18–22). Our discovery that mAbs PRC5 and PRP7, which recognize discontinuous conformation-dependent epitopes, react with wild type PrP on Western blots was also surprising. However, our findings indicate that, subsequent to denaturing treatments, PrP readily refolds during or after Western blotting to a structure amenable to detection by conformation-dependent antibodies. Although it is widely assumed that discontinuous epitopes are generally irreversibly destroyed under the denaturing conditions used for SDS-PAGE, positive reactions on Western blots with antibodies recognizing discontinuous epitopes are not without precedent (44).
Because PRC5, PRC7, and 6H4 also react with immunoblotted PrP(27–30), our results demonstrate that PK-resistant PrP must also maintain the structural determinants required for recognition by these conformation-dependent mAbs. Our findings therefore indicate that, following denaturation, PrPC and PrP(27–30) re-nature into a common structure amenable to detection by conformation-dependent mAbs However, recent studies using mass spectrometry analysis coupled with hydrogen-deuterium exchange indicate that PrPSc consists of β-strands, relatively short turns, and/or loops, and no α-helical structure, which represents a conformation radically different from PrPC (45). Our results therefore also suggest that the conformation of renatured PrP(27–30) is structurally more akin to PrPC than to PrPSc from which it is derived.
PRC mAbs Discriminate PrP Polymorphisms and PrP Glycotypes
Epitopes of two of the four PRC mAbs are composed of PrP residues that are polymorphic and influence disease susceptibility in sheep and deer. PRC5 reactivity is dependent on the sheep Ala/Val polymorphism at residue 136. Susceptibility of sheep to classical scrapie is strongly associated with amino acid polymorphism at this residue, as well as polymorphisms at residues 154 and 171 (46). Although previous studies have shown mAbs to be capable of distinguishing OvPrP-Q171 and OvPrP-R171 allotypes (47, 48), the direct involvement of residue 171 in these epitopes was not demonstrated, and in the case of glycosylation-dependent antibodies, the effect was indirectly influenced by N-linked glycan occupancy of OvPrP (47). Recognition by PRC1 is also dependent on a known PrP polymorphism. Detection of deer PrP was inhibited when His replaced Gln at residue 95 but not when Ser replaced Gly at adjacent residue 96 (deer PrP numberings). These amino acid variations correspond to naturally occurring polymorphisms affecting susceptibility of white tail deer to CWD (49).
Monoclonal antibody PRC1 also fails to react with PrP primary structures containing three instead of two Gly residues immediately downstream of mouse PrP Gln-90. Our studies indicate that an additional Gly sterically hinders PRC1 recognition of its epitope, which includes Gln at mouse PrP residue 90/deer PrP residue 95. Other than deer harboring the H95 polymorphism, monoclonal antibody PRC1 has excellent reactivity with cervid PrP, and this specificity is therefore likely to be useful for studies on CWD. Like other mAbs with epitopes located at the boundary between the octarepeat region and the protease-resistant core of PrP(27–30) that have been used for molecular strain typing (13, 50–52), PRC1 may be useful for differentiating biochemically distinct forms of PrP(27–30) associated with different strains.
In addition to its conformational dependence, PRC7 immunoreactivity is also dependent of the state of glycosylation of PrP. Because PRC antibodies were derived from mice immunized with bacterially derived RecPrP, an N-linked glycan is unlikely to contribute to the PRC7 epitope. Instead, the preferential binding of PRC7 to under-glycosylated PrP suggests that occupancy of one of the two PrP N-linked glycosylation sites precludes antibody binding to the fully glycosylated protein, making it immunologically silent to PRC7. A similar mechanism has been suggested to explain the properties of previously isolated glycosylation-dependent antibodies (13, 19, 47).
