Background: Grhl2 regulates cell-junction gene transcription in several epithelia but has not been fully characterized in lungs.
Results: In lung epithelial cells GRHL2 regulates cell-cell interaction genes, collective cell migration, and Nkx2-1 transcription. Conversely, NKX2-1 regulates transcription of Grhl2.
Conclusion: A Grhl2- and Nkx2-1-positive transcriptional loop coordinates morphogenesis and differentiation of lung epithelial cells.
Significance: This regulatory loop reinforces normal lung epithelial cell identity.
Keywords: Cell Adhesion, Cell Junctions, Cell Migration, Cell-Cell Interaction, Development, DNA Transcription, Gene Regulation, Lung, Transcription/Developmental Factors, Cell Shape
Abstract
The Grainyhead family of transcription factors controls morphogenesis and differentiation of epithelial cell layers in multicellular organisms by regulating cell junction- and proliferation-related genes. Grainyhead-like 2 (Grhl2) is expressed in developing mouse lung epithelium and is required for normal lung organogenesis. The specific epithelial cells expressing Grhl2 and the genes regulated by Grhl2 in normal lungs are mostly unknown. In these studies we identified the NK2-homeobox 1 transcription factor (Nkx2-1) as a direct transcriptional target of Grhl2. By binding and transcriptional assays and by confocal microscopy we showed that these two transcription factors form a positive feedback loop in vivo and in cell lines and are co-expressed in lung bronchiolar and alveolar type II cells. The morphological changes observed in flattening lung alveolar type II cells in culture are associated with down-regulation of Grhl2 and Nkx2-1. Reduction of Grhl2 in lung epithelial cell lines results in lower expression levels of Nkx2-1 and of known Grhl2 target genes. By microarray analysis we identified that in addition to Cadherin1 and Claudin4, Grhl2 regulates other cell interaction genes such as semaphorins and their receptors, which also play a functional role in developing lung epithelium. Impaired collective cell migration observed in Grhl2 knockdown cell monolayers is associated with reduced expression of these genes and may contribute to the altered epithelial phenotype reported in Grhl2 mutant mice. Thus, Grhl2 functions at the nexus of a novel regulatory network, connecting lung epithelial cell identity, migration, and cell-cell interactions.
Introduction
During lung organogenesis, the progenitor cell layer lining the developing airways maintains its epithelial characteristics while proliferating and differentiating into multiple cell types to form a functional lung (1). Genes that establish and maintain cell polarity and cell junctions must be coordinately regulated with cell-specific differentiation programs to generate multiple lung epithelial cell types (2–4).
In Drosophila, the CP2 family transcription factor Grainyhead (Grh) contributes to tracheal organogenesis by regulating the size of the apical membrane and morphogenesis of tracheal epithelial cells (5). In vertebrates, three homologs of Grh, Grainyhead like-1, -2, and -3 (Grhl1, Grhl2, and Grhl3), share high similarity in their DNA binding sequence and in the carboxyl terminal dimerization region but display a different pattern of expression in the lung and other epithelial tissues (6). Previous in situ hybridization analyses indicate that Grhl2 is the only family member that is highly expressed in distal lung epithelium throughout development, although the particular cells expressing Grhl2 have not been identified nor has its functional role in lung epithelium. Grhl1 and Grhl3, in contrast, are expressed in proximal lung epithelium until E15.5, but later their expression is reduced in bronchi/bronchioles and is undetectable in the alveolar lung epithelium (6).
Grhls seem to have conserved functions controlling cell shape, cell growth, cell proliferation, and cell fate (7–14). They maintain epithelial cell characteristics by regulating cell-cell junction genes including the desmosomal cadherin Desmoglein-1 (Dsg1) by Grhl1 and Claudin1 (Cldn1) and Occludin (Ocdn) by Grhl3. Recently, E-cadherin (Cdh1) and Cldn4 were identified as direct transcriptional targets of Grhl2. Grhl2 null mutant mice die by embryonic day E11.5 (15) due to defects in neural tube closure and defective apical junction complex composition in epithelial tissues. Expression patterns of Cdh1 and Cldn4 were drastically reduced in foregut endoderm and otic epithelium as well as in the surface ectoderm, indicating that apical junction genes are regulated in vivo by Grhl2. Due to the early lethality of Grhl2 null mutants, studies at later stages of development were not possible. Similarly, N-ethyl-N-nitrosourea (ENU)4-generated Grhl2 mutant mice die by E12.5 due to defects in neural tube closure and heart development (16). Apical junction gene expression in epithelial organs was also reduced. A few embryos that survived to E18.5 had smaller lungs, disorganized epithelial apical junctions, and collapsed alveolar sacs, suggesting a functional role for Grhl2 in lung development and regulation of lung epithelial genes.
Herein we identify genes regulated by Grhl2 in lung epithelial cells and provide evidence for a novel positive transcriptional feedback loop between Grhl2 and the homeobox transcription factor Nkx2-1 in embryonic lung. The critical role of Nkx2-1 in regulating epithelial cell proliferation and differentiation and of Grhl2 in regulating cell-cell interactions and epithelial structure suggest that the Grhl2-Nkx2-1-positive feedback loop may serve to differentiate and reinforce distal lung epithelial phenotypes.
EXPERIMENTAL PROCEDURES
Cell Lines and Tissues
E10 is a spontaneously immortalized adult mouse lung epithelial cell line (17) provided by Dr. A. Malkinson (University of Colorado) and Dr. R. J. Ruch (University of Toledo). MLE15 (18) is an adult mouse lung epithelial cell line provided by Dr. J. A. Whitsett (Cincinnati Children's Hospital Medical Center). Cells were cultured in conditions described previously (19). Lung tissues used in chromatin immunoprecipitation analysis and immunohistochemistry were dissected from CD1 mice (Charles River Laboratories). All experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee at Boston University Medical Center in accordance with National Institute of Health guidelines.
Isolation and Culture of Murine Alveolar Type II Cells
Cells were isolated from SFTPC-Gfp mice provided by Dr. J. K. Heath (University of Birmingham, Birmingham, UK) (20) using a previously described method (21) with some modifications. SFTPC-Gfp mice were anesthetized, and tracheae were exposed and cannulated with a 20-gauge luer stub adapter. Lungs were perfused with 10 ml of 1× Phosphate Buffered Saline (PBS) via the pulmonary artery, and 1–2 ml of dispase (2.4 units/ml, Roche Diagnostics, #14435800) was rapidly instilled through the tracheal cannula followed by 0.5 ml of low-melting agarose solution warmed at 55 °C. Lungs were immediately covered with ice for 2 min to gel the agarose, removed from the animals, and then incubated in dispase II/collagenase A (Roche Diagnostics) (2.4 units/ml/0.1%) solution for 45 min at 37 °C. After this incubation, lung tissues were treated with DNase 1 (0.025 mg/ml, Qiagen) on ice for 5 min. Cells in suspension were then filtered through a 100-μm filter and centrifuged at 800 rpm for 6 min at 4 °C. Cells were resuspended with 1 ml of RBC lysis buffer (Invitrogen) for 90 s before adding DMEM (Invitrogen) without serum (6 ml for each lung). FBS (Invitrogen) (0.5 ml) was added to the bottom of the tube, and cells were centrifuged at 800 rpm for 5 min at 4 °C. The pellet was resuspended in PF10 (PBS supplemented with 10% FBS), and cells were filtered just before sorting in a MoFlow instrument (Dako Cytomation) at the Boston University School of Medicine flow cytometry core. Cells with high GFP and high side-scatter signals were either collected in RLT buffer (RNAeasy kit, Qiagen) for RNA isolation or in DMEM medium supplemented with 20% FBS for culturing on fibronectin-coated or plain 24-well culture plates (Corning).
RNA Purification and Real Time RT (qRT)-PCR
Total RNA was isolated from mouse lung tissue, sorted cells, and cell lines using the RNAeasy kit (Qiagen) and treated with DNase1 (Qiagen). Isolated RNA was reverse-transcribed (RT) using TaqMan Reverse Transcription Reagents (Applied Biosystems) following the manufacturer's protocols. The StepOnePlus system (Applied Biosystems) and commercially available TaqMan gene expression assays (Applied Biosystems) were used for qRT-PCR analyses (supplemental Table S1). Reactions were performed with TaqMan PCR master mix or Fast TaqMan PCR master mix (Applied Biosystems). Relative expression levels normalized to Gapdh were determined using the comparative 2−ΔΔCT method.
Plasmid Construction
Full-length Grhl2 cDNA was subcloned from the pGADT7-HA-Grhl2 vector provided by Dr. Bogi Andersen (University of California, Irvine, CA). Briefly, the Grhl2 cDNA was amplified by PCR using primers 5′-CAA GCG GCC GCC ATG TCA CAA GAG TCG GAC-3′ and 5′CGC TGA TGG AGA TCT GAG GAT CCA TTC-3′ that contain Not1 and BamH1 adaptors, respectively. This fragment was inserted in place of the dsRed gene in the dual promoter-reporter lentiviral plasmid, pCMV-dsred-UBC-Gfp (22), to generate pCMV-Grhl2-UBC-Gfp. The construct was verified by restriction enzyme digestion and by sequencing. This Grhl2 vector was used in overexpression experiments in MLE15 cells. The genomic DNA fragment containing the 5′ end region of the mouse Nkx2-1 gene (−339 to −2230 bp from the second ATG site) (supplemental Fig. S1) was generated by PCR using genomic DNA from mouse 129/Ola ES cells, cloned into the pCR-BluntII-TOPO shuttle plasmid, and then subcloned into KpnI and HindIII sites of pGL3 basic vector (Promega). The −350-bp fragment of the Nkx2-1 proximal promoter (−3 to −352 bp from the second ATG site) was generated by PCR and cloned in the pGL3 basic vector (Promega) (supplemental Fig. S1). The constructs were verified by sequencing and were identified as −2kbNkx2-1Luc and −0.35kbNkx2-1Luc. Two fragments in the first intron of the Grhl2 gene that bind NKX2-1 protein (region H (high binding) and region L (low binding) (Fig. 6A)) were amplified by PCR from genomic DNA, cloned in pGEM T vector (Promega), and subcloned in the Kpn1 and SacI sites 5′ to the Sv40 promoter in the pGL3 promoter vector (Promega) (supplemental Fig. S1). The constructs were verified by sequencing and identified as H-Grhl2 Sv40Luc and L-Grhl2 SVv40Luc.
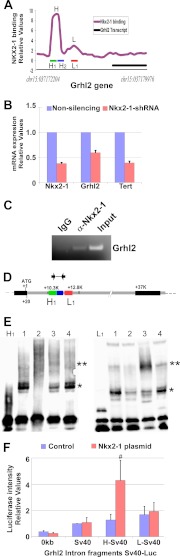
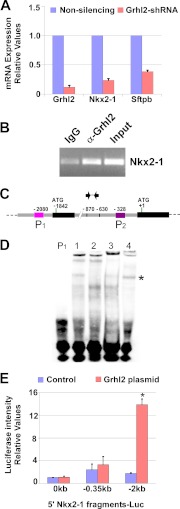
FIGURE 6.
