Background: The Rad52-type mitochondrial recombination protein Mgm101 forms large oligomeric rings, but its functional implication is poorly understood.
Results: Mutations that either disassemble the rings or induce the formation of super-stable macroscopic filaments affect Mgm101 function.
Conclusion: A properly configured ring structure is essential for Mgm101 function in vivo.
Significance: This finding helps the better understanding of how Mgm101 promotes homologous recombination in mitochondria.
Keywords: Bioenergetics, Mitochondria, Mitochondrial DNA, Molecular Genetics, Yeast Genetics
Abstract
Mgm101 is a Rad52-type recombination protein of bacteriophage origin required for the repair and maintenance of mitochondrial DNA (mtDNA). It forms large oligomeric rings of ∼14-fold symmetry that catalyze the annealing of single-stranded DNAs in vitro. In this study, we investigated the structural elements that contribute to this distinctive higher order structural organization and examined its functional implications. A pair of vicinal cysteines, Cys-216 and Cys-217, was found to be essential for mtDNA maintenance. Mutations to the polar serine, the negatively charged aspartic and glutamic acids, and the hydrophobic amino acid alanine all destabilize mtDNA in vivo. The alanine mutants have an increased propensity of forming macroscopic filaments. In contrast, mutations to aspartic acid drastically destabilize the protein and result in unstructured aggregates with severely reduced DNA binding activity. Interestingly, the serine mutants partially disassemble the Mgm101 rings into smaller oligomers. In the case of the C216S mutant, a moderate increase in DNA binding activity was observed. By using small angle x-ray scattering analysis, we found that Mgm101 forms rings of ∼200 Å diameter in solution, consistent with the structure previously established by transmission electron microscopy. We also found that the C216A/C217A double mutant tends to form broken rings, which likely provide free ends for seeding the growth of the super-stable but functionally defective filaments. Taken together, our data underscore the importance of a delicately maintained ring structure critical for Mgm101 activity. We discuss a potential role of Cys-216 and Cys-217 in regulating Mgm101 function and the repair of damaged mtDNA under stress conditions.
Introduction
The MGM101 gene of Saccharomyces cerevisiae is essential for the maintenance of the mitochondrial genome (1–3). It encodes a positively charged protein of 247 amino acids associated with the mitochondrial nucleoids (4–6). The specific association of Mgm101 homologs with mitochondrial nucleoids has also been reported in other eukaryotic species (7). Several lines of evidence suggest that Mgm101 may participate in the recombinational repair of mitochondrial DNA (mtDNA). First, mtDNA in temperature-sensitive mutants of mgm101 cultured under semi-permissive conditions are hyper-sensitive to DNA-damaging agents, including ultraviolet and γ-ray irradiations, and hydrogen peroxide (5). Recombination is a well established pathway for the repair of double strand DNA breaks. Second, it has been shown that defects in Mgm101 function can be suppressed by hyper-recombinogenic DNA sequences. Although Mgm101 is essential for the stability of the wild-type (ρ+) mtDNA, the maintenance of the highly repeated hypersuppressive ρ− genomes was found to be independent of Mgm101 (3). These genomes contain the GC-rich ori sequences that are known to be highly recombinogenic (8, 9). A high concentration of the recombinogenic elements may bypass the requirement for an Mgm101-catalyzed recombination process. Third, our recent studies have shown that Mgm101 shares biochemical and structural similarities with the Rad52 family proteins (10), which are known to be involved in homologous recombination.
Homologous recombination is a highly conserved molecular process that has primarily evolved for the repair of double strand breaks and stalled or collapsed replication forks. The Rad52-type proteins participate in several recombination processes (11–13). In a canonical recombination pathway, Rad52, or its bacterial ortholog, RecO, binds to processed 3′ ssDNA4 tails and recruits the core ATP-dependent recombinases, including Rad51 in the eukaryotes and RecA in bacteria. It is Rad51 or RecA that forms the helical nucleoprotein filaments and initiates strand invasion and homologous pairing within duplex DNA templates. In this respect, Rad52 functions as a recombination mediator for Rad51. In a later stage of the recombination process, Rad52 acts as a ssDNA-annealing protein, which is critical for the capture of the second DNA end at the recombination site to generate Holliday junctions (14–16). Finally, the ssDNA annealing property confers on Rad52 the ability to catalyze recombination in a RecA/Rad51-independent manner. Following the resection of dsDNA breaks by exonucleases, the exposed homologous ssDNA can be directly annealed by Rad52. This process is proposed to initiate by successive interactions between two separate Rad52-ssDNA complexes until the formation of a homologous and stable duplex (17). The operation mode of single strand annealing generally leads to deletions of the resected sequences present between directly repeated regions (18).
The Rad52-type single strand DNA-annealing proteins are widespread in the temperate bacteriophages (19–21). These phage proteins can promote homologous recombination through the single strand annealing mode without causing deletions in the genome. For instance, the Redβ protein in phage λ initiates strand annealing by preferentially targeting on the lagging strand of a replication fork (22, 23), which leads to the error-free recombinational repair of the phage DNA.
Mgm101 is structurally more close to the viral proteins rather than to the eukaryotic Rad52, although they all possess a single strand annealing activity. Unlike Rad52, Mgm101 and its bacteriophage counterparts do not contain an extended C terminus required for interaction with Rad51/RecA (24, 25). It is therefore unlikely that they act as mediators for the Rad51/RecA-type recombinases. Despite very limited similarity in their primary sequences, Mgm101 shares similar structural organization with its viral orthologs. Mgm101, Redβ, and Erf from the bacteriophages λ and P22, RecT from the prophage rac, and Sak from the lactococcal phage ul36 all form large homo-oligomeric rings of 10–14-fold symmetry in vitro (10, 19, 26, 27). This is in contrast with the native Rad52, which mainly forms heptameric rings (28). Like Redβ, Mgm101 also forms highly compressed helical filaments in the absence of DNA (10, 19), which is not seen with the eukaryotic Rad52. When bound to ssDNA, Mgm101 forms condensed nucleoprotein filaments like Sak from the lactococcal phage ul36. These observations suggest that Mgm101 may functionally operate like the bacteriophage proteins and catalyze recombination by the single strand annealing mode. It may cooperate with other proteins like Mhr1, the single strand-binding protein Rim1, and the MRX complex to promote the recombinational repair of double strand breaks in mtDNA (29–33).
The functional implication of the higher order structural organization of the Rad52-type proteins is poorly understood. By taking advantage of the genetic versatility of the yeast system, we investigated structural elements that determine and possibly regulate the higher order structural organization of Mgm101. In this study, we characterized Mgm101 variants with mutations in a pair of vicinal cysteines that are conserved among Mgm101 homologs. The mutant proteins are all defective in maintaining mtDNA in vivo, concomitant with altered higher order structural organization in vitro. The data support the notion that the maintenance of a properly configured ring structure of Mgm101 is crucial for its in vivo functionality. We will discuss the possibility that potential modifications of the specific cysteine residues may be important for activating mtDNA transactions in response to oxidative damage or other stresses in mitochondria.
EXPERIMENTAL PROCEDURES
Growth Media, Yeast Strains, and Plasmid Construction
Complete (YP) and minimal medium (YNB) were prepared with 2% dextrose (D) or 2% glycerol plus 2% ethanol (GE). The temperature-sensitive mgm101 mutant used for complementation test was M2915-7C (MATa, ade2, leu2, his4, ura3, mgm101-1ts). The mutant alleles of Cys-216 and Cys-217 were generated by in vitro site-directed mutagenesis using the plasmid pCXJ22-MGM101 as a template. For complementation test in an mgm101 null background, the mutant alleles were cloned into the integrative vector, pUC-URA3/4. After linearizing with the restriction enzyme EcoRV that is located in the URA3 gene, the plasmids were integrated into the ura3 locus of the diploid strain CS1638/1 (MATa/MATα, ade2/ade2, his3/his3, trp1/trp1, leu2/leu2, ura3/ura3, +/mgm101Δ::kan) by selection for Ura+ transformants. After sporulation and tetrad dissection, spores carrying mgm101Δ::kan and the mutant mgm101 alleles were identified and tested for respiratory growth on glycerol plus ethanol medium by replica plating.