PRC mAbs Expand the Range of Options for PrP Epitope Manipulation
The involvement of specific amino acids in mAb recognition has, to date, only been defined for a handful of mAbs, in particular 3F4 (53) and L42 (54). One application for which such information has proved useful is epitope tagging of PrP molecules. Previous studies showed that PrP molecules engineered to include amino acid residues required for 3F4 binding could be converted to PrPSc in murine cell lines, chronically infected with adapted scrapie prions (24). Whereas such an approach is useful for assessing the effects of amino acid alterations on PrPSc formation, it is not without drawbacks, because 3F4 epitope inclusion can lead to interfering effects on PrPSc formation (55). In addition, the exact composition of the 3F4 epitope has been recently debated (56). Stable expression of PRC epitope-mutated PrP constructs in RK13 cells not only allowed us to confirm the requirement of those residues for recognition by various PRC mAbs but also to assess the effects of substitutions on conversion of PrPSc. Because RK13 cells support the replication of prions from a variety of species (29, 30, 57), we were able to assess the effects of these mutations on PrPSc formation in the context of multiple PrP primary structures. We found that mutation of amino acids constituting various PRC mAb epitopes had different effects on the abilities of mouse and cervid PrP to be converted to PrPSc (summarized in Table 1), which is consistent with previous observations that structural requirements for PrPC conversion to protease-resistant PrP vary between PrP primary structures (58). Such information expands the available options for introducing new or eliminating existing PrP epitopes, while preserving the ability of mutated PrPC to convert to PrPSc. For example, removal of one of three Gly residues after residue 90 preserved the ability of mutated mouse PrP to convert to PrPSc and facilitated recognition by PRC1. Similarly, mutation of mouse PrP at residues 158 and 154 preserved conversion to PrPSc and abolished recognition by mAbs PRC5 and PRC7, respectively. Such manipulations allow, for example, newly converted PrPSc to be distinguished from PrPSc in the inoculum and provide novel means of assessing the kinetics and cell biology of PrPSc formation.
In conclusion, we used a directed molecular evolution approach to create shuffled genes expressing novel PrP epitopes as immunogens. To our knowledge, these studies are the first to describe the involvement of specific amino acids in functional, discontinuous, and conformation-dependent anti-PrP mAb epitopes. We show that perturbations of the correct tertiary structure of the PrP globular domain, either by disulfide bond reduction or by the introduction of conformation-disrupting deletion mutants, affect the topology of a variety of conformational epitopes. We also show that conformation-dependent PrP epitopes readily reform following conditions of harsh denaturation, and that both PrPC and PrP(27–30) re-nature into a structure amenable to detection by conformation-dependent mAbs. The epitopes of two of the four new antibodies described here are composed of residues that are polymorphic in certain species and affect disease susceptibility in those hosts. Because of their dependence on these polymorphisms for reactivity, PRC1 and PRC5 represent unique immunological tools to assess the conversion and pathogenic effects of individual sheep and deer allotypes.4 Finally, the mapping and mutational analyses of PrP mAb epitopes described here not only expand the repertoire of available options for tagging PrP in Tg mice and transfected cell lines, but they also provide new information about the role of these amino acids in conversion of PrPC to PrPSc.
Acknowledgments
We thank Tanya Seward for technical help during the early phase of these studies; Richard Bessen and Charles Merrill for critical reading of the manuscript; Stanley Prusiner for the kind gift of mAbs D18, R1, and HumP; Adriano Aguzzi for the kind gift POM2 mAb, and Prionics for mAb 6H4.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 NS040334 and PO1 AI077774-015261 (to G. C. T.) and T32 DA022738 (to V. K., R. A., and G. C. T.).

This article contains supplemental Experimental Procedures and Figs. 1 and 2.
E. Saijo and G. C. Telling, manuscript in preparation.
- CWD
- chronic wasting disease
- PrP
- prion protein
- PrPC
- cellular form of the prion protein
- PrPSc
- scrapie form of the prion protein
- mAb
- monoclonal antibody
- PK
- proteinase K
- Tg
- transgenic
- RK13
- rabbit kidney epithelial cells
- PNGase F
- peptide:N-glycosidase F
- βME
- β-mercaptoethanol
- SPR
- surface plasmon resonance
- Rec
- recombinant.