NKX2-1 binds to Grhl2 intronic regions and increases transcription. A, shown is the binding pattern of NKX2-1 protein to the Grhl2 intron identified in a global ChIP-on-chip analysis of NKX2-1 binding profiles in E11.5 mouse lung epithelium (23). H and L identify the binding regions; H1, H2, and L1 identify the probes used in EMSAs. B, shown is qRT-PCR analysis of Nkx2-1, Grhl2, and Tert expression in MLE15 cells infected with Nkx2-1-shRNA or non-silencing-shRNA. The data set was normalized against Gapdh (n = 3). C, a representative ChIP-PCR analysis shows NKX2-1 binding to the Grhl2 intron. ChIP was performed with chromatin from E11.5 day lung bud immunoprecipitated with NKX2-1-specific antibody or its isotype control. Immunoprecipitated and input DNA fragments were amplified using primers corresponding to the Grhl2 intron. D, shown is a scheme of the Grhl2 gene depicting, first, and second exons (black boxes) and first intron (gray box). Numbers are relative to the ATG (+1). Green (H1), blue (H2), and red boxes (L1) represent probes used in EMSAs. Arrows indicate oligonucleotides used in ChIP-PCR analyses. E, left panel, EMSA analysis shows binding of MLE15 nuclear proteins to probe H1 (lane 1), competition with 100-fold unlabeled probe (lane 2), super-shift of the complex using NKX2-1 antibody (lane 3), and control with IgG. The single asterisk marks the H1-protein complex. The double asterisk marks the super-shifted complex. Right panel, EMSA analysis shows binding of MLE15 nuclear proteins to probe L1 (lane 1), competition with 100-fold unlabeled probe (lane 2), super-shift of complex formation using Nkx2-1 antibody (lane 3), and control with IgG. The single asterisk marks the L1-protein complex. The double asterisk marks the super-shifted complex. F, MLE15 cells were co-transfected with empty luciferase plasmid (pGL3; 0-Luc-plasmid), Sv40 promoter-luciferase (Sv40Luc), Grhl2 intron fragment H inserted 5′ to Sv40-luciferase plasmid (H-Sv40Luc), or Grhl2 intron fragment L inserted 5′ to Sv40-luciferase plasmid (L-Sv40Luc) with CMV-Nkx2-1 or pCMV plasmids. Firefly luciferase was normalized to Renilla luciferase levels (n = 5). Data represent the mean ± S.E. # indicates p ≤ 0.05.
Lentivirus Production and Transduction
To knock down Grhl2 gene expression, a replication incompetent, vesicular stomatitis virus G-pseudotyped lentiviral vector that contained a Gfp tracking reporter gene along with either a non-targeting control shRNA (RHS4346) or the shRNA targeting Grhl2 (V2LMM_20130) was employed (Open Biosystems). Lentiviruses were packaged and concentrated as described previously (19). MLE15 cell infections with lentiviral vectors were performed for 16 h in the presence of Polybrene (5 μg/ml) at a multiplicity of infection of 50. Stable cell lines were generated by puromycin dihydrochloride (5 μg/ml, Sigma) selection for 6 days. Cells were harvested after selection and analyzed for transduction efficiency and viability by flow cytometry analysis of GFP expression and exclusion of propidium iodide. Knockdown of Grhl2 in the infected population was quantified at the mRNA level by qRT-PCR and at the protein level by immunostaining and Western blot analyses. Similarly, Grhl2 was overexpressed in E10 cells by transduction of packaged bicistronic construct pCMV-Grhl2-UBC-Gfp, and cell morphology was compared with the cells transfected with pCMV-dsred-UBC-Gfp. Nkx2-1 gene expression knockdown was performed as described previously (19) using a mixture of three lentiviral clones (TRCN0000020449, TRCN0000020450, and TRCN0000086264 Open Biosystems) targeting Nkx2-1 mRNA of mouse, rat, and human origin.
Chromatin Immunoprecipitation Assays (ChIP)-PCR
Mouse lung buds (5–7 per reaction) were dissected from E11.5 day embryos and fixed in 1% formaldehyde in 1× PBS at room temperature for 15 min as described previously (23). Samples were sonicated using a Branson 450 dismembrator to achieve a chromatin fragment size of 500–2000 bp. To immunoprecipitate chromatin fragments, we incubated the samples (equal amount of DNA) at 4 °C overnight with rabbit GRHL2 (HPA00482, Sigma) or rabbit NKX2-1 (07-601, Millipore-Upstate) antibodies. Control experiments were performed with the corresponding IgG isotype (Santa Cruz Biotechnology) to determine nonspecific binding. Pre-absorbed protein A/G beads (Santa Cruz Biotechnology) were used to immunoprecipitate chromatin-antibody complexes. Equal volumes of immunoprecipitated DNA solution, 10% of the input DNA fragments, and genomic DNA were amplified by PCR using oligonucleotides spanning 228 bp of the proximal promoter of Nkx2-1 (sense primer, 5′-GCA CAC TCT TTT GGT GGT GA-3′; antisense primer, 5′-GCA ACC AAC TTG GGG AGT TA-3′) and 224 bp of the Grhl2 first intron (sense primer, 5′-GGG TTA CGT GGC TGC TTC A-3′; antisense primer, 5′-CGT CAG GTT GCT AAG GGC A-3′). Binding of GRHL2 to the Cldn4 promoter and to the Cdh1 enhancer region was also determined with samples immunoprecipitated using the rabbit GRHL2 antibody and quantified by qPCR on StepOnePlus equipment using oligonucleotides (IDT) previously described (15). Data were normalized to the input and compared with IgG control.
Electrophoretic Mobility Shift Assays (EMSA)
The DNA probes (supplemental Fig. S1) were annealed as described previously (24) and labeled with the Biotin 3′ End DNA labeling kit (Thermo Scientific). The assay was carried out using the Thermo Light Shift Chemiluminescent EMSA kit (Thermo Scientific). Samples were run in 5% TBE acrylamide gels (Bio-Rad) and detected using the Chemiluminescent Nucleic Acid Detection Module (Thermo Scientific). MLE15 cells nuclear proteins were extracted as described previously (24). 10 μg of protein was used per reaction. All binding reactions were carried out at room temperature. For competition experiments, unlabeled oligonucleotides (100-fold) were incubated with proteins for 10 min before the addition of the labeled oligonucleotide. For super shift analysis, samples were preincubated with 5 μl of NKX2-1 antibody (ab76013, Abcam), GRHL2 antibody (HPA004820, Sigma), or nonspecific rabbit IgG (Santa Cruz, sc-2027).
Immunocytochemistry
MLE15 cells were grown in conditions described previously on glass coverslips to reach 70–80% confluence, fixed with 4% formaldehyde in 1× PBS, and processed for immunostaining. For GRHL2 nuclear staining, MLE15 cells were treated with 0.01% Triton X-100 (Fisher) in 1× PBS, blocked with 1× PBS containing 2% FBS for 30 min, and incubated overnight at 4 °C with rabbit GRHL2 antibody (HPA00482, Sigma; 1:500 dilution). After three washes with 1× PBS, cells were incubated with secondary anti-rabbit FITC-conjugated antibody (Invitrogen, 1:2000 dilution) for 1 h at room temperature. Images were taken on a Leica inverted microscope or on a Leitz Aristoplan microscope. For phalloidin staining, cells were processed in a similar fashion and stained with Alexa Fluor 594 Phalloidin (Invitrogen) according to the manufacturer's protocol. Confocal imaging was performed using a Zeiss 710 microscope. Images were processed using Metamorph microscopy image analysis software to generate maximum projection image and orthogonal images of MLE15 stained with phalloidin.
Immunohistochemistry
E9.5 whole embryos as well as E15.5, E18.5, and adult dissected lungs were fixed in freshly prepared 4% paraformaldehyde in 1× PBS, pH 7.4, at 4 °C for 16 h. These samples were embedded in paraffin following standard processing with ethanol dehydration as described previously (25). Sections (6 μm) of these tissues were deparaffinized and hydrated by standard methods. Antigen retrieval was performed using an Antigen Unmasking solution (Vector Laboratories, Inc.). Endogenous peroxidase was quenched with 3% H2O2 in methanol for 15 min. Blocking was performed with 2% normal goat serum in 1× PBS for 1 h at room temperature. The tissues were incubated with monoclonal rabbit NKX2-1 antibody (ab76013, Abcam, 1:500 dilution) for 16 h at 4 °C or rabbit GRHL2 antibody (HPA00482, Sigma, 1:250 dilution) in 1× PBS + 0.2% Triton X-100 (PBST) and washed with 1× PBS (5 min, twice). Antibody binding was detected with the Vectastain Elite ABC kit (Vector Laboratories) that includes incubation with biotinylated tyramide signal amplification (TSA) reagent (PerkinElmer Life Sciences) and diaminobenzidine as substrate. Images were taken using a Leitz Aristoplan microscope.
For GRHL2 immunofluorescence analyses and double staining for NKX2-1 and CDH1, 6-μm tissue sections were depariffinized at 60 °C for 1 h and hydrated by incubation in HistoChoice Clearing Agent (Sigma) for 6 min followed by incubations in progressive ethanol dilutions as previously described. Antigen retrieval was performed in antigen unmasking solution (Vector Laboratories) and heating in a microwave (8 min on high power and 17 min on low power). The slides were then cooled down to room temperature for 1 h. Permeabilization was performed by incubation in 1× TN buffer (20 mm Tris-HCl, pH 7.4, 150 mm NaCl) containing 0.5% Triton X-100 at room temperature for 1 h, blocked in 3% BSA in 1× TN containing 0.5% Tween 20 at room temperature for 1 h, incubated with GRHL2 antibody (HPA00482, Sigma, 1:350) or monoclonal NKX2-1 antibody (Seven Hills, 8G7G3-1, 1:350) or CDH1 antibody (BD Biosciences, 610182, 1:500) in 1× TNT (1× PBS containing 5% goat serum, 0.2% BSA, and 0.05% Tween 20) at 4 °C overnight, washed in 1× TNT for 5 min, and then exposed to Alexa Fluor-labeled antibodies. We used Alexa Fluor 488 goat anti-rabbit IgG (H+L) (A11008, Invitrogen, 1:200 dilution) in 1× TNT for 5 min and Alexa Fluor 647 goat anti-mouse IgG (H+L) (A21235, Invitrogen, 1:200). Sections were washed in 1× TNT for 5 min and mounted under coverslips with Prolong Gold with DAPI (Invitrogen). Confocal images were collected using a Zeiss 710 microscope. Autofluorescence from the red blood cells was removed by spectral imaging. The images were then processed with Zen 2009 LE software (Carl Zeiss).
Transient Co-transfection and Luciferase Assays
These assays were performed in MLE15 cells as described previously (19). Cells were co-transfected with empty pGL3 vector, −0.35kbNkx2-1Luc or −2kbNkx2-1Luc and pCMV-dsred-UBC-Gfp or pCMV-Grhl2-UBC-Gfp by the DEAE-dextran/chloroquine method (24). pGL3 promoter vector Sv40Luc, H-Sv40Luc, or L-Sv40Luc were co-transfected with pCMV empty vector or pCMV-Nkx2-1 expression construct provided by Dr. Roberto Di Lauro (Stazione Zoologica A. Dohrn, Napoli, Italy). In all experiments renilla luciferase was used to normalize for transfection efficiency. Firefly and renilla luciferase activities were measured after 48 h with the dual luciferase system (Promega).
Gene Array Experiments
All procedures were performed at Boston University Microarray Resource Facility following protocols described in the GeneChip® Whole Transcript (WT) Sense Target Labeling Assay Manual (Affymetrix). Samples were analyzed on the GeneChip Mouse Gene Array 1.0ST (Affymetrix). Data were analyzed using the Robust Multi-Array Analysis (RMA) algorithm (26, 27). Differential expression between experimental and control samples was examined using Welch's t test and a false discovery rate (FDR) correction (28). Genes with a p ≤ 0.02 and an FDR adjusted p value < 0.3 were considered to be differentially expressed. Data have been deposited in the GEO repository, accession number GSE 40729.
