Background: Human cholesteryl ester transfer protein (CETP) transfers cholesteryl esters from high-density to low-density lipoprotein particles.
Results: Crystallographic, mutagenesis, and biochemical studies illuminated inhibition mechanisms of CETP by torcetrapib and a structurally distinct compound, ((2R)-3-{[4-(4-chloro-3-ethylphenoxy)pyrimidin-2-yl][3-(1,1,2,2-tetrafluoroethoxy)benzyl]amino}-1,1,1-trifluoropropan-2-ol.
Conclusion: These small molecules inhibit CETP through blocking its lipid tunnel.
Significance: Potential polar interactions at compound binding site may be utilized in design of inhibitors with improved physical properties.
Keywords: Cardiovascular Disease, Crystallography, HDL, Low Density Lipoprotein (LDL), Medicinal Chemistry, Cholesteryl Ester Transfer Protein, Inhibitor Complex
Abstract
Human plasma cholesteryl ester transfer protein (CETP) transports cholesteryl ester from the antiatherogenic high-density lipoproteins (HDL) to the proatherogenic low-density and very low-density lipoproteins (LDL and VLDL). Inhibition of CETP has been shown to raise human plasma HDL cholesterol (HDL-C) levels and is potentially a novel approach for the prevention of cardiovascular diseases. Here, we report the crystal structures of CETP in complex with torcetrapib, a CETP inhibitor that has been tested in phase 3 clinical trials, and compound 2, an analog from a structurally distinct inhibitor series. In both crystal structures, the inhibitors are buried deeply within the protein, shifting the bound cholesteryl ester in the N-terminal pocket of the long hydrophobic tunnel and displacing the phospholipid from that pocket. The lipids in the C-terminal pocket of the hydrophobic tunnel remain unchanged. The inhibitors are positioned near the narrowing neck of the hydrophobic tunnel of CETP and thus block the connection between the N- and C-terminal pockets. These structures illuminate the unusual inhibition mechanism of these compounds and support the tunnel mechanism for neutral lipid transfer by CETP. These highly lipophilic inhibitors bind mainly through extensive hydrophobic interactions with the protein and the shifted cholesteryl ester molecule. However, polar residues, such as Ser-230 and His-232, are also found in the inhibitor binding site. An enhanced understanding of the inhibitor binding site may provide opportunities to design novel CETP inhibitors possessing more drug-like physical properties, distinct modes of action, or alternative pharmacological profiles.
Introduction
Extensive epidemiological and preclinical mechanistic and intervention studies suggest that low HDL-C5 levels, in addition to high LDL-C levels, are powerful predictors of cardiovascular risks and that increasing plasma HDL-C levels may be beneficial (1). By shuttling cholesteryl ester from HDL to LDL, CETP decreases plasma HDL-C levels and increases VLDL cholesterol (VLDL-C) and subsequently LDL cholesterol (LDL-C) levels (2, 3). In addition to cholesteryl esters, CETP also transfers triglycerides and phospholipids between plasma lipoproteins. The suggestion that inhibition of CETP could provide a lipid profile with reduced atherosclerotic risk first arose >20 years ago (4). Epidemiological studies as well as transgenic animal models strongly suggest a role of CETP in the development of atherosclerosis and that blocking CETP could be beneficial (5–7). For example, populations with a mutation that leads to CETP deficiency have a lower incidence of cardiovascular diseases (5).
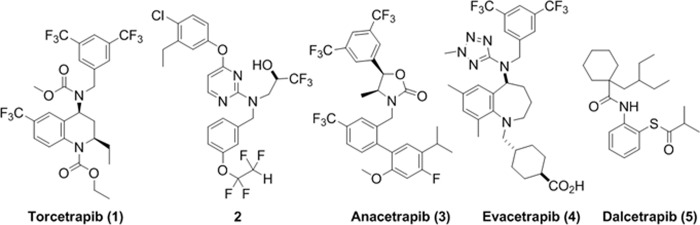
Inhibition of CETP by small molecule drugs to raise human plasma HDL-C levels has been actively pursued as an approach to prevent cardiovascular diseases (8–11). Despite the success of statins, which lower LDL-C levels significantly, cardiovascular disease is still the leading cause of mortality worldwide claiming ∼17 million lives each year (12). Many CETP inhibitors have been developed, with four having reached late stage clinical trials (see Fig. 1): torcetrapib (1) (13), anacetrapib (3, MK-859) (14, 15), evacetrapib (4, LY2484595) (16), and dalcetrapib (5, R1658, also known as JTT-705) (17). Torcetrapib, the first CETP inhibitor to advance to late stage clinical trials, showed a substantial effect on plasma lipoprotein levels, raising HDL-C and lowering LDL-C. However, torcetrapib also exhibited adverse effects in a cardiovascular events trial in combination with atorvastatin and was found to increase the risk of mortality and morbidity over atorvastatin alone (18–21). Dalcetrapib did not show the same adverse effects but was not potent enough to demonstrate clinical efficacy (22). The other CETP inhibitors named in Fig. 1 are being actively tested in later stage clinical trials (23–25); their effects on disease outcome remain unknown (26–29).
FIGURE 1.
Chemical structures of the CETP inhibitors described in this work.
Torcetrapib (1) is a representative of the tetrahydroquinoline (THQ, Fig. 1) series of inhibitors with a very potent CETP IC50 of 50 nm, measured in whole human plasma (30). Anacetrapib (3) contains the triad of trifluoromethyl groups found in torcetrapib but also has a distinct biaryl moiety (Fig. 1). Evacetrapib (4) contains a homologated core of torcetrapib and the 3,5-bis-trifluoromethylbenzyl group but also a methyl tetrazole and cyclohexane carboxylic acid side chain. Dalcetrapib (5) is more distinct from the fluorine-containing structures above, with an ortho-thio-anilide core (Fig. 1) and only modest CETP inhibitory activity in the human plasma assay (IC50, 9 μm), although it relies on forming a covalent disulfide linkage with CETP rather than reversible binding as in the above compounds. In a phase III clinical trial, torcetrapib demonstrated an increase of 72.1% in HDL-C and a decrease of 24.9% in LDL-C (18). In the same clinical trial, torcetrapib was also shown to increase systolic blood pressure and serum bicarbonate and decrease serum potassium at a daily dose of 60 mg (18). It is still not clear whether the increased risk associated with torcetrapib is due to its off-target effects. On the other hand, anacetrapib, dalcetrapib, and evacetrapib have been shown to increase HDL-C and lower LDL-C without increasing blood pressure (14–17, 23–25).
Illuminating the interactions of these small molecule compounds with CETP at the atomic level may further our understanding of how CETP facilitates lipid transfer, shed light onto how these compounds inhibit CETP, and provide insight into drug design. Previously, we reported the crystal structure of the holo-CETP structure (3). The holo-protein structure reveals a long hydrophobic tunnel in which four lipid molecules are found to bind. Based on the structure we proposed a mechanism by which the lipids transfer through this hydrophobic tunnel between HDL and LDL (3). A recent report of single particle electron microscopy imaging experiments indicate that CETP bridges a ternary complex with its N-terminal domain penetrating into HDL particles and its C-terminal domain interacting with LDL or VLDL particles, supporting the tunnel mechanism for neutral cholesteryl ester transfer and illuminating the direction of lipid transfer (31).
To study the molecular basis of CETP inhibition, holo-CETP crystals were produced using previously described methods (3) and soaked with saturated solutions of torcetrapib or a structurally distinct inhibitor ((2R)-3-{[4-(4-chloro-3-ethylphenoxy)pyrimidin-2-yl][3-(1,1,2,2-tetrafluoroethoxy)benzyl]amino}-1,1,1-trifluoropropan-2-ol), herein called compound 2 (Fig. 1, IC50 of 37 nm in the whole human plasma inhibition assay) to produce complex crystals. The inhibitors were observed unambiguously in the omit (2Fo − Fc) and (Fo − Fc) electron density maps generated from x-ray diffraction data collected at synchrotron sources (Fig. 2). Both CETP-inhibitor complex structures have been fully refined, with the torcetrapib complex structure having the higher resolution of 2.6 Å (Table 1).
FIGURE 2.
Qualities of electron density maps for inhibitors. a, the refined model of torcetrapib (stick model, with carbon atoms colored magenta, oxygen colored red, nitrogen colored blue, and fluorine colored light blue) embedded in the initial 2Fo − Fc omit map contoured at 1.2σ (colored in gray). Protein carbon atoms are colored green, and cholestryl ester carbon atoms are colored yellow. Cys-13 is highlighted. b, the refined model of compound 2 embedded in the initial Fo − Fc omit map contoured at 3σ.
TABLE 1.
Data collection and refinement statistics
| Torcetrapib | Compound 2 | |
|---|---|---|
| Data collection | ||
| Space group | P212121 | P212121 |
| Cell dimensions | a = 69.6, b = 69.3, and c = 188.2 Å; α, β, and γ = 90° | a = 68.8, b = 69.9, c = 187.1 Å; α, β, and γ = 90° |
| Resolution (Å)a | 50.0–2.6 (2.67–2.6) | 50–3.1 (3.21–3.1) |
| Rmerge | 0.083 (0.34) | 0.111 (0.513) |
| I/σ | 18.5 (2.4) | 12.1 (2.1) |
| Completeness (%) | 79.2 (30.9) | 76.1 (30.6) |
| Redundancy | 4.0 (1.6) | 5.0 (2.2) |
| Refinement | ||
| Resolution (Å) | 50–2.6 | 50–3.1 |
| No. of reflections | 21,810 | 11,592 |
| Rwork/Rfree | 0.213/0.259 | 0.198/0.239 |
| No. of atoms | ||
| Protein | 3,740 | 3,700 |
| Ligand/ion | 239 | 227 |
| Water | 110 | 32 |
| r.m.s.d bond lengths (Å)b | 0.009 | 0.008 |
| r.m.s.d bond angles | 1.3° | 1.2° |
| Ramachandran plot favored, allowed, & disallowed (%) | 95.7, 99.6, and 0.6 | 92.6, 99.6, and 0.4 |
a Statistics in the highest resolution shell are shown in parentheses.
b r.m.s.d., root mean square deviation.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Detailed descriptions on DNA constructs, protein expression, and purifications have been reported previously (3). The human CETP construct (1–476, sequence numbering starts at the first amino acid of the mature protein after signal peptide removed) used for crystallization in this study contains five point mutations, C1A, N88D, C131A, N240D, and N341D, to eliminate heterogeneous post-translational modifications on the protein surface to facilitate protein crystallization (3). Protein expression was carried out in Chinese hamster ovary cell line DG44. CETP was purified through an immobilized monoclonal antibody column (the monoclonal antibody was immobilized on CNBr-activated Sepharose Fast Flow resin, GE Healthcare), a hydrophobic interaction column (Butyl-650 Toyopearl M) (Toshoh Haas, Montgomeryville, PA), and an anion exchange column (Q Sepharose Fast Flow, GE Healthcare) (3). Two CETP peaks eluted from the anion exchange column were further purified through the same hydrophobic interaction column described above (3). The purified mutant CETP has similar activity as the wild type protein. The typical protein yield after final purification is >10 mg protein per liter of expression media, with purity >95%.
Crystal Soaking and Structure Determination
Crystallization of the holo-CETP has been reported previously (3). Briefly, the holo-CETP crystals were obtained by hanging drop vapor diffusion, using 10 mg ml−1 protein in a buffer of 20 mm Tris, pH 8.0, 250 mm NaCl, and 1 mm EDTA mixed 1:1 with a well solution of 0.1 m HEPES (pH 7.5), 0.2 m MgCl2, and 27–35% (w/v) PEG 400 at 4 °C. Note that neither cholesteryl ester nor phospholipid was added during purification and crystallization, but they were detected in purified protein sample using mass spectrum analysis and observed in the holo-CETP crystal structure (3). A large number of holo-CETP crystals were soaked at 4 °C in mother liquors containing saturate concentrations of inhibitors, 0.1% β-octylglucoside, 0.2 m MgCl2, 0.1 m HEPES buffer at pH 7.5 and 30% polyethylene glycol 400 for days. The crystals were then cooled directly in liquid nitrogen before data collection.
Crystallographic data sets were collected at the 17-ID beamline of the Advanced Photon Source at the Argonne National Laboratory (Chicago, IL). Diffraction data were processed with the program suite HKL-2000 (32), whereas the structure solution and refinement were carried out using the CCP4 program suite (33). The starting CETP model was derived from the holo-CETP structure (Protein Data Bank code 2OBD), excluding the bound lipids and solvent molecules, and the manual model building was carried out using program COOT (34). The inhibitor-bound structures have been refined satisfactorily. The diffraction data collection and final refinement statistics are listed in Table 1. Probably due to prolonged soaking that was necessary to observe inhibitors, all crystals suffered from loss of resolution and anisotropic diffractions. The torcetrapib-CETP complex crystal has a higher resolution. Its data were complete to 2.8 Å, and partially complete between 2.6 to 2.8 Å due to anisotropic diffraction pattern (Table 1).
Mutagenesis of CETP
Mutant CETP cDNAs were cloned into a modified version of pSecTag2/Hygro containing N-terminal His6 and V5 tags (Invitrogen). HEK293S cells were cultured and transfected with the cDNAs as described previously (35). Medium from transfected cells was collected and concentrated. With the aid of GeneTools software (Syngene), CETP yields were normalized by Western blot analysis. The cDNAs of C13A, R201A, H232A, and F263A mutants were engineered by overlapping PCR using a wild-type DNA template and mutagenic primer. These CETP mutants were purified as described.
Activity and Inhibition Assay of CETP
The activity of CETP and its mutants was measured using a cholesterol transfer assay (30). In this assay, the dual-labeled [3H]triolein and [14C]cholesteryl oleate, and 200 ng of CETP was used. The amount of [3H,14C]LDL was 6.7 nmol per assay, whereas HDL was 10.5 nmol per assay. The HDL contained a 2:1 mix of HDL2 and HDL3 fractions. CETP inhibition assays were performed using equal amounts of the wild-type and mutant protein samples (35, 36).
Synthesis of Torcetrapib and Compound 2
The chemical synthesis and characterizations of torcetrapib and compound 2 have been reported previously (37, 38). The characterization and purities of compounds are done using mass spectrum and NMR.
RESULTS
Torcetrapib Occupies the N-terminal Pocket of the CETP Tunnel
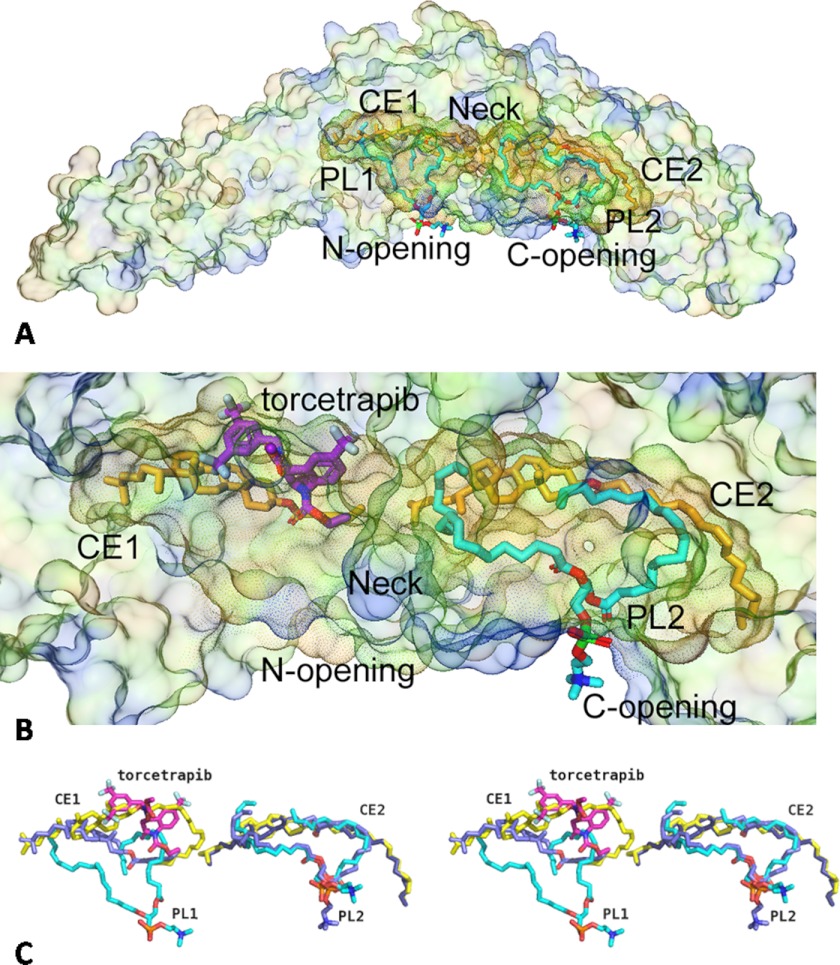
The holo-CETP structure revealed an exceptionally long hydrophobic tunnel made of two large pockets (the N-terminal pocket and the C-terminal pocket, herein called the N-pocket and the C-pocket, respectively), connected by a narrower neck (3). Point mutations narrowing the neck blocked triglyceride transfer and severely hindered cholesteryl ester transfer, suggesting a molecular mechanism involving neutral lipids transiting through the tunnel via the neck (3). Each binding pocket was occupied by one cholesteryl ester and one phospholipid molecule (CE1, PL1, CE2, and PL2), respectively, with the cholesteryl esters deeply buried and the phospholipids plugging the tunnel openings toward solvent (Fig. 3a). This tunnel mechanism is further supported by the recently reported single particle electron microscopy imaging studies of the CETP complex with HDL and LDL (31). In the structures of the CETP-inhibitor complexes, which were obtained from soaking the holo-CETP crystals that contain one copy of CETP in the asymmetric unit, one inhibitor was found in each protein, which provided molecular level validation of the 1:1 stoichiometry and non-competitive inhibition model previously suggested through traditional biochemical methods (30).
FIGURE 3.
CETP inhibitors occupy the N-terminal pocket of CETP lipid binding tunnel. a, in holo-CETP, the N- (N) and C-terminal (C) pockets of the lipid tunnel are occupied by one cholesterol ester molecule and one phospholipid molecule (carbon atoms are colored cyan) each (labeled CE1, PL1, CE2, and PL2, respectively). Two pockets are connected via a narrower neck (highlighted). The outer surface of CETP is shown in light, transparent color, and the inside surface of the lipid binding tunnel is shown in darker colors. b, an enlarged view of the lipid tunnel of CETP in CETP-torecetrapib complex. Torcetrapib (carbon atoms are colored magenta) occupies only the N-terminal pocket of CETP, shifting CE1 and displacing PL1. c, stereo view of overlay of ligands in the holo-CETP and torcetrapib-CETP complex (carbon atoms are in light blue), to show that the inhibitor shifts the cholesterol molecule and displaces the phospholipid in the N-terminal pocket of CETP, but lipids remain bound in the C-terminal pocket.
Torcetrapib occupies a volume of ∼12 Å × 12 Å × 7 Å within the N-terminal pocket of the CETP tunnel (Fig. 3b). In the CETP-inhibitor complex, no electron density was observed for the phospholipid molecule (PL1) in the N-pocket of the holo-CETP crystals, although there was still a large space left in the N-pocket after inhibitor binding (Fig. 3b), suggesting that the inhibitor entered the tunnel through the N-terminal opening and displaced the bound lipid during crystal soaking. Torcetrapib may weaken phospholipid binding because it has some overlaps with the oleyl chain of PL1 but does not necessarily exclude phospholipid from binding in vivo in the space left in the presence of the inhibitor because of the flexibility of phospholipids. The cholesteryl ester in the N-terminal pocket (CE1), cohabiting with the bound inhibitor, is observed in a drastically different conformation, with the CE1 ester moiety shifted by >10 Å from its position in holo-CETP (Fig. 3c). CE1, specially its oleoyl tail, appears to be less ordered in the inhibitor complex than in the holo-CETP (Fig. 3c).
In contrast, the overall structures of CETP protein and the bound lipids in the C-terminal side of the tunnel (CE2 and PL2) remain undisturbed upon torcetrapib binding, although the phosphate head group of PL2 does occupy slightly different positions in different crystal structures, most probably due to its flexible nature (Fig. 3, b and c). Our results suggest that torcetrapib binding physically interferes with PL1 binding and forces CE1 into a position that is presumably unfavorable for lipid transfer by blocking the narrow passage. Indeed, torcetrapib locates at a position that is near the narrow neck of the hydrophobic tunnel of CETP and can restrict the lipid flow through the tunnel (Fig. 3b). This mechanism supports the proposed tunnel-based lipid transfer model in which all neutral lipid transfers pass through this neck, and it is also consistent with the effects of CETP inhibitors (30).
All Torcetrapib-CETP Interactions Are Hydrophobic
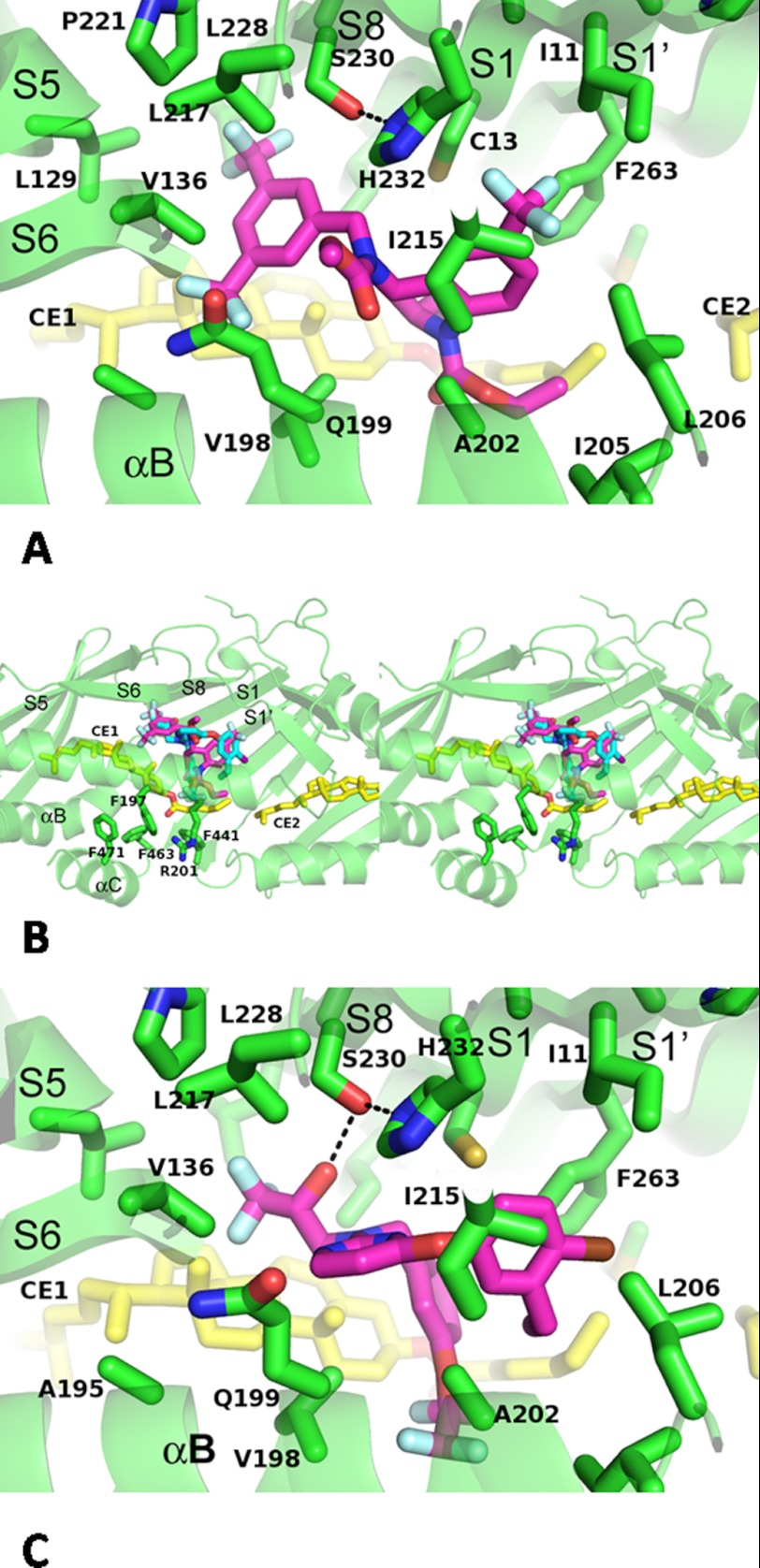
Torcetrapib interacts with residues located on the β-strands of S1, S5, S6, S8, and S1′, and α-helix B, all in the N-terminal half of CETP except for the S1′ strand (Fig. 4a). The S1′ strand contains Phe-263, which is at the neck of the hydrophobic tunnel. All these interactions are of a hydrophobic nature, and the entire N-terminal pocket is observed to be highly hydrophobic, which is consistent with both the highly lipophilic nature of neutral lipid substrates and the high log partition coefficients (logP) for the potent inhibitors. However, there are also three polar residues in the center of the inhibitor binding site (Fig. 4a), Gln-199, Ser-230, and His-232. The hydrophilic atoms of these residues either point away from the inhibitor (Gln-199) or interact with each other (Ser-230 and His-232) and do not form hydrogen bonds with the inhibitor.
FIGURE 4.
CETP-inhibitor interactions. a, binding interactions of torcetrapib with CETP. Protein main chains are shown as secondary structures cross-out. Side chains that interact with torcetrapib are shown as stick models and labeled. Secondary structures are named according to Ref. 3. b, stereo view of overlay of torcetrapib (magenta-colored carbon atoms) and compound 2 (carbon atoms colored cyan) in the N-terminal pocket of CETP. The N-terminal opening is highlighted side chains of surrounding residues. c, interactions between compound 2 and CETP. Hydrogen bonds are shown as dashes.
The THQ core of torcetrapib binds near the narrowing of the CETP tunnel, forming hydrophobic interactions with Phe-263 (Fig. 4a), which forms part of the neck of the CETP tunnel (3). The ethyl group on the THQ makes hydrophobic contacts with the oleoyl tail of CE1, which was incompletely modeled due to its disorder. The 6-trifluoromethyl group on the THQ projects deeply into the N-terminal pocket, occupying a sub-pocket formed by side chains of Ile-11, Cys-13, Ile-215, and the aromatic faces of His-232 and Phe-263 (Fig. 4a). This interaction involving the trifluoromethyl group explains the structure-activity relationship observed in other THQ-containing analogs: removing the 6-trifluoromethyl group from the THQ results in a greater than 18-fold loss in activity. One of the trifluoromethyl groups from the bis(trifluoromethyl)benzyl group inserts between the cholesterol rings of CE1 and Val-136 and Gln-199, and the second trifluoromethyl group occupies a another sub-pocket that is defined by the side chains of Leu-129, Val-136, Val-198, Pro-221, and Leu-228 (Fig. 4a).
The interactions between torcetrapib and CE1 indicate that the latter is an integral part of the inhibitor binding site and should be considered in inhibitor design. Indeed, replacing the 2-ethyl group on the THQ with bulkier groups, such as cyclopropyl or isopropyl, led to 1.5- or 1.9-fold lower potencies, respectively.
The methyl carbamate occupies a third sub-pocket formed by Val-198, Gln-199, Ala-202, and Ile-215. The ethyl carbamate points toward the opening of the N-terminal pocket, which is shielded by Arg-201 and aromatic residues Phe-197, Phe-441, Phe-463, and Phe-471 (Fig. 4b).
Potentially, a polar substituent could be introduced to reach out of the N-terminal opening and disrupt the flexible C-terminal helix that is important to CETP activity (3) or to interact with the side chain of Arg-201. Additional polar groups could also be incorporated to interact with the side chains of Gln-199, Ser-230, and His-232, which are close to torcetrapib. Such a polar substituent may not decrease the binding energy of CETP inhibitors if a favorable hydrogen bond can be formed with those residues and could help their aqueous solubility and free fractions in plasma, which are generally very low in this series and require non-traditional formulations for good oral absorption (39).
Binding Mode of Compound 2
Compound 2 is a potent, structurally distinct CETP inhibitor with unique functional groups, a trifluoropropan-2-ol and a tetrafluoroethoxyphenyl. It was designed in an effort to improve the overall solubility and free fraction of the CETP inhibitors by adding hydrophilic groups (hydroxyl and pyrimidine ring in this case) before structural information was available. Unfortunately, the aqueous solubility of compound 2 is still low, with a >8 calculated logP. However, its slightly more polar nature did not decrease its binding affinity (IC50 = 12 nm, serum-free) or its activity in human plasma (IC50 = 37 nm).
Despite the large structure differences, compound 2 occupies the same binding site as torcetrapib in the N-terminal pocket of CETP, with the physicochemically equivalent groups overlaying well in the two structures (Fig. 4b). The chlorophenoxyl group of compound 2 is closest to the neck of the hydrophobic tunnel of CETP (Fig. 4b) and occupies the same pocket as the THQ core trifluoromethyl of torcetrapib. The pyrimidine of compound 2 overlays the methyl carbamate of torcetrapib, occupying the sub-pocket formed by Val-198, Gln-199, Ala-202, and Ile-215 and making π-π interactions with His-232 side chain. Although the trifluoropropan-2-ol binds to the same sub-pocket as the second trifluoromethyl of the bis(trifluoromethyl)benzyl in torcetrapib, compound 2 lacks a group equivalent to the first trifluoromethyl group that interacts with the ring of CE1 and Val-136 and Gln-199.
The most distinct interaction between compound 2 and CETP is the hydrogen bond between the hydroxyl and the side chain of Ser-230 (Fig. 4C). Such a hydrogen bond could compensate the dehydration penalty of burying a hydrophilic group and preserve the binding affinity of inhibitors. The phenyl moiety of the tetrafluoroethoxylbenzyl group superimposes with the THQ 2-ethyl in torcetrapib, interacting with the tail of CE1, whereas the tetrafluoroethoxyl group extends further toward the N-terminal opening and is in van der Waals contact distances to the side chains of Arg-201 and Phe-441 (Fig. 4b). Further extension of this group may provide another option for solubility improvement.
Mutagenesis Studies of CETP Inhibition
To further validate the inhibitor binding modes observed in our crystal structures, four single point mutants, C13A, R201A, H232A, and F263A, were produced and tested in the in vitro inhibition assay under serum-free conditions. The specific lipid transfer activities of the mutants are mostly similar to that of the WT protein, with H232A being slightly (70%) more active than WT (Table 2).
TABLE 2.
Activity and inhibition of the wild type CETP (WT) and C13A, R201A, H232A, and F263A mutants (serum-free)
| Transfer activity (Relative CE) | Torcetrapib (IC50) | Compound 2 (IC50) | |
|---|---|---|---|
| nm | nm | ||
| WT | 1.0 | 4.3 | 13 |
| C13A | 0.9 | 44 | 11 |
| R201A | 1.1 | 12 | 45 |
| H232A | 1.7 | 220 | Inactive |
| F263A | 1.1 | 35 | 14 |
Torcetrapib is shown to inhibit the transfer activity of the wild-type protein with an IC50 of 4.3 nm serum-free (50 nm with serum), compared with serum-free IC50 of 44 nm, 12 nm, 220 nm, and 35 nm for C13A, R201A, H232A, and F263A, respectively (Table 2). Cys-13, His-232, and Phe-263 are in direct contact with the bound inhibitor (Fig. 4a), explaining the 8–50-fold IC50 increase for these mutants (Table 2). Among these mutations, H232A has the largest influence on the IC50 of torcetrapib, presumably because it not only loses a direct interaction with the quinoline of torcetrapib, but also makes Ser-230 lose its hydrogen bond partner (Fig. 4c) and thus the binding site less hydrophobic. The Arg-201 side chain does not interact directly with torcetrapib in the crystal structure, and accordingly, only a small effect (<3-fold) is seen with the R201A mutant.
In contrast to torcetrapib, the tetrafluoroethoxyl group of compound 2 makes a direct hydrophobic contact with the side chain of Arg-201 (Fig. 4b), explaining the weaker potency of this compound for R201A mutant (Table 2). H232A mutation produces the most dramatic decrease in potency for compound 2, making it essentially inactive (Table 2). A H232A mutation removes the hydrogen bond between His-232 and Ser-230, which makes the latter not positioned to accept the hydrogen bond from the hydroxyl of compound 2. It is not entirely clear why C13A and F263A mutations have minimum effects on the potency of compound 2. Compound 2 might be able to maintain similar potency at the binding sites of C13A or F263A mutants with a rotational adjustment of the chloro-ethylphenoxy group in combination with shifts in the CE binding mode, which would not be unusual given the nonspecific nature of these hydrophobic interactions. Further structural work might be needed to fully resolve this question.
DISCUSSION
The mechanism for the adverse effects of torcetrapib is still not entirely clear, but the combination of the highly lipophilic nature (cLogP = 7.6), the extremely low aqueous solubility and high protein binding of torcetrapib might increase the chances of off-target interactions. The high lipophilicity of torcetrapib and its molecular weight (>500) constitute two violations of the “Lipinski's Rule-of-5” (40).
Based on our CETP-inhibitor complex structures, it may be possible to incorporate more hydrophilic groups that interact with Ser-230, His-232, or Gln-199, which are located at the interior of the inhibitor binding pocket, or Arg-201, at the entrance, to improve compound solubility while maintaining binding affinities. For compound 2, the formation of a hydrogen bond between the sole polar hydroxyl group and Ser-230 demonstrates that polar interactions can be compatible with high potency (Fig. 4c). Similarly, polar groups may be attached to the ethyl carbamate of torcetrapib or the tetrafluoroethoxyl group of compound 2 via a spacer (13). In such a binding mode, the polar groups would point toward the opening of the N-terminal pocket. In our modeling exercise, the cyclohexanecarboxylic acid of evacetrapib indeed points toward the opening of the N-terminal pocket.
The chemical structure of JTT-705 is completely different from that of torcetrapib, anacetrapib, and evacetrapib (Fig. 1). Chemical analysis and mutagenesis studies have shown that the thioester of JT-705 can be readily hydrolyzed to produce a free thiol and form a disulfide bond to Cys-13 (17), possibly leading to a slow off-rate for the covalently attached moiety. Results from a recent study indicate that JTT-705 inhibition of CETP is time-dependent (41). We were not able to soak JTT-705 into the holo-CETP crystal, probably due to the weak activity (IC50 of 9 μm) (17), and lack of chemical reactivity under our soaking conditions. In our structures, the Cys-13 free thiol is sandwiched between the side chains of His-232 and Phe-263. All three residues spread at the depth of the N-terminal pocket and make critical interactions with inhibitors. It is therefore tempting to model the JTT-705 binding mode, with its thiol covalently linked to Cys-13 and the phenyl ring in aromatic stacking interactions with Phe-263, a residue on the passage from the N-terminal pocket to the neck. JT-705, the smallest of the four compounds (Fig. 1), should occupy the least volume in the N-terminal pocket but still large enough to clog the N-terminal pocket and hinder the binding and transfer of neutral lipids.
Due to the chemical similarities between torcetrapib, anacetrapib, and evacetrapib (Fig. 1), it is reasonable to speculate that anacetrapib and evacetrapib adopt binding modes similar to that of torcetrapib, which is further supported by additional structures of CETP in complex with similar compounds.6 Therefore, our results suggest that all current clinical trial compounds, anacetrapib, dalcetrapib (JTT-705), and evacetrapib, bind in the CETP N-terminal pocket and inhibit lipid transfer in a mechanism similar to that of torcetrapib. This conclusion is supported by the experimental results that torcetrapib, anacetrapib, and dalcetrapib are competitive in CETP binding (41).
In surface plasmon resonance and gel shift experiments, torcetrapib (and compound 2) was found to enhance CETP binding to HDL particles, a model that could explain its ability to block neutral lipid and phospholipid transfer (30). It was further speculated that the increased HDL binding may be linked to the adverse clinical effects of torcetrapib (29, 42). Our structure revealed that torcetrapib and compound 2, similar to neutral lipids, are buried deep inside the tunnel rather than bridging the CETP-HDL interface. Indeed, anacetrapib and dalcetrapib have also been found to promote the formation of CETP-HDL complex (41), suggesting that tunnel blocking by inhibitors slow cholesteryl ester transfer and the subsequent HDL dissociation.
Unlike torcetrapib, in clinical trials anacetrapib and dalcetrapib do not increase the blood pressure (14). Furthermore, the effect of blood pressure increase by torcetrapib was shown not to be linked to CETP inhibition but instead to increased levels of aldosterone (43). Alternatively, the adverse clinic effects of torcetrapib may be completely due to its interactions with targets yet to be determined, and these off-targets are not likely shared by all CETP inhibitors.
Plasma HDL is a very complex biological system and plays multiple physiological roles (44–47). Although the blood pressure side effects of torcetrapib could be addressed with current compounds (14–17, 23–25), there is still an urgent need for CETP inhibitors with distinct pharmacological properties and modes of action to fully test the target via clinical trials and to provide treatments for the growing population of cardiovascular disease patients worldwide. Our crystal structure should help these efforts. In addition to ideas outlined above, other parts of the CETP tunnel such as the C-terminal pocket could be potentially targeted for inhibitor design as well. Finally, once the molecular basis for CETP-lipoprotein interaction is better defined, further opportunities may exist for designing novel CETP inhibitors.
Acknowledgments
We thank Tim Subashi, Kim McGrath, Kim Stutzman-Engwall, Yang Cong, George Karam, and Peter Dorff for various reagents and Meihua Tu, Bruce Lefker, Mary Didiuk, Gregory Chang, Cheryl Hayward, Mark Bamberger, John Thompson, Lee Morehouse, David Cunningham, David Hepworth, Andrew Seddon, and Ron Wester for valuable discussions. We also thank Kieran Geoghegan for proofreading the manuscript.
The atomic coordinates and structure factors (codes 4EWS and 4F2A) have been deposited in the Protein Data Bank (http://wwpdb.org/).
S. Liu, A. Mistry, J. M. Reynolds, D. B. Lloyd, M. C. Griffor, D. A. Perry, R. B. Ruggeri, R. W. Clark, and X. Qiu, unpublished material.
- HDL-C
- HDL cholesterol
- CETP
- cholesteryl ester transfer protein
- THQ
- tetrahydroquinoline
- LDL-C
- LDL-cholesterol
- VLDL
- very low-density lipoprotein.
REFERENCES
- 1. Bruckert E., Hansel B. (2007) HDL-c is a powerful lipid predictor of cardiovascular diseases. Int. J. Clin. Pract. 61, 1905–1913 [DOI] [PubMed] [Google Scholar]
- 2. Barter P. J., Brewer H. B., Jr., Chapman M. J., Hennekens C. H., Rader D. J., Tall A. R. (2003) Cholesteryl ester transfer protein: a novel target for raising HDL and inhibiting atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 23, 160–167 [DOI] [PubMed] [Google Scholar]
- 3. Qiu X., Mistry A., Ammirati M. J., Chrunyk B. A., Clark R. W., Cong Y., Culp J. S., Danley D. E., Freeman T. B., Geoghegan K. F., Griffor M. C., Hawrylik S. J., Hayward C. M., Hensley P., Hoth L. R., Karam G. A., Lira M. E., Lloyd D. B., McGrath K. M., Stutzman-Engwall K. J., Subashi A. K., Subashi T. A., Thompson J. F., Wang I. K., Zhao H., Seddon A. P. (2007) Crystal structure of cholesteryl ester transfer protein reveals a long tunnel and four bound lipid molecules. Nat. Struct. Mol. Biol. 14, 106–113 [DOI] [PubMed] [Google Scholar]
- 4. Brown M. L., Inazu A., Hesler C. B., Agellon L. B., Mann C., Whitlock M. E., Marcel Y. L., Milne R. W., Koizumi J., Mabuchi H. (1989) Molecular basis of lipid transfer protein deficiency in a family with increased high-density lipoproteins. Nature 342, 448–451 [DOI] [PubMed] [Google Scholar]
- 5. Inazu A., Brown M. L., Hesler C. B., Agellon L. B., Koizumi J., Takata K., Maruhama Y., Mabuchi H., Tall A. R. (1990) Increased high-density lipoprotein levels caused by a common cholesteryl-ester transfer protein gene mutation. N. Engl. J. Med. 323, 1234–1238 [DOI] [PubMed] [Google Scholar]
- 6. Agellon L. B., Walsh A., Hayek T., Moulin P., Jiang X. C., Shelanski S. A., Breslow J. L., Tall A. R. (1991) Reduced high density lipoprotein cholesterol in human cholesteryl ester transfer protein transgenic mice. J. Biol. Chem. 266, 10796–10801 [PubMed] [Google Scholar]
- 7. Sugano M., Makino N., Sawada S., Otsuka S., Watanabe M., Okamoto H., Kamada M., Mizushima A. (1998) Effect of antisense oligonucleotides against cholesteryl ester transfer protein on the development of atherosclerosis in cholesterol-fed rabbits. J. Biol. Chem. 273, 5033–5036 [DOI] [PubMed] [Google Scholar]
- 8. Linsel-Nitschke P., Tall A. R. (2005) HDL as a target in the treatment of atherosclerotic cardiovascular disease. Nat. Rev. Drug. Discov. 4, 193–205 [DOI] [PubMed] [Google Scholar]
- 9. Dullaart R. P., Dallinga-Thie G. M., Wolffenbuttel B. H., van Tol A. (2007) CETP inhibition in cardiovascular risk management: a critical appraisal. Eur. J. Clin. Invest. 37, 90–98 [DOI] [PubMed] [Google Scholar]
- 10. Joy T., Hegele R. A. (2008) Is raising HDL a futile strategy for atheroprotection? Nat. Rev. Drug. Discov. 7, 143–155 [DOI] [PubMed] [Google Scholar]
- 11. Cannon C. P. (2011) High-density lipoprotein cholesterol as the Holy Grail. Jama 306, 2153–2155 [DOI] [PubMed] [Google Scholar]
- 12. World Health Organization (2008) The Atlas of Heart Disease and Stroke, World Health Organization, Geneva, Switzerland [Google Scholar]
- 13. Ruggeri R. B. (2005) Cholesteryl ester transfer protein: pharmacological inhibition for the modulation of plasma cholesterol levels and promising target for the prevention of atherosclerosis. Curr. Top Med. Chem. 5, 257–264 [DOI] [PubMed] [Google Scholar]
- 14. Krishna R., Anderson M. S., Bergman A. J., Jin B., Fallon M., Cote J., Rosko K., Chavez-Eng C., Lutz R., Bloomfield D. M., Gutierrez M., Doherty J., Bieberdorf F., Chodakewitz J., Gottesdiener K. M., Wagner J. A. (2007) Effect of the cholesteryl ester transfer protein inhibitor, anacetrapib, on lipoproteins in patients with dyslipidaemia and on 24-h ambulatory blood pressure in healthy individuals: two double-blind, randomised placebo-controlled phase I studies. Lancet 370, 1907–1914 [DOI] [PubMed] [Google Scholar]
- 15. Ali A., Napolitano J. M., Deng Q., Lu Z., Sinclair P. J., Taylor G. E., Thompson C. F., Quraishi N. (July 1, 2005) U. S. Patent 2006/0040999 A1
- 16. Cao G., Beyer T. P., Zhang Y., Schmidt R. J., Chen Y. Q., Cockerham S. L., Zimmerman K. M., Karathanasis S. K., Cannady E. A., Fields T., Mantlo N. B. (2011) Evacetrapib is a novel, potent, and selective inhibitor of cholesteryl ester transfer protein that elevates HDL cholesterol without inducing aldosterone or increasing blood pressure. J. Lipid Res. 52, 2169–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Okamoto H., Yonemori F., Wakitani K., Minowa T., Maeda K., Shinkai H. (2000) A cholesteryl ester transfer protein inhibitor attenuates atherosclerosis in rabbits. Nature 406, 203–207 [DOI] [PubMed] [Google Scholar]
- 18. Barter P. J., Caulfield M., Eriksson M., Grundy S. M., Kastelein J. J., Komajda M., Lopez-Sendon J., Mosca L., Tardif J. C., Waters D. D., Shear C. L., Revkin J. H., Buhr K. A., Fisher M. R., Tall A. R., Brewer B. (2007) Effects of torcetrapib in patients at high risk for coronary events. N. Engl. J. Med. 357, 2109–2122 [DOI] [PubMed] [Google Scholar]
- 19. Nissen S. E., Tardif J. C., Nicholls S. J., Revkin J. H., Shear C. L., Duggan W. T., Ruzyllo W., Bachinsky W. B., Lasala G. P., Lasala G. P., Tuzcu E. M., and ILLUSTRATE Investigators (2007) Effect of torcetrapib on the progression of coronary atherosclerosis. N. Engl. J. Med. 356, 1304–1316 [DOI] [PubMed] [Google Scholar]
- 20. Kastelein J. J., van Leuven S. I., Burgess L., Evans G. W., Kuivenhoven J. A., Barter P. J., Revkin J. H., Grobbee D. E., Riley W. A., Shear C. L., Duggan W. T., Bots M. L. (2007) Effect of torcetrapib on carotid atherosclerosis in familial hypercholesterolemia. N. Engl. J. Med. 356, 1620–1630 [DOI] [PubMed] [Google Scholar]
- 21. Bots M. L., Visseren F. L., Evans G. W., Riley W. A., Revkin J. H., Tegeler C. H., Shear C. L., Duggan W. T., Vicari R. M., Grobbee D. E., Kastelein J. J. (2007) Torcetrapib and carotid intima-media thickness in mixed dyslipidaemia (RADIANCE 2 study): a randomized, double-blind trial. Lancet 370, 153–160 [DOI] [PubMed] [Google Scholar]
- 22. Hooper A. J., Burnett J. R. (2012) Dalcetrapib, a cholesteryl ester transfer protein modulator. Expert Opin. Investig. Drugs 21, 1427–1432 [DOI] [PubMed] [Google Scholar]
- 23. Vergeer M., Kastelein J. J. (2008) Anacetrapib: new hope for cholesteryl ester transfer protein inhibitors in the treatment of dyslipidemia. Nat. Clin. Pract. Cardiovasc Med. 5, 302–303 [DOI] [PubMed] [Google Scholar]
- 24. Stein E. A., Roth E. M., Rhyne J. M., Burgess T., Kallend D., Robinson J. G. (2010) Safety and tolerability of dalcetrapib (RO4607381/JTT-705): results from a 48-week trial. Eur. Heart. J. 31, 480–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nicholls S. J., Brewer H. B., Kastelein J. J., Krueger K. A., Wang M. D., Shao M., Hu B., McErlean E., Nissen S. E. (2011) Effects of the CETP inhibitor evacetrapib administered as monotherapy or in combination with statins on HDL and LDL cholesterol: a randomized controlled trial. Jama 306, 2099–2109 [DOI] [PubMed] [Google Scholar]
- 26. Tall A. R., Yvan-Charvet L., Wang N. (2007) The failure of torcetrapib: was it the molecule or the mechanism? Arterioscler. Thromb. Vasc. Biol. 27, 257–260 [DOI] [PubMed] [Google Scholar]
- 27. Tall A. R. (2007) CETP inhibitors to increase HDL cholesterol levels. N. Engl. J. Med. 356, 1364–1366 [DOI] [PubMed] [Google Scholar]
- 28. Joy T., Hegele R. A. (2009) The end of the road for CETP inhibitors after torcetrapib? Curr. Opin. Cardiol. 24, 364–371 [DOI] [PubMed] [Google Scholar]
- 29. Hamilton J. A., Deckelbaum R. J. (2007) Crystal structure of CETP: new hopes for raising HDL to decrease risk of cardiovascular disease? Nat. Struct. Mol. Biol. 14, 95–97 [DOI] [PubMed] [Google Scholar]
- 30. Clark R. W., Ruggeri R. B., Cunningham D., Bamberger M. J. (2006) Description of the torcetrapib series of cholesteryl ester transfer protein inhibitors, including mechanism of action. J. Lipid Res. 47, 537–552 [DOI] [PubMed] [Google Scholar]
- 31. Zhang L., Yan F., Zhang S., Lei D., Charles M. A., Cavigiolio G., Oda M., Krauss R. M., Weisgraber K. H., Rye K. A., Pownall H. J., Qiu X., Ren G. (2012) Structural basis of transfer between lipoproteins by cholesteryl ester transfer protein. Nat. Chem. Biol. 8, 342–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Otwinowski Z., Borek D., Majewski W., Minor W. (2003) Multiparametric scaling of diffraction intensities. Acta Crystallogr. A 59, 228–234 [DOI] [PubMed] [Google Scholar]
- 33. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 34. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lloyd D. B., Reynolds J. M., Cronan M. T., Williams S. P., Lira M. E., Wood L. S., Knight D. R., Thompson J. F. (2005) Novel variants in human and monkey CETP. Biochim. Biophys. Acta 1737, 69–75 [DOI] [PubMed] [Google Scholar]
- 36. Lloyd D. B., Lira M. E., Wood L. S., Durham L. K., Freeman T. B., Preston G. M., Qiu X., Sugarman E., Bonnette P., Lanzetti A., Milos P. M., Thompson J. F. (2005) Cholesteryl ester transfer protein variants have differential stability but uniform inhibition by torcetrapib. J. Biol. Chem. 280, 14918–14922 [DOI] [PubMed] [Google Scholar]
- 37. Damon D. B., Dugger R. W., Scott R. W. (April 18, 2003) U. S. Patent 6706881
- 38. Chang G., Garigipati R. S., Lefker B., Perry D. A., Zeng D. (January 3, 2007) U. S. Patent 7919506
- 39. Gumkowski M. J., Franco L., Murdande S. B., Perlman M. E. (June 19, 2002) U. S. Patent 6962931
- 40. Lipinski C. A., Lombardo F., Dominy B. W., Feeney P. J. (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Del. Rev. 46, 3–26 [DOI] [PubMed] [Google Scholar]
- 41. Ranalletta M., Bierilo K. K., Chen Y., Milot D., Chen Q., Tung E., Houde C., Elowe N. H., Garcia-Calvo M., Porter G., Eveland S., Frantz-Wattley B., Kavana M., Addona G., Sinclair P., Sparrow C., O'Neill E. A., Koblan K. S., Sitlani A., Hubbard B., Fisher T. S. (2010) Biochemical characterization of cholesteryl ester transfer protein inhibitors. J. Lipid Res. 51, 2739–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Howes L. G., Kostner K. (2007) The withdrawal of torcetrapib from drug development: implications for the future of drugs that alter HDL metabolism. Expert Opin. Investig. Drugs 16, 1509–1516 [DOI] [PubMed] [Google Scholar]
- 43. Forrest M. J., Bloomfield D., Briscoe R. J., Brown P. N., Cumiskey A. M., Ehrhart J., Hershey J. C., Keller W. J., Ma X., McPherson H. E., Messina E., Peterson L. B., Sharif-Rodriguez W., Siegl P. K., Sinclair P. J., Sparrow C. P., Stevenson A. S., Sun S. Y., Tsai C., Vargas H., Walker M., 3rd, West S. H., White V., Woltmann R. F. (2008) Torcetrapib-induced blood pressure elevation is independent of CETP inhibition and is accompanied by increased circulating levels of aldosterone. Br J. Pharmacol. 154, 1465–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Barter P. J., Nicholls S., Rye K. A., Anantharamaiah G. M., Navab M., Fogelman A. M. (2004) Antiinflammatory properties of HDL. Circ. Res. 95, 764–772 [DOI] [PubMed] [Google Scholar]
- 45. Yvan-Charvet L., Matsuura F., Wang N., Bamberger M. J., Nguyen T., Rinninger F., Jiang X. C., Shear C. L., Tall A. R. (2007) Inhibition of cholesteryl ester transfer protein by torcetrapib modestly increases macrophage cholesterol efflux to HDL. Arterioscler. Thromb. Vasc. Biol. 27, 1132–1138 [DOI] [PubMed] [Google Scholar]
- 46. Vaisar T., Pennathur S., Green P. S., Gharib S. A., Hoofnagle A. N., Cheung M. C., Byun J., Vuletic S., Kassim S., Singh P., Chea H., Knopp R. H., Brunzell J., Geary R., Chait A., Zhao X. Q., Elkon K., Marcovina S., Ridker P., Oram J. F., Heinecke J. W. (2007) Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J. Clin. Invest. 117, 746–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pecheniuk N. M., Deguchi H., Elias D. J., Xu X., Griffin J. H. (2006) Cholesteryl ester transfer protein genotypes associated with venous thrombosis and dyslipoproteinemia in males. J. Thromb. Haemost. 4, 2080–2082 [DOI] [PubMed] [Google Scholar]