REFERENCES
- 1. Stahl N., Borchelt D. R., Hsiao K., Prusiner S. B. (1987) Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell 51, 229–240 [DOI] [PubMed] [Google Scholar]
- 2. Viles J. H., Cohen F. E., Prusiner S. B., Goodin D. B., Wright P. E., Dyson H. J. (1999) Copper binding to the prion protein. Structural implications of four identical cooperative binding sites. Proc. Natl. Acad. Sci. U.S.A. 96, 2042–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Riek R., Hornemann S., Wider G., Billeter M., Glockshuber R., Wüthrich K. (1996) NMR structure of the mouse prion protein domain PrP(121–231). Nature 382, 180–182 [DOI] [PubMed] [Google Scholar]
- 4. Chen S. G., Teplow D. B., Parchi P., Teller J. K., Gambetti P., Autilio-Gambetti L. (1995) Truncated forms of the human prion protein in normal brain and in prion diseases. J. Biol. Chem. 270, 19173–19180 [DOI] [PubMed] [Google Scholar]
- 5. Yadavalli R., Guttmann R. P., Seward T., Centers A. P., Williamson R. A., Telling G. C. (2004) Calpain-dependent endoproteolytic cleavage of PrPSc modulates scrapie prion propagation. J. Biol. Chem. 279, 21948–21956 [DOI] [PubMed] [Google Scholar]
- 6. Bendheim P. E., Barry R. A., DeArmond S. J., Stites D. P., Prusiner S. B. (1984) Antibodies to a scrapie prion protein. Nature 310, 418–421 [DOI] [PubMed] [Google Scholar]
- 7. Barry R. A., Kent S. B., McKinley M. P., Meyer R. K., DeArmond S. J., Hood L. E., Prusiner S. B. (1986) Scrapie and cellular prion proteins share polypeptide epitopes. J. Infect. Dis. 153, 848–854 [DOI] [PubMed] [Google Scholar]
- 8. Büeler H., Fischer M., Lang Y., Bluethmann H., Lipp H. P., DeArmond S. J., Prusiner S. B., Aguet M., Weissmann C. (1992) Normal development and behavior of mice lacking the neuronal cell-surface PrP protein. Nature 356, 577–582 [DOI] [PubMed] [Google Scholar]
- 9. Prusiner S. B., Groth D., Serban A., Koehler R., Foster D., Torchia M., Burton D., Yang S. L., DeArmond S. J. (1993) Ablation of the prion protein (PrP) gene in mice prevents scrapie and facilitates production of anti-PrP antibodies. Proc. Natl. Acad. Sci. U.S.A. 90, 10608–10612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Antonyuk S. V., Trevitt C. R., Strange R. W., Jackson G. S., Sangar D., Batchelor M., Cooper S., Fraser C., Jones S., Georgiou T., Khalili-Shirazi A., Clarke A. R., Hasnain S. S., Collinge J. (2009) Crystal structure of human prion protein bound to a therapeutic antibody. Proc. Natl. Acad. Sci. U.S.A. 106, 2554–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kanyo Z. F., Pan K. M., Williamson R. A., Burton D. R., Prusiner S. B., Fletterick R. J., Cohen F. E. (1999) Antibody binding defines a structure for an epitope that participates in the PrPC → PrPSc conformational change. J. Mol. Biol. 293, 855–863 [DOI] [PubMed] [Google Scholar]
- 12. Khalili-Shirazi A., Kaisar M., Mallinson G., Jones S., Bhelt D., Fraser C., Clarke A. R., Hawke S. H., Jackson G. S., Collinge J. (2007) β-PrP form of human prion protein stimulates production of monoclonal antibodies to epitope 91–110 that recognize native PrPSc. Biochim. Biophys. Acta 1774, 1438–1450 [DOI] [PubMed] [Google Scholar]
- 13. Polymenidou M., Moos R., Scott M., Sigurdson C., Shi Y. Z., Yajima B., Hafner-Bratkovic I., Jerala R., Hornemann S., Wuthrich K., Bellon A., Vey M., Garen G., James M. N., Kav N., Aguzzi A. (2008) The POM monoclonals. A comprehensive set of antibodies to nonoverlapping prion protein epitopes. PLoS ONE 3, e3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jones M., McLoughlin V., Connolly J. G., Farquhar C. F., MacGregor I. R., Head M. W. (2009) Production and characterization of a panel of monoclonal antibodies against native human cellular prion protein. Hybridoma 28, 13–20 [DOI] [PubMed] [Google Scholar]