RESULTS
NKX2-1 and GRHL2 Protein Expression Patterns Overlap in Developing Lung Epithelium
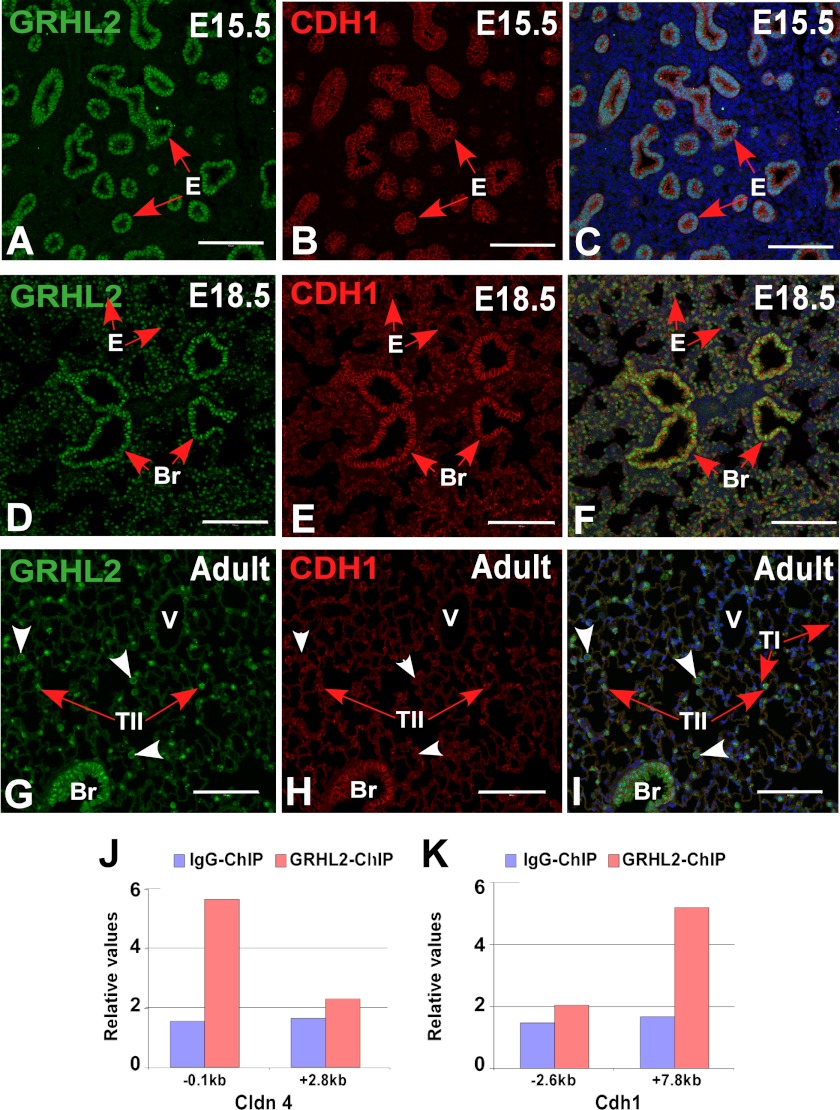
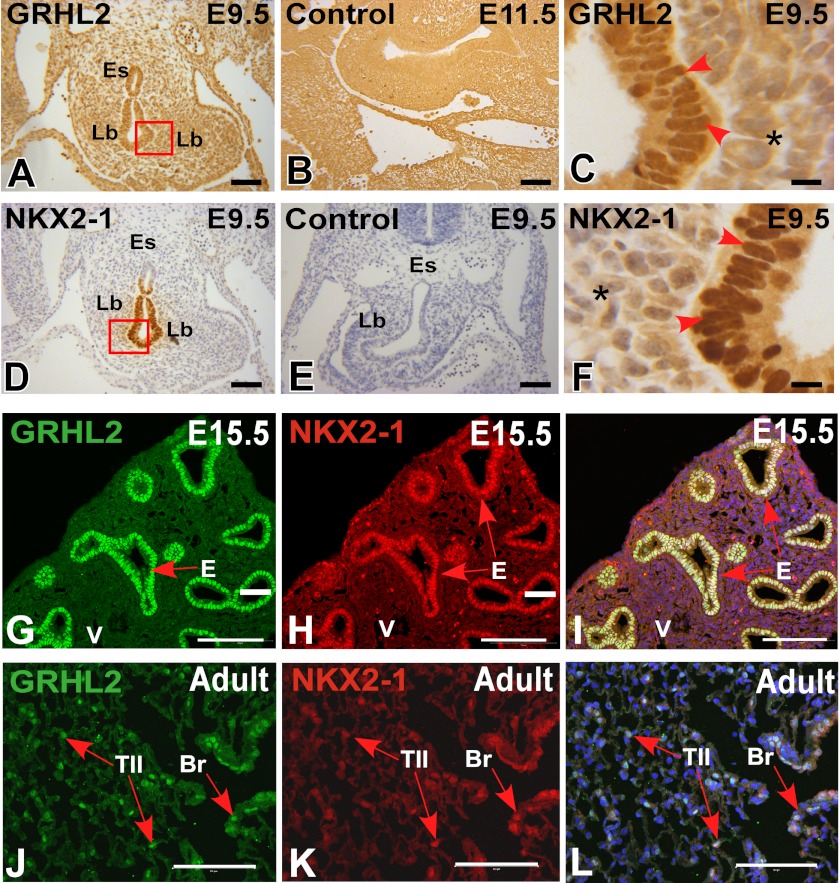
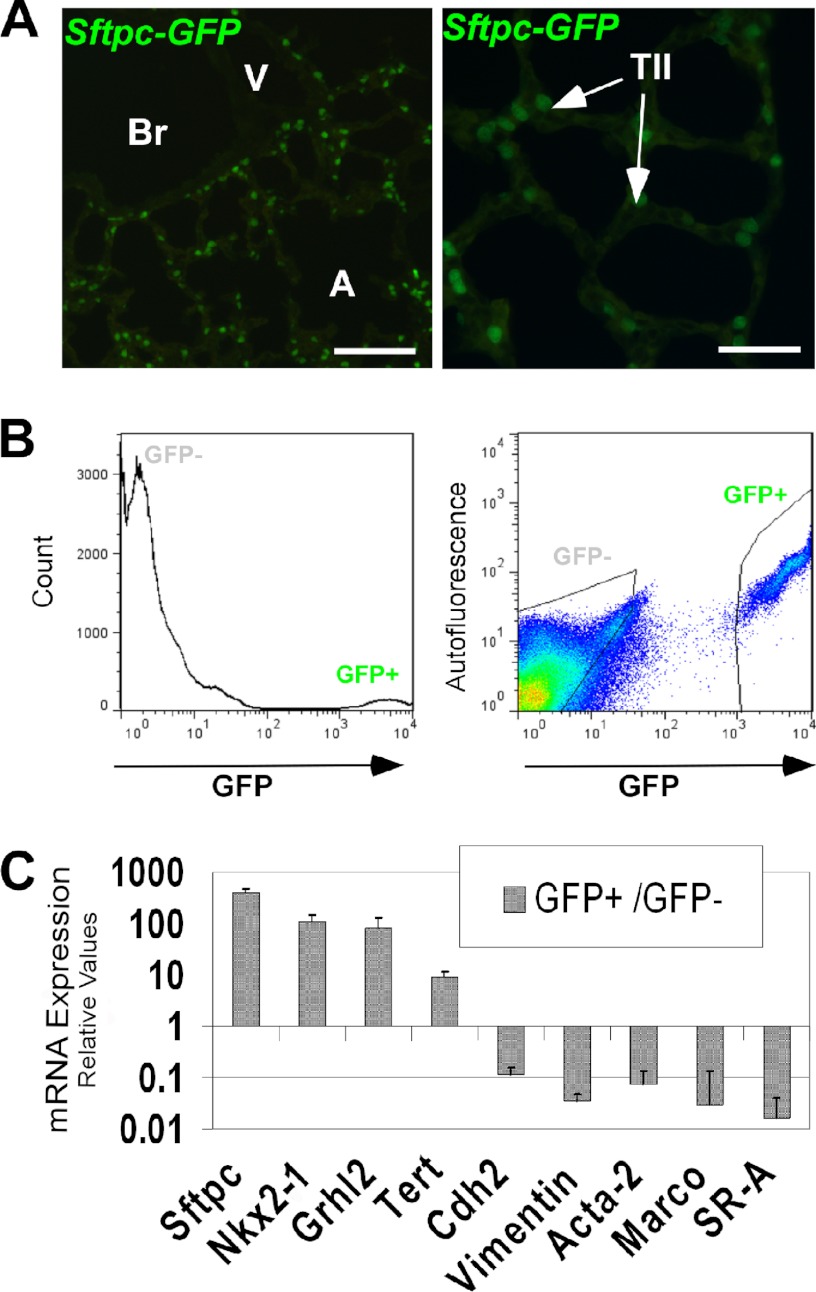
We characterized the expression pattern of GRHL2 protein by immunohistochemistry and immunofluorescence at different stages of lung development. GRHL2 protein was detected in the lung epithelium at E9.5 and E15.5 and in bronchiolar and alveolar epithelial cells at E18.5 and adult in a pattern similar to the expression pattern of the message RNA (6) and of its target Cdh1 (Fig. 1, A–I). To evaluate whether GRHL2 protein expressed in the lung epithelium binds in vivo to the same targets identified in a kidney cell line (15), we performed ChIP-PCR analyses using E11.5 mouse lungs. GRHL2 binds to the proximal promoter region (−0.1 kb) of Cldn4 but not to the intronic region (+2.8 kb) (Fig. 1J) and to the intronic region (+7.8 kb) of the Cdh1 gene but not to the proximal promoter region (−2.6 kb) (Fig. 1K). We compared the patterns of expression of GRHL2 and the essential lung transcription factor NKX2-1. At E9.5, when mouse lung morphogenesis begins, GRHL2 has a wider field of expression than NKX2-1. Although the expression of both proteins overlaps in the forming lung buds, GRHL2 can be detected in the dorsal and ventral esophagus cells, whereas NKX2-1 is only detected in the ventral cells (Fig. 2, A–F). By immunofluorescence and confocal microscopy, we found considerable overlap in the expression of GRHL2 and NKX2-1 in E15.5 (Fig. 2, G–I) and adult lungs (Fig. 2, J–L). Similar expression patterns were also detected in primary lung epithelial cells. Surfactant protein C-Gfp (SFTPC-Gfp) mouse lungs (20) (Fig. 3A) have been used previously to isolate by fluorescence-activated flow cytometry enriched populations of alveolar type II epithelial cells. Using this method, we obtained from total lung cell suspensions ∼5% high fluorescence-intensity cells (Fig. 3B). GFP-positive cells have higher expression levels of Nkx2-1 and Grhl2 than GFP-negative cells, as determined by qRT-PCR (Fig. 3C). Other markers of epithelial type II cells, such as Sftpc and Cdh1, were detected at higher levels in GFP-positive cells, whereas levels of markers of alveolar type I cells (Podoplanin (Pdpn), also known as T1α), mesenchymal cells (Cadherin2 (Cdh2), Vimentin (Vim), α-smooth muscle actin-2 (Acta2)) and of macrophages (macrophage receptor with collagenous structure (Marco)) were low.
FIGURE 1.
Expression and binding patterns of GRHL2 in mouse lung development. Immunofluorescence co-localization analyses of GRHL2 and one of its known targets CDH1 in days E15.5 (A–C) and E18.5 (D–F) and in adult lung (G–I) are shown. ChIP-qPCR analysis of GRHL2 binding to Cldn4 regulatory regions (−0.1 and +2.8 kb) (J) and to Cdh1 regulatory regions (−2.6 and +7.8 kb) (K) in vivo in E11.5 mouse lung. Bars, A–I, 100 μm. E, epithelium; Br, bronchi/bronchioles; V, blood vessel; TII, alveolar type II cells; TI, alveolar type I cells.
FIGURE 2.