Expression and Purification of Mutant Mgm101
Mgm101 and its cysteine mutants were expressed and purified from Escherichia coli by the MBP-tagging strategy as described previously (10). Briefly, cell lysate was first treated with DNase I for 2 h on ice to reduce contaminant DNA. The salt concentration was adjusted to 500 mm, and the MBP fusion proteins were purified with amylose resin at 4 °C. The mutant Mgm101 proteins were then released from MBP fusions after cleavage with the PrescissionTM protease. Cation exchange chromatography on a Bio-ScaleTM Mini Macro-Prep High S cartridge (Bio-Rad) allowed the separation of Mgm101 from MBP. The proteins were further purified by size exclusion chromatography on a calibrated Superose 6 prep grade column equilibrated in buffer GF (20 mm MOPS, pH 7.0, 150 mm NaCl, 5 mm β-mercaptoethanol, 1 mm EDTA, 0.2 mm phenylmethanesulfonyl fluoride). Peak fractions were analyzed by SDS-PAGE, concentrated, and stored at −80 °C.
Biochemical Analysis of Mgm101
The oligomerization state of Mgm101 was examined by size exclusion chromatography on a calibrated Superose 6 column equilibrated with buffer GF. For electrophoretic mobility shift assay (EMSA), the oligonucleotide HS40AT-F (5′-TTAATATTTAATAATATAATCAATAAATAATATTATAATAATATAATATAA-3′) was end-labeled with [γ-32P]ATP and used as a probe. Recombinant proteins were mixed with 1.25 × 10−3 μm of the probe in the binding buffer of 20 mm Tris-HCl, pH 8.0, 100 mm KCl, 5 mm MgCl2, 1 mm EDTA, 10% glycerol, 1 mm DTT, 50 μg/ml BSA, 1 mm PMSF and incubated at room temperature for 30 min. The samples were then loaded onto a 6% native acrylamide gel in 0.5× TBE buffer at 4 °C. Transmission electron microscopy was carried out using a JEM-2100 transmission electron microscope (JEOL) operating at 200 kV as described previously (10). For long term negative stain electron microscopy analysis, the protein was stored at 4 °C in buffer GF. For testing NEM accessibility to cysteines in Mgm101, the protein was equilibrated in alkylation buffer (20 mm MOPS, 1 mm EDTA, 0.2 mm PMSF) and concentrated to 4.2 mg/ml in a 50-kDa molecular mass cutoff spin concentrator (Vivaspin). Mgm101 (1 mg/ml) was then treated with 3 mm NEM dissolved in alkylation buffer at room temperature for 40 min. The alkylation reaction was stopped with 15 mm cysteine. The alkylated protein was then buffer-changed by a spin column (G-25 resin) equilibrated in 40 mm ammonium bicarbonate, diluted 1:3 into 50% acetonitrile and 0.1% formic acid, and analyzed by an electrospray Q-TOF mass spectrometer.
SAXS Data Collection and Analysis
SAXS experiments were conducted at the F2 beamline at the Cornell High Energy Synchrotron Source (CHESS) (Cornell University, Ithaca, NY). Purified Mgm101 and Mgm101C216A,C217A were analyzed in 20 mm MOPS, pH 7.0, buffer containing 150 mm NaCl, 1 mm EDTA, 5 mm β-mercaptoethanol, and 0.2 mm PMSF. Data were collected using an ADSC Quantum 1 detector, with wavelength = 1.2524 Å, at 4 °C. Data acquisition was performed by taking two successive 300-s frames of each 30-μl sample. The protein buffer was analyzed under the same conditions for buffer subtraction of the raw data. The sample-to-detector distance was set at 1467 mm, which yielded a q-range of 0.00814–0.275 Å−1. Guinier plots of the data at four different concentrations (from 0.625 to 5 mg/ml) were linear at small angles (34) and showed very similar scattering patterns. No significant time-dependent protein aggregation or radiation damage was detected under these experimental conditions. After initial data integration and background subtraction, SAXS data were analyzed using the ATSAS program package (35). The radius of gyration (Rg), the pair distance distribution function [P(r)], and the maximum intermolecular distance (Dmax) of the protein were calculated using the program GNOM (36). The program DAMMIF was used to generate 10 ab initio low resolution bead models using simulated annealing of a single-phase dummy atom model. These models were averaged and filtered using the program DAMAVER, yielding final models with average normalized discrepancy (NSD) values of 1.08 and 0.985 for Mgm101 and Mgm101C216A,C217A, respectively, which are consistent with NSD values observed for model calculations of ring or disk-shaped objects (37). The bead model was converted to a volumetric bead map using the pdb2vol kernel convolution utility from the Situs Program Package, and snap shots were taken using Chimera (University of California, San Francisco).
RESULTS
Substitution by Alanine Revealed That Cys-216 and Cys-217 Are Critical for Mgm101 Function in Vivo
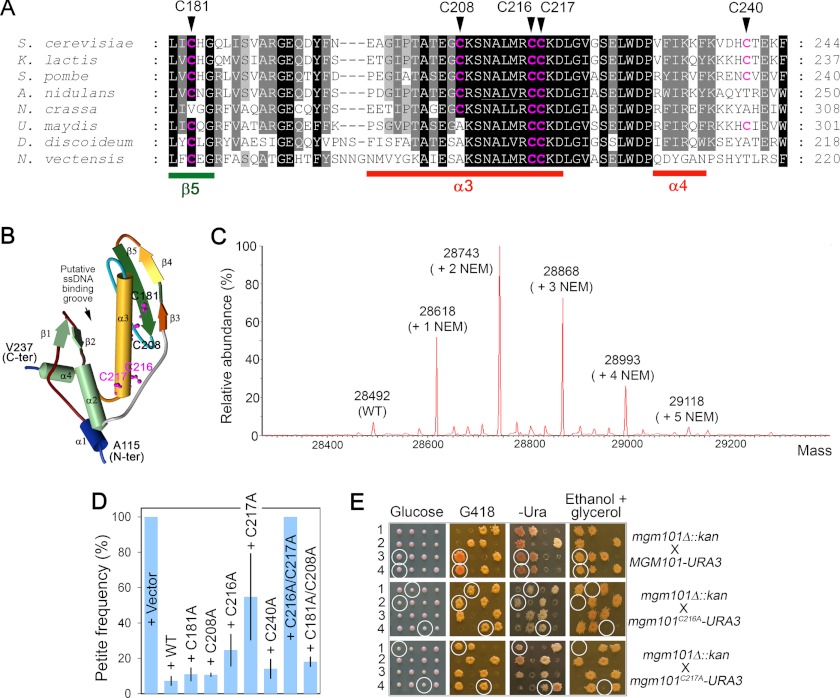
A salient feature of Mgm101 is the formation of large oligomeric rings of ∼14-fold symmetry with a molecular mass of ∼400 kDa. We asked how the ring structure is maintained, how this structure affects the mtDNA maintenance function of Mgm101, and whether the ring structure is actively regulated under stress conditions that increase mtDNA damage. In this study, we mainly focused on the cysteine residues that may potentially be involved in the redox-based remodeling of the Mgm101 rings. Mgm101 contains five cysteines. Cys-181, Cys-208, and Cys-240 are not absolutely conserved through evolution, whereas Cys-216 and Cys-217 are remarkably conserved among Mgm101 homologs from yeasts, filamentous fungi, and lower animals (e.g. Nematostella vectensis) (Fig. 1A). By using the Phyre2 (Protein Homology/analogy Recognition Engine) program (38), the core domain of Mgm101 (Ala-115–Val-237) can be modeled to the N-terminal ssDNA binding domain of the human Rad52 protein with an overall 97% confidence (39, 40). In this structural model, Cys-216 and Cys-217 are projected at the end of the α-helix 3, which is situated at the bottom of the putative ssDNA binding groove (Fig. 1B). All five cysteines seemed to be accessible to thiol-modifying agents. By mass spectrometry, modified Mgm101 with the addition of 1–5 NEM molecules was readily identifiable (Fig. 1C).