- 15. Stanker L. H., Serban A. V., Cleveland E., Hnasko R., Lemus A., Safar J., DeArmond S. J., Prusiner S. B. (2010) Conformation-dependent high affinity monoclonal antibodies to prion proteins. J. Immunol. 185, 729–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Williamson R. A., Peretz D., Pinilla C., Ball H., Bastidas R. B., Rozenshteyn R., Houghten R. A., Prusiner S. B., Burton D. R. (1998) Mapping the prion protein using recombinant antibodies. J. Virol. 72, 9413–9418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li R., Liu T., Wong B. S., Pan T., Morillas M., Swietnicki W., O'Rourke K., Gambetti P., Surewicz W. K., Sy M. S. (2000) Identification of an epitope in the C terminus of normal prion protein whose expression is modulated by binding events in the N terminus. J. Mol. Biol. 301, 567–573 [DOI] [PubMed] [Google Scholar]
- 18. Khalili-Shirazi A., Quaratino S., Londei M., Summers L., Tayebi M., Clarke A. R., Hawke S. H., Jackson G. S., Collinge J. (2005) Protein conformation significantly influences immune responses to prion protein. J. Immunol. 174, 3256–3263 [DOI] [PubMed] [Google Scholar]
- 19. Khalili-Shirazi A., Summers L., Linehan J., Mallinson G., Anstee D., Hawke S., Jackson G. S., Collinge J. (2005) PrP glycoforms are associated in a strain-specific ratio in native PrPSc. J. Gen. Virol. 86, 2635–2644 [DOI] [PubMed] [Google Scholar]
- 20. Cordes H., Bergström A. L., Ohm J., Laursen H., Heegaard P. M. (2008) Characterization of new monoclonal antibodies reacting with prions from both human and animal brain tissues. J. Immunol. Methods 337, 106–120 [DOI] [PubMed] [Google Scholar]
- 21. Harmeyer S., Pfaff E., Groschup M. H. (1998) Synthetic peptide vaccines yield monoclonal antibodies to cellular and pathological prion proteins of ruminants. J. Gen. Virol. 79, 937–945 [DOI] [PubMed] [Google Scholar]
- 22. Yuan J., Kinter M., McGeehan J., Perry G., Kneale G., Gambetti P., Zou W. Q. (2005) Concealment of epitope by reduction and alkylation in prion protein. Biochem. Biophys. Res. Commun. 326, 652–659 [DOI] [PubMed] [Google Scholar]
- 23. Stemmer W. P. (1994) Rapid evolution of a protein in vitro by DNA shuffling. Nature 370, 389–391 [DOI] [PubMed] [Google Scholar]
- 24. Scott M. R., Köhler R., Foster D., Prusiner S. B. (1992) Chimeric prion protein expression in cultured cells and transgenic mice. Protein Sci. 1, 986–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kurt T. D., Telling G. C., Zabel M. D., Hoover E. A. (2009) Trans-species amplification of PrP(CWD) and correlation with rigid loop 170N. Virology 387, 235–243 [DOI] [PubMed] [Google Scholar]
- 26. Borchelt D. R., Davis J., Fischer M., Lee M. K., Slunt H. H., Ratovitsky T., Regard J., Copeland N. G., Jenkins N. A., Sisodia S. S., Price D. L. (1996) A vector for expressing foreign genes in the brains and hearts of transgenic mice. Genet. Anal. 13, 159–163 [DOI] [PubMed] [Google Scholar]
- 27. Crameri A., Raillard S. A., Bermudez E., Stemmer W. P. (1998) DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature 391, 288–291 [DOI] [PubMed] [Google Scholar]
- 28. Angers R. C., Kang H. E., Napier D., Browning S., Seward T., Mathiason C., Balachandran A., McKenzie D., Castilla J., Soto C., Jewell J., Graham C., Hoover E. A., Telling G. C. (2010) Prion strain mutation determined by prion protein conformational compatibility and primary structure. Science 328, 1154–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bian J., Napier D., Khaychuck V., Angers R., Graham C., Telling G. (2010) Cell-based quantification of chronic wasting disease prions. J. Virol. 84, 8322–8326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vilette D., Andreoletti O., Archer F., Madelaine M. F., Vilotte J. L., Lehmann S., Laude H. (2001) Ex vivo propagation of infectious sheep scrapie agent in heterologous epithelial cells expressing ovine prion protein. Proc. Natl. Acad. Sci. U.S.A. 98, 4055–4059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Korth C., Stierli B., Streit P., Moser M., Schaller O., Fischer R., Schulz-Schaeffer W., Kretzschmar H., Raeber A., Braun U., Ehrensperger F., Hornemann S., Glockshuber R., Riek R., Billeter M., Wüthrick K., Oesch B. (1997) Prion (PrPSc)-specific epitope defined by a monoclonal antibody. Nature 390, 74–77 [DOI] [PubMed] [Google Scholar]
