Background: The role of HSSTnT in cardiac muscle regulation is unknown.
Results: HSSTnT isoforms regulate Ca2+ sensitivity and maximal force of contraction in skinned fibers in a different fashion compared with HCTnT3.
Conclusion: HSSTnT isoforms despite being homologues of CTnT display distinct functional properties.
Significance: The HSSTnT isoforms, when expressed in the heart, would play a specific role in desensitizing the myofilament to Ca2+.
Keywords: Calcium-binding Proteins, Heart, Protein Complexes, Skeletal Muscle, Troponin, ATPase Activity, Slow Skeletal Troponin T, Circular Dichroism, Proteolysis, Skinned Fiber
Abstract
Human slow skeletal troponin T (HSSTnT) shares a high degree of homology with cardiac TnT (CTnT). Although the presence of HSSTnT has not been confirmed in the heart at the protein level, detectable levels of HSSTnT mRNA have been found. Whether HSSTnT isoforms are expressed transiently remains unknown. Because transient re-expression of HSSTnT may be a potential mechanism of regulating function, we explored the effect of HSSTnT on the regulation of cardiac muscle. At least three HSSTnT isoforms have been found to exist in slow skeletal muscle: HSSTnT1 (+exons 5 and 12), HSSTnT2 (+exon 5, −exon 12), and HSSTnT3 (−exons 5 and 12). Another isoform, HSSTnT hypothetical (Hyp) (−exon 5, +exon 12), has only been found at the mRNA level. Compared with HCTnT3 (adult isoform), Tn complexes containing HSSTnT1, -2, and -3 did not alter the actomyosin ATPase activation and inhibition in the presence and absence of Ca2+, respectively. HSSTnTHyp was not evaluated as it did not form a Tn complex under a variety of conditions. Porcine papillary skinned fibers displaced with HSSTnT1, -2, or -3 and reconstituted with human cardiac troponin I and troponin C (HCTnI·TnC) complex showed a decrease in the Ca2+ sensitivity of force development and an increase in maximal recovered force (HSSTnT1 and -3) compared with HCTnT3. In contrast, HSSTnTHyp showed an increase in the Ca2+ sensitivity of force development. This suggests that re- or overexpression of specific SSTnT isoforms might have therapeutic potential in the failing heart because they increase the maximal force of contraction. In addition, circular dichroism and proteolytic digestion experiments revealed structural differences between HSSTnT isoforms and HCTnT3 and that HSSTnT1 is more susceptible to calpain and trypsin proteolysis than the other HSSTnTs. Overall, HSSTnT isoforms despite being homologues of CTnT may display distinct functional properties in muscle regulation.
Introduction
The thin filament regulatory system consists of the troponin complex and tropomyosin (Tm).2 Tm has a double-stranded α-helical coiled-coil structure that non-covalently associates with actin, and the interaction between actin and myosin occurs at a much slower rate in the absence of Ca2+. Cardiac troponin (CTn) consists of three subunits: troponin C (CTnC), which binds Ca2+; troponin I (CTnI), which inhibits muscle contraction; and troponin T (CTnT), whose function is to link Tn to Tm. Cardiac contraction is activated when the intracellular free Ca2+ concentration rises. Consequently, Ca2+ binds to the N-terminal regulatory site of CTnC, inducing conformational changes that expose a pocket of hydrophobic residues that are then able to form new tight interactions with TnI (1–3). The strengthened TnC-TnI interaction is transmitted to TnT and promotes the translocation of the Tn·Tm complex away from the outer domain of the actin filaments, thus allowing cyclic interactions between myosin heads (S1) and actin. The myosin head, an actin-activated, Mg2+-ATPase-dependent motor, binds to actin and undergoes a power stroke, the phenomenon responsible for generating contractile force in muscle (4).
Three TnT genes have been identified: cardiac TnT (CTnT), fast skeletal TnT (FSTnT), and slow skeletal TnT (SSTnT). Each TnT gene transcript can be alternatively spliced to generate multiple isoforms (5). Three human SSTnT (HSSTnT) isoforms have been described, and this is due to alternative splicing of exons 5 and 12 (of a total of 14 exons) of the SSTnT gene that generates a limited number of isoforms (5–7). A high degree of diversity between FSTnT and CTnT can result from alternative splicing of the TnT gene, which can affect interactions between TnT and Tm (8) and the contractile properties of the cardiac system (9). Skinned cardiac fibers reconstituted with the human CTnT (HCTnT) isoform 1 (fetal) had a greater Ca2+ sensitivity of force development, whereas the HCTnT isoform 4 (fetal and adult diseased isoform) had a lower Ca2+ sensitivity of force development when compared with the HCTnT isoform 3 (adult)-reconstituted fibers (9). The possibility that TnT isoforms differ in their regulatory function is further supported by additional biochemical and physiological studies (10–13). The existence of multiple HSSTnT isoforms also suggests that they may play a functional role in regulating muscle physiology.
The SSTnT isoforms are abundantly expressed in skeletal muscle during development and in slow fibers from adult skeletal muscle (14), and the cardiac TnT gene is expressed throughout cardiogenesis. However, recent reports have shown that SSTnT is also transiently expressed during mouse (15, 16) and human (17) heart development. Interestingly, SSTnT mRNA is up-regulated in patients with end stage heart failure (17). However, the structural properties of SSTnT isoforms and their distinct effect on muscle regulation have never been explored. Because little is known about the role of SSTnT in adult heart and cardiac disease, we thought to investigate the functional consequences of HSSTnT isoforms on cardiac muscle regulation. Knowledge of the action of SSTnT isoforms in the heart is important because there is evidence of their transient expression as the heart becomes dysfunctional that may provide beneficial changes in function similar to the induction of the fetal gene program where α-myosin switches to β-myosin in the diseased mouse ventricle (18, 19).
Functional properties of sarcomeric variants have long been studied in divergent systems to further understand differences in their regulatory action. The function of FSTnT in the cardiac background has been studied in animal models (20, 21). Transgenic mice overexpressing FSTnT in the heart showed a reduction in cardiac function and diminished contractility, which led to the development of a pathologic phenotype (20). Skinned fiber measurements demonstrated that transgenic hearts overexpressing the FSTnT isoform were less tolerant of acidic pH conditions, manifested as a large decrease in Ca2+ sensitivity and maximal force at pH 6.5 (21). In another study, FSTnT exchanged into cardiac skinned fibers resulted in a lower maximal force and ATPase rate and a faster rate constant of tension redevelopment (22). These data demonstrate that FSTnT and CTnT although related as homologues modulate and regulate muscle contraction differently.
Here, we show that cardiac skinned fiber preparations that were displaced with HSSTnT isoforms demonstrated decreased Ca2+ sensitivity compared with fibers displaced with HCTnT3. In addition, the maximal force was found to be increased in fibers that were displaced with HSSTnT isoforms 1 and 3 compared with HCTnT. These results suggest that SSTnT isoforms possess distinct physical properties among themselves and also when compared with CTnT. These factors are likely to contribute to differences in their direct role in desensitizing the cardiac myofilament for Ca2+ (HSSTnT1 > HSSTnT2 = HSSTnT3) and their ability to regulate the amount of maximal force developed by cardiac fibers. In addition, the re- or overexpression of specific SSTnT isoforms might have therapeutic potential in the failing heart because they increase the maximal force of contraction.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of Human SSTnT
Human skeletal muscle poly(A)+ RNA from Clontech was utilized for RT-PCR using primers designed to amplify HSSTnT1. The complete nucleotide sequence of the cloned cDNA corresponded to the HSSTnT isoform 2 (+exon 5, −exon 12). The other HSSTnT isoforms were made using a sequential overlapping PCR method. The HSSTnT2 was used as a template for PCR with primers designed to introduce or delete the 33- or 48-bp regions, resulting in various combinations that are known as isoforms 1 (+exons 5 and 12) and 3 (−exons 5 and 12). The HSSTnT produced with the potential combination of alternatively spliced exons (−exon 5, +exon 12) has not been described previously and was also created by sequential overlapping PCR. This HSSTnT isoform (−exon 5, +exon 12) is called HSSTnT hypothetical (HSSTnTHyp). Screening of a human muscle cDNA library by Samson et al. (7) showed the presence of three HSSTnT isoforms and strongly suggested the expression of a fourth isoform. This hypothetical HSSTnT isoform found by RT-PCR and described in this report may be the fourth isoform proposed by Samson et al. (7). All subcloned DNA sequences were inserted into the expression plasmid pET3d and sequenced to verify the correct sequences prior to expression and purification. The expression and purification of the HSSTnT isoforms followed our standard laboratory protocol for HCTnT3. Briefly, the HSSTnT isoforms were expressed in Escherichia coli BL21 (DE3) CodonPlus bacterial cells (Stratagene). The bacterially expressed HSSTnT isoforms were passed over a fast flow S-Sepharose column and eluted with a linear gradient of 0–0.6 m KCl (4 °C) in a buffer containing 50 mm Tris-HCl (pH 7.0), 6 m urea, 2 mm EDTA, and 1 mm DTT. The fractions containing HSSTnT were dialyzed against 50 mm Tris-HCl (pH 7.8), 6 m urea, 1 mm EDTA, and 1 mm DTT; loaded onto a fast flow Q-Sepharose column; and eluted using a 0–0.6 m KCl gradient (4 °C). The purest fractions (>97% pure protein) analyzed by SDS-PAGE were pooled and immediately stored at −80 °C.
Cloning, Expression, and Purification of Human CTnT, CTnI, and CTnC
cDNAs cloned in our laboratory from human cardiac tissue were used for the expression and purification of CTnT, CTnI, and CTnC (23). Briefly, the CTnT, CTnI, and CTnC were expressed in E. coli BL21 (DE3) CodonPlus bacterial cells (Stratagene). The bacterially expressed HCTnT3 was purified according to a well established laboratory protocol and as described above for HSSTnT isoforms. The bacterially expressed CTnI was first purified by column chromatography on a fast flow S-Sepharose column at 4 °C and eluted with a linear KCl gradient of 0–0.6 m in a buffer as described above. Semipure CTnI was dialyzed against a solution containing 50 mm Tris-HCl (pH 7.5), 1 m NaCl, 2 mm CaCl2, and 1 mm DTT and loaded onto a CTnC affinity column. Pure HCTnI was eluted using a gradient of 0–1 mm EDTA and 0–6 m urea. The bacterially expressed CTnC was passed over a Q-Sepharose column and eluted with a linear gradient of 0–0.6 m KCl (4 °C) in a buffer as described above. The purest TnC fractions were dialyzed into 50 mm Tris-HCl (pH 7.5), 1 mm CaCl2, 1 mm MgCl2, 50 mm NaCl, and 1 mm DTT; loaded onto a phenyl-Sepharose column after the addition of 0.5 m ammonium sulfate; and eluted with Ca2+-free buffer containing 50 mm Tris-HCl (pH 7.5), 1 mm EDTA, and 1 mm DTT at room temperature. Collected fractions of CTnT, CTnI, and CTnC were run on SDS-PAGE, and the fractions containing the purest most concentrated fractions were pooled, yielding >97% pure protein. The purified CTnT and CTnI were immediately stored at −80 °C, and the CTnC was dialyzed exhaustively against 5 mm NH4HCO3 and then lyophilized. All steps described above were performed at 4 °C unless otherwise indicated.
Formation of Troponin Complexes
The HSSTnT isoforms were first concentrated using an Amicon stirred ultrafiltration cell (Millipore Corp.). The protein concentration of the individual subunits was determined using the Coomassie Plus kit (Pierce), and then the subunits were mixed in a 3.0:1.2:1.0 TnT:TnI:TnC molar ratio. After 1 h, the complexes were successively dialyzed against decreasing concentrations of urea and NaCl. The initial buffer contained 50 mm Tris-HCl (pH 8.0), 6 m urea, 1 m NaCl, 3 mm CaCl2, 2.5 mm MgCl2, 1 mm DTT, and 0.1 mm phenylmethanesulfonyl fluoride (PMSF). The following two dialysis buffers contained 4 and 2 m urea with 1 m NaCl. After that, the concentration of NaCl was decreased to 1, 0.8, 0.6, 0.3, 0.15, and 0.1 m consecutively. The last dialysis was repeated twice, and the Tris-HCl concentration in the buffer was decreased to 10 mm, and CaCl2 was excluded. The excess precipitated HSSTnT and HCTnI that accumulated during complex formation were removed by centrifugation. Proper stoichiometry was verified by SDS-PAGE before storage of troponin complexes at −80 °C. The following troponin complexes were formed for this study: HCTnT3·HCTnI·HCTnC, HSSTnT1·HCTnI·HCTnC, HSSTnT2·HCTnI·HCTnC, and HSSTnT3·HCTnI·HCTnC. Interestingly, HSSTnTHyp did not form complexes with HCTnI·HCTnC under the conditions utilized for Tn complex formation.
Actin-Tm-Tn-activated Myosin ATPase Activity Assay (Inhibition and Activation)
Porcine cardiac myosin, rabbit skeletal F-actin, and porcine cardiac Tm were prepared as described previously (24). The protein concentrations used for actomyosin ATPase assays were 0.6 μm porcine cardiac myosin, 3.5 μm rabbit skeletal F-actin, 1 μm porcine cardiac tropomyosin, and 0–2 μm preformed Tn complexes as described above. The final ionic strength of the reactions was ∼150 mm when considering ionic contributions obtained from the various protein buffers. The ATPase inhibitory assay was performed in a 0.1-ml reaction mixture of 3.4 mm MgCl2, 0.13 μm CaCl2, 1.5 mm EGTA, 3.5 mm ATP, 1 mm DTT, and 11.5 mm MOPS (pH 7.0) at 25 °C. The ATPase activation assay was performed using the same 0.1-ml buffer mixture with 3.3 mm MgCl2 and 1.7 mm CaCl2. The ATPase reaction was initiated with the addition of ATP and quenched after 20 min using trichloroacetic acid to a final concentration of 35%. The assay proteins that precipitated were removed by centrifugation. The inorganic phosphate concentration (released by ATP hydrolysis) in the supernatant was determined according to the method developed by Fiske and Subbarow (25).
Skinned Fiber Studies
Fiber Preparation
The porcine heart was collected from a local slaughterhouse and washed using Krebs solution (140 mm NaCl, 4 mm KCl, 1.8 mm CaCl2, 1 mm MgCl2, 1.8 mm NaH2PO4, 5.5 mm glucose, and 50 mm HEPES (pH 7.4) sparged with oxygen for 1.5 h). Strips of left ventricle papillary muscle were extracted and incubated overnight in a pCa 8.0 solution containing 50% glycerol and 1% Triton X-100 at −20 °C and thereafter transferred to a similar replacement solution without Triton X-100 and stored up to 2 weeks at −20 °C.
Steady State and Calcium Dependence of Force Development
Freshly isolated fibers were attached to tweezer clips connected to a force transducer. The fibers were submerged in a 1.3-ml cuvette containing pCa 8.0 solution (10−8 m Ca2+, 1 mm Mg2+, 7 mm EGTA, 2.5 mm MgATP2−, 20 mm MOPS (pH 7.0), 20 mm creatinine phosphate, and 15 units/ml creatine phosphokinase; ionic strength, 150 mm). To ensure complete removal of the sarcoplasmic reticulum, the fibers were incubated in pCa 8.0 solution containing 1% Triton X-100 for 30 min prior to the experiment. The length and diameter of each fiber were recorded. After extensive washing with pCa 8.0 solution to remove all the residual Triton X-100, the fibers were tested for steady state force development in a pCa 4.0 solution (same as pCa 8.0 solution except the [Ca2+] was 10−4 m) followed by relaxation in the pCa 8.0 solution. Once relaxed, the fibers were exposed to solutions of increasing [Ca2+] (from pCa 8.0 to 4.0). The various pCa solutions were calculated using a computer program (pCa calculator) developed in our laboratory (26). Data were analyzed using the following equation: Percent change in force = 100 × [Ca2+]n/([Ca2+]n + [Ca2+50]n) where “[Ca2+50]” is the free [Ca2+] that produces 50% force and “n” is the Hill coefficient.
SSTnT Preparation for Fiber Studies
The HSSTnT isoforms were slowly thawed on ice and sequentially dialyzed in buffers containing decreasing concentrations of urea and KCl. The HSSTnT isoforms and HCTnT (control) were stored at −80 °C in a final buffer containing 10 mm MES (pH 6.2), 0.5 m KCl, 1 mm DTT, and 0.1 mm PMSF.
Tn-displaced Skinned Cardiac Fibers
To determine the effects of the four different HSSTnT isoforms on the various Ca2+-dependent parameters of muscle contraction, the endogenous porcine CTn was displaced with the HSSTnT isoforms of interest. Once the fiber was mounted and tested for calcium responsiveness, the fibers were relaxed and submerged in HSSTnT solution (3 × 1-h incubation at room temperature with fresh HSSTnT). Before incubating the fiber with HSSTnT, the protein was diluted 1:1 using the same buffer as described above but without KCl, which reduces the protein and salt concentration by half. The final HSSTnT protein concentration varied from 0.1 to 0.25 mg/ml depending on the isoform. The HCTnT3 was adjusted to the same concentration as the HSSTnT isoforms. This procedure results in the loss of Ca2+-activated force, and the fibers begin to exercise unregulated force irrespective of the [Ca2+]. To restore Ca2+-regulated force development after TnT incubation, fibers were incubated with a preformed CTnI·CTnC complex (30 μm in relaxing solution) for ∼1 h. Upon reconstitution with the CTnI·CTnC binary complex, the fibers gradually relaxed. Once the fibers fully relaxed, the calcium-regulated force was restored.
Digestion Studies
The degradation of HSSTnT isoforms and HCTnT3 by calpain I (a cytoplasmic, micromolar Ca2+-requiring thiol proteinase) and trypsin was investigated. Equivalent amounts of HSSTnT isoforms (1.2 μm each) were incubated with 1.2 units of calpain I (from porcine erythrocytes; Calbiochem) in the presence of 50 mm Tris-HCl (pH 7.5), 1 mm EDTA, 400 mm NaCl, 2 mm CaCl2, and 0.5 mm DTT in a total volume of 100 μl. HSSTnT isoforms were also incubated with trypsin (Promega) at a protein:trypsin ratio of 1:1000 in 50 mm Tris-HCl (pH 7.5), 1 mm EDTA, 400 mm NaCl, and 0.5 mm DTT. Proteolysis was initiated with the addition of calpain or trypsin. Both calpain and trypsin digestions were carried out at 37 °C, and portions of the samples were removed (25 μl) and heated with SDS sample buffer at different time points. Samples incubated in the absence of calpain or trypsin were utilized as controls. Samples were then run on 12% acrylamide gels, stained with Coomassie Blue G-250, and imaged using a Bio-Rad Versadoc 4000MP. Band intensities were quantified using Quantity One software from Bio-Rad. Each trypsin and calpain digestion study was repeated a minimum of three times.
Circular Dichroism Measurements
Far-UV circular dichroism (CD) spectra were collected using a 1-mm-path quartz cell in a Jasco J-720 spectropolarimeter. Spectra were recorded at 195–250 nm with a bandwidth of 1 nm, speed of 50 nm/min, and resolution of 0.5 nm at room temperature (∼20 °C). Ten scans were averaged, and no numerical smoothing was applied. The optical activity of the buffer was subtracted from relevant protein spectra. Mean residue ellipticity ([θ]MRE in degrees·cm2/dmol) for the spectra was calculated utilizing the same Jasco system software and the following equation: [θ]MRE = [θ]/(10 × Cr × l) where [θ] is the measured ellipticity in millidegrees, Cr is the mean residue molar concentration, and l is the path length in cm (27). Protein concentrations were determined by the Coomassie Plus kit (Pierce) using bovine serum albumin as a standard. The experimental protein concentration for the HSSTnT isoforms and HCTnT was 0.2 mg/ml. The CD experiments were performed using the following buffer conditions: 10 mm potassium phosphate buffer, 1 m KCl, and 1 mm DTT (added fresh each time). The salt concentration was maintained at a high level to prevent protein precipitation during data collection.
Statistical Analysis
The experimental results were reported as x ± S.E. and analyzed for significance using Student's t test at p < 0.05 (paired or unpaired depending on the experimental design).
RESULTS
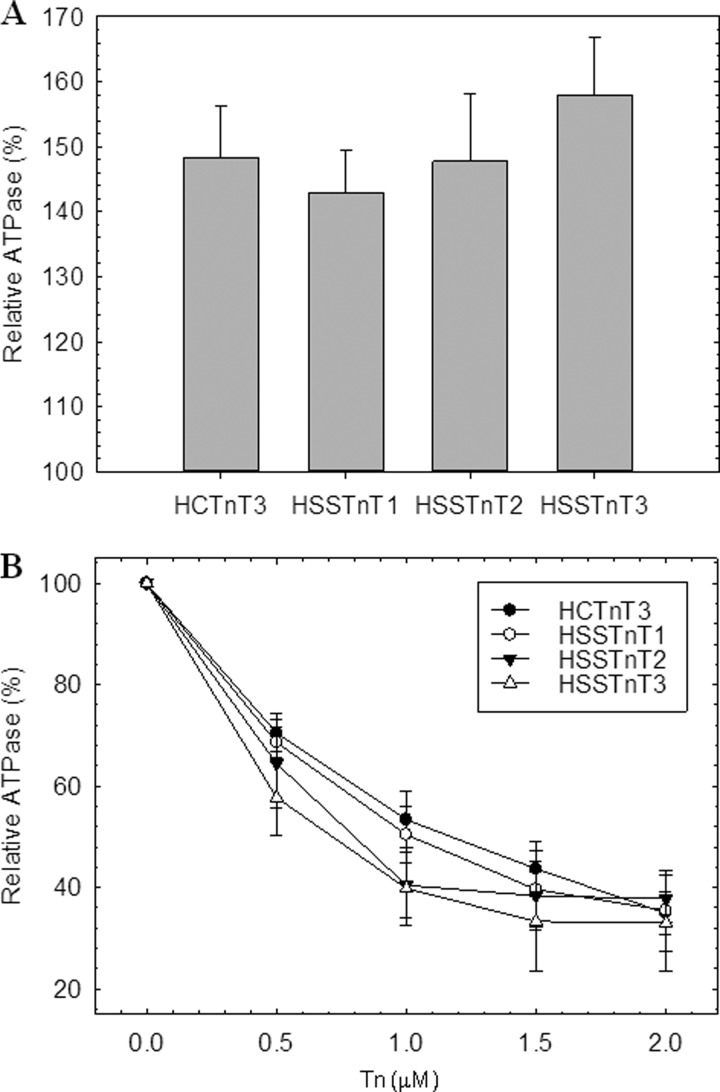
We evaluated the capability of the slow skeletal Tn complexes to activate or inhibit actomyosin ATPase activity. Three different slow skeletal Tn complexes were made (HSSTnT1·CTnI·CTnC, HSSTnT2·CTnI·CTnC, and HSSTnT3·CTnI·CTnC), and these were compared with the cardiac Tn complex containing the adult isoform CTnT3 (HCTnT3·CTnI·CTnC). The HSSTnTHyp did not form a Tn complex. Several different protocols were tested (i.e. varying the pH and ionic strength), but we were unable to obtain a Tn complex with this isoform. Fig. 1A illustrates the differing abilities of the slow skeletal Tn complexes to activate the ATPase activity in the presence of Ca2+. The actomyosin ATPase activity in the absence of Tn complex was considered to be the basal activity level for both activation and inhibition and was set as 100%. No statistically significant differences were observed between slow skeletal Tn complexes when compared with cardiac Tn complex at 1 μm Tn. We observed a trend of the HSSTnT isoforms 1–3 to increase the actomyosin ATPase activity; however, the difference between them was not significant. Fig. 1B shows the entire inhibition curve of the ATPase activity by increasing concentrations of the Tn complex at low Ca2+ levels (pCa 8.5). Several concentrations of Tn complexes containing either the HSSTnT isoforms or HCTnT3 were utilized; these did not show any differences in their ability to inhibit ATPase activity.
FIGURE 1.
Regulation of actomyosin ATPase by Tn complex. A, activation of actomyosin ATPase activity determined at 1 μm troponin complex in the presence of Ca2+. B, inhibition of actomyosin ATPase activity at increasing ratios of troponin complex in the absence of Ca2+. The assay conditions were as described under “Experimental Procedures.” The myosin ATPase activity that occurs in the absence of Tn complexes is considered 100% ATPase activity. Each point represents an average of six experiments, each performed in triplicate, and is expressed as mean ± S.E. Error bars represent S.E.
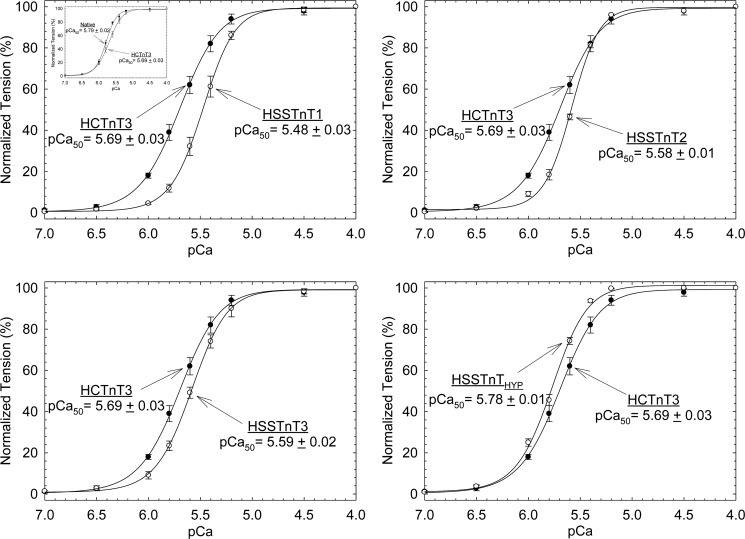
To determine the effects of HSSTnT isoforms on the biophysical and contractile properties of cardiac muscle, various HSSTnT isoforms were exchanged into porcine papillary skinned fiber preparations. Fibers were treated with exogenous HSSTnT isoforms or HCTnT3 (control) at protein concentrations ranging from 0.15 to 0.25 mg/ml depending on the isoform and reconstituted with human CTnI·CTnC complex, and the Ca2+ sensitivity of force development was then measured as a function of increasing Ca2+ concentration (see “Experimental Procedures” for details). As shown in Fig. 2, fibers displaced with HSSTnT1, -2, and -3 showed a decrease in the Ca2+ sensitivity of force development compared with the HCTnT3 (adult isoform of CTnT). The HSSTnT 1 (pCa50 = 5.48 ± 0.03) had the highest rightward shift (ΔpCa50 = −0.21) in Ca2+ sensitivity of force development when compared with the HCTnT3 (pCa50 = 5.69 ± 0.03). The HSSTnT2 and -3 isoforms produced a less pronounced decrease in the Ca2+ sensitivity of force development than HSSTnT1 but had statistically significant differences when compared with the HCTnT3 with pCa50 values of 5.58 ± 0.01 and 5.59 ± 0.02, respectively. Although HSSTnTHyp (found only at the mRNA level) did not form a Tn complex in solution, once it was within the thin filament (skinned fibers) it was able to displace the native troponin complex, to associate with/reconstitute TnI and TnC, and regulate contraction. The HSSTnTHyp isoform produced a completely distinct result. The Ca2+ sensitivity of force development was increased, which contrasts with the functional phenotype found for the other HSSTnT isoforms. After the HSSTnT isoforms were incorporated into the fibers and reconstituted with human CTnI·CTnC complex, the Hill coefficient values showed a tendency to increase but were not statistically different when compared with the HCTnT3. This suggests that the presence of SSTnT isoforms in the heart does not modify the cooperativity of the thin filament. The mean values of pCa50 and Hill coefficient (nH) for each condition are summarized in Table 1. The inset of Fig. 2, upper left panel, shows that porcine fibers treated with HCTnT3 and reconstituted with human CTnI·CTnC complex have a decreased Ca2+ sensitivity of force development. This effect has been reported several times by our laboratory (9, 23, 28).
FIGURE 2.
Normalized pCa-force relationship in skinned cardiac muscle fibers. Each skinned muscle preparation was treated with HCTnT or HSSTnT to displace the endogenous troponin complex and reconstituted with the HCTnI·HCTnC binary complex. The Ca2+ dependence of force development was measured in each preparation after whole troponin complex reconstitution at pH 7.0. Data are plotted as a comparison between HCTnT3 and HSSTnT isoforms, are reported as the average of several experiments, and expressed as mean ± S.E. The inset in the left top panel shows the difference between native and HCTnT3 displaced fibers. Error bars represent S.E.
TABLE 1.
Summary of pCa-force relationship curves in fibers displaced with HSSTnT and reconstituted with cardiac TnI·TnC complex at pH 7
ΔpCa50 = HSSTnT pCa50 − HCTnT3 pCa50.
| HSSTnT | pCa50 | Hill coefficient, nH | ΔpCa50 | Force recovery | Ca2+-unregulated force | No. of experiments |
|---|---|---|---|---|---|---|
| % | % | |||||
| HCTnT3 | 5.69 ± 0.03 | 2.24 ± 0.10 | 56.9 ± 2.6 | 94.8 ± 3.3 | 4 | |
| HSSTnT1 | 5.48 ± 0.03a | 2.78 ± 0.17 | −0.21 | 76.0 ± 5.4a | 71.9 ± 1.4a | 4 |
| HSSTnT2 | 5.58 ± 0.01a | 3.17 ± 0.11 | −0.11 | 68.5 ± 4.0 | 94.8 ± 2.3 | 4 |
| HSSTnT3 | 5.59 ± 0.02a | 2.53 ± 0.22 | −0.10 | 73.1 ± 3.3a | 90.2 ± 6.8 | 5 |
| HSSTnTHyp | 5.78 ± 0.01a | 2.58 ± 0.19 | +0.09 | 63.4 ± 2.2 | 93.1 ± 3.3 | 6 |
a p < 0.05 compared with HCTnT3.
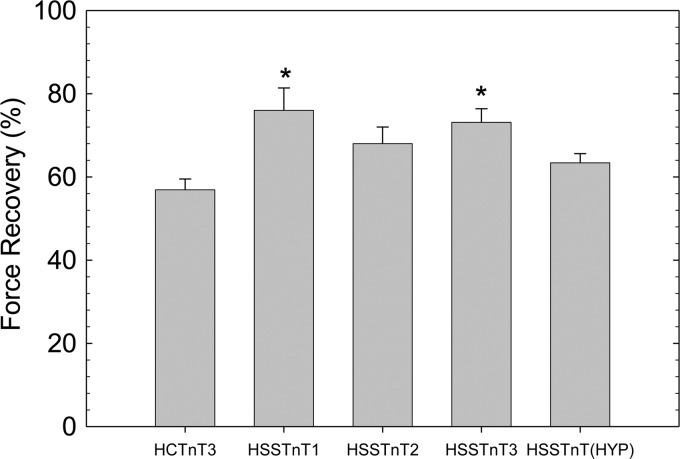
After displacement and reconstitution, the maximal force obtained at high Ca2+ concentrations (pCa 4.0) for each fiber was evaluated. This force was measured relative to the initial maximal force of the skinned fibers before TnT treatment. In Fig. 3, we observed a statistically significant increase in the maximal force of recovery by fibers displaced with the HSSTnT1 and -3 isoforms compared with HCTnT3. However, fibers containing the HSSTnT2 and Hyp isoforms do not appear to be significantly different compared with adult HCTnT3.
FIGURE 3.
Effect of HSSTnT isoforms on Ca2+-activated maximal force at pH 7.0. The relative Ca2+-activated maximal force values of HSSTnT isoforms were compared with that of HCTnT3. *, p < 0.05. Values are reported as mean ± S.E. Error bars represent S.E.
To address the possibility that HSSTnT isoforms may not properly incorporate into the skinned fibers, we measured the unregulated tension at a low Ca2+ concentration (pCa 8.0) immediately after incubation with TnT. When TnT is incorporated into the fibers and displaces the whole native CTn complex, the inhibitory subunit (TnI) is removed, and the fiber becomes unregulated as it is unable to block myosin and actin interactions even at low Ca2+ concentrations. This parameter can be correlated with the extent of endogenous CTn complex displacement. Only HSSTnT1 showed a diminished ability to displace the CTn native complex compared with HCTnT3. The mean values of unregulated tension are shown in Table 1.
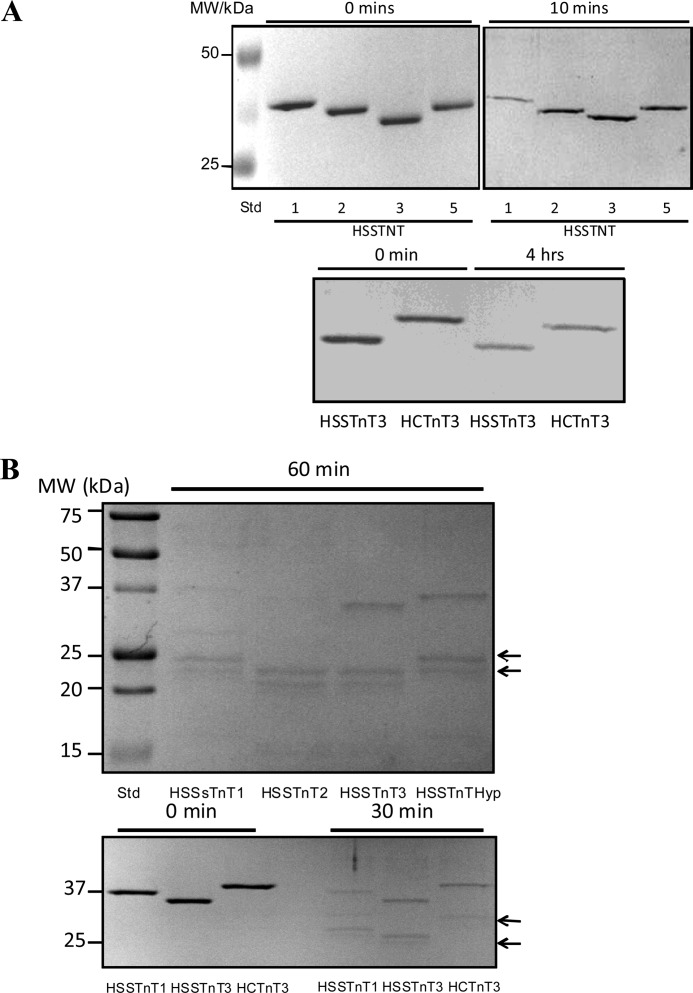
Calpains have been suggested to be the initial proteolytic enzymes involved in the degradation of myofibrils, making them more susceptible to other proteases (29). To investigate the possibility that the different HSSTnT isoforms show different susceptibilities to calpain, we compared the degradation of HSSTnT isoforms by calpain I. Calpain I-mediated degradation of HSSTnT1 was significantly faster than that of HSSTnT2, HSSTnT3, or HSSTnTHyp (Fig. 4A). In separate experiments, the degradation of HSSTnT3 and HCTnT3 by calpain I were compared (Fig. 4A), and both were found to be degraded to a similar extent. Sensitivity to limited trypsin proteolysis is often utilized as a probe of structure-function relationships. We investigated the digestion of HSSTnT isoforms by trypsin and observed that HSSTnT1 and HSSTnT2 were both proteolyzed significantly faster than HSSTnT3 or HSSTnTHyp (Fig. 4B). HSSTnT3 and HCTnT3 were both similar with respect to the rate of degradation by trypsin. In trypsin-treated samples, lower molecular mass protein bands were observed by SDS-PAGE. These lower molecular mass bands were similar for HSSTnT1 and -2. Although the estimated molecular masses of HSSTnTs were 30–33 kDa, they run on SDS-PAGE as 35–38-kDa bands due to the highly charged N terminus in these proteins (17 glutamic acids in the first 30 N-terminal residues) that binds atypically to SDS. These results suggest that HSSTnT1 may be more susceptible to calpain cleavage than the other HSSTnTs.
FIGURE 4.
Proteolytic susceptibility of human slow skeletal TnT isoforms to calpain I and trypsin. A, the upper panel shows SDS-PAGE of 1 μg of HSSTnT isoforms 1, 2, 3, and Hyp at 0 and 10 min after addition of calpain I (0.12 unit). The lower panel shows SDS-PAGE of 1 μg of HSSTnT3 and HCTnT3 at 0 and 4 h after addition of calpain I (0.048 unit). B, the upper panel shows SDS-PAGE of 1 μg of each HSSTnT isoform 60 min after addition of trypsin (1:1000 protein:trypsin ratio). The lower panel shows SDS-PAGE of 1 μg of HSSTnT1, -3, and HCTnT3 at 0 and 30 min after addition of trypsin (1:1000 protein:trypsin ratio). Arrows indicate proteolytic fragments.
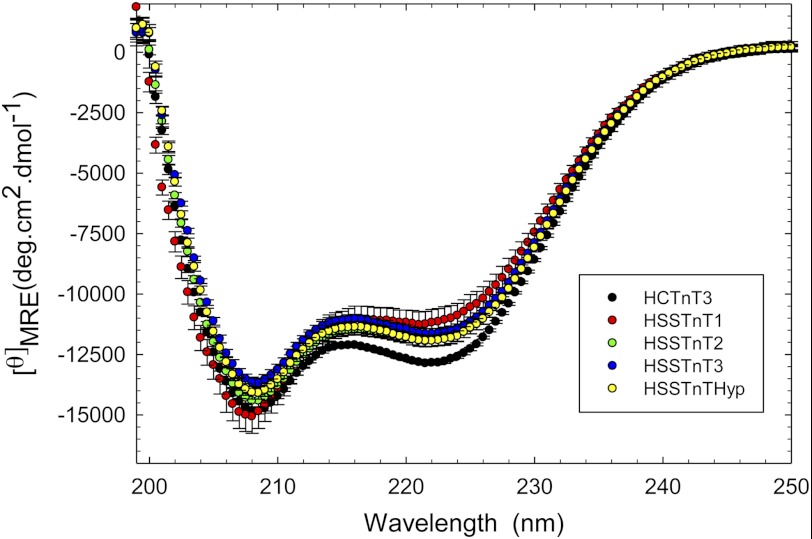
To address whether significant differences existed between the structures of the HSSTnT isoforms and HCTnT3, CD experiments were performed, and the amount of α-helix and β-sheet was recorded at 222 and 208 nm, respectively. The HSSTnT isoforms displayed lower α-helical content compared with HCTnT3. However, at 208 nm, only HSSTnT3 and HSSTnTHyp showed changes in structure compared with HCTnT3 (Fig. 5). The mean residue ellipticity (MRE) values at 222 and 208 nm are shown in Table 2.
FIGURE 5.
Circular dichroism spectra of human slow skeletal TnT isoforms. Far-UV CD spectra were recorded at 195–250 nm utilizing a 1-mm-path quartz cell in a Jasco J-720 spectropolarimeter at room temperature (20 °C). To prevent protein precipitation during the experimental time frame, the buffer contained 1 m KCl, and the protein concentration was 0.2 mg/ml. The spectra shown are the average of six independent measurements. For each independent measurement, 10 scans were averaged, and no numerical smoothing was applied. The optical activity of the buffer was subtracted from relevant protein spectra. Mean residue ellipticity ([θ]MRE in degrees (deg)·cm2/dmol) for the spectra was calculated utilizing the same Jasco system software and the following equation: [θ]MRE = [θ]/(10 × Cr × l) where [θ] is the measured ellipticity in millidegrees, Cr is the mean residue molar concentration, and l is the path length in cm. Error bars represent S.E. Additional details of the buffer conditions and machine setup are described under “Experimental Procedures.”
TABLE 2.
Summary of circular dichroism results for HSSTnT isoforms and HCTnT3
Error is reported as ±S.E. n, number of experiments; deg, degrees.
| TnT | [θ]222 nm | [θ]208 nm | n |
|---|---|---|---|
| deg·cm2·dmol−1 | deg·cm2·dmol−1 | ||
| HCTnT3 | −12,851.82 ± 74.06 | −14,837.18 ± 90.98 | 6 |
| HSSTnT1 | −11,191.22 ± 549.22a | −15,069.30 ± 690.70 | 6 |
| HSSTnT2 | −11,937.00 ± 234.79a | −14,422.73 ± 240.33 | 6 |
| HSSTnT3 | −11,655.38 ± 226.01a | −13,660.70 ± 224.25a | 6 |
| HSSTnTHyp | −11,937.78 ± 228.66a | −14,075.27 ± 259.41a | 6 |
a p < 0.05 compared with HCTnT3.
DISCUSSION
This study focused on the role of HSSTnT isoforms and their ability to regulate cardiac muscle function. In contrast to CTnT, which has four isoforms produced by alternative splicing of exons 4 and 5 located in the N terminus, HSSTnT has different isoforms produced by alternative splicing of the two exons located in two different domains (exon 5 located in the N terminus and exon 12 located in the C terminus) (9). The genetic control of the expression of specific isoforms appears more complex for TnT and may be regulated according to the contractile demands of the heart under certain conditions.
By using recombinant fragments of TnT, Reinach and co-workers (30) have shown that in the absence of TnI and TnC the first 191 amino acids of chicken skeletal TnT (TnT1) were able to activate the S1-actin-tropomyosin Mg2+-ATPase activity. This finding was irrespective of the [Ca2+]; whereas wild type TnT was unable to contribute to its activation (30). Furthermore, the authors identified a region between residues 77 and 191 as a domain of FSTnT that interacts with Tm and actin. It is likely that this domain is responsible for the activation of actomyosin ATPase activity. Here, we show that the utilization of different human slow skeletal TnT isoforms in the cardiac CTnI·CTnC complex did not significantly affect the activation of the actomyosin ATPase. These results demonstrate that the region that controls the activity of the actomyosin ATPase may lie outside of residues 24–34 (exon 5) of HSSTnT. We observed a trend of increased ATPase activation: HSSTnT isoform 1 < 2 < 3; however, these changes were not statistically significant compared with HCTnT3-containing complexes. No significant changes were observed in the inhibition of the actomyosin ATPase activity at different concentrations of Tn complex. In addition, there were no significant differences in the inhibition of Ca2+-unregulated tension upon reconstitution with CTnI·CTnC (data not shown). After CTn displacement and incorporation of HSSTnT isoforms or HCTnT3, the fiber becomes Ca2+-unregulated due to the absence of the inhibitory subunit (CTnI) and capable of generating tension in a solution without Ca2+ (pCa 8.0). At this point, the fiber is incubated with the binary complex (CTnI·CTnC) to recover the Ca2+ regulation of muscle contraction. These measurements further indicate that these isoforms do not alter the parameters of relaxation in skinned fibers. In addition, we were unable to obtain the Tn complex containing HSSTnTHyp; therefore, no ATPase data were generated with this isoform. However, we were able to incorporate HSSTnTHyp into the skinned fiber as shown by the Ca2+ sensitivity data, which argues that the protein, although insoluble in vitro under physiological conditions, is able to associate with the thin filament in skinned fibers. The fiber is able to reconstitute the troponin complex after TnT exchange under physiological in situ conditions. This suggests that the other proteins of the sarcomere lattice, i.e. tropomyosin, actin, and myosin, may assist with the incorporation of HSSTnTHyp into the thin filament. It is likely that the interaction between tropomyosin and HSSTnTHyp enhances the ability of HSSTnTHyp to bind to the CTnI·CTnC complex.
The results suggest that the SSTnT isoforms, when expressed in the heart, would play a specific role in desensitizing the myofilament to Ca2+. In addition, it appears that they have an isoform-specific effect on the maximal force developed by cardiac fibers. These functional effects were shown by incorporation of the HSSTnT isoforms into skinned porcine cardiac fibers and would likely determine the role that SSTnT plays in the developing and adult heart. The existence of SSTnT in the heart is supported by previous studies that have shown that SSTnT is transiently expressed during mouse cardiogenesis (15, 16). It has also been found that both cardiac and skeletal muscle can dynamically regulate the expression patterns of sarcomeric proteins and isoforms that are commonly found in either muscle type. Barton et al. (31) used in situ hybridization and real time quantitative PCR techniques to show that the SSTnT gene shows transient and regional expression in sham-induced hypertrophic rats. Despite this finding, they did not show the presence of SSTnT at the protein level. It is not surprising that there could be an overlap in the regulation of skeletal and cardiac muscle in the expression of slow skeletal isoforms in cardiac muscle as a result of development or as a stress response (32).
HSSTnT1 and -3 had a statistically significant increase in the recovered maximal force in cardiac skinned fibers compared with HCTnT3. However, HSSTnT2 displayed a trend toward increasing maximal force that was not statistically significant. The desensitization to Ca2+ and increase in maximal force shown in this study when HSSTnT isoforms were incorporated into skinned cardiac porcine fibers illustrate the beneficial effects of re-expression of SSTnT in the diseased heart. For example, re-expression of SSTnT isoforms in the sham-induced hypertrophic rat model may be used to compensate for the increased contractile status (Ca2+ sensitivity) of the myocardium that often accompanies the hypertrophic changes seen in familial hypertrophic cardiomyopathy (17). In the studies outlined above, the changes in SSTnT expression may act as a compensatory mechanism that decreases Ca2+ sensitivity in the diseased heart. The increased force recovery seen in the HSSTnT1 and -3 may assist in restoring function to the compromised heart. There was no significant change in cooperativity of Ca2+ activation in the skinned fiber studies compared with HCTnT3, indicating that the alternative splicing may not greatly affect the interaction of HSSTnT isoforms with cardiac Tm.
However, the CD studies suggest that structural differences exist between the HSSTnT isoforms when compared with HCTnT3 as they all demonstrated a decreased amount of α-helical content (see Table 1, 222 nm). This could be due in part to the absence of exons 15–17 in the HSSTnT isoforms that are normally found in CTnT3. The HSSTnT isoform 3 and Hyp are both missing exon 5, which may explain their decreased amount of β-sheet content (see Table 1, 208 nm). The amount of β-sheet content may correspond to the presence or absence of exon 5 because the HSSTnT isoforms 1 and 2, which contain exon 5, have the highest amount of β-sheet content compared with the HSSTnT isoform 3 and Hyp. The large increase in α-helical content appears to have an impact on the Ca2+ sensitivity of force development as isoform 1 has the largest rightward shift in Ca2+ sensitivity with a ΔpCa50 of −0.21. This is consistent with studies conducted in HCTnT where exon 5 was shown to be an important determinant of Ca2+ sensitivity because removal of exon 5 in CTnT isoforms 3 and 4 caused a pronounced decrease in Ca2+ sensitivity compared with CTnT isoforms 1 and 2 (including exon 5) (9). The loss of α-helical secondary structure may contribute to an increased susceptibility to proteolysis that can be partially correlated to the data obtained from the proteolytically mediated degradation studies. It was observed that the HSSTnTHyp isoform underwent much faster proteolysis by trypsin. Although it is uncertain when and where each isoform is expressed, the HSSTnT1 isoform has the highest molecular weight and is the isoform normally described in normal adult slow muscle fibers (33). Charcot-Marie-Tooth disease (a group of inherited peripheral polyneuropathies caused by various neuronal defects) has been found to be associated with a significant up-regulation of the low molecular weight HSSTnT isoform in human skeletal muscle (possibly either HSSTnT2 or -3) (33). This study also showed differences between the high and low molecular weight SSTnT isoforms with respect to their binding affinity for troponin I and tropomyosin (using antibodies against different regions of troponin). The faster proteolytic digestion rates for HSSTnT1 when compared with the other HSSTnT isoforms and HCTnT3 may possibly be due to the lower α-helical content of this protein relative to the other isoforms.
What remains to be determined is exactly how the expression of muscle protein isoforms is controlled in the heart and skeletal muscle. It has been observed that CTnC isoforms can be transiently expressed in skeletal muscle during development and expressed in such disease states as Duchenne muscular dystrophy (34). It appears that expression of skeletal TnT and CTnT isoforms is dynamic and may be interrelated in that both sets of isoforms can be expressed in the alternative muscle type. The CTnT skeletal muscle re-expression theory was explored on the basis that denervation induces abnormal CTnT expression in skeletal muscle (35, 36). This previous study was intended to explore mechanisms that increase the expression of muscle isoforms in alternative muscle types and argues that there may be additional types of regulation of protein expression between muscles. In conclusion, the present study outlines the functional consequences of SSTnT isoforms introduced into the cardiac system and provides novel findings that begin to unravel their physiological impact in the regulation of cardiac muscle. Finally, re- or overexpression of specific SSTnT isoforms might have therapeutic potential in the failing heart since they increase the maximal force of contraction.
This work was supported, in whole or in part, by National Institutes of Health Grants HL096819 (to A. V. G.), AR050199, and HL042325 (to J. D. P.).
- Tm
- tropomyosin
- Tn
- troponin
- SSTnT
- slow skeletal troponin T
- CTn
- cardiac troponin
- FSTnT
- fast skeletal TnT
- HSS
- human slow skeletal
- Hyp
- hypothetical
- HCTn
- human cardiac troponin
- MRE
- mean residue ellipticity.
REFERENCES
- 1. Zot A. S., Potter J. D. (1987) Structural aspects of troponin-tropomyosin regulation of skeletal muscle contraction. Annu. Rev. Biophys. Biophys. Chem. 16, 535–559 [DOI] [PubMed] [Google Scholar]
- 2. Farah C. S., Reinach F. C. (1995) The troponin complex and regulation of muscle contraction. FASEB J. 9, 755–767 [DOI] [PubMed] [Google Scholar]
- 3. Takeda S., Yamashita A., Maeda K., Maéda Y. (2003) Structure of the core domain of human cardiac troponin in the Ca2+-saturated form. Nature 424, 35–41 [DOI] [PubMed] [Google Scholar]
- 4. Gordon A. M., Homsher E., Regnier M. (2000) Regulation of contraction in striated muscle. Physiol. Rev. 80, 853–924 [DOI] [PubMed] [Google Scholar]
- 5. Perry S. V. (1998) Troponin T: genetics, properties and function. J. Muscle Res. Cell Motil. 19, 575–602 [DOI] [PubMed] [Google Scholar]
- 6. Gahlmann R., Troutt A. B., Wade R. P., Gunning P., Kedes L. (1987) Alternative splicing generates variants in important functional domains of human slow skeletal troponin T. J. Biol. Chem. 262, 16122–16126 [PubMed] [Google Scholar]
- 7. Samson F., Mesnard L., Mihovilovic M., Potter T. G., Mercadier J. J., Roses A. D., Gilbert J. R. (1994) A new human slow skeletal troponin T (TnTs) mRNA isoform derived from alternative splicing of a single gene. Biochem. Biophys. Res. Commun. 199, 841–847 [DOI] [PubMed] [Google Scholar]
- 8. Pearlstone J. R., Smillie L. B. (1981) Identification of a second binding region on rabbit skeletal troponin-T for α-tropomyosin. FEBS Lett. 128, 119–122 [DOI] [PubMed] [Google Scholar]
- 9. Gomes A. V., Guzman G., Zhao J., Potter J. D. (2002) Cardiac troponin T isoforms affect the Ca2+ sensitivity and inhibition of force development. Insights into the role of troponin T isoforms in the heart. J. Biol. Chem. 277, 35341–35349 [DOI] [PubMed] [Google Scholar]
- 10. Tobacman L. S., Lee R. (1987) Isolation and functional comparison of bovine cardiac troponin T isoforms. J. Biol. Chem. 262, 4059–4064 [PubMed] [Google Scholar]
- 11. Schachat F. H., Diamond M. S., Brandt P. W. (1987) Effect of different troponin T-tropomyosin combinations on thin filament activation. J. Mol. Biol. 198, 551–554 [DOI] [PubMed] [Google Scholar]
- 12. McCall S. J., Nassar R., Malouf N. N., Saunders A. J., Oakeley A. E., Henderson P. M., Solaro R. J., Pielak G. J., Alexander K. A., Anderson P. A. (2006) Development and cardiac contractility: cardiac troponin T isoforms and cytosolic calcium in rabbit. Pediatr. Res. 60, 276–281 [DOI] [PubMed] [Google Scholar]
- 13. Anderson P. A., Malouf N. N., Oakeley A. E., Pagani E. D., Allen P. D. (1991) Troponin T isoform expression in humans. A comparison among normal and failing adult heart, fetal heart, and adult and fetal skeletal muscle. Circ. Res. 69, 1226–1233 [DOI] [PubMed] [Google Scholar]
- 14. Sabry M. A., Dhoot G. K. (1991) Identification of and pattern of transitions of cardiac, adult slow and slow skeletal muscle-like embryonic isoforms of troponin T in developing rat and human skeletal muscles. J. Muscle Res. Cell Motil. 12, 262–270 [DOI] [PubMed] [Google Scholar]
- 15. Krishan K., Morgan M. J., Zhao W., Dhoot G. K. (2000) Slow troponin T mRNA in striated muscles is expressed in both cell type and developmental stage specific manner. J. Muscle Res. Cell Motil. 21, 527–536 [DOI] [PubMed] [Google Scholar]
- 16. Wang Q., Reiter R. S., Huang Q. Q., Jin J. P., Lin J. J. (2001) Comparative studies on the expression patterns of three troponin T genes during mouse development. Anat. Rec. 263, 72–84 [DOI] [PubMed] [Google Scholar]
- 17. Cullen M. E., Dellow K. A., Barton P. J. (2004) Structure and regulation of human troponin genes. Mol. Cell. Biochem. 263, 81–90 [DOI] [PubMed] [Google Scholar]
- 18. Lompre A. M., Schwartz K., d'Albis A., Lacombe G., Van Thiem N., Swynghedauw B. (1979) Myosin isoenzyme redistribution in chronic heart overload. Nature 282, 105–107 [DOI] [PubMed] [Google Scholar]
- 19. Izumo S., Lompré A. M., Matsuoka R., Koren G., Schwartz K., Nadal-Ginard B., Mahdavi V. (1987) Myosin heavy chain messenger RNA and protein isoform transitions during cardiac hypertrophy. Interaction between hemodynamic and thyroid hormone-induced signals. J. Clin. Investig. 79, 970–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang Q. Q., Feng H. Z., Liu J., Du J., Stull L. B., Moravec C. S., Huang X., Jin J. P. (2008) Co-expression of skeletal and cardiac troponin T decreases mouse cardiac function. Am. J. Physiol. Cell Physiol. 294, C213–C222 [DOI] [PubMed] [Google Scholar]
- 21. Nosek T. M., Brotto M. A., Jin J. P. (2004) Troponin T isoforms alter the tolerance of transgenic mouse cardiac muscle to acidosis. Arch. Biochem. Biophys. 430, 178–184 [DOI] [PubMed] [Google Scholar]
- 22. Chandra M., Tschirgi M. L., Rajapakse I., Campbell K. B. (2006) Troponin T modulates sarcomere length-dependent recruitment of cross-bridges in cardiac muscle. Biophys. J. 90, 2867–2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pinto J. R., Parvatiyar M. S., Jones M. A., Liang J., Potter J. D. (2008) A troponin T mutation that causes infantile restrictive cardiomyopathy increases Ca2+ sensitivity of force development and impairs the inhibitory properties of troponin. J. Biol. Chem. 283, 2156–2166 [DOI] [PubMed] [Google Scholar]
- 24. Lang R., Gomes A. V., Zhao J., Housmans P. R., Miller T., Potter J. D. (2002) Functional analysis of a troponin I (R145G) mutation associated with familial hypertrophic cardiomyopathy. J. Biol. Chem. 277, 11670–11678 [DOI] [PubMed] [Google Scholar]
- 25. Fiske C. H., Subbarow Y. (1925) The colorimetric determination of phosphorus. J. Biol. Chem. 66, 375–400 [Google Scholar]
- 26. Dweck D., Reyes-Alfonso A., Jr., Potter J. D. (2005) Expanding the range of free calcium regulation in biological solutions. Anal. Biochem. 347, 303–315 [DOI] [PubMed] [Google Scholar]
- 27. Szczesna D., Ghosh D., Li Q., Gomes A. V., Guzman G., Arana C., Zhi G., Stull J. T., Potter J. D. (2001) Familial hypertrophic cardiomyopathy mutations in the regulatory light chains of myosin affect their structure, Ca2+ binding, and phosphorylation. J. Biol. Chem. 276, 7086–7092 [DOI] [PubMed] [Google Scholar]
- 28. Szczesna D., Zhang R., Zhao J., Jones M., Guzman G., Potter J. D. (2000) Altered regulation of cardiac muscle contraction by troponin T mutations that cause familial hypertrophic cardiomyopathy. J. Biol. Chem. 275, 624–630 [DOI] [PubMed] [Google Scholar]
- 29. Goll D. E., Thompson V. F., Taylor R. G., Christiansen J. A. (1992) Role of the calpain system in muscle growth. Biochimie 74, 225–237 [DOI] [PubMed] [Google Scholar]
- 30. Oliveira D. M., Nakaie C. R., Sousa A. D., Farah C. S., Reinach F. C. (2000) Mapping the domain of troponin T responsible for the activation of actomyosin ATPase activity. Identification of residues involved in binding to actin. J. Biol. Chem. 275, 27513–27519 [DOI] [PubMed] [Google Scholar]
- 31. Barton P. J., Felkin L. E., Koban M. U., Cullen M. E., Brand N. J., Dhoot G. K. (2004) The slow skeletal muscle troponin T gene is expressed in developing and diseased human heart. Mol. Cell. Biochem. 263, 91–97 [DOI] [PubMed] [Google Scholar]
- 32. Fredericks S., Degens H., McKoy G., Bainbridge K., Collinson P. O., Coulton G., Elmahdi H., Holt D. W. (2007) Effect of denervation on the content of cardiac troponin-T and cardiac troponin-I in rat skeletal muscle. Clin. Biochem. 40, 423–426 [DOI] [PubMed] [Google Scholar]
- 33. Larsson L., Wang X., Yu F., Höök P., Borg K., Chong S. M., Jin J. P. (2008) Adaptation by alternative RNA splicing of slow troponin T isoforms in type 1 but not type 2 Charcot-Marie-Tooth disease. Am. J. Physiol. Cell Physiol. 295, C722–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bodor G. S., Survant L., Voss E. M., Smith S., Porterfield D., Apple F. S. (1997) Cardiac troponin T composition in normal and regenerating human skeletal muscle. Clin. Chem. 43, 476–484 [PubMed] [Google Scholar]
- 35. Saggin L., Gorza L., Ausoni S., Schiaffino S. (1990) Cardiac troponin T in developing, regenerating and denervated rat skeletal muscle. Development 110, 547–554 [DOI] [PubMed] [Google Scholar]
- 36. Ricchiuti V., Apple F. S. (1999) RNA expression of cardiac troponin T isoforms in diseased human skeletal muscle. Clin. Chem. 45, 2129–2135 [PubMed] [Google Scholar]