FIGURE 1.
Cys-216 and Cys-217 are critical for mtDNA maintenance in vivo. A, sequence similarities between Mgm101 homologs from fungi (Saccharomyces cerevisiae, Kluyveromyces lactis, Schizosaccharomyces pombe, Aspergillus nidulans, Neurospora crassa and Ustilago maydis), Dictyostelium discoideum and lower animals represented by Nematostella vectensis. The cysteines residues are highlighted in red. B, solid ribbon structure of Mgm101(115–237) modeled on the N-terminal ssDNA binding domain of human Rad52 (Protein Data Bank code 1H2I). The projected positions of Cys-181, Cys-208, Cys-216, and Cys-217 are represented by scaled balls and sticks. C, mass spectrometry analysis showing the modification of Mgm101 by NEM. D, stability of the ρ+ genome in the mgm101-1ts mutant expressing the various mutant alleles of MGM101. The mgm101-1ts mutant was transformed with a centromeric vector expressing the indicated alleles. Ura+ transformants were plated on YPD medium and incubated at 35 °C. Petite colonies were scored as white colonies in contrast to the red ρ+ colonies due to the presence of the ade2 mutation in the host strain. The data are the averaged petite frequency of at least three independent transformants. Error bars represent standard deviations. E, meiotic analysis showing that mgm101C216A and mgm101C217A failed to complement the null mgm101Δ::kan allele for respiratory growth on ethanol plus glycerol medium at 30 °C. Four complete tetrads independently segregating the null mgm101Δ::kan and the mgm101C216A or mgm101C217A allele were dissected on complete glucose medium. The meiotic segregants were then replica-plated onto G418, −Ura, and ethanol plus glycerol medium. G418R marks the presence of the null mgm101Δ::kan allele. Ura+ marks the mgm101C216A and mgm101C217A alleles. Circled are the meiotic spores that cosegregate mgm101Δ::kan with mgm101C216A or mgm101C217A.
We next substituted the five cysteines with alanine. The mutant alleles were first examined for their mtDNA maintenance activity in the temperature-sensitive mgm101-1ts mutant. As shown in Fig. 1D, transformants carrying the control vector are completely converted into petite colonies at the nonpermissive temperature (35 °C). Petite formation was suppressed by expressing the mgm101C181A, mgm101C208A, and mgm101C240A alleles, suggesting that these three cysteines are not critical for mtDNA maintenance. Interestingly, substitution with alanine at Cys-216 and Cys-217 significantly affected the capability of the protein in suppressing mgm101-1ts. Moreover, the mgm101C216A/C217A double mutant was found to completely lose the complementing activity. The additive effect suggests that the action of Cys-216 and Cys-217 does not involve the formation of a vicinal disulfide bond. Furthermore, it is unlikely that Cys-216 and Cys-217 affect Mgm101 function by forming disulfide bonds with Cys-181 or Cys-208, as a C181A/C208A double mutant has little effect on mtDNA stability.
We found that the mgm101C216A and mgm101C217A alleles exhibited a much more severe defect when tested in an mgm101 null mutant (Fig. 1E). After chromosomal integration into a diploid strain in which one copy of the wild-type MGM101 is disrupted by the insertion of the kan (G418R) marker, meiotic segregants expressing the mutant alleles (marked by URA3) in the mgm101Δ::kan background failed to show respiratory growth on the nonfermentable ethanol plus glycerol medium. The data indicate that Cys-216 and Cys-217 are crucial for Mgm101 function in vivo. The mild phenotype of the C216A and C217A alleles in the mgm101-1ts mutant may result from an inter-subunit complementation with the temperature-sensitive allele. Cells expressing only mgm101C181A, mgm101C208A, and mgm101C240A were found to be respiratory-competent (data not shown).
Oligomerization and Filamentation by the Alanine Variants
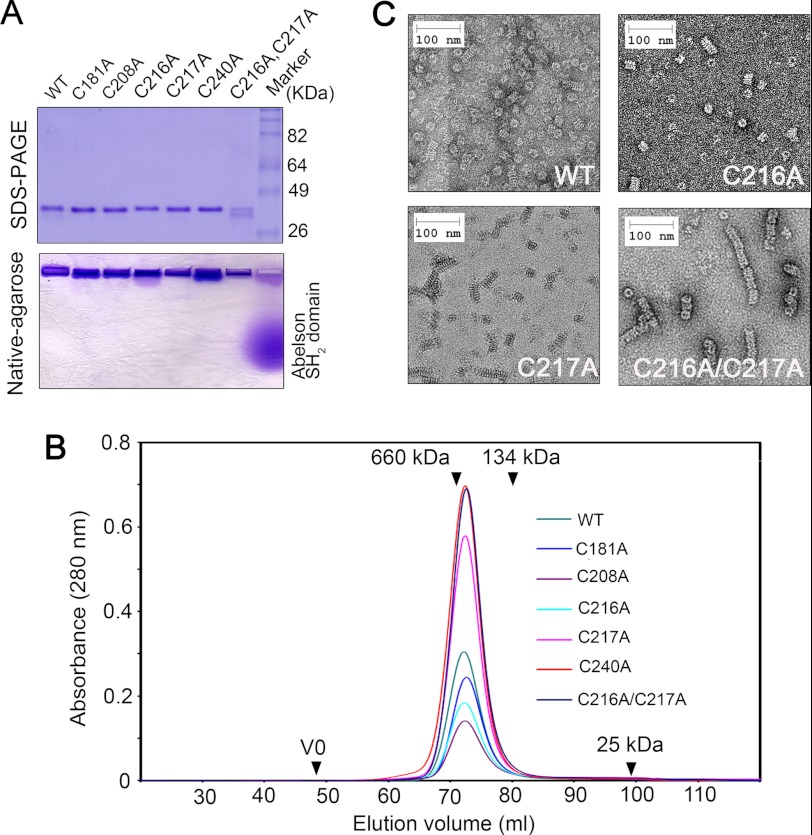
By using the MBP-tagging strategy, we expressed and purified the five alanine variants as well as the C216A/C217A double mutant from E. coli. All the proteins were expressed at high yields and remained soluble after cation exchange and size exclusion chromatography. Like the wild-type Mgm101, the mutant proteins could hardly migrate into a native agarose gel, suggesting that they all form large oligomers (Fig. 2A). In size exclusion chromatography, the mutant proteins were indistinguishable from the wild type. They all formed monodisperse peaks corresponding to a molecular mass of ∼400 kDa (Fig. 2B). It can therefore be concluded that the substitution of the five cysteines by alanine does not significantly affect the gross oligomeric organization of Mgm101.
FIGURE 2.
Characterization of the mutant Mgm101 with the substitution of the five cysteines by alanine. A, upper panel, SDS-PAGE showing the purified Mgm101, Mgm101C181A, Mgm101C208A, Mgm101C216A, Mgm101C217A, Mgm101C240A, and Mgm101C216A/C217A. Lower panel, native agarose gel electrophoresis showing that Mgm101 and its mutant variants migrate poorly toward the cathode. 10 μg of purified proteins were loaded on the gel. 10 μg of the positively charged Abelson Src homology 2 domain was included as control. B, size exclusion chromatography of purified Mgm101 variants on a Superose 6 column. V0, void volume. C, negative stain transmission electron microscopy showing that the C216A/C217A double mutant forms extended filaments after 4 weeks of incubation at 4 °C.
By using negative stain electron microscopy, we found that differences between the mutant and the wild-type proteins became obvious after 4 weeks of incubation at 4 °C. The C217A and C216A mutants appeared to have slightly increased tendency of forming short filaments relative to the wild type (Fig. 2C). Our previous studies have shown that the Mgm101 filaments are not formed by simple stacking of the rings and that there is clearly a condensed helical structure in these macromolecules (10). Interestingly, the C216A/C217A double mutant formed extensive filaments under the same conditions (Fig. 2C). This observation suggests that C216A and C217A may have subtle effect on the structural organization of the Mgm101 rings, which cannot be detected in freshly prepared samples. The additive effect between C216A and C217A on the higher order structure correlates with the synthetic functional deficiency in vivo (see Fig. 1D). It appears that the engagement in filament growth is unfavorable for Mgm101 activity in vivo. The detected differences in filament growth do not seem to have dramatic effect on the ssDNA binding activity in vitro. The fresh and aged samples of Mgm101 have a Kd of 235.12 ± 75.69 and 222.83 ± 63.82 nm, whereas the fresh and aged Mgm101C216A/C217A have a Kd of 248.73 ± 103.10 and 251.72 ± 93.41 nm, respectively.
Substitutions with Aspartic and Glutamic Acids Lead to the Aggregation and Functional Inactivation of Mgm101
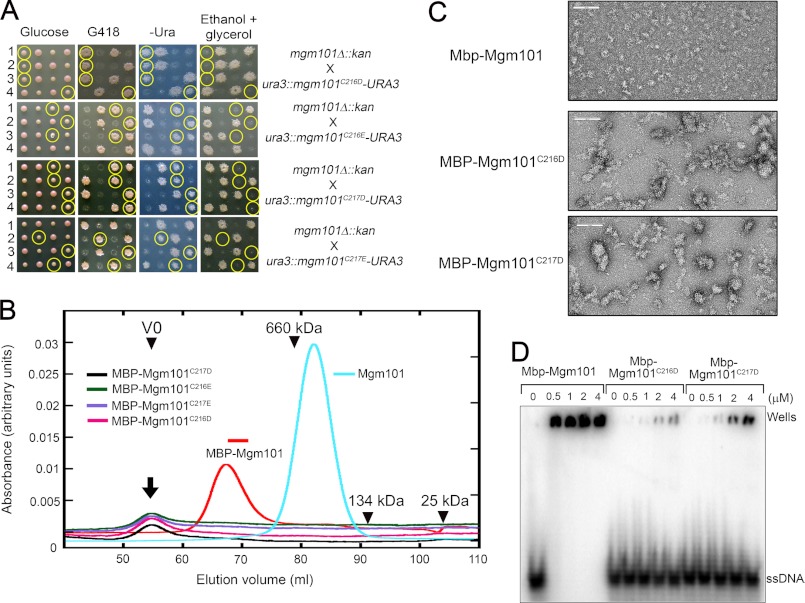
Based on their critical role for Mgm101 function in vivo and the formation of the super-stable structures by the alanine substitutions at Cys-216 and Cys-217, we hypothesized that this vicinal cysteine pair may be important for regulating the higher order structural organization of the protein. Modifications of Cys-216 and/or Cys-217 by processes such as oxidation may introduce negative charges that remodel the Mgm101 rings into a conformation favorable for DNA binding. To test whether the introduction of charged amino acids in these sites affects ring stability, we mutated Cys-216 and Cys-217 to the negatively charged aspartic and glutamic acids. Surprisingly, we found that the C216D, C217D, C216E, and C217E mutants are all defective in mtDNA maintenance. The mutant alleles failed to complement the null allele of MGM101 in the meiotic assay (Fig. 3A).
FIGURE 3.
Characterization of the mutant Mgm101 with the substitution of Cys-216 and Cys-217 by aspartic and glutamic acids. A, meiotic analysis showing that the mgm101C216D, mgm101C216E, mgm101C217D, and mgm101C217E alleles are defective in complementing the null mgm101Δ::kan allele for respiratory growth on ethanol plus glycerol medium at 30 °C. Details are described as in Fig. 1E. Circled are the meiotic spores that cosegregate mgm101Δ::kan with the mutant alleles. B, size exclusion chromatography of purified MBP-Mgm101C216D, MBP-Mgm101C216E, MBP-Mgm101C217D, and MBP-Mgm101C217E. The MBP-Mgm101 fusion and the wild-type Mgm101 are included as controls. C, negative stain transmission electron microscopy showing that MBP-Mgm101C216D and Mgm101C217D form unstructured aggregates, in contrast to MBP-Mgm101 that forms rather homogeneous particles. The scale bar, 100 nm. D, electrophoretic mobility shift assays showing that MBP-Mgm101C216D and MBP-Mgm101C217D have a weak DNA binding activity compared with MBP-Mgm101.
We prepared the C216D, C217D, C216E, and C217E mutant proteins in an MBP-fused form and attempted to characterize their biochemical properties. It was found that none of the mutant proteins could be recovered after releasing from MBP by proteolytic cleavage, suggesting that the mutant proteins are unstable in solution. We then decided to directly characterize the fusion proteins and to have their biochemical properties compared with the wild-type control. By using size exclusion chromatography, MBP-Mgm101 typically eluted as a monodisperse peak of ∼940 kDa (Fig. 3B). In contrast, all four fusion proteins with the aspartic and glutamic acid substitutions eluted in the void volume, which suggests the formation of larger molecular species. By negative stain electron microscopy, we found that MBP-Mgm101 forms relatively homogeneous globular particles with a diameter of ∼30 nm (Fig. 3C). No clear ring structure was visible, which is normally seen with untagged Mgm101. The tagging by MBP, which has a molecular mass of 42 kDa compared with 28 kDa for Mgm101, likely masks the central channel of Mgm101. More importantly, we observed that the mutant fusions, as represented by MBP-Mgm101C216D and MBP-Mgm101C217D indeed formed irregularly shaped aggregates. Although MBP-Mgm101 retains a robust ssDNA binding activity, with a Kd of 460.97 ± 75.66 nm compared with 234.32 ± 97.83 nm for Mgm101, the activity of MBP-Mgm101C216D and MBP-Mgm101C217D is reduced by 73- and 38-fold, with Kd of 33.43 ± 26.28 and 17.48 ± 5.25 μm, respectively (Fig. 3D). These data clearly indicate that the introduction of the acidic amino acids at Cys-216 or Cys-217 drastically alters the conformation of the protein, which affects the maintenance of a stable ring structure and reduces protein stability and DNA binding activity.
Mutations to Serine at Cys-216 and Cys-217 Partially Disassemble the Mgm101 Rings
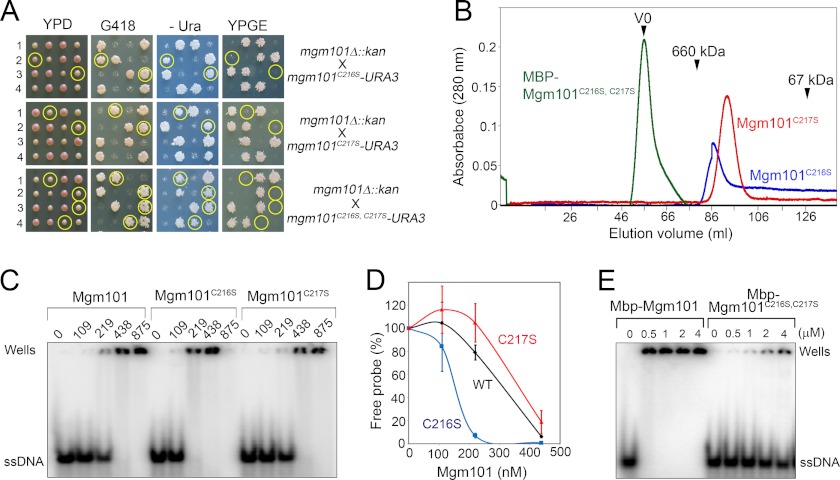
The drastic effect of the acidic amino acids on the Mgm101 ring structure prompted us to mutate Cys-216 and Cys-217 to serine, which is polar but uncharged. We assumed that a polar amino acid may partially disassemble the rings. Again, we found that these mutations severely affected mtDNA maintenance in vivo when nascent spores expressing the mutant alleles are grown on YPD medium (Fig. 4A). Respiratory-competent cells can be recovered from the C216S spores only when they were dissected on nonfermentable carbon sources (data not shown). We prepared the two mutant proteins and both remained stable in solution. Size exclusion chromatography revealed that the C216S mutant forms a peak of ∼400 kDa corresponding to ∼14 subunit oligomers as seen in the wild-type protein (Fig. 4B). In addition, a significant portion of the C216S mutant protein is present in a continuous tail of smaller sizes, which can be interpreted as partially disassembled Mgm101 rings/oligomers. The C217S mutant forms a major peak slightly smaller than 400 kDa, but the presence of the continuous tail, which is suggestive of ring disassembly, is also visible. We have not been able to recover the Mgm101C216S/C217S double mutant in a soluble form. Drastic structural alterations by the double mutation are obvious even at its MBP-fused form, which eluted in the void volume in size exclusion chromatography (Fig. 4B). These data suggest that the C216S and C217S mutations can partially disassemble the Mgm101 rings. The C216S and C217S mutations synergistically destabilize the protein and cause aggregation.
FIGURE 4.
Ring disassembly and ssDNA binding activity of the C216S and C217S mutants. A, meiotic analysis showing that spores expressing only mgm101C216S, mgm101C217S, or mgm101C216S/C217S become respiratory-deficient after dissection on YPD. Details are described as in Fig. 1E. Circled are the meiotic spores that cosegregate mgm101Δ::kan with the mutant alleles. B, size exclusion chromatography of purified Mgm101C216S, Mgm101C217S, and MBP-Mgm101C216S/C217S. C, electrophoretic mobility shift assays showing the ssDNA binding activity of Mgm101C216S and Mgm101C217S. Note that at high Mgm101/ssDNA ratios, the protein-DNA complexes formed by Mgm101C216S moved toward the anode (data not shown) instead of being trapped in the wells. D, graphic representation of the results from C. Error bars represent standard deviations of three independent experiments. E, electrophoretic mobility shift assays showing the ssDNA binding activity of MBP-Mgm101C216S/C217S.
EMSA revealed that the C217S mutant has an ssDNA binding activity close to that of the wild-type protein, with a Kd of 318.26 ± 157.97 nm compared with 234.32 ± 97.83 nm for the control. The C216S mutant seems to have a moderate increase in DNA binding, with a Kd value of 96.74 ± 40.79 nm (Fig. 4, C and D). As expected, the MBP-Mgm101C216S/C217S aggregates bind poorly to DNA (Fig. 4E), with a Kd of 8.91 ± 2.58 μm compared with 460.97 ± 75.66 nm for the MBP-Mgm101 control. In summary, subtle changes in the ring organization such as in the case of C216S seem to stimulate moderately DNA binding activity, whereas excessive structural changes in the C216S/C217S double mutant has the opposite effect.
Structural Analysis of Mgm101 and Its Mutant Variants by SAXS
To gain further insight into the structural dynamics of Mgm101, we used small angle x-ray scattering to establish the molecular envelope of Mgm101 and its mutant variants. As the aspartic and glutamic acid mutants are unstable in solution and the serine mutants form heterogeneous oligomeric species (see Fig. 4B), these proteins were precluded from the SAXS analysis. Our efforts were therefore focused on potential structural differences between the wild-type and the over-filamenting C216A/C217A double mutant. The C216A and C217A single mutants were not included in the analysis because of their subtle difference from the wild type with regard to the filamentation phenotype.
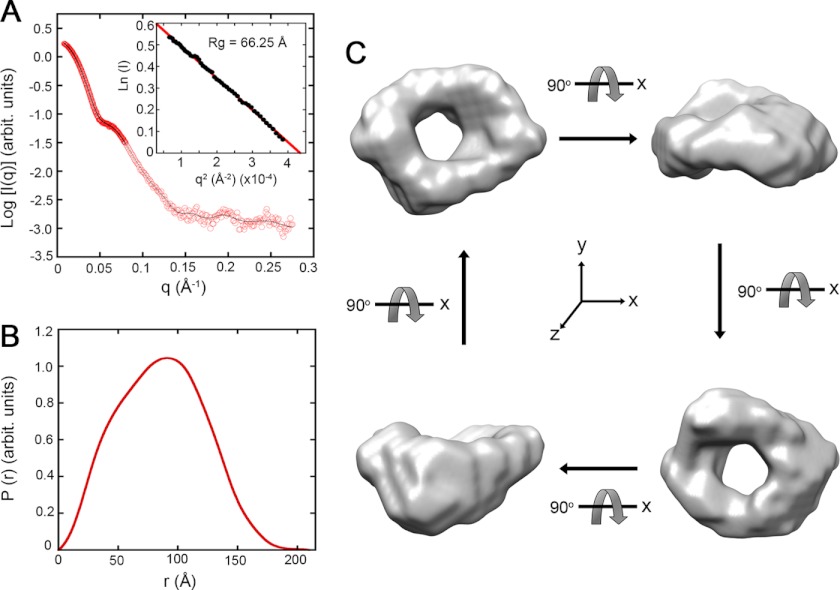
Fig. 5A shows the characteristic x-ray scattering profile of freshly prepared Mgm101 after solvent subtraction. No significant inter-particle interference and protein aggregation in the Mgm101 solution was observed as reflected by the linear relationship in the Guinier region of the small angle data (Fig. 5A, inset). The radius of gyration (Rg) of Mgm101 was found to be 66.25 ± 0.49 Å, and the pair distance distribution function (P(r)) was generated by the indirect Fourier transformation method using the program GNOM (Fig. 5B) (36). The calculated P(r) displayed an excellent total estimate score of 0.98. The maximum intermolecular distance (Dmax) of Mgm101 was estimated to be ∼210 Å, which is close to the diameter of ∼200 Å for the Mgm101 rings deduced by transmission electron microscopy (10).
FIGURE 5.
Solution structure of Mgm101 determined by SAXS. A, scattering intensity I(q) of Mgm101 at 5 mg/ml as a function of the scattering vector q. Experimental data were fitted with the GNOM program (fit, solid line). The inset shows the linear fit of the scattering data in the Guinier region. B, pair distance distribution (P(r)) of Mgm101. C, surface envelope model of Mgm101 generated from SAXS data. The surface envelope is an average of 10 independent models generated by the program DAMAVER. The four structures shown are 90° rotations of the same model generated with the software Chimera.