- 32. Safar J. G., Scott M., Monaghan J., Deering C., Didorenko S., Vergara J., Ball H., Legname G., Leclerc E., Solforosi L., Serban H., Groth D., Burton D. R., Prusiner S. B., Williamson R. A. (2002) Measuring prions causing bovine spongiform encephalopathy or chronic wasting disease by immunoassays and transgenic mice. Nat. Biotechnol. 20, 1147–1150 [DOI] [PubMed] [Google Scholar]
- 33. Raviprakash K., Apt D., Brinkman A., Skinner C., Yang S., Dawes G., Ewing D., Wu S. J., Bass S., Punnonen J., Porter K. (2006) A chimeric tetravalent dengue DNA vaccine elicits neutralizing antibody to all four virus serotypes in rhesus macaques. Virology 353, 166–173 [DOI] [PubMed] [Google Scholar]
- 34. Apt D., Raviprakash K., Brinkman A., Semyonov A., Yang S., Skinner C., Diehl L., Lyons R., Porter K., Punnonen J. (2006) Tetravalent neutralizing antibody response against four dengue serotypes by a single chimeric dengue envelope antigen. Vaccine 24, 335–344 [DOI] [PubMed] [Google Scholar]
- 35. Dupuy L. C., Locher C. P., Paidhungat M., Richards M. J., Lind C. M., Bakken R., Parker M. D., Whalen R. G., Schmaljohn C. S. (2009) Directed molecular evolution improves the immunogenicity and protective efficacy of a Venezuelan equine encephalitis virus DNA vaccine. Vaccine 27, 4152–4160 [DOI] [PubMed] [Google Scholar]
- 36. Du S. X., Xu L., Zhang W., Tang S., Boenig R. I., Chen H., Mariano E. B., Zwick M. B., Parren P. W., Burton D. R., Wrin T., Petropoulos C. J., Ballantyne J. A., Chambers M., Whalen R. G. (2011) A directed molecular evolution approach to improved immunogenicity of the HIV-1 envelope glycoprotein. PLoS ONE 6, e20927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nazor K. E., Kuhn F., Seward T., Green M., Zwald D., Pürro M., Schmid J., Biffiger K., Power A. M., Oesch B., Raeber A. J., Telling G. C. (2005) Immunodetection of disease-associated mutant PrP, which accelerates disease in GSS transgenic mice. EMBO J. 24, 2472–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Petsch B., Müller-Schiffmann A., Lehle A., Zirdum E., Prikulis I., Kuhn F., Raeber A. J., Ironside J. W., Korth C., Stitz L. (2011) Biological effects and use of PrPSc- and PrP-specific antibodies generated by immunization with purified full-length native mouse prions. J. Virol. 85, 4538–4546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Biasini E., Seegulam M. E., Patti B. N., Solforosi L., Medrano A. Z., Christensen H. M., Senatore A., Chiesa R., Williamson R. A., Harris D. A. (2008) Non-infectious aggregates of the prion protein react with several PrPSc-directed antibodies. J. Neurochem. 105, 2190–2204 [DOI] [PubMed] [Google Scholar]
- 40. Morel N., Simon S., Frobert Y., Volland H., Mourton-Gilles C., Negro A., Sorgato M. C., Créminon C., Grassi J. (2004) Selective and efficient immunoprecipitation of the disease-associated form of the prion protein can be mediated by nonspecific interactions between monoclonal antibodies and scrapie-associated fibrils. J. Biol. Chem. 279, 30143–30149 [DOI] [PubMed] [Google Scholar]
- 41. Laver W. G., Air G. M., Webster R. G., Smith-Gill S. J. (1990) Epitopes on protein antigens. Misconceptions and realities. Cell 61, 553–556 [DOI] [PubMed] [Google Scholar]
- 42. Zanusso G., Farinazzo A., Prelli F., Fiorini M., Gelati M., Ferrari S., Righetti P. G., Rizzuto N., Frangione B., Monaco S. (2004) Identification of distinct N-terminal truncated forms of prion protein in different Creutzfeldt-Jakob disease subtypes. J. Biol. Chem. 279, 38936–38942 [DOI] [PubMed] [Google Scholar]
- 43. Sasamori E., Suzuki S., Kato M., Tagawa Y., Hanyu Y. (2010) Characterization of discontinuous epitope of prion protein recognized by the monoclonal antibody T2. Arch. Biochem. Biophys. 501, 232–238 [DOI] [PubMed] [Google Scholar]
- 44. Zhou Y. H., Chen Z., Purcell R. H., Emerson S. U. (2007) Positive reactions on Western blots do not necessarily indicate the epitopes on antigens are continuous. Immunol. Cell Biol. 85, 73–78 [DOI] [PubMed] [Google Scholar]