Co-localization of GRHL2 and NKX2-1 in mouse lung development. Shown are immunohistochemical analyses of GRHL2 at E9.5 (A) and its corresponding control (B) and GRHL2 expression in the nuclei of lung epithelial cells (C). Immunohistochemical analyses of NKX2-1 protein expression in E9.5 mouse embryos (D) and its corresponding control (E) are shown. F, NKX2-1 expression in the nuclei of lung epithelial cells (F). Immunofluorescence co-localization of NKX2-1 and GRHL2 proteins in day E15.5 lung (G–I) and in adult lung (J–L). Lb, lung buds; Es, esophagus; E, epithelium; V, blood vessel; Br, bronchi/bronchioles; TII, alveolar type II cells; *, negative mesenchymal cell nuclei. Bars A, B, D, and E = 50 μm, C and F = 5 μm, and G–L = 100 μm.
FIGURE 3.
Grhl2 expression in alveolar type II cells. A, fluorescence microscopy analyses show the expression pattern of GFP in the distal lung epithelium of an adult 3.7 kb-SFTPC-Gfp transgenic mouse used for alveolar type II isolation. Left panel bar = 50 μm. Br, bronchioles; A, alveoli. Right panel bar = 25 μm. TII, alveolar type II cells. B, cell sorting of GFP-positive cells from 3.7KbSFTPC-Gfp lungs. A histogram shows that ∼5% of the cells are GFP-positive (left panel); gates used to collect GFP-positive (GFP+) and GFP-negative (GFP−) cells (right panel) are shown. C, shown is a gene expression profile of GFP+ cells isolated from 3.7KbSFTPC-Gfp lungs. Relative quantities of different cell-type markers in GFP+ cells compared with GFP- cells are shown. The data sets were normalized against Gapdh (n = 3). Data represent the mean ± S.D.
Grhl2 Regulates NKX2-1in Lung Epithelial Cells
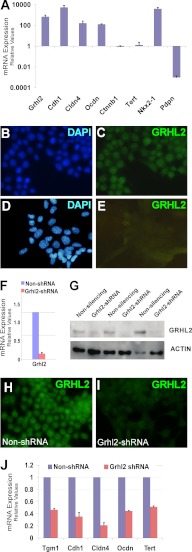
To analyze the regulatory network of the Grhl2 gene, we used two mouse lung epithelial cell lines. MLE15 are cuboidal cells expressing several alveolar epithelial type II cell-specific genes, like Sftpb and Nkx2-1, whereas E10 are squamous cells expressing alveolar epithelial type I genes like Pdpn and Caveolin1 (Cav1). The mRNA expression levels of Grhl2, Cdh1, Cldn4, and Ocdn in MLE15 are 2–3 orders of magnitude higher than the levels in E10 cells (Fig. 4A). Immunostaining results corroborated the high expression level of GRHL2 protein in MLE15 compared with E10 cells (Fig. 4, B–E). MLE15 cells express high levels of Grhl2, whereas Grhl1 and Grhl3 are low or undetected by qRT-PCR (data not shown), making this cell system attractive for studying genes regulated only by Grhl2. We infected MLE15 cells with Grhl2-shRNA lentivirus. Reduction of Grhl2 mRNA by >80% (Fig. 4F) results in 69 ± 12% lower GRHL2 protein levels determined by Western blot and immunofluorescence (Fig. 4, G–I). Grhl2 target genes are also down-regulated (Fig. 4J). Nkx2-1 mRNA and its downstream target, surfactant protein B (Sftpb) (Fig. 5A), were significantly reduced. To determine whether Nkx2-1 is a direct target of GRHL2 in vivo, we performed ChIP with E11.5 lungs (Fig. 5B). A GRHL2 antibody was used to immunoprecipitate chromatin fragments bound by GRHL2. Immunoprecipitated DNA fragments were amplified with specific primers for an Nkx2-1 regulatory region. A higher signal of the PCR product in GRHL2 over isotype control indicates that GRHL2 binds specifically to the Nkx2-1 promoter in lung epithelium at E11.5. We also performed EMSAs to further characterize binding of GRHL2 to three regions containing putative GRHL2 binding sites. These sites were selected based on the conserved sequence of Grhl family member binding sites in mammals and other organisms (supplemental Fig. S1). Three regions were tested (Fig. 5C), P1 located −2230 bp from the second ATG (+1) and upstream of the first ATG (at −1842 bp), P2 located −328 bp from the second ATG (+1), and P3. P1 forms a complex with MLE15 nuclear proteins under the conditions tested (Fig. 5D). This complex can be competed by a 100-fold excess of unlabeled oligonucleotide. The addition of GRHL2 antibody but not of IgG interferes with complex formation, indicating specific binding of GRHL2 protein. No DNA-protein complexes were detected with P2 and P3. To determine the transcriptional response of these two Nkx2-1 promoter regions to GRHL2 overexpression, we co-transfected luciferase constructs containing −0.35 or −2 kb of the Nkx2-1 5′-flanking region with a Grhl2 expression vector (CMV-Grhl2-UBC-Gfp) into MLE15 cells (Fig. 5E). A 14-fold increase in the level of luciferase activity with the 2-kb fragment containing the P1 region supports a direct role of GRHL2 in regulation of Nkx2-1.
FIGURE 4.
Down-regulation of Grhl2 affects expression of its target genes. A, qRT-PCR analysis of the expression levels of Grhl2, its potential targets, and type I epithelial cell marker Pdpn in lung epithelial MLE15 and E10 cells is shown. The data sets were normalized against Gapdh (n = 3). Data represent the mean ± S.D. Immunofluorescence analysis of GRHL2 expression in MLE15 cells (B and C) and in E10 cells (D and E) is shown. F, qRT-PCR expression analysis of Grhl2 in MLE15 cells infected with Grhl2-shRNA V2LMM_20130 (Open Biosystems) or non-silencing-shRNA. Data set was normalized against Gapdh (n = 3). Data represent the mean ± S.D. G, the Western blot shows down-regulation of GRHL2 protein in MLE15 cells by shRNA in three independent transductions. Data were normalized to β-actin. H–I, immunofluorescence analysis of GRHL2 protein expression in MLE15 cells infected with Grhl2 shRNA or non-silencing control is shown. J, by qRT-PCR analysis, we determined that knockdown of Grhl2 in MLE15 cells lowers the expression of its target genes. The data sets were normalized against Gapdh (n = 3). Data represent the mean ± S.D.
FIGURE 5.
GRHL2 binds to Nkx2-1 promoter and activates its transcription. A, qRT-PCR expression analysis of Nkx2-1 and its target Sftpb in MLE15 cells infected with Grhl2-shRNA or non-silencing-shRNA. Data sets were normalized against Gapdh (n = 3). Data represent the mean ± S.D. B, ChIP-PCR analysis shows GRHL2 binding to the Nkx2-1 promoter in the embryonic lung. ChIP was performed with chromatin from E11.5 day lung buds immunoprecipitated with GRHL2-specific antibody or its isotype control. Immunoprecipitated and input DNA fragments were amplified using primers corresponding to Nkx2-1. C, a scheme of the Nkx2-1 gene depicting 5′ regulatory regions, first and second exons (black boxes), and first intron (gray boxes). Numbers are relative to the second ATG (+1). Pink (P1) and purple boxes (P2) represent probes used in EMSAs. Arrows indicate oligonucleotides used in ChIP-PCR analyses. D, EMSA analysis shows binding of MLE15 nuclear proteins to probe P1 (lane 1), competition with 100-fold unlabeled probe (lane 2), interference of complex formation using GRHL2 antibody (lane 3), and control with IgG. The asterisk marks the P1-protein complex. E, the Nkx2-1 promoter is activated in the presence of exogenous GRHL2. MLE15 cells were transfected with either empty luciferase plasmid (pGL3; 0-Luc-plasmid), Nkx2-1 promoter luciferase plasmids −0.35kbNkx2-1Luc or −2.1KbNkx2-1Luc, and co-transfected with CMV-Grhl2-UBC-Gfp or CMV-dsred-UBC-Gfp plasmids. Firefly luciferase was normalized to renilla luciferase levels (n = 2–3). Data represent the mean ± S.D. The asterisk indicates p ≤ 0.05.
NKX2-1 Regulates Grhl2 in Lung Epithelial Cells
In a previous study we analyzed direct transcriptional targets of NKX2-1 in developing lung by ChiP-on-chip (23). In that analysis we observed that NKX2-1 binds to a DNA region within the first intron of the Grhl2 gene (Fig. 6A), suggesting a regulatory loop between Nkx2-1 and Grhl2. To test this hypothesis, we investigated whether Nkx2-1 directly regulates Grhl2 in lung epithelial cells. Nkx2-1 mRNA was reduced in MLE15 cells by transduction of Nkx2-1 shRNA lentivirus (19) (Fig. 6B). Levels of Grhl2 mRNA expression and of one of its regulated genes, the telomerase reverse transcriptase gene (Tert) were reduced by ∼50 and ∼70%, respectively. Reduction of Nkx2-1 mRNA results in changes in cell phenotype as cells tend to move apart, reducing the appearance of cell clusters (supplemental Fig. S2). To confirm whether Grhl2 is a direct target of NKX2-1 in vivo, we performed ChIP with E11.5 lungs (Fig. 6C). A NKX2-1 antibody was used to immunoprecipitate chromatin fragments bound by NKX2-1. Immunoprecipitated DNA fragments were amplified with specific primers spanning Grhl2 regulatory regions. A higher signal of the PCR product in NKX2-1 over isotype control indicates that NKX2-1 binds specifically to the Grhl2 intronic region in lung epithelium at E11.5. We also performed EMSAs to further characterize binding of NKX2-1 to three regions of the Grhl2 intron containing putative NKX2-1 binding sites. These sites were selected based on the conserved core sequence of NKX2-1 (CTTG/CAAG) binding site (supplemental Fig. S1). Three regions were tested, H1 and H2, corresponding to peak H highly bound by NKX2-1, and L1, corresponding to peak L weakly bound by NKX2-1 (Fig. 6, A and D). Two probes (H1 and L1) form complexes with MLE15 nuclear proteins under the conditions tested (Fig. 6E). These complexes can be competed by a 100-fold excess of unlabeled oligonucleotide. The addition of NKX2-1 antibody but not of IgG reduces the mobility of the complexes, super-shifting the bands and indicating specific binding of NKX2-1 protein. No DNA-protein complexes were detected with H2. To determine the transcriptional response of these two Grhl2 intronic regions to Nkx2-1 overexpression, we co-transfected Sv40 promoter-pGL3 luciferase constructs (Sv40Luc) containing fragments H or L of the Grhl2 intron 5′ upstream of the Sv40 minimal promoter with an Nkx2-1 expression vector (pCMV-Nkx2-1) into MLE15 cells. A 4-fold increase in the level of luciferase activity with the H fragment supports a direct role of Nkx2-1 in regulation of Grhl2 (Fig. 6F). Fragment L, although bound by NKX2-1, does not increase the luciferase activity in response to Nkx2-1 overexpression.
Grhl2 and Its Known Targets Are Down-regulated When Isolated Type II Cells Flatten in Culture
Isolated lung alveolar type II cells cultured on fibronectin-coated dishes are an ex vivo model that mimics many of the plasticity features associated with the transition from a type II alveolar epithelial cell phenotype into a type I-like phenotype. These features include changes in cell morphology, from cuboidal to flat, a decrease in expression of well characterized type II markers, and an increase in expression of the type I cell alveolar epithelial markers (19, 29, 30).
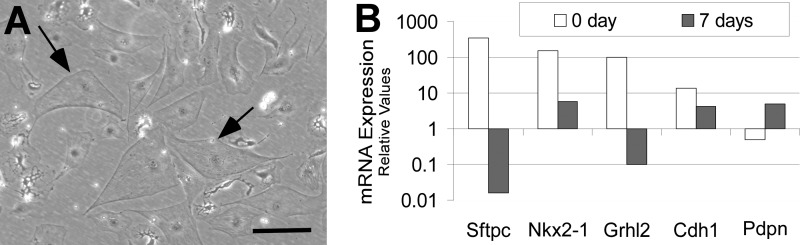
GFP-positive cells isolated by cell sorting from SFTPC-Gfp adult mouse lungs were cultured on fibronectin-coated plates for 7 days (Fig. 7A). We observed down-regulation of the type II markers Sftpc and Nkx2-1 and up-regulation of the type I cell marker Pdpn (Fig. 7B). The expression level of Grhl2 was substantially down-regulated, and its downstream target Cdh1 was down-regulated 3.2-fold. This suggested either a functional role for Grhl2 in maintaining the cuboidal cell shape of type II epithelial cells or conversely that a change in cell shape regulates the expression of Grhl2.
FIGURE 7.
Grhl2 is down-regulated as alveolar type II cells flatten in culture. A, shown are changes in the cell morphology of sorted GFP-positive cells upon culturing on fibronectin. Bar = 10 μm. B, gene expression analyses by qRT-PCR of Grhl2 type I and type II marker genes in freshly isolated versus cultured alveolar type II cells are shown.
Grhl2 Regulates Lung Epithelial Cell Shape, Migration, and Expression Levels of Cell-Cell Interaction Genes
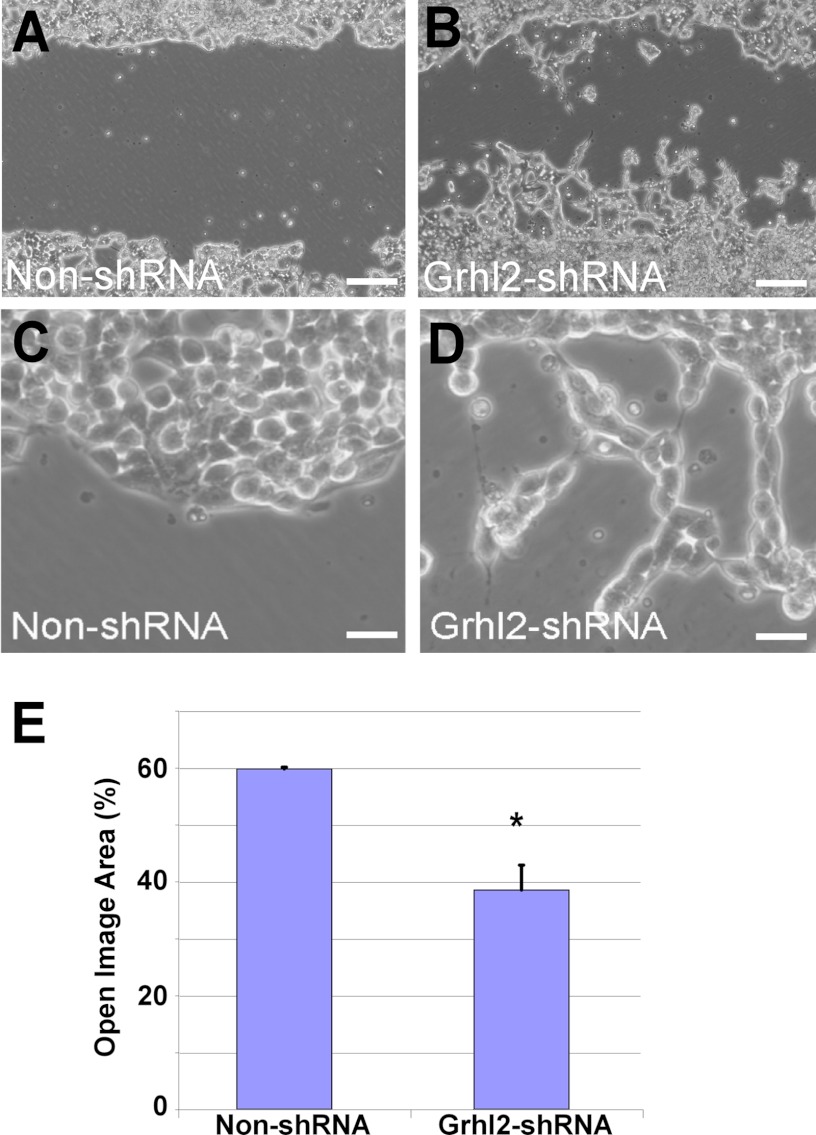
We further evaluated whether changes in Grhl2 levels were a cause or an effect of the alteration in epithelial cell shape using MLE15 and E10 cell lines (19). To determine the effect of Grhl2 on the regulation of genes involved in cell shape, we used MLE15 cells infected with lentivirus expressing Grhl2-shRNA or a non-silencing shRNA and investigated the change in cell morphology and in expression patterns of selected cell junction genes. Reduced levels of Grhl2 relaxed the cuboidal morphology of MLE15 cells into an expanded cell phenotype and reorganize F-actin in the apical and basal sides of the cells (Fig. 8, A–D), suggesting that GRHL2 may directly or indirectly regulate cell-cell interaction and cytoskeletal genes or their arrangement. Confocal x-z orthogonal images of phalloidin-stained cells (Fig. 8, E and F) showed a drastic reorganization of F-actin, leading to relaxation of the cuboidal shape. Conversely, mis-expression of CMV-Grhl2-UBC-Gfp in E10 cells (supplemental Fig. S3) changes the squamous and elongated phenotype of E10 cells into a cuboidal phenotype favoring formation of small cell clusters. We tested the effect of reduced Grhl2 levels on the migratory behavior of MLE15 cell monolayers upon induced injury. Scratch assays of confluent and serum-starved MLE15 cells infected with non-silencing control or Grhl2-shRNA showed that lower levels of Grhl2 alter migration patterns, particularly the collective migration of the epithelial layer, as cells detach from each other and move in smaller clusters (Fig. 9, A–D). Scratch healing analysis using Tscratch software indicated that cells with reduced levels of Grhl2 closed ∼30% more of the wound than control cells (Fig. 9E) (31). Reduction in the expression of cell junction genes in Grhl2-shRNA-infected cells could contribute to the different migration pattern of these cells.
FIGURE 8.
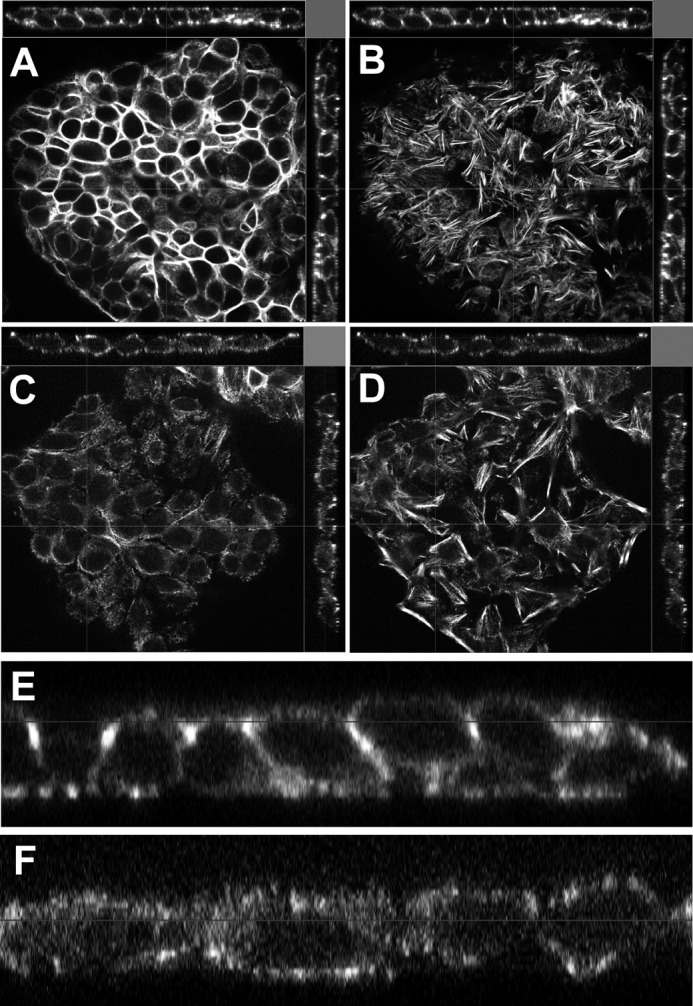
Reduced Grhl2 expression affects morphology of lung epithelial cells. Confocal orthogonal images of phalloidin stained MLE15 cells transduced with non-silencing shRNA (non-shRNA) apical XY image (A), basal XY image (B), or with Grhl2-shRNA lentivirus apical XY image (C) and basal XY image (D). Bar = 20 μm. Shown is zoom-in of the orthogonal XZ image of MLE15 transduced with non-silencing shRNA (E) and of the orthogonal XZ image of MLE15 transduced with Grhl2 shRNA (F).
FIGURE 9.
Reduced Grhl2 expression affects migration pattern of lung epithelial cells. Shown is representative wound healing image of MLE15 cells transduced with non-silencing shRNA (non-shRNA) (A) or with Grhl2-shRNA lentivirus 48 h after scratch (B). Bar = 50 μm. Knockdown of Grhl2 alters migration of MLE15 cells in wound healing scratch assays. C and D, higher magnification images show the difference in migratory pattern with reduced levels of Grhl2. Bar = 10 μm. E, quantification of the scratch healing using Tscratch software (n = 3) is shown. Data represent the mean ± S.E. The asterisk indicates p ≤ 0.05.
Identification of Downstream Targets of GRHL2 in Lung Epithelial Cells
To identify additional downstream genes regulated by GRHL2 in lung epithelial cells, we performed cDNA microarray analyses in MLE15 cells infected with Grhl2-shRNA and compared with a non-silencing control. Among the probe sets represented in the gene expression microarray, we found 286 down-regulated and 218 up-regulated genes in Grhl2-shRNA cells compared with control cells (p < 0.02; FDR adjusted p value <0.3, -fold change >1.25). Grhl2 knockdown leads to down-regulation of genes that could be direct or indirect targets of GRHL2 in lung epithelial cells. To explore the potential biological functions of the down-regulated genes in Grhl2 knockdown cells, annotations were derived from GO biological processes, and significant annotated functional groups were evaluated using GATHER (32). Among the top functional groups of genes containing more than 10 genes and a Bayes factor >1 were “morphogenesis” and “lipid metabolism,” both processes involved in organ development and in the regulation of cell shape (data not shown).
Among the genes involved in morphogenesis there were several semaphorins, a repulsion-attraction family of genes expressed in lung epithelium (33, 34). This prompted us to further evaluate changes in expression within known families of cell-cell interaction genes. We determined that several members of the semaphorin family and their corresponding receptors as well as Notch and Ephrin genes were reduced when Grhl2 levels were lower (Table 1). Many of these genes are expressed in the lung epithelium in a pattern similar to Grhl2 at E14.5 (35).
TABLE 1.
Selected cell-cell interaction and other lung genes regulated by Grhl2 in MLE15 cells
(p < 0.02; FDR adjusted p value <0.3, fold-change >1.25.
| Symbol | Description | -Fold change | p value | FDR | Lung expression E14.5 | Probeset |
|---|---|---|---|---|---|---|
| Fabp5 | Fatty acid-binding protein 5 | −3.1 | 0.0014 | 0.19 | 10585699 | |
| Fgb | Fibrinogen β chain | −2.6 | 0.0058 | 0.23 | 10498981 | |
| Hapln1 | Hyaluronan and proteoglycan link protein 1 | −2.3 | 0.0003 | 0.17 | 10406519 | |
| Klf12 | Kruppel-like factor 12 | −2.1 | 0.0083 | 0.25 | 10422013 | |
| Cldn8 | Claudin 8 | −1.9 | 0.0061 | 0.24 | E + | 10440647 |
| Sema3a | Semaphorin 3A | −1.5 | 0.0003 | 0.17 | 10519717 | |
| Sema3c | Semaphorin 3C | −1.4 | 0.0051 | 0.23 | E + + + | 10519886 |
| Cldn4 | Claudin 4 | −1.3 | 0.0471 | 0.39 | E + + + | 10534395 |
| Efna5 | Ephrin A5 | −1.3 | 0.0008 | 0.17 | 10452419 | |
| Sema6d | Semaphorin 6D | −1.3 | 0.0033 | 0.21 | 10475544 | |
| Notch1 | Notch gene homolog 1 | −1.3 | 0.0124 | 0.27 | E + + + | 10481056 |
| Sftpd | Surfactant associated protein D | −1.3 | 0.0114 | 0.27 | E + | 10419096 |
| Nrp2 | Neuropilin 2 | −1.2 | 0.0064 | 0.24 | E + | 10346843 |
| Efna1 | Ephrin A1 | −1.2 | 0.0035 | 0.22 | E + + + | 10499536 |
| Plxnb1 | Plexin B1 | −1.2 | 0.0011 | 0.19 | E + + + | 10589368 |
| Plxna2 | Plexin A2 | −1.2 | 0.0002 | 0.15 | 10352867 | |
| Grhl2 | Grainyhead-like 2 | −1.2 | 0.0602 | 0.42 | E + + + | 10423770 |
| Ephb3 | Eph receptor B3 | −1.2 | 0.0060 | 0.24 | 10434559 | |
| Notch2 | Notch gene homolog 2 | −1.2 | 0.0053 | 0.23 | 10494595 | |
| Plxna4 | Plexin A4 | −1.2 | 0.0047 | 0.23 | 10543802 | |
| Plxna1 | Plexin A1 | −1.2 | 0.0113 | 0.27 | 10546184 | |
| Enpp2 | Ectonucleotide pyrophosphatase/phosphodiesterase 2 | 2.2 | 0.0066 | 0.24 | 10428619 | |
| Scgb3a1 | Secretoglobin, family 3A, member 1 | 2.4 | 0.0018 | 0.19 | E + + | 10375608 |
| Hddc2 | HD domain containing 2 | 2.5 | 0.0054 | 0.23 | 10362394 | |
| Bex1 | Brain expressed gene 1 | 2.9 | 0.0005 | 0.17 | 10606868 |
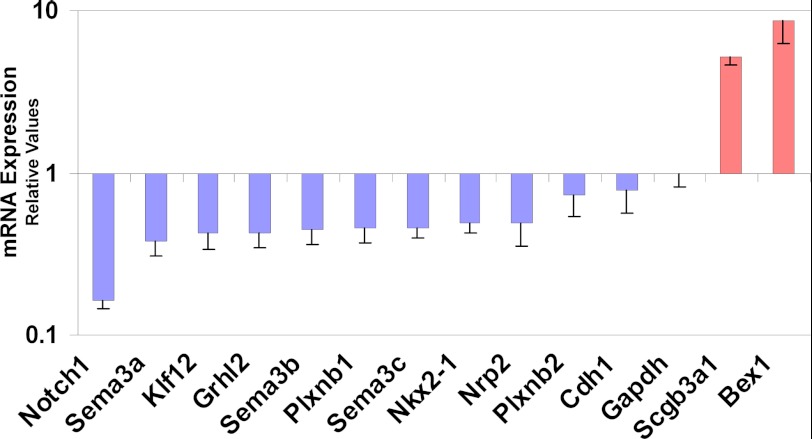
We validated by qRT-PCR arrays (Applied Biosystems) 13 of 20 genes selected from the microarray analysis (Fig. 10). These included the cell-cell interaction genes Sema3b, Sema3c, and their receptor Nrp2, the cell fate specification gene Notch1, and the Kruppel-like factor 12 (Klf12). Sema3a, Plxnb1, and Plxnb2, although validated by qRT-PCR, are expressed in mesenchymal or subepithelial cells during lung development (34).
FIGURE 10.
Down-regulation of Grhl2 affects the expression of other cell-cell interaction genes. Genes involved in cell-cell interaction identified in the microarray analysis were validated by qRT-PCR. 13 of 20 genes selected were confirmed (n = 2). Data represent the mean ± S.D.
DISCUSSION
We studied GRHL2-mediated gene regulation in the lung and explored the molecular mechanisms by which GRHL2 may regulate epithelial characteristics and differentiation of alveolar cells. We present evidence that GRHL2 directly regulates the key lung transcription factor Nkx2-1. Notably, Grhl2 and Nkx2-1 bind to each other's promoter in vivo, forming a positive feedback regulatory loop. Because Nkx2-1 is critical for distal lung cell differentiation and Grhl2 regulates cell morphogenesis, we propose that these two factors sustain each other's expression to maintain epithelial cell features during differentiation of distinct lung epithelial cell phenotypes.
Because of the larger expression domain of GRHL2 in the foregut endoderm at E9.5 and the overlapping patterns thereafter, we initially hypothesized that GRHL2 regulates Nkx2-1 expression. In vivo and in vitro experiments proved that GRHL2 binds to a particular region of the Nkx2-1 promoter and activates its transcriptional activity. Similarly, NKX2-1 protein binds to the first intron of the Grhl2 gene and regulates its expression. Although the identification of the particular cis-element in these regulatory regions will require further deletion and mutation analyses, the results presented indicate the presence of a regulatory loop connecting these transcription factors during mouse lung epithelial cell differentiation.
Positive transcriptional feedback loops have been described during pattern formation in development of several organisms such as zebrafish and mouse. These loops restrict transcriptional activity to give rise to different cell types in specific spatial and temporal domains or to reinforce initial cell fate decisions, providing robustness in gene expression to resist variations of external signals (36). The factors involved in any positive feedback loop are not necessarily involved in initiation of their own expression (36). In early Grhl2 ENU mutant lungs (16), the expression of Nkx2-1 is not altered, suggesting that the initial transcription does not depend on Grhl2 alone. Other members of the Grhl family present in the foregut endoderm, such as Grhl1 and/or Grhl3, or other transcription factors may initiate Nkx2-1 expression. Later in gestation, when Grhl2 is the only member of the Grhl family expressed at high levels in the distal lung epithelium, the effect of its specific loop may be more evident. The few viable E18.5 Grhl2 ENU mutant fetuses were reported to have smaller lungs and disorganized cell junctions (16). It is possible that at E18.5, the absence of Grhl2 may affect expression levels or patterns of Nkx2-1 and result in altered epithelial cell fates, although this has not been evaluated. Nkx2-1−/− mice, in contrast, have an abnormal lung with rudimentary main bronchi and an absence of peripheral lung structures (37, 38). Despite the importance of Nkx2-1 in lung morphogenesis, there is limited information concerning the transcriptional regulation of Nkx2-1 (39–41) or the restriction of its expression to lung bronchiolar or alveolar type II cells. These findings add Grhl2 as a new factor in the Nkx2-1 transcriptional network controlling lung development.
The alveolar epithelium includes cuboidal type II and flattened type I cells. These two types of epithelial cells with distinct morphological characteristics and functions lie adjoined to each other in the distal lung to form the alveoli. During lung development, any alteration in the differentiation of type I and type II epithelial cells may cause an aberrant alveolar morphology, which in turn may lead to respiratory failure at birth. Similarly, during lung injury, alterations in alveolar structure compromise the respiratory capacity and may lead to disease (42). GRHL2 protein is co-expressed with NKX2-1 in alveolar type II cells. Conversely, flattened cells facing the alveolar lumen that have an elongated nucleus and are negative for NKX2-1-likely type I cells- are also negative for GRHL2. Our analysis of GRHL2 function in primary alveolar type II cells and lung cell lines showed that there is a link between GRHL2 expression and cell morphogenesis, migration, and gene expression. Changes in type II alveolar cell morphology, as they flatten in culture, is accompanied by a reduction in Grhl2 expression. This reduction could be a cause or a consequence of the changes in cell shape. To address this issue, we down-regulated the expression of Grhl2 in the lung epithelial cell line MLE15. Changes in the levels of expression of cell-cell interaction genes and differences in the pattern of collective cell migration of the epithelial layer indicate that Grhl2 down-regulation is a cause rather than a consequence of any of these changes. Importantly, changes in cell shape and migration in MLE15 cells reflects mostly the effect of Grhl2 in those processes, as these cells do not express detectable levels of Grhl1 or Grhl3. In contrast, in the E18.5 Grhl2 ENU or null mutant models, the presence of Grhl1 or Grhl3 in lung epithelium in early development may compensate for the lack of Grhl2, masking the effects of individual members of this family of transcription factors. Nevertheless, the lung phenotype of the few Grhl2 ENU mutant mice that survived to E18.5 supports the hypothesis that Grhl2 participates in lung epithelial cell differentiation. Due to the lethality of homozygous Grhl2 mutations in mice, the ultimate role of Grhl2 in alveolar sac morphogenesis will need to be evaluated in vivo by conditional ablation of Grhl2 expression in type II cells. These experiments are beyond the scope of the present report but will allow the validation of the functional role of Grhl2 in vivo.
GRHL2 directly regulates transcription of Cdh1 in lung bud epithelium. NKX2-1 also regulates Cdh1 expression in a human adenocarcinoma cell line (43), although this regulation could be direct or indirect. Our finding that Nkx2-1 forms a feed-forward regulatory loop with Grhl2 supports an indirect link between Nkx2-1 and Cdh1 in the lung epithelial layer as it differentiates into distinct cell types to form a functional respiratory tree. Indeed, Grhl2 and Nkx2-1 are among the top 25 candidate genes identified in a microarray analysis of genes involved in EMT through regulation of Cdh1 (44). The maintenance of the regulatory loop between Grhl2 and Nkx2-1 would thus be expected to favor an epithelial phenotype by preventing the differentiation of epithelial cells into mesenchymal cells, such as myofibroblasts, in fibrotic lung diseases.
Grhl2 knockdown affects cell migration after scratch-wounding in culture as well as the expression of Tert in lung epithelial MLE15 cells. This evidence together with the reported regulation of RhoGEF19 by GRHL2 (7), the impaired injury/repair process in the cochlea in a family segregating DFNA28, the progressive hearing loss associated with a GRHL2 mutation (45), and the down-regulation of Grhl2 observed in microarray analyses of hyperoxia injured lung (GEO GDS248 hyperoxic lung injury) suggest that Grhl2 plays an important role in the lung injury/repair process. Grhls also regulate proliferation of some human carcinoma cell lines (46). Tert, a gene regulated by GRHL2 (47, 48) in humans, is crucial in lung morphogenesis during development (49) as well as in injury/repair in mice and humans (50). These data and the co-localization reported for Tert in type II cells lead us to infer that GRHL2 may also regulate Tert in mouse lung alveolar cells. Genetic mutations in hTERT within human families predispose them to idiopathic pulmonary fibrosis (51–53), pointing to a potential role of GRHL2 in lung fibrosis through regulation of Tert and possibly other lung epithelial genes.
Microarray analyses revealed novel genes regulated by GRHL2 in lung epithelial cells. Expression of several members of the semaphorin family and their corresponding receptors were reduced when Grhl2 levels were down-regulated. Although the regulation of these genes by Grhl2 could be direct or indirect, the change in expression of attraction-repulsion genes further supports the role of Grhl2 in balancing collective epithelial cell movements and differentiation. As Grhl2, semaphorins are also important for the development of different organs, neural cell migration (16, 54), angiogenesis, tumorigenesis, and the immune response (34, 55). Previous analysis of the role of semaphorins in lung development suggested that semaphorins may control branching morphogenesis and promote epithelial proliferation (33, 34). Therefore, we propose that through regulation of these genes, Grhl2 contributes to lung organogenesis.
Notably, we also showed that Grhl2 regulates expression of Klf12, a Kruppel-like transcription factor that directly represses expression of the helix-loop-helix transcription factor AP-2α (56). AP-2α, in turn, directly regulates expression of Cdh1 (57). Although both GRHL2 and AP-2α are positive regulators of Cdh1, experiments in Grhl2 or AP-2α mutant embryos indicated that these factors do not regulate each other's expression in neural tissues (16). Our findings suggest that Grhl2 can directly activate Cdh1 expression or inhibit Cdh1 expression by activating Klf12; these are potential mechanisms for fine-tuning Cdh1 expression levels to allow simultaneous epithelial layer migration and cell differentiation. Down-regulation of Grhl2 in MLE15 cells also caused reduced expression of genes such as Nkx2-1, Sftpb, and surfactant protein D (Sftpd) and increased the expression of a tracheal/bronchial gene secretoglobin 3a1 (Sctgb3a1) (58), suggesting a role for GRHL2 in proximal-distal cell differentiation in lung development. The early lethality of Grhl2 null mutant embryos limits further in vivo lung analyses.
The Grhl2-Nkx2-1-positive feedback loop, the cell-cell interaction genes regulated by GRHL2, and the Nkx2-1 direct targets that we recently identified in lung development (23) expand the knowledge about the regulatory networks in distal lung epithelium. The Grhl2-Nkx2-1 loop may be conserved in lung development and disease similarly to the conservation of Nkx2-1 regulatory patterns in development and tumors (23). Nkx2-1 haploinsufficiency in humans results in abnormal airway and alveolar morphogenesis (59). In lung tumors, Nkx2-1 controls cell differentiation and limits metastatic potential (60, 61). It is possible that through the Grhl2-Nkx2-1-positive feedback, loop Nkx2-1 regulates Cdh1 and other cell-cell interaction genes controlling cell metastatic potential.
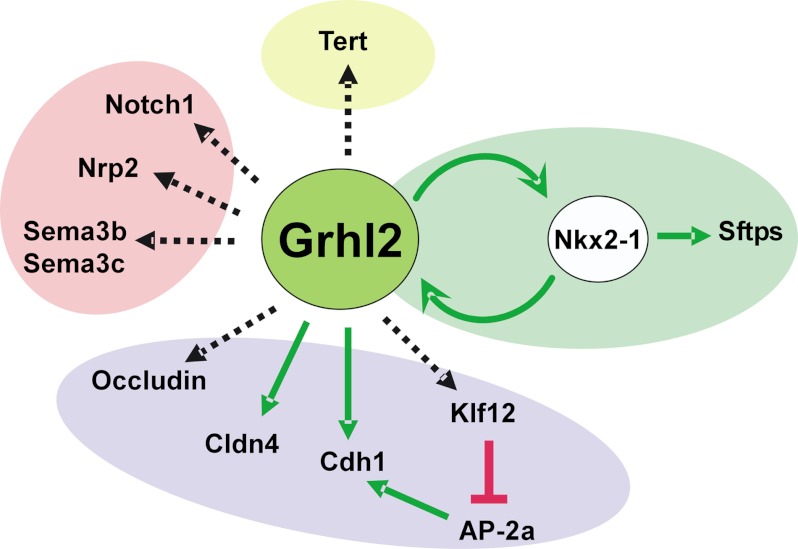
In summary, we identified novel cell-cell interaction genes regulated by Grhl2 that are known to participate in lung morphogenesis and cell differentiation. Most importantly, we identified a positive transcriptional feedback loop between Grhl2 and Nkx2-1 that mutually and positively affects their expression levels (Fig. 11). We, therefore, propose that the Grhl2-Nkx2-1-positive feedback loop serves to reinforce distal lung epithelial gene expression, epithelial layer integrity, and maintenance of particular epithelial cell fates.
FIGURE 11.
Summary of genes regulated by GRHL2 identified in these studies. GRHL2 regulates lung cell differentiation genes (Nkx2-1 feed forward loop, green oval), cell junction genes (purple oval), and cell-cell interaction genes (pink oval) as well as Tert. Solid green or red lines represent direct regulation. Dotted black arrows represent direct or indirect regulation.
Acknowledgments
We thank Dr. Jerome Brody and Dr. Wellington Cardoso at Boston University for thoughtful suggestions about the project and the manuscript. We thank Dr. Barbara Driscoll, Sonia Navarro, and Tessa Hyland at Children's Hospital, Los Angeles, and Prof. Dr. Kai M. Schmidt-Ott and Katharina Walentin at Max-Delbrück-Center for Molecular Medicine, Charité-Universitätsmedizin Berlin for help with immunofluorescence analysis protocol in lung sections. We thank Dr. G. Esteban Fernandez and the Cellular Imaging Core at Hospital, Los Angeles for help with confocal and spectral imaging and Dr. Marc Lenburg and Jacqui Milton from the Clinical and Translational Science Institute Bioinformatics Support Group at Boston University for microarray data analysis (UL1 RR025771).
This work was supported, in whole or in part, by National Institutes of Health Grant R01 HL0833034 (NHLBI; to M. I. R., D. N. K., and S. V.) and in part by P01 HL47049 (NHLBI; to M. I. R. and D. N. K.). This work was also supported by the California Institute of Regenerative Medicine TG2-01168 (to S. V. and D. W.).

This article contains supplemental Table S1 and Figs. S1–S3.
Data have been deposited in the GEO repository, accession number GSE 40729.
- ENU
- N-ethyl-N-nitrosourea
- qRT-PCR
- quantitative real-time PCR
- FDR
- false discovery rate.
REFERENCES
- 1. Warburton D., El-Hashash A., Carraro G., Tiozzo C., Sala F., Rogers O., De Langhe S., Kemp P. J., Riccardi D., Torday J., Bellusci S., Shi W., Lubkin S. R., Jesudason E. (2010) Lung organogenesis. Curr. Top. Dev. Biol. 90, 73–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cardoso W. V., Lü J. (2006) Regulation of early lung morphogenesis. Questions, facts, and controversies. Development 133, 1611–1624 [DOI] [PubMed] [Google Scholar]
- 3. Morrisey E. E., Hogan B. L. (2010) Preparing for the first breath. Genetic and cellular mechanisms in lung development. Dev. Cell 18, 8–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Warburton D., Bellusci S. (2004) The molecular genetics of lung morphogenesis and injury repair. Paediatr. Respir. Rev. 5, S283–S287 [DOI] [PubMed] [Google Scholar]
- 5. Hemphälä J., Uv A., Cantera R., Bray S., Samakovlis C. (2003) Grainy head controls apical membrane growth and tube elongation in response to Branchless/FGF signalling. Development 130, 249–258 [DOI] [PubMed] [Google Scholar]
- 6. Auden A., Caddy J., Wilanowski T., Ting S. B., Cunningham J. M., Jane S. M. (2006) Spatial and temporal expression of the Grainyhead-like transcription factor family during murine development. Gene Expr. Patterns 6, 964–970 [DOI] [PubMed] [Google Scholar]
- 7. Boglev Y., Wilanowski T., Caddy J., Parekh V., Auden A., Darido C., Hislop N. R., Cangkrama M., Ting S. B., Jane S. M. (2011) The unique and cooperative roles of the Grainy head-like transcription factors in epidermal development reflect unexpected target gene specificity. Dev. Biol. 349, 512–522 [DOI] [PubMed] [Google Scholar]
- 8. Gustavsson P., Copp A. J., Greene N. D. (2008) Grainyhead genes and mammalian neural tube closure. Birth Defects Res. A Clin. Mol. Teratol. 82, 728–735 [DOI] [PubMed] [Google Scholar]
- 9. Janicke M., Renisch B., Hammerschmidt M. (2010) Zebrafish grainyhead-like1 is a common marker of different non-keratinocyte epidermal cell lineages, which segregate from each other in a Foxi3-dependent manner. Int. J. Dev. Biol. 54, 837–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tao J., Kuliyev E., Wang X., Li X., Wilanowski T., Jane S. M., Mead P. E., Cunningham J. M. (2005) BMP4-dependent expression of Xenopus Grainyhead-like 1 is essential for epidermal differentiation. Development 132, 1021–1034 [DOI] [PubMed] [Google Scholar]
- 11. Wilanowski T., Caddy J., Ting S. B., Hislop N. R., Cerruti L., Auden A., Zhao L. L., Asquith S., Ellis S., Sinclair R., Cunningham J. M., Jane S. M. (2008) Perturbed desmosomal cadherin expression in grainy head-like 1-null mice. EMBO J. 27, 886–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yu Z., Bhandari A., Mannik J., Pham T., Xu X., Andersen B. (2008) Grainyhead-like factor Get1/Grhl3 regulates formation of the epidermal leading edge during eyelid closure. Dev. Biol. 319, 56–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yu Z., Lin K. K., Bhandari A., Spencer J. A., Xu X., Wang N., Lu Z., Gill G. N., Roop D. R., Wertz P., Andersen B. (2006) The Grainyhead-like epithelial transactivator Get-1/Grhl3 regulates epidermal terminal differentiation and interacts functionally with LMO4. Dev. Biol. 299, 122–136 [DOI] [PubMed] [Google Scholar]
- 14. Yu Z., Mannik J., Soto A., Lin K. K., Andersen B. (2009) The epidermal differentiation-associated Grainyhead gene Get1/Grhl3 also regulates urothelial differentiation. EMBO J. 28, 1890–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Werth M., Walentin K., Aue A., Schönheit J., Wuebken A., Pode-Shakked N., Vilianovitch L., Erdmann B., Dekel B., Bader M., Barasch J., Rosenbauer F., Luft F. C., Schmidt-Ott K. M. (2010) The transcription factor grainyhead-like 2 regulates the molecular composition of the epithelial apical junctional complex. Development 137, 3835–3845 [DOI] [PubMed] [Google Scholar]
- 16. Pyrgaki C., Liu A., Niswander L. (2011) Grainyhead-like 2 regulates neural tube closure and adhesion molecule expression during neural-fold fusion. Dev. Biol. 353, 38–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Porter S. E., Dwyer-Nield L. D., Malkinson A. M. (2001) Regulation of lung epithelial cell morphology by cAMP-dependent protein kinase type I isozyme. Am. J. Physiol. Lung Cell Mol. Physiol. 280, L1282–L1289 [DOI] [PubMed] [Google Scholar]
- 18. Wikenheiser K. A., Vorbroker D. K., Rice W. R., Clark J. C., Bachurski C. J., Oie H. K., Whitsett J. A. (1993) Production of immortalized distal respiratory epithelial cell lines from surfactant protein C/simian virus 40 large tumor antigen transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 90, 11029–11033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cao Y., Vo T., Millien G., Tagne J. B., Kotton D., Mason R. J., Williams M. C., Ramirez M. I. (2010) Epigenetic mechanisms modulate thyroid transcription factor 1-mediated transcription of the surfactant protein B gene. J. Biol. Chem. 285, 2152–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lo B., Hansen S., Evans K., Heath J. K., Wright J. R. (2008) Alveolar epithelial type II cells induce T cell tolerance to specific antigen. J. Immunol. 180, 881–888 [DOI] [PubMed] [Google Scholar]
- 21. Kim C. F., Jackson E. L., Woolfenden A. E., Lawrence S., Babar I., Vogel S., Crowley D., Bronson R. T., Jacks T. (2005) Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell 121, 823–835 [DOI] [PubMed] [Google Scholar]
- 22. Wilson A. A., Kwok L. W., Hovav A. H., Ohle S. J., Little F. F., Fine A., Kotton D. N. (2008) Sustained expression of α1-antitrypsin after transplantation of manipulated hematopoietic stem cells. Am. J. Respir. Cell Mol. Biol. 39, 133–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tagne J. B., Gupta S., Gower A. C., Shen S. S., Varma S., Lakshminarayanan M., Cao Y., Spira A., Volkert T. L., Ramirez M. I. (2012) Genome-wide analyses of Nkx2–1 binding to transcriptional target genes uncover novel regulatory patterns conserved in lung development and tumors. PLoS One 7, e29907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ramirez M. I., Rishi A. K., Cao Y. X., Williams M. C. (1997) TGT3, thyroid transcription factor I, and Sp1 elements regulate transcriptional activity of the 1.3-kilobase pair promoter of T1α, a lung alveolar type I cell gene. J. Biol. Chem. 272, 26285–26294 [DOI] [PubMed] [Google Scholar]
- 25. Williams M. C., Cao Y., Hinds A., Rishi A. K., Wetterwald A. (1996) T1α protein is developmentally regulated and expressed by alveolar type I cells, choroid plexus, and ciliary epithelia of adult rats. Am. J. Respir. Cell Mol. Biol. 14, 577–585 [DOI] [PubMed] [Google Scholar]
- 26. Irizarry R. A., Bolstad B. M., Collin F., Cope L. M., Hobbs B., Speed T. P. (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Irizarry R. A., Hobbs B., Collin F., Beazer-Barclay Y. D., Antonellis K. J., Scherf U., Speed T. P. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 [DOI] [PubMed] [Google Scholar]
- 28. Benjamini Y., Drai D., Elmer G., Kafkafi N., Golani I. (2001) Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 125, 279–284 [DOI] [PubMed] [Google Scholar]
- 29. Borok Z., Danto S. I., Lubman R. L., Cao Y., Williams M. C., Crandall E. D. (1998) Modulation of t1α expression with alveolar epithelial cell phenotype in vitro. Am. J. Physiol. 275, L155–L164 [DOI] [PubMed] [Google Scholar]
- 30. Olsen C. O., Isakson B. E., Seedorf G. J., Lubman R. L., Boitano S. (2005) Extracellular matrix-driven alveolar epithelial cell differentiation in vitro. Exp. Lung Res. 31, 461–482 [DOI] [PubMed] [Google Scholar]
- 31. Gebäck T., Schulz M. M., Koumoutsakos P., Detmar M. (2009) TScratch. A novel and simple software tool for automated analysis of monolayer wound healing assays. Biotechniques 46, 265–274 [DOI] [PubMed] [Google Scholar]
- 32. Chang J. T., Nevins J. R. (2006) GATHER. A systems approach to interpreting genomic signatures. Bioinformatics 22, 2926–2933 [DOI] [PubMed] [Google Scholar]
- 33. Ito T., Kagoshima M., Sasaki Y., Li C., Udaka N., Kitsukawa T., Fujisawa H., Taniguchi M., Yagi T., Kitamura H., Goshima Y. (2000) Repulsive axon guidance molecule Sema3A inhibits branching morphogenesis of fetal mouse lung. Mech. Dev. 97, 35–45 [DOI] [PubMed] [Google Scholar]
- 34. Kagoshima M., Ito T. (2001) Diverse gene expression and function of semaphorins in developing lung. Positive and negative regulatory roles of semaphorins in lung branching morphogenesis. Genes Cells 6, 559–571 [DOI] [PubMed] [Google Scholar]
- 35. Visel A., Thaller C., Eichele G. (2004) GenePaint.org. An atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 32, D552–D556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meinhardt H., Gierer A. (2000) Pattern formation by local self-activation and lateral inhibition. Bioessays 22, 753–760 [DOI] [PubMed] [Google Scholar]
- 37. Minoo P., Su G., Drum H., Bringas P., Kimura S. (1999) Defects in tracheoesophageal and lung morphogenesis in Nkx2.1(−/−) mouse embryos. Dev. Biol. 209, 60–71 [DOI] [PubMed] [Google Scholar]
- 38. Yuan B., Li C., Kimura S., Engelhardt R. T., Smith B. R., Minoo P. (2000) Inhibition of distal lung morphogenesis in Nkx2.1(−/−) embryos. Dev. Dyn. 217, 180–190 [DOI] [PubMed] [Google Scholar]
- 39. Das A., Acharya S., Gottipati K. R., McKnight J. B., Chandru H., Alcorn J. L., Boggaram V. (2011) Thyroid transcription factor-1 (TTF-1) gene. Identification of ZBP-89, Sp1, and TTF-1 sites in the promoter and regulation by TNF-α in lung epithelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 301, L427–L440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li C., Ling X., Yuan B., Minoo P. (2000) A novel DNA element mediates transcription of Nkx2.1 by Sp1 and Sp3 in pulmonary epithelial cells. Biochim. Biophys. Acta 1490, 213–224 [DOI] [PubMed] [Google Scholar]
- 41. Shaw-White J. R., Bruno M. D., Whitsett J. A. (1999) GATA-6 activates transcription of thyroid transcription factor-1. J. Biol. Chem. 274, 2658–2664 [DOI] [PubMed] [Google Scholar]
- 42. Crosby L. M., Waters C. M. (2010) Epithelial repair mechanisms in the lung. Am. J. Physiol. Lung Cell Mol. Physiol. 298, L715–L731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Saito R. A., Watabe T., Horiguchi K., Kohyama T., Saitoh M., Nagase T., Miyazono K. (2009) Thyroid transcription factor-1 inhibits transforming growth factor-β-mediated epithelial-to-mesenchymal transition in lung adenocarcinoma cells. Cancer Res. 69, 2783–2791 [DOI] [PubMed] [Google Scholar]
- 44. Shimamura T., Imoto S., Shimada Y., Hosono Y., Niida A., Nagasaki M., Yamaguchi R., Takahashi T., Miyano S. (2011) A novel network profiling analysis reveals system changes in epithelial-mesenchymal transition. PLoS One 6, e20804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Peters L. M., Anderson D. W., Griffith A. J., Grundfast K. M., San Agustin T. B., Madeo A. C., Friedman T. B., Morell R. J. (2002) Mutation of a transcription factor, TFCP2L3, causes progressive autosomal dominant hearing loss, DFNA28. Hum. Mol. Genet. 11, 2877–2885 [DOI] [PubMed] [Google Scholar]
- 46. Tanaka Y., Kanai F., Tada M., Tateishi R., Sanada M., Nannya Y., Ohta M., Asaoka Y., Seto M., Shiina S., Yoshida H., Kawabe T., Yokosuka O., Ogawa S., Omata M. (2008) Gain of GRHL2 is associated with early recurrence of hepatocellular carcinoma. J. Hepatol. 49, 746–757 [DOI] [PubMed] [Google Scholar]
- 47. Chen W., Dong Q., Shin K. H., Kim R. H., Oh J. E., Park N. H., Kang M. K. (2010) Grainyhead-like 2 enhances the human telomerase reverse transcriptase gene expression by inhibiting DNA methylation at the 5′-CpG island in normal human keratinocytes. J. Biol. Chem. 285, 40852–40863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kang X., Chen W., Kim R. H., Kang M. K., Park N. H. (2009) Regulation of the hTERT promoter activity by MSH2, the hnRNPs K and D, and GRHL2 in human oral squamous cell carcinoma cells. Oncogene 28, 565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Driscoll B., Buckley S., Bui K. C., Anderson K. D., Warburton D. (2000) Telomerase in alveolar epithelial development and repair. Am. J. Physiol. Lung Cell Mol. Physiol. 279, L1191–L1198 [DOI] [PubMed] [Google Scholar]
- 50. Fridlender Z. G., Cohen P. Y., Golan O., Arish N., Wallach-Dayan S., Breuer R. (2007) Telomerase activity in bleomycin-induced epithelial cell apoptosis and lung fibrosis. Eur. Respir. J. 30, 205–213 [DOI] [PubMed] [Google Scholar]
- 51. Alder J. K., Chen J. J., Lancaster L., Danoff S., Su S. C., Cogan J. D., Vulto I., Xie M., Qi X., Tuder R. M., Phillips J. A., 3rd, Lansdorp P. M., Loyd J. E., Armanios M. Y. (2008) Short telomeres are a risk factor for idiopathic pulmonary fibrosis. Proc. Natl. Acad. Sci. U.S.A. 105, 13051–13056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Armanios M. Y., Chen J. J., Cogan J. D., Alder J. K., Ingersoll R. G., Markin C., Lawson W. E., Xie M., Vulto I., Phillips J. A., 3rd, Lansdorp P. M., Greider C. W., Loyd J. E. (2007) Telomerase mutations in families with idiopathic pulmonary fibrosis. N. Engl. J. Med. 356, 1317–1326 [DOI] [PubMed] [Google Scholar]
- 53. Tsakiri K. D., Cronkhite J. T., Kuan P. J., Xing C., Raghu G., Weissler J. C., Rosenblatt R. L., Shay J. W., Garcia C. K. (2007) Adult-onset pulmonary fibrosis caused by mutations in telomerase. Proc. Natl. Acad. Sci. U.S.A. 104, 7552–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Osborne N. J., Begbie J., Chilton J. K., Schmidt H., Eickholt B. J. (2005) Semaphorin/neuropilin signaling influences the positioning of migratory neural crest cells within the hindbrain region of the chick. Dev. Dyn. 232, 939–949 [DOI] [PubMed] [Google Scholar]
- 55. Potiron V., Roche J. (2005) Class 3 semaphorin signaling. The end of a dogma. Sci. STKE 2005, pe24. [DOI] [PubMed] [Google Scholar]
- 56. Roth C., Schuierer M., Günther K., Buettner R. (2000) Genomic structure and DNA binding properties of the human zinc finger transcriptional repressor AP-2rep (KLF12). Genomics 63, 384–390 [DOI] [PubMed] [Google Scholar]
- 57. Schwartz B., Melnikova V. O., Tellez C., Mourad-Zeidan A., Blehm K., Zhao Y. J., McCarty M., Adam L., Bar-Eli M. (2007) Loss of AP-2α results in deregulation of E-cadherin and MMP-9 and an increase in tumorigenicity of colon cancer cells in vivo. Oncogene 26, 4049–4058 [DOI] [PubMed] [Google Scholar]
- 58. Reynolds S. D., Reynolds P. R., Pryhuber G. S., Finder J. D., Stripp B. R. (2002) Secretoglobins SCGB3A1 and SCGB3A2 define secretory cell subsets in mouse and human airways. Am. J. Respir. Crit. Care Med. 166, 1498–1509 [DOI] [PubMed] [Google Scholar]
- 59. Galambos C., Levy H., Cannon C. L., Vargas S. O., Reid L. M., Cleveland R., Lindeman R., deMello D. E., Wert S. E., Whitsett J. A., Perez-Atayde A. R., Kozakewich H. (2010) Pulmonary pathology in thyroid transcription factor-1 deficiency syndrome. Am. J. Respir. Crit. Care Med. 182, 549–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kwei K. A., Kim Y. H., Girard L., Kao J., Pacyna-Gengelbach M., Salari K., Lee J., Choi Y. L., Sato M., Wang P., Hernandez-Boussard T., Gazdar A. F., Petersen I., Minna J. D., Pollack J. R. (2008) Genomic profiling identifies TITF1 as a lineage-specific oncogene amplified in lung cancer. Oncogene 27, 3635–3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Winslow M. M., Dayton T. L., Verhaak R. G., Kim-Kiselak C., Snyder E. L., Feldser D. M., Hubbard D. D., DuPage M. J., Whittaker C. A., Hoersch S., Yoon S., Crowley D., Bronson R. T., Chiang D. Y., Meyerson M., Jacks T. (2011) Suppression of lung adenocarcinoma progression by Nkx2-1. Nature 473, 101–104 [DOI] [PMC free article] [PubMed] [Google Scholar]