To generate the envelope structure of Mgm101, bead models from 10 independent ab initio simulations were averaged and filtered on the basis of occupancy (Fig. 5C). The final model clearly shows a ring structure with a large central channel consistent with the results from negative staining electron microscopy (10). The envelope structure also shows that the surface of Mgm101 rings is not completely flat. The precise nature of the protruding structures remains unclear. One possibility is that the large N-terminal domain of Mgm101 may be unevenly distributed on the ring surface because of its structural flexibility.
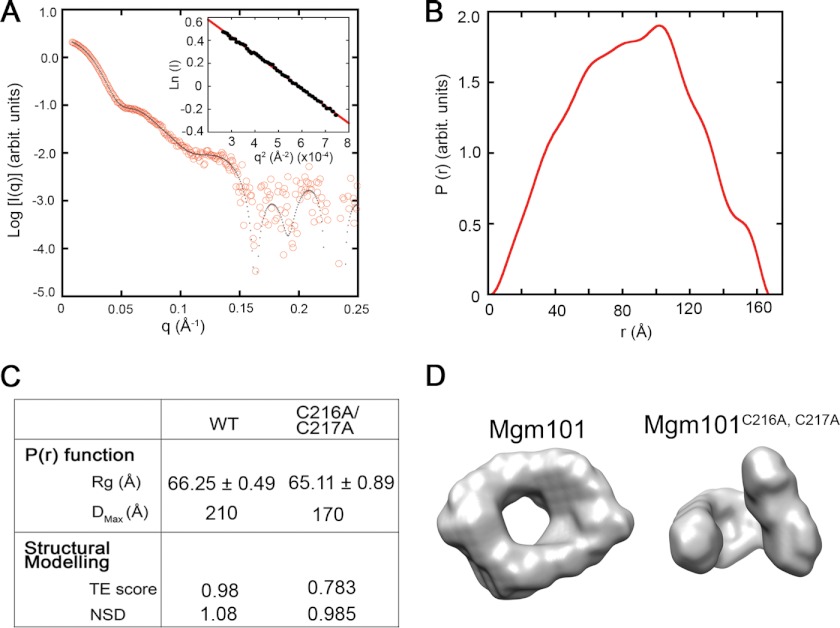
The freshly prepared C216A/C217A double mutant showed an x-ray scattering profile very similar to that of the wild-type Mgm101 (Fig. 6, A and B). As summarized in Fig. 6C, the mutant protein has a slightly lower radius of gyration (Rg). It has a Dmax of 170 Å, compared with 210 Å for the wild type. Structural modeling of the mutant produced high quality total estimate score and NSD value as seen with the wild type. Interestingly, instead of being a closed ring like the wild-type protein, Mgm101C216A/C217A seems to adopt a “broken-washer” conformation. It is possible that this specific conformation provides the free ends for the growth of filaments during the extended incubation as detected by negative stain electron microscopy (see Fig. 2D).
FIGURE 6.
Solution structure of Mgm101C216A/ C217A in comparison with the wild type. A, scattering intensity I(q) of Mgm101C216A/C217A as a function of the scattering vector q. Experimental data were fitted with the GNOM program (fit, solid line). The inset shows the linear fit of the scattering data in the Guinier region. B, pair distance distribution (P(r)) of Mgm101C216A/C217A. C, comparison of SAXS data between Mgm101 and Mgm101C216A/C217A. D, surface envelope model of Mgm101C216A/C217A generated from SAXS data in comparison with that of Mgm101.
DISCUSSION
Single strand annealing proteins in the Rad52 superfamily are present across all domains of life. The prototypical members of this protein family are probably those from the bacteriophages, as represented by Redβ and Erf from the bacteriophages λ and P22, RecT from the prophage rac, and Sak from the lactococcal phage ul36. These proteins functionally promote recombination by single strand annealing, an operation mode independent of the RecA/Rad51 recombinase. This is particularly the case for the phage proteins and the Rad59-type proteins in eukaryotes that lack an identifiable RecA/Rad51-interacting domain (41). Structurally, the Rad52 family proteins are all predicted to have a typical β-β-β-α fold (β3-β4-β5-α3 in Fig. 1B). Another characteristic feature is that these proteins all form homo-oligomeric rings of 10–14-fold symmetry (19, 26, 27). We recently found that the Mgm101 protein in yeast mitochondria share biochemical, structural, and functional similarities with the Rad52-type recombination proteins (10). Similar proteins were also identified in mitochondria from plants and lower animals (10, 42, 43). Like its bacteriophage counterparts, Mgm101 forms oligomeric rings. The Mgm101 rings have an ∼14-fold symmetry with a diameter of ∼200 Å as revealed by negative stain electron microscopy. In this study, we have been able to establish the molecular envelope of Mgm101 by SAXS analysis (Fig. 5). This provides the first solution structure of the Rad52-type recombination proteins. SAXS analysis is commonly used to establish macromolecular assemblies, and it is particularly useful to provide complementary evidence to other low resolution structural analyses established by techniques, including electron microscopy. The SAXS models of Mgm101 and Mgm101C216A/C217A have NSD values of 1.08 and 0.985, respectively. The NSD value evaluates the overall variance among 10 or more independently calculated models and discards models that lie outside an acceptable discrepancy. Typically, shape reconstructions of flat or disc-like objects are more difficult, and give relatively higher NSD values than for globular objects (37). Based on the NSD values and the available data from analysis by electron microscopy, we consider that the established Mgm101 envelopes are reasonably acceptable, although caution needs to be taken when building any ab initio structural models. The SAXS analysis confirms the presence of a large central channel of ∼60 Å within the Mgm101 ring. The ring formation appears to be an inherent property of the protein under physiological conditions rather than an artifact of negative staining.
The molecular determinants and functional implications of ring formation by the Rad52-type strand annealing proteins are largely unclear. The essential role of Mgm101 for mtDNA maintenance has allowed us to address these questions by combining in vivo functional complementation and in vitro biochemical analysis of the mutant proteins. We identified a vicinal cysteine pair, Cys-216/Cys-217, that plays a crucial role in modulating higher order structural organization and the in vivo function of Mgm101. Mutations to alanine accelerated the formation of macromolecular filaments in vitro. Whether Mgm101 forms filaments in vivo is a challenging question for future studies. SAXS analysis supported a broken ring structure in the freshly prepared Mgm101C216A/C217A. This characteristic structure could provide the free ends for Mgm101 polymerization and the formation of helical filaments. The cysteine to alanine mutations may contort the monomer-to-monomer binding interface, and the accumulation of 14 contortions impairs complete ring formation. We previously reported a 50-Å helical pitch of the Mgm101 filaments supporting our hypothesis of an altered monomer interface (10). However, substitutions of Cys-216 and Cys-217 by the uncharged but polar serine partially disassembled Mgm101 into oligomers of lower molecular mass. Substitution of Cys-216 and Cys-217 with the negatively charged aspartic and glutamic acids drastically destabilized Mgm101, as manifested by the formation of unstructured large aggregates. These data suggest that the ring structure of Mgm101 needs to be delicately maintained for its in vivo function. We favor the model that in contrast to the hydrophobic alanine the polar serine may mildly inhibit polymerization. As a consequence, the mutant Mgm101 forms smaller oligomers with or without a full ring structure. Substitutions of Cys-216 and Cys-217 by aspartic and glutamic acids may severely alter the conformation of Mgm101. This may destabilize the protein in solution leading to aggregate formation. Thus, Cys-216 and Cys-217 are likely located at positions critical for modulating the dynamics of higher order structures. Substitutions by the hydrophobic and small alanine engage the protein into a conformation favoring polymerization and filamentation, whereas the introduction of polar or charged amino acids tends to disassemble or destabilize the oligomers.
All the substitution mutants so far studied, including alanine, serine, aspartic, and glutamic acids, rapidly lose mtDNA. This is not surprising for the aspartic acid mutants as they form aggregates incapable of binding to DNA. However, the DNA binding activity of the rings formed by the alanine variants is little affected compared with the wild type. Moreover, ssDNA binding is moderately increased in the C216S mutant. Based on their DNA binding activity, it would be reasonably expected that the alanine and serine alleles may be functionally active in mtDNA maintenance. Contrary to this expectation, we found that these mutants rapidly lose mtDNA. This raises the question as to whether Mgm101 with the unmodified cysteines at the positions Cys-216 and Cys-217 is active in vivo. It is possible that these cysteines need specific post-translational modifications in mitochondria, which reconfigures the ring structure and stimulates the DNA binding activity. In support of this hypothesis, it is important to note that the presence of the vicinal Cys-216 and Cys-217 pair is unique to the Mgm101 homologs in mitochondria. These residues are not conserved in Rad52 family members from bacteriophages and eukaryotic cells. Put differently, these sites do not have to be cysteines to fulfill the Rad52-type function. By replacing these two amino acids with cysteines during evolution, Mgm101 might have acquired a novel property. The DNA binding activity of the protein with unmodified cysteines, just like the alanine and serine variants, may be too low to be effective in vivo. Subtle modifications to the cysteines may be required to activate DNA binding and mtDNA repair function. The crystal structure of the single strand annealing domain of Rad52, homologous to Mgm101 and its bacteriophage orthologs, have been previously solved (39, 40). It was proposed that ssDNA binds to the outer rim of the undecameric rings formed by Rad52. Our previous work has shown that once bound to ssDNA, the Mgm101 rings become undetectable (10). A similar situation has been observed with the Sak protein from the lactococcal phage ul36 (27). It is likely that DNA binding may be initiated by ring-ssDNA interaction. Specific modification of Cys-216 and Cys-217 may be necessary to facilitate the mobilization of Mgm101 and to form nucleoprotein filaments competent for homology search and strand annealing. At this stage, we cannot exclude the possibility that modifications may be critical for activating an event upstream or downstream of DNA binding.
Among the Mgm101 variants, C216S was found to have a moderate increase in ssDNA binding activity. The C216S allele also significantly disassembles the ring structure, which is yet not sufficiently severe for inducing aggregation. Furthermore, C216S is also the only mutant that retains a residual activity in mtDNA maintenance when the cells are maintained on a nonfermentable carbon source (data not shown). The increased DNA binding activity and the partial retention of in vivo function in the C216S mutant suggest that an adequately introduced electrophoretic polarity is in favor of weakening ring rigidity and enhancing Mgm101 function. Substitution by serine at Cys-217 is probably too weak to be effective. A synergistic effect between C216S and C217S may occur, which introduces drastic changes and renders the protein unstable like the aspartic and glutamic acid mutants. Together with the accessibility of the cysteine to NEM treatment, we speculate that Cys-216 and/or Cys-217 may be oxidized in response to oxidative stress to negatively charged derivatives (e.g. sulfenic acid). The modified products may be more effective than serine in weakening inter-subunit interaction and allowing the mobilization of the protein for DNA binding. In an attempt to test this model, we used various thiol-modifying agents to see whether they increase the DNA binding activity of Mgm101. We found that neither hydrogen peroxide nor phenylarsine oxide, which cross-links vicinal thiol groups, directly stimulates DNA binding activity in vitro. S-Nitrosylation using S-nitrosoglutathione as a nitric oxide donor also failed to stimulate DNA binding activity (data not shown). These observations suggest that a redox mediator (e.g. peroxiredoxin, metal ions, etc.) may be required for oxidizing Mgm101 and for channeling electrons to hydrogen peroxide or other reactive oxygen species in vivo. The ultimate capture of the cysteine modifications would need the identification of the putative mediator, the in vitro reconstitution of the electron transfer events, and the identification of the modified cysteines by mass spectrometry or by x-ray crystallography. In this regard, it is also important to note that cysteine is subject to many other forms of modifications, which includes deprotonation, sulfuration, S-thiolation, metal thiolation, acetylation, prenylation, etc. (44). Further studies are necessary to clarify how Cys-216 and Cys-217 are exactly modified in mitochondria and whether this mechanism is used as a switch for regulating the mtDNA repair function of Mgm101 under stress conditions.
Acknowledgments
We thank the Cornell High Energy Synchrotron Source (CHESS) and the Macromolecular Diffraction at CHESS (MacCHESS) facility for beam time; Richard Gillilan for advice in SAXS data collection and analysis; Stewart Loh for the use of FPLC, and Jeung-Hoi (Jenny) Ha for help in the analysis of DNA binding data. CHESS is supported by the National Science Foundation and NIGMS, National Institutes of Health, and the MacCHESS resource is supported by NCRR, National Institutes of Health.
This work was supported, in whole or in part, by National Institutes of Health Grants R01AG023731 and R01GM058600 (to X. J. C. and S. W.).
- ssDNA
- single-stranded DNA
- MBP
- maltose-binding protein
- NEM
- N-ethylmaleimide
- SAXS
- small angle x-ray scattering
- NSD
- normalized spatial discrepancy.
REFERENCES
- 1. Chen X. J., Guan M. X., Clark-Walker G. D. (1993) MGM101, a nuclear gene involved in maintenance of the mitochondrial genome in Saccharomyces cerevisiae. Nucleic Acids Res. 21, 3473–3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Clark-Walker G. D., Chen X. J. (1996) A vital function for mitochondrial DNA in the petite-negative yeast Kluyveromyces lactis. Mol. Gen. Genet. 252, 746–750 [DOI] [PubMed] [Google Scholar]
- 3. Zuo X. M., Clark-Walker G. D., Chen X. J. (2002) The mitochondrial nucleoid protein, Mgm101p, of Saccharomyces cerevisiae is involved in the maintenance of rho+ and ori/rep-devoid petite genomes but is not required for hypersuppressive rho− mtDNA. Genetics 160, 1389–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zuo X., Xue D., Li N., Clark-Walker G. D. (2007) A functional core of the mitochondrial genome maintenance protein Mgm101p in Saccharomyces cerevisiae determined with a temperature-conditional allele. FEMS Yeast Res. 7, 131–140 [DOI] [PubMed] [Google Scholar]
- 5. Meeusen S., Tieu Q., Wong E., Weiss E., Schieltz D., Yates J. R., Nunnari J. (1999) Mgm101p is a novel component of the mitochondrial nucleoid that binds DNA and is required for the repair of oxidatively damaged mitochondrial DNA. J. Cell Biol. 145, 291–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kaufman B. A., Newman S. M., Hallberg R. L., Slaughter C. A., Perlman P. S., Butow R. A. (2000) In organello formaldehyde cross-linking of proteins to mtDNA. Identification of bifunctional proteins. Proc. Natl. Acad. Sci. U.S.A. 97, 7772–7777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Itoh K., Izumi A., Mori T., Dohmae N., Yui R., Maeda-Sano K., Shirai Y., Kanaoka M. M., Kuroiwa T., Higashiyama T., Sugita M., Murakami-Murofushi K., Kawano S., Sasaki N. (2011) DNA packaging proteins Glom and Glom2 coordinately organize the mitochondrial nucleoid of Physarum polycephalum. Mitochondrion 11, 575–586 [DOI] [PubMed] [Google Scholar]
- 8. Zinn A. R., Pohlman J. K., Perlman P. S., Butow R. A. (1988) In vivo double strand breaks occur at recombinogenic G + C-rich sequences in the yeast mitochondrial genome. Proc. Natl. Acad. Sci. U.S.A. 85, 2686–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gaillard C., Strauss F., Bernardi G. (1980) Excision sequences in the mitochondrial genome of yeast. Nature 283, 218–220 [DOI] [PubMed] [Google Scholar]
- 10. Mbantenkhu M., Wang X., Nardozzi J. D., Wilkens S., Hoffman E., Patel A., Cosgrove M. S., Chen X. J. (2011) Mgm101 is a Rad52-related protein required for mitochondrial DNA recombination. J. Biol. Chem. 286, 42360–42370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kowalczykowski S. C., Dixon D. A., Eggleston A. K., Lauder S. D., Rehrauer W. M. (1994) Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 58, 401–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. San Filippo J., Sung P., Klein H. (2008) Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77, 229–257 [DOI] [PubMed] [Google Scholar]
- 13. Symington L. S. (2002) Role of RAD52 epistasis group genes in homologous recombination and double strand break repair. Microbiol. Mol. Biol. Rev. 66, 630–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shi I., Hallwyl S. C., Seong C., Mortensen U., Rothstein R., Sung P. (2009) Role of the Rad52 amino-terminal DNA binding activity in DNA strand capture in homologous recombination. J. Biol. Chem. 284, 33275–33284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sugiyama T., Kantake N., Wu Y., Kowalczykowski S. C. (2006) Rad52-mediated DNA annealing after Rad51-mediated DNA strand exchange promotes second ssDNA capture. EMBO J. 25, 5539–5548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nimonkar A. V., Sica R. A., Kowalczykowski S. C. (2009) Rad52 promotes second-end DNA capture in double-stranded break repair to form complement-stabilized joint molecules. Proc. Natl. Acad. Sci. U.S.A. 106, 3077–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rothenberg E., Grimme J. M., Spies M., Ha T. (2008) Human Rad52-mediated homology search and annealing occurs by continuous interactions between overlapping nucleoprotein complexes. Proc. Natl. Acad. Sci. U.S.A. 105, 20274–20279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pâques F., Haber J. E. (1999) Multiple pathways of recombination induced by double strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63, 349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Passy S. I., Yu X., Li Z., Radding C. M., Egelman E. H. (1999) Rings and filaments of β protein from bacteriophage λ suggest a superfamily of recombination proteins. Proc. Natl. Acad. Sci. U.S.A. 96, 4279–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Iyer L. M., Koonin E. V., Aravind L. (2002) Classification and evolutionary history of the single strand annealing proteins, RecT, Redβ, ERF, and RAD52. BMC Genomics 3, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lopes A., Amarir-Bouhram J., Faure G., Petit M. A., Guerois R. (2010) Detection of novel recombinases in bacteriophage genomes unveils Rad52, Rad51, and Gp2.5 remote homologs. Nucleic Acids Res. 38, 3952–3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lim S. I., Min B. E., Jung G. Y. (2008) Lagging strand-biased initiation of red recombination by linear double-stranded DNAs. J. Mol. Biol. 384, 1098–1105 [DOI] [PubMed] [Google Scholar]
- 23. Poteete A. R. (2008) Involvement of DNA replication in phage λ Red-mediated homologous recombination. Mol. Microbiol. 68, 66–74 [DOI] [PubMed] [Google Scholar]
- 24. Sung P. (1997) Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem. 272, 28194–28197 [DOI] [PubMed] [Google Scholar]
- 25. New J. H., Sugiyama T., Zaitseva E., Kowalczykowski S. C. (1998) Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature 391, 407–410 [DOI] [PubMed] [Google Scholar]
- 26. Poteete A. R., Sauer R. T., Hendrix R. W. (1983) Domain structure and quaternary organization of the bacteriophage P22 Erf protein. J. Mol. Biol. 171, 401–418 [DOI] [PubMed] [Google Scholar]
- 27. Ploquin M., Bransi A., Paquet E. R., Stasiak A. Z., Stasiak A., Yu X., Cieslinska A. M., Egelman E. H., Moineau S., Masson J. Y. (2008) Functional and structural basis for a bacteriophage homolog of human RAD52. Curr. Biol. 18, 1142–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stasiak A. Z., Larquet E., Stasiak A., Müller S., Engel A., Van Dyck E., West S. C., Egelman E. H. (2000) The human Rad52 protein exists as a heptameric ring. Curr. Biol. 10, 337–340 [DOI] [PubMed] [Google Scholar]
- 29. Ling F., Makishima F., Morishima N., Shibata T. (1995) A nuclear mutation defective in mitochondrial recombination in yeast. EMBO J. 14, 4090–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ling F., Morioka H., Ohtsuka E., Shibata T. (2000) A role for MHR1, a gene required for mitochondrial genetic recombination, in the repair of damage spontaneously introduced in yeast mtDNA. Nucleic Acids Res. 28, 4956–4963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shibata T., Ling F. (2007) DNA recombination protein-dependent mechanism of homoplasmy and its proposed functions. Mitochondrion 7, 17–23 [DOI] [PubMed] [Google Scholar]
- 32. Van Dyck E., Foury F., Stillman B., Brill S. J. (1992) A single-stranded DNA-binding protein required for mitochondrial DNA replication in S. cerevisiae is homologous to E. coli SSB. EMBO J. 11, 3421–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kalifa L., Quintana D. F., Schiraldi L. K., Phadnis N., Coles G. L., Sia R. A., Sia E. A. (2012) Mitochondrial genome maintenance. Roles for nuclear nonhomologous end-joining proteins in Saccharomyces cerevisiae. Genetics 190, 951–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guinier A. (1939) La diffraction des rayons X aux très petits angles. Application l'étude de phénomènes ultramicroscopiques. Ann. Phys. (Paris) 12, 161–237 [Google Scholar]
- 35. Petoukhov M. V., Svergun D. I. (2007) Analysis of x-ray and neutron scattering from biomacromolecular solutions. Curr. Opin. Struct. Biol. 17, 562–571 [DOI] [PubMed] [Google Scholar]
- 36. Svergun D. I. (1992) Determination of the regularization parameter indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 37. Volkov V. V., Svergun D. I. (2004) Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36, 860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web. A case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 39. Kagawa W., Kurumizaka H., Ishitani R., Fukai S., Nureki O., Shibata T., Yokoyama S. (2002) Crystal structure of the homologous-pairing domain from the human Rad52 recombinase in the undecameric form. Mol. Cell 10, 359–371 [DOI] [PubMed] [Google Scholar]
- 40. Singleton M. R., Wentzell L. M., Liu Y., West S. C., Wigley D. B. (2002) Structure of the single-strand annealing domain of human RAD52 protein. Proc. Natl. Acad. Sci. U.S.A. 99, 13492–13497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bai Y., Symington L. S. (1996) A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 10, 2025–2037 [DOI] [PubMed] [Google Scholar]
- 42. Samach A., Melamed-Bessudo C., Avivi-Ragolski N., Pietrokovski S., Levy A. A. (2012) Identification of plant RAD52 homologs and characterization of the Arabidopsis thaliana RAD52-like genes. Plant Cell 23, 4266–4279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Janicka S., Kuhn K., Le Ret M., Bonnard G., Imbault P., Augustyniak H., Gualberto J. M. (2012) A RAD52-like single-stranded DNA-binding protein affects mitochondrial DNA repairs by recombination. Plant J., in press [DOI] [PubMed] [Google Scholar]
- 44. Nagahara N., Matsumura T., Okamoto R., Kajihara Y. (2009) Protein cysteine modifications. 1) Medical chemistry for proteomics. Curr. Med. Chem. 16, 4419–4444 [DOI] [PubMed] [Google Scholar]