- 45. Smirnovas V., Baron G. S., Offerdahl D. K., Raymond G. J., Caughey B., Surewicz W. K. (2011) Structural organization of brain-derived mammalian prions examined by hydrogen-deuterium exchange. Nat. Struct. Mol. Biol. 18, 504–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Goldmann W. (2008) PrP genetics in ruminant transmissible spongiform encephalopathies. Vet. Res. 39, 30. [DOI] [PubMed] [Google Scholar]
- 47. Moudjou M., Treguer E., Rezaei H., Sabuncu E., Neuendorf E., Groschup M. H., Grosclaude J., Laude H. (2004) Glycan-controlled epitopes of prion protein include a major determinant of susceptibility to sheep scrapie. J. Virol. 78, 9270–9276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bilheude J. M., Brun A., Morel N., Díaz San Segundo F., Lecroix S., Espinosa J. C., González L., Steele P., Grassi J., Andréoletti O., Torres J. M. (2007) Discrimination of sheep susceptible and resistant to transmissible spongiform encephalopathies by a haplotype-specific monoclonal antibody. J. Virol. Methods 145, 169–172 [DOI] [PubMed] [Google Scholar]
- 49. Johnson C., Johnson J., Clayton M., McKenzie D., Aiken J. (2003) Prion protein gene heterogeneity in free-ranging white-tailed deer within the chronic wasting disease affected region of Wisconsin. J. Wildl. Dis. 39, 576–581 [DOI] [PubMed] [Google Scholar]
- 50. Stack M. J., Chaplin M. J., Clark J. (2002) Differentiation of prion protein glycoforms from naturally occurring sheep scrapie, sheep-passaged scrapie strains (CH1641 and SSBP1), bovine spongiform encephalopathy (BSE) cases, and Romney and Cheviot breed sheep experimentally inoculated with BSE using two monoclonal antibodies. Acta Neuropathol. 104, 279–286 [DOI] [PubMed] [Google Scholar]
- 51. Thuring C. M., Erkens J. H., Jacobs J. G., Bossers A., Van Keulen L. J., Garssen G. J., Van Zijderveld F. G., Ryder S. J., Groschup M. H., Sweeney T., Langeveld J. P. (2004) Discrimination between scrapie and bovine spongiform encephalopathy in sheep by molecular size, immunoreactivity, and glycoprofile of prion protein. J. Clin. Microbiol. 42, 972–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Polymenidou M., Stoeck K., Glatzel M., Vey M., Bellon A., Aguzzi A. (2005) Coexistence of multiple PrPSc types in individuals with Creutzfeldt-Jakob disease. Lancet Neurol. 4, 805–814 [DOI] [PubMed] [Google Scholar]
- 53. Kascsak R. J., Rubenstein R., Merz P. A., Tonna-DeMasi M., Fersko R., Carp R. I., Wisniewski H. M., Diringer H. (1987) Mouse polyclonal and monoclonal antibody to scrapie-associated fibril proteins. J. Virol. 61, 3688–3693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vorberg I., Buschmann A., Harmeyer S., Saalmüller A., Pfaff E., Groschup M. H. (1999) A novel epitope for the specific detection of exogenous prion proteins in transgenic mice and transfected murine cell lines. Virology 255, 26–31 [DOI] [PubMed] [Google Scholar]
- 55. Priola S. A., Caughey B., Race R. E., Chesebro B. (1994) Heterologous PrP molecules interfere with accumulation of protease-resistant PrP in scrapie-infected murine neuroblastoma cells. J. Virol. 68, 4873–4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zou W. Q., Langeveld J., Xiao X., Chen S., McGeer P. L., Yuan J., Payne M. C., Kang H. E., McGeehan J., Sy M. S., Greenspan N. S., Kaplan D., Wang G. X., Parchi P., Hoover E., Kneale G., Telling G., Surewicz W. K., Kong Q., Guo J. P. (2010) PrP conformational transitions alter species preference of a PrP-specific antibody. J. Biol. Chem. 285, 13874–13884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Courageot M. P., Daude N., Nonno R., Paquet S., Di Bari M. A., Le Dur A., Chapuis J., Hill A. F., Agrimi U., Laude H., Vilette D. (2008) A cell line infectable by prion strains from different species. J. Gen. Virol. 89, 341–347 [DOI] [PubMed] [Google Scholar]
- 58. Priola S. A., Chabry J., Chan K. (2001) Efficient conversion of normal prion protein (PrP) by abnormal hamster PrP is determined by homology at amino acid residue 155. J. Virol. 75, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]