Background: Pathogenic Pseudomonas aeruginosa strains produce a colicin M-like bacteriocin exhibiting peptidoglycan lipid II-degrading activity.
Results: We have determined the crystal structure of the Pseudomonas aeruginosa PaeM bacteriocin and functionally characterized its C-terminal activity domain.
Conclusion: This study highlights structural plasticity of the active site of this enzyme family.
Significance: The PaeM pyocin could potentially be exploited as antibacterial agent.
Keywords: Bacterial Toxins, Peptidoglycan, Phosphodiesterases, Protein Structure, Pseudomonas aeruginosa, X-ray Crystallography, Bacteriocin, Colicin M, Lipid II
Abstract
Colicin M (ColM) is the only enzymatic colicin reported to date that inhibits cell wall peptidoglycan biosynthesis. It catalyzes the specific degradation of the lipid intermediates involved in this pathway, thereby provoking lysis of susceptible Escherichia coli cells. A gene encoding a homologue of ColM was detected within the exoU-containing genomic island A carried by certain pathogenic Pseudomonas aeruginosa strains. This bacteriocin (pyocin) that we have named PaeM was crystallized, and its structure with and without an Mg2+ ion bound was solved. In parallel, site-directed mutagenesis of conserved PaeM residues from the C-terminal domain was performed, confirming their essentiality for the protein activity both in vitro (lipid II-degrading activity) and in vivo (cytotoxicity against a susceptible P. aeruginosa strain). Although PaeM is structurally similar to ColM, the conformation of their active sites differs radically; in PaeM, residues essential for enzymatic activity and cytotoxicity converge toward a same pocket, whereas in ColM they are spread along a particularly elongated active site. We have also isolated a minimal domain corresponding to the C-terminal half of the PaeM protein and exhibiting a 70-fold higher enzymatic activity as compared with the full-length protein. This isolated domain of the PaeM bacteriocin was further shown to kill E. coli cells when addressed to the periplasm of these bacteria.
Introduction
Numerous strains of Escherichia coli secrete colicins to kill competitors belonging to the same or closely related bacterial species (1–3). Most of the colicins characterized to date either possess a pore-forming activity targeting the cytoplasmic membrane or a nuclease activity degrading specifically rRNA, tRNA, or chromosomal DNA (1). Colicin M (ColM)4 is unique as it is the only colicin known to interfere with cell-wall peptidoglycan biosynthesis (4, 5). Indeed, it was demonstrated to be an enzyme (phosphodiesterase) catalyzing the specific degradation of the peptidoglycan lipid II intermediate, thereby provoking the arrest of the synthesis of this essential cell-wall polymer and ultimately cell lysis (6). ColM cleaves the bond connecting the undecaprenyl and pyrophosphoryl groups in the lipid II structure, a reaction releasing undecaprenol and 1-pyrophospho-MurNAc(-pentapeptide)-GlcNAc products, which could not be directly reused for synthesis of the polymer (6).
To enter into susceptible cells and reach its target (lipid II) located in the outer layer of the cytoplasmic membrane, ColM first binds to the outer membrane protein receptor FhuA (7) and then parasitizes the TonB/ExbB/ExbD import machinery (1, 8). The recent discovery that the in vivo activity of ColM was also dependent upon a specific periplasmic protein, FkpA, from the targeted cells, further complicated the story (9). E. coli mutants affected in either of the latter proteins are, therefore, fully resistant to ColM. All colicins show a similar three-domain structural organization, each of these domains playing a specific role in the mode of action of the colicin, the N-terminal and central domains being required for toxin translocation and receptor binding steps, respectively, and the C-terminal domain carrying the toxic activity (1). As is the case for colicinogenic strains in general, ColM-producing strains are protected against the toxin they secrete by concomitant expression of a specific immunity protein. The mechanism of action of the ColM immunity protein, Cmi, whose x-ray structure has recently been solved (10, 11), remains unknown.
Genes encoding proteins that exhibit similarity to E. coli ColM were recently identified in the genomes of Pseudomonas aeruginosa, Pseudomonas syringae, Pseudomonas fluorescens, Burkholderia spp., and Pectobacterium carotovorum species (12, 13). Only a limited number of strains from these species harbored these genes, which were often localized within pathogenicity islands. In particular, the P. aeruginosa homologue was located in the 80-kb exoU-containing genomic island A, a large horizontally acquired genetic element and virulence determinant (14). These ColM homologues exhibited significant sequence homology (35–45% identity) in the C-terminal region, roughly the second half of these proteins, which corresponds to the catalytic domain. In contrast, no homology was detected in their N-terminal regions, which presumably participate in receptor binding and import (12, 15). The translocation/reception domain sequences of ColM-like bacteriocins from the phytopathogenic P. carotovorum bacterium (pectocins M1 and M2) present extensive similarity to those of plant ferredoxins. It was further demonstrated that these domains are used to parasitize an iron uptake system from the targeted bacteria (13). This shows that a ColM-like killing domain can be fused through evolution to domains of different folds to hijack specific import machineries. The ColM-like proteins from P. aeruginosa (PaeM), P. syringae (PsyM), and P. fluorescens (PflM) were purified, and their lipid II-degrading activity was confirmed. All these enzymes require Mg2+ for activity and cleave lipid II at the same position as ColM. In vivo assays showed that these ColM homologues have narrow antibacterial spectra (12). For instance, despite its high catalytic activity (600-fold higher kcat/Km ratio as compared with E. coli ColM), PaeM did not show any cytotoxic effect toward E. coli cells, suggesting a failure for this toxin to parasitize the ColM reception and import systems. However, PaeM exerted a bacteriostatic effect on certain P. aeruginosa strains chosen among reference strains and clinical isolates (12).
The x-ray structure of ColM, recently determined by Zeth et al. (16), revealed a very compact fold lacking the well individualized domains observed in all other colicins. We report here the crystal structure of the ColM-like protein (PaeM) from P. aeruginosa as well as the functional characterization of its activity domain. This structure highlights major differences within the active site, which could explain why the PaeM enzyme exhibits much higher specific enzymatic activity toward lipid II than its ColM homologue.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Growth Conditions
The E. coli strains DH5α (Invitrogen) and XL1-blue (Stratagene) were used as the hosts for propagation of plasmids and site-directed mutagenesis experiments, respectively. Strains C43(DE3) (Avidis) and Rosetta were used for the production of PaeM proteins. The P. aeruginosa strains JJ692 and DET08 were described earlier (12). The construction of pMLD245, a pET2160 derivative plasmid allowing high level expression of the PaeM protein with a C-terminal six-histidine tag (Arg-Ser-His6 extension) was previously described (12). The pREP4groESL plasmid allowing overproduction of the bacterial chaperones was obtained from K. Amrein (17), and the two cloning vectors pET2130 and pET2160 were described previously (12). Unless otherwise noted, cells were grown in 2YT medium (18) at 37 °C. Ampicillin, kanamycin, and chloramphenicol were used at 100, 50, and 25 μg/ml, respectively. Growth was monitored at 600 nm with a Shimadzu UV-1601 spectrophotometer.
Molecular Biology Techniques
Polymerase chain reaction (PCR) amplification of genes was performed in a Bio-med Thermocycler 60 apparatus (B. Braun) using Expand High Fidelity polymerase (Roche Applied Science). DNA fragments were purified using the Wizard PCR Preps DNA purification kit (Promega), and standard procedures for DNA digestion, ligation, and agarose gel electrophoresis were used (19). Plasmid isolation was carried out by the alkaline lysis method, and E. coli cells were transformed with plasmid DNA by the method of Dagert and Ehrlich (20).
Construction of Expression Plasmids
Site-directed mutagenesis of the C-terminal His6-tagged PaeM protein was performed directly on the pMLD245 expression plasmid (12) by using the QuikChange II XL mutagenesis kit. Pairs of complementary oligonucleotides used for introducing mutations in the gene sequence are shown in Table 1. Truncated variants of PaeM lacking the N-terminal domain were also generated; the corresponding gene sequences were amplified using PaeM-Δ1-xxx (where 1-xxx represents deleted residues) and PaeM-rev primers (Table 1), and the resulting PCR fragments were cut by BamHI and HindIII and then cloned into pET2130. The sequences of cloned inserts and mutagenesis products were verified by DNA sequencing.
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequencea |
|---|---|
| Cloning | |
| PaeM-Δ1–126 | CGCGGGATCCCCTAGCTGGGATCGTCCTAAAATAGATACG (BamHI) |
| PaeM-Δ1–133 | CGCGGGATCCATAGATACGTTCAATGGCGGGGTCTACACG (BamHI) |
| PaeM-rev | CCTGAAGCTTTTAACCAGAAATATTTACAGGGATAGC (HindIII) |
| Site-directed mutagenesis | |
| W54K | CTATGTGAACGGTGATAAGGAAAAACCTCTAC |
| E55A | GAACGGTGATTGGGCAAAACCTCTACTCGC |
| W54K/E55A | CTATGTGAACGGTGATAAGGCAAAACCTCTACTCG |
| A238S | CGTGGGGAGATCCGTAGCTATGATGATCTG |
| D241A | CCGTGCATATGATGCTCTGTATGACTTC |
| Y243A | GCATATGATGATCTGGCTGACTTCAATCCTTC |
| D244A | GATGATCTGTATGCCTTCAATCCTTCAAATC |
| D240A/D241A | GAGATCCGTGCATATGCAGCACTGTATGACTTC |
| D241A/D244A | GCATATGATGCACTGTATGCATTCAATCC |
| N249A | GACTTCAATCCTTCAGCTCACAGAACCGAAAC |
| H250A | CAATCCTTCAAATGCCAGAACCGAAACTGC |
| R251A | CCTTCAAATCACGCAACCGAAACTGCTG |
a Restriction sites (in parentheses) in oligonucleotide sequences are shown in bold. Mutations introduced in site-directed mutagenesis oligonucleotides are underlined (for each mutation, one pair of forward and reverse complementary primers was used; only the sequence of the forward primer is shown here). In names of oligonucleotides used for generating truncated PaeM protein variants, Δ1-xxx indicates the deleted protein residues (in each case, the remaining sequence is fused to a N-terminal MH6GS tag).
Protein Production and Purification
For expression of full-length PaeM proteins (wild-type and mutants), C43(DE3)(pMLD245) cells were grown at 37 °C in 2YT-ampicillin medium (1-liter cultures). When the culture optical density (A600 nm) reached 1.0, isopropyl-β-d-thiogalactopyranoside was added (1 mm), and growth was continued for 3 h at 37 °C. Truncated forms of PaeM protein were expressed in the presence of chaperones by using Rosetta(pLysS)(pREP4groESL) as the host strain. This strain carrying plasmids pMLD362 (PaeM-Δ1–126) or pMLD363 (PaeM-Δ1–133) was grown at 37 °C as described above, but at A600 nm = 1.0 the temperature was lowered to 15 °C before the addition of isopropyl-β-d-thiogalactopyranoside (1 mm), and cells were incubated overnight at this temperature. In all cases cells were harvested, washed with 40 ml of cold 20 mm potassium phosphate buffer, pH 7.4, containing 0.5 mm MgCl2 and 0.1% 2-mercaptoethanol (buffer A), resuspended in 12 ml of the same buffer, and disrupted by sonication (Bioblock Vibracell sonicator, model 72412). The resulting suspension was centrifuged at 4 °C for 20 min at 200,000 × g in a TL100 Beckman centrifuge, and the supernatant was stored at −20 °C. The PaeM mutants W54K, E55A, and W54K/E55A formed inclusion bodies when expressed as described for the wild-type protein. They were solubilized and renaturated from these inclusion bodies as follows. After sonication, the suspension was centrifuged at 4 °C for 10 min at 7000 × g to collect the insoluble material, and this pellet was suspended in 2 ml of 8 m urea for 1 h at 4 °C and centrifuged again. The proteins were finally refolded by a 50-fold dilution of the supernatant in buffer A before purification.
His6-tagged proteins were purified on nickel-nitrilotriacetate (Ni2+-NTA)-agarose under native conditions. Crude soluble protein extracts obtained as described above were incubated with 1.5 ml of polymer pre-equilibrated in buffer A containing 20 mm imidazole for 1 h at 4 °C. The polymer was washed extensively with buffer A containing 20 and 40 mm imidazole, and proteins were eluted using increasing concentrations of imidazole (100, 200, and 300 mm). Purified protein fractions were pooled, concentrated by ultrafiltration (10K Amicon Ultra-15 centrifugal filter, Millipore), and dialyzed overnight against 100 volumes of buffer A. Final preparations were stored at −20 °C after the addition of 10% glycerol. For crystallization trials, the His6-tagged full-length PaeM protein eluted from the Ni2+-NTA polymer was further subjected to a S75 size-exclusion chromatography column (GE Healthcare) previously equilibrated in 20 mm Tris-HCl, pH 7.5, 200 mm NaCl, and 10 mm 2-mercaptoethanol.
SDS-PAGE analysis of proteins was performed as described previously (21). Protein concentrations were determined by using the Bradford procedure (22) as well as quantitative amino acid analysis with a Hitachi model L8800 analyzer (ScienceTec) after hydrolysis of samples for 24 h at 105 °C in 6 m HCl containing 0.05% 2-mercaptoethanol. For Western blot and immunodetection, proteins were transferred from the gels onto nitrocellulose membranes, and blots were developed using 5-bromo-4-chloro-3-indolyl phosphate and nitro blue tetrazolium. Anti-PaeM polyclonal antibodies produced in rat (GeneCust, Dudelange, Luxembourg) were used at 1:100 dilutions.
Lipid II Hydrolase Activity
The enzymatic activity of PaeM and of its different variants was tested in a reaction mixture (10 μl) consisting of 100 mm Tris-HCl, pH 7.5, 20 mm MgCl2, 150 mm NaCl, 2.3 μm [14C]radiolabeled lipid II (140 Bq), and 0.2% n-dodecyl-β-d-maltoside. Unless otherwise noted, the meso-diaminopimelic acid-containing lipid II from E. coli was used as the substrate in this assay. Lipids II from other bacterial species were also tested to analyze the substrate specificity of the PaeM protein. These lipids were obtained by a chemo-enzymatic procedure, as previously described (23). The reaction was initiated by the addition of the purified protein (from 2 to 10 μg in 5 μl of buffer A), and incubated for 30 min at 37 °C with shaking (Thermomixer, Eppendorf). The reaction was stopped by heating at 100 °C for 1 min and then analyzed by thin-layer chromatography on LK6D silica gel plates (Whatman) using 1-propanol/ammonium hydroxide/water (6:3:1; v/v/v) as a mobile phase. The radioactive spots were visualized with a Storm 860 PhosphorImager and located and quantified with a radioactivity scanner (Rita Star, Raytest Isotopenmeβgeräte GmbH, Straubenhardt, Germany). Under these conditions the radiolabeled substrate (lipid II) and product (1-pyrophospho-MurNAc[pentapeptide]-GlcNAc) migrated with Rf values of 0.7 and 0.3, respectively.
Antibacterial Activity
2YT top agar (3 ml) was inoculated with the P. aeruginosa PaeM-susceptible DET08 strain (108 cells) and poured onto 2YT agar plates. 2-μl samples of the purified PaeM proteins (wild-type and mutated forms) were then spotted onto the surface of the top agar, and growth inhibition was observed after 24 h of incubation at 37 °C. The antibacterial activity of these proteins was also similarly tested against some Gram-positive species: Staphylococcus aureus (strain RN4220), Enterococcus faecalis (strain JH2–2), and Enterococcus faecium (strain D344). The cytotoxic effect of these different proteins was tested on the E. coli strain BW25113 under previously described conditions (15) that allow the introduction of proteins from the external milieu into the periplasm while preserving cell integrity (the so called “bypass” assay).
Crystallization and Structure Determination
Crystals of P. aeruginosa PaeM protein were obtained at 19 °C by mixing equal volumes of protein (15–25 mg/ml in 20 mm Tris-HCl, pH 7.5, 200 mm NaCl, and 10 mm 2-mercaptoethanol) with a crystallization buffer composed of either 100 mm HEPES, pH 7.5, and 2 m sodium formate (formate-form) or 100 mm Tris-HCl, pH 8.5, 200 mm MgCl2, and 30% (w/v) PEG 4000 (Mg2+-form). For data collection, crystals were transferred into a cryoprotectant solution consisting of crystallization buffer provided with progressively higher ethylene glycol concentrations up to 30%. The datasets were collected on beam line Proxima-1 (SOLEIL, St-Aubin, France). Crystals from the Mg2+-form diffracted up to 1.7 Å resolution, whereas those from the formate-form diffracted more weakly (2.4 Å resolution). The structure was determined by the multiple-wavelength anomalous dispersion method using the anomalous signal extracted from x-ray data collected on selenomethionine-substituted protein crystals from the Mg2+-form. Diffraction data were recorded at the wavelengths corresponding to the peak, edge, and remote of the selenium fluorescence spectrum. Data were processed using the XDS package (24). The space group was P212121 with one molecule per asymmetric unit. All the expected selenium sites (two per monomer) were found with the program SHELXD in the 41–3 Å resolution range (25). Refinement of the selenium atom positions, phasing, and density modification were performed with the program SHARP (26). The quality of the experimental phases allowed automatic building of more than 95% of the protein residues using the ARP/WARP software (27). This model was then refined against the dataset collected at the selenium edge using PHENIX (28) and then rebuilt with COOT (29). In the structure of PaeM obtained with MgCl2 as a crystallization reagent, a strong peak in the Fo − Fc electron density map was located near the Asp-241 side chain and was attributed to an Mg2+ ion based on the following elements. First, this peak was only present when the structure was refined against the dataset obtained from crystals grown at a high concentration of Mg2+ ions (200 mm) in the crystallization solution but not in the formate-form. Second, the distances to the center of this peak (between 2.2 and 2.4 Å) and the octahedral configuration of the surrounding atoms corresponded to a typical coordination sphere of an Mg2+ ion. Third, the refined B-factor values for either a water molecule or an Mg2+ ion modeled at this position were in favor of Mg2+. The structure was also refined to 2.4 Å resolution using the dataset collected from crystal of the formate-form using the same protocol.
All the PaeM residues (from Val-2 to Ser-291, the N-terminal methionine being excised after expression in E. coli according to mass spectrometry analysis (12)) as well as the first two histidine residues from the C-terminal His tag, a magnesium ion, 433 water molecules, and 12 ethylene glycol molecules from the cryoprotectant solution were modeled into electron density maps using the Mg2+-form data. The structure refined against the dataset collected from formate-form crystals contained all the same protein residues as the structure refined at high resolution as well as 112 water molecules. Statistics on data processing and structure refinement are gathered in Table 2.
TABLE 2.
Statistics for data processing and structure refinement
Values in parentheses are for highest resolution shell. r.m.s.d., root mean square deviation.
| Formate form | SeMet (Mg2+ form) |
|||
|---|---|---|---|---|
| Peak | Inflexion | Remote | ||
| Data | ||||
| Resolution (Å) | 50-2.38 (2.53-2.38) | 41-1.7 (1.74-1.7) | 41-1.7 (1.74-1.7) | 41-1.7 (1.74-1.7) |
| Wavelength (Å) | 0.97918 | 0.97918 | 0.97934 | 0.97779 |
| Space group | P212121 | P212121 | ||
| Cell parameters | a = 42.9Å; b = 72.5 Å; c = 100 Å | a = 42.9Å; b = 71.5 Å; c = 99.9 Å | ||
| Total number of reflections | 73,769 | 130,459 | 145,934 | 147,906 |
| Total number of unique reflections | 22,837 | 62,510 | 63,194 | 63,857 |
| Rsym (%)a | 8.4 (23.8) | 7.3 (41.8) | 7.5 (44.9) | 7.5 (43.6) |
| Completeness (%) | 95.7 (83.5) | 95.6 (98.4) | 96.7 (98.8) | 97.7 (98.6) |
| I/σ(I) | 12.7 (5.2) | 8.6 (2.2) | 9.1 (2.2) | 9.3 (2.2) |
| Redundancy | 3.2 | 2 | 2.3 | 2.3 |
| Refinement | ||||
| Resolution (Å) | 50-2.4 | 41-1.7 | ||
| R/Rfree (%)b | 17.0/24.3 | 17.9/21.4 | ||
| r.m.s.d. bonds (Å) | 0.006 | 0.005 | ||
| r.m.s.d. angles (°) | 0.898 | 0.933 | ||
| B factor (Å2) | ||||
| Protein | 27.3 | 14.8 | ||
| Magnesium | 11.3 | |||
| Water | 30 | 29.7 | ||
| Ethylene glycol | 32.5 | |||
| PDB code | 4G76 | 4G75 | ||
a Rsym = ΣhΣi|Ihi − 〈Ih〉|/ΣhΣhiIhi, where Ihi is the ith observation of the reflection h, and 〈Ih〉 is the mean intensity of reflection h.
b Rfactor = I£ ‖Fo| − |Fc‖/‖Fo|. Rfree was calculated with a small fraction (5%) of randomly selected reflections.
Materials and Chemicals
Isopropyl-β-d-thiogalactopyranoside was obtained from Eurogentec, and Ni2+-NTA-agarose was from Qiagen. Antibiotics and reagents were from Sigma. DNA ligase and restriction enzymes were obtained from New England Biolabs, and DNA purification kits were from Promega and Macherey-Nagel. The QuikChange II XL kit used for site-directed mutagenesis experiments was from Stratagene. Oligonucleotide synthesis and DNA sequencing was performed by Eurofins MWG Operon.
RESULTS AND DISCUSSION
Expression of PaeM by P. aeruginosa Strains
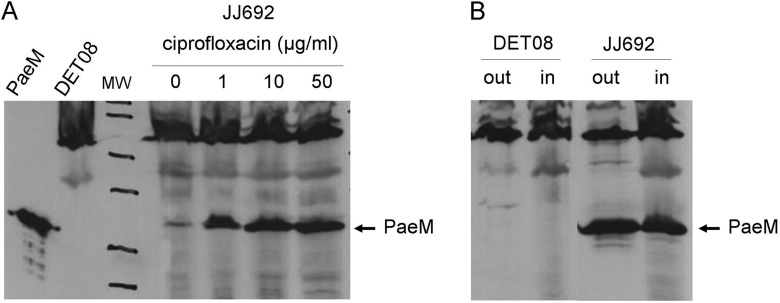
The exoU-containing genomic island A that is found in the genome of many pathogenic strains of P. aeruginosa (strain JJ692, for instance) contains a gene encoding a ColM homologue (14) that we have named PaeM. This protein was previously purified and shown to exhibit a high lipid II-degrading activity (12). It was inactive against all strains of E. coli tested, but it exerted a bacteriostatic effect on certain P. aeruginosa strains, including clinical isolates (strain DET08, for instance) (12). To verify that P. aeruginosa strains carrying this gene indeed produced the protein, antibodies were raised against PaeM, and crude extracts from the two strains JJ692 and DET08 were analyzed by SDS-PAGE and Western blot. The DET08 strain, which does not contain the gene, was used as a control. As shown in Fig. 1A, a faint band corresponding to PaeM was detected only in crude extracts from the strain JJ692. As the expression of ColM and other colicins is known to be primarily under SOS control in E. coli (1), we tested whether the expression of PaeM in P. aeruginosa could also be induced under particular stress conditions. The JJ692 strain was treated by ciprofloxacin, an antibiotic known to induce the SOS response by causing DNA damage. This treatment indeed resulted in a significant stimulation of gene expression, as judged by the accumulation of PaeM protein in the cell content (Fig. 1A). Whether this bacteriocin was released by the strain in the external growth medium was also questioned. As shown in Fig. 1B, the PaeM protein was detected in both the cell content and the external growth medium of the strain JJ692. These data clearly demonstrated the functionality of this gene and the effective production and release of the bacteriocin by P. aeruginosa strains under physiological growth conditions. The mode of excretion of ColM, PaeM, and their homologues is unknown as, in contrast to many other bacteriocins, they are not expressed together with a dedicated lysis protein. Therefore, whether excretion results from a lysis of a small fraction of the cell population or whether it occurs through a specific mechanism still remains to be determined.
FIGURE 1.
Production and excretion of PaeM. A, the P. aeruginosa JJ692 strain was grown and treated for 2 h by ciprofloxacin at the indicated concentrations. Crude cell protein extracts were prepared and analyzed by SDS-PAGE, and the PaeM protein was detected by Western-blot, as detailed in “Experimental Procedures.” Purified PaeM protein and a crude extract from strain DET08 were also analyzed as controls (two lanes on the left). B, the P. aeruginosa DET08 (PaeM-susceptible) and JJ692 (PaeM-producing) strains were grown and treated for 2 h by ciprofloxacin at 10 μg/ml. Cells were harvested, and the presence of the PaeM protein was searched for both in the external growth medium (out) and crude cell extracts (in) fractions.
Substrate Specificity of PaeM
The structure of peptidoglycan and consequently that of its lipid II intermediate varies to some extent in the bacterial world, in particular in the peptide moiety (30). ColM was previously shown to hydrolyze lipids II from E. coli, E. faecalis, E. faecium, and S. aureus that contain either meso-diaminopimelic acid (E. coli) or l-Lys (Gram-positive species) at position 3 of the peptide as well as (for the latter species) additional amino acid residues branched on the Lys residue: l-Ala and l-Ala-l-Ala (E. faecalis), d-iso-Asn (E. faecium), Gly1 to Gly5 (S. aureus) (23). The substrate specificity of PaeM toward these different forms of lipid II was tested. All of them were hydrolyzed at a similar rate (less than 10% deviation as compared with E. coli lipid II), showing that the catalytic efficiency of PaeM was neither affected by the nature of the residue at the third position of the stem peptide nor by the length and composition of the side chain. It was noteworthy that bulky groups (l-Ala2, Gly5) did not hamper the enzyme activity.
Overall Protein Structure
Milligram quantities of PaeM protein (15–20 mg per liter of culture) were purified and subsequently used for crystallization experiments (see “Experimental Procedures” for details). The structure of the PaeM protein was solved using the multiple-wavelength anomalous dispersion method from crystals of selenomethionine-labeled protein. Two radically different crystallization conditions yielded diffracting crystals in the same space group (P212121). Crystals obtained in the presence of Mg2+ ions (200 mm MgCl2), a cation necessary for the enzymatic activity of ColM and its homologues (6, 12), diffracted to significantly higher resolution than those obtained in the absence of MgCl2. The PaeM crystal structure was refined against datasets collected from crystals grown in both conditions. The resulting structures were virtually identical (root mean square deviation value of 0.2 Å). The only significant difference between these two structures was the presence of a Mg2+ ion bound to the PaeM active site in the crystals grown in the presence of MgCl2 (see below). Hence, unless stated, we will mostly discuss the PaeM structure from the Mg2+-form, which has been refined at the highest resolution (1.7 Å).
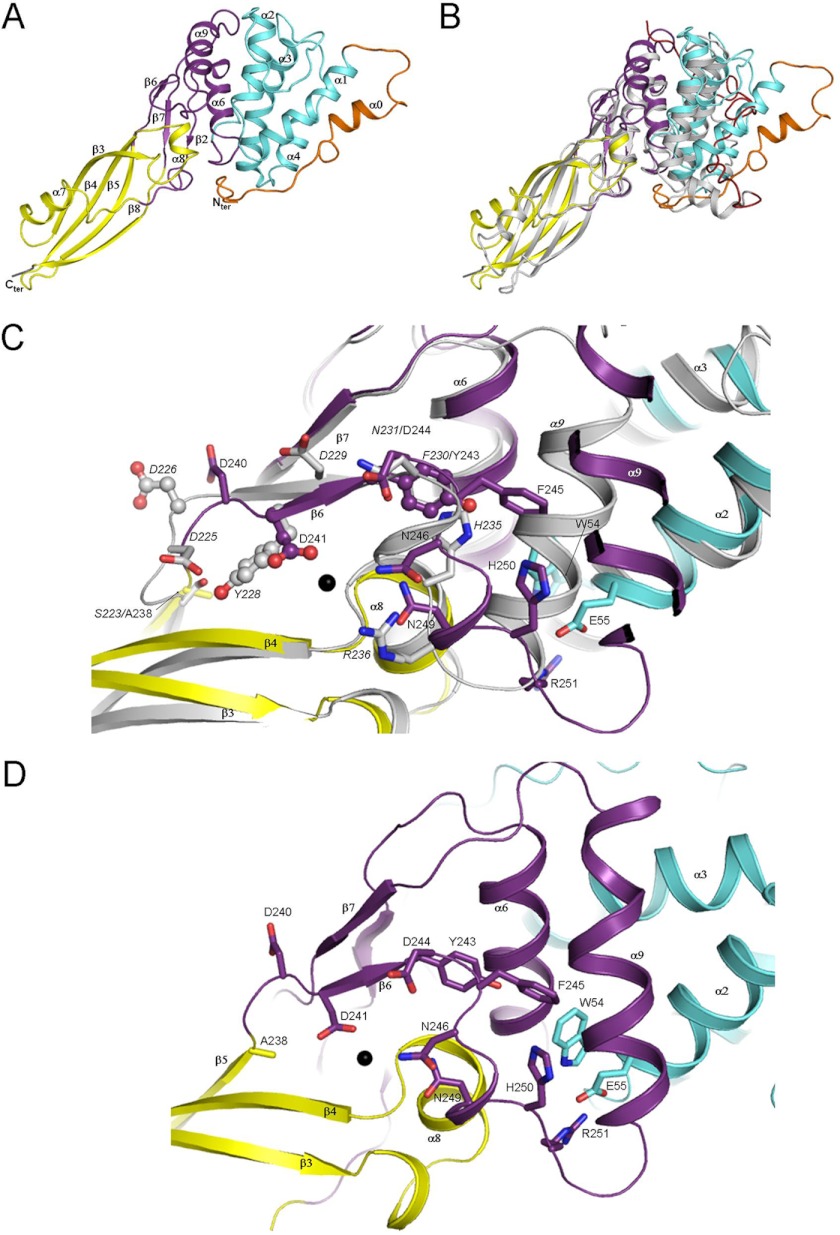
PaeM is an α/β protein that adopts an elongated cylinder-like shape (90 Å long and 35 Å wide). The overall structure is similar to that of E. coli ColM (Fig. 2, A and B, and Ref. 16). Based on both the crystal structure and enzymatic assays of truncated forms of ColM, the translocation, receptor binding, and cytotoxic domains of ColM are delineated by residues 1–35, 36–121, and 122–272, respectively (15, 16). In PaeM, the region encompassing residues Ser-33–Thr-136 (colored in cyan in Fig. 2A) folds as a four-helix bundle (helices α1 to α4) similarly to the receptor binding domain from ColM (root mean square deviation of 2.3 Å over 61 Cα atoms and 11% sequence identity). However, although helices α1 and α2 are packed in an antiparallel manner in PaeM, helix α1 is almost perpendicular to α2 in ColM (Fig. 2B). The structure of the PaeM region including residues Phe-137 to Ser-291 is very similar to the catalytic domain of ColM (root mean square deviation of 2.9 Å over 138 Cα atoms and 23% sequence identity). This domain can be subdivided into two subdomains. One subdomain (residues Pro-170–Ala-238 and Gly-281–Ser-291, colored in yellow in Fig. 2A) adopts an opened β-barrel formed by strands β3, β4, β5, and β8, with helix α7 running into the barrel and an additional helix (α8) on top of this domain and at the interface with the second subdomain. The other subdomain (residues Phe-137–Lys-169 and Tyr-239–Pro-280, colored in purple in Fig. 2A) is formed by two α-helices (α6 and α9) packed onto a three-stranded β-sheet (strands β2, β6, and β7). The largest structural difference between the PaeM and ColM catalytic domains originates from a 10° variation in the angle between the subdomains (Fig. 2B). More significant differences are observed between PaeM (residues Val-2 to Pro-32) and ColM (residues Met-1 to Pro-35) N-terminal regions (colored in orange and red, respectively, in Fig. 2, A and B), which correspond to the translocation domain. In ColM, this region adopts an extended conformation that wraps around the receptor binding domain, allowing the N terminus to interact with the C-terminal end of helix α9 from the catalytic domain (Fig. 2B). In PaeM, this region contains an α-helix (α0) that packs against helix α1 and otherwise adopts an extended conformation that runs in an opposite direction compared with the ColM translocation domain (Fig. 2B).
FIGURE 2.
Structure of PaeM. A, shown is a ribbon representation of the PaeM structure. The translocation and receptor binding domains are colored in orange and cyan, respectively. The catalytic domain is colored in yellow with the small subdomain in purple. The part of the His tag present at the C-terminal end of PaeM and defined in the 2 Fo − Fc electron density map is shown in gray. B, superimposition of ColM onto PaeM is shown. PaeM is colored according to the same color code as panel A. The ColM translocation domain is shown in red, and the remaining domains are in gray. C, shown is a comparison of PaeM (same color code as panel A) and ColM (gray) active sites. Residues discussed in the manuscript are shown as sticks and/or ball and sticks. Residues and secondary structure elements from ColM are labeled in italics. The Mg2+ ion is depicted as a black sphere. D, shown is a detailed representation of the PaeM active site (same color code as panel C).
Active Site Structure
Extensive site-directed mutagenesis studies identified residues Asp-226, Tyr-228, Asp-229, His-235, and Arg-236 from ColM as extremely important for both in vitro catalytic activity and in vivo cytotoxicity (15, 31–33). These residues are conserved in PaeM and are concentrated on the protein surface (Figs. 2 and 3), constituting the putative active site. They form a particularly large and atypical active site located around strand β6 (Fig. 3). Comparison of the active site regions in ColM and PaeM reveals a striking difference in their conformation (Fig. 2, C and D). Indeed, in both structures of PaeM obtained with or without MgCl2, the 240DDLYDF245 peptide slips by 6.5 Å compared with the corresponding peptide (225DDKYDF230) in ColM. This slip results from a more extended conformation of the loop connecting strands β5 and β6 compared with ColM. As a result, Tyr-243 from PaeM does not superimpose onto Tyr-228 from ColM, as would be expected from sequence comparison, but coincides with Phe-230 from ColM (Fig. 2, C and D). Importantly, Asp-241 from PaeM, which is the homologue of the catalytic Asp-226 from ColM, slips and flips over by 180° and superposes with ColM Tyr-228. In the structure of PaeM obtained with MgCl2, a strong peak in the Fo − Fc electron density map was located near the carboxylate of Asp-241 and was attributed to an Mg2+ ion (see “Experimental Procedures” for details). The comparison of the PaeM structures in the absence or presence of the Mg2+ ion does not show important changes. Only the Asp-241 side chain slightly rotates upon Mg2+ binding to complete the coordination sphere. This indicates that the slip of the active site peptide observed in PaeM compared with ColM is not induced by Mg2+ binding but is inherent to PaeM. It is noteworthy that recombinant PaeM is much more active (600-fold) than recombinant ColM (12). We suspect that our PaeM structure corresponds to a “pre-formed” active conformation and that the active site configuration of ColM represented an inactive or less active form. However, the His-250 and Arg-251 side chains, which correspond to the critical residues His-235 and Arg-236 of ColM, are not oriented toward the putative substrate binding site but face the interior of the protein (Fig. 2, C and D), suggesting that these residues may reorient upon catalysis. Hence, we speculate that the active site will only adopt its fully active conformation upon binding of the substrate. It has been hypothesized that ColM must be unfolded and refolded with the aid of a periplasmic chaperone with cis/trans peptidyl-proline isomerase activity to acquire its cytotoxic activity (9, 34). We can also assume that PaeM follows a similar scenario that ends up with a fully active conformation of the enzyme.
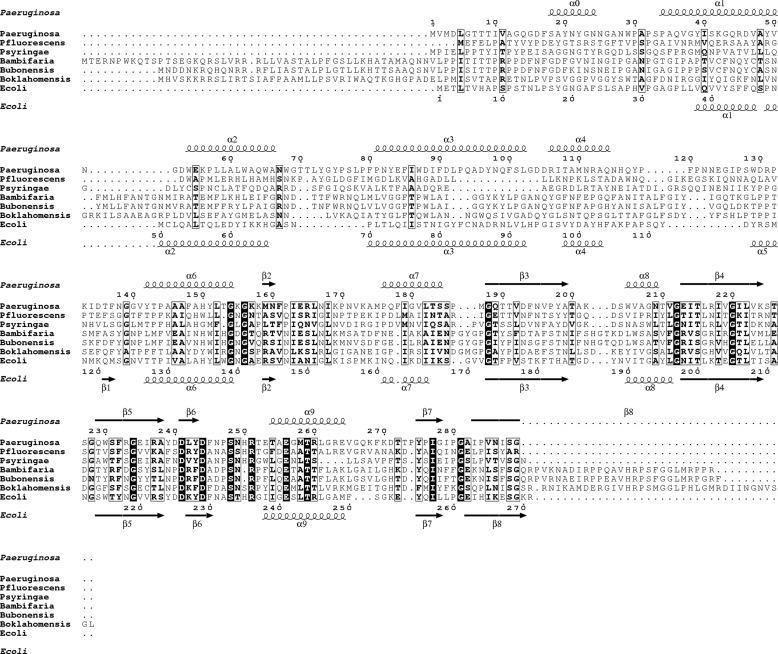
FIGURE 3.
Sequence alignment of PaeM orthologues. Strictly conserved residues are in white on a black background. Partially conserved amino acids are boxed. Secondary structure elements present in the crystal structures of PaeM and ColM are shown above and below the alignment, respectively. The aligned PaeM orthologues sequences are from Pseudomonas aeruginosa (Paeruginosa), Pseudomonas fluorescens (Pfluorescens), Pseudomonas syringae (Psyringae), Burkholderia ambifaria (Bambifaria), Burkholderia ubonensis (Bubonensis), Burkholderia oklahomensis (Boklahomensis), and Escherichia coli (Ecoli).
Localization of the PaeM Active Site
The kinetic parameters of the purified wild-type PaeM protein (His-tagged form) were previously determined and compared with those of ColM (12). Although these two proteins had a quite similar Km value for lipid II (about 45 μm), the kcat value of PaeM appeared much higher than that of ColM (32 versus 0.055 min−1), revealing a ∼600-fold greater catalytic efficiency of the P. aeruginosa homologue. The important structural differences observed in the active site of PaeM and ColM may explain this discrepancy. Site-directed mutagenesis experiments were now performed to more precisely delineate the active site of PaeM and to identify amino acid residues that are essential for the catalytic process. Based on alignments of ColM-like proteins, several putative active site residues were previously identified whose role in substrate binding or catalysis in ColM was questioned (15). In particular, five residues were shown to be essential for ColM activity, namely Asp-226, Tyr-228, Asp-229, His-235, and Arg-236. Mutagenesis of the corresponding PaeM residues (Asp-241, Tyr-243, Asp-244, His-250, and Arg-251) was performed (Table 3). Both a dramatic decrease (by 95–99%) of the in vitro lipid II-degrading activity and a loss of cytotoxicity toward the DET08 strain were observed for the D241A, D244A, Y243A, and R251A mutants. The perfect correlation observed between the in vitro (enzymatic activity) and in vivo (P. aeruginosa growth inhibition) effects of these different wild-type and mutant PaeM proteins confirmed that the cytotoxicity of PaeM was related to its catalytic properties of degradation of the lipid II peptidoglycan precursor. Compared with all single mutants that still exhibited a very low level of residual activity in vitro, the double mutant D241A/D244A had no detectable activity, showing that these aspartate residues play a major role in catalysis. The H250A mutation only yielded a limited reduction of in vitro activity and had no effect on the cytotoxicity of PaeM. This is in contrast with the homologous His-235 residue of ColM that was found critical for activity. This histidine residue is conserved in ColM homologues from E. coli and Pseudomonas species but not those from Burkholderia spp. As judged from the structure of PaeM, no other amino acid residue was identified that could play the potential role of ColM His-235. These data suggest that the catalytic mechanism, which is expected to be identical for these different homologues, does not involve a histidine residue. As discussed previously, PaeM residues His-250 and Arg-251 are localized on a loop that sticks out of the active site center. This correlates well with the fact that His-250 is not essential. However, residue Arg-251 of PaeM and the homologous Arg-236 of ColM were both found critical for activity, suggesting that this loop may still undergo a structural change to reorient Arg-251 toward the active site center. In ColM, the substitution of the Ser-223 residue by an alanine was shown to result in a 1.5-fold activity increase (15). According to the ColM structure, this serine side chain forms a hydrogen bond with the Tyr-228 hydroxyl group, whose side chain is oriented away from the active site (Fig. 2C). Based on the conformation observed in our PaeM structure, we propose that the S223A mutation in ColM releases this Tyr-228 constrained conformation, thereby facilitating the slip of strand β6 concomitant with the movement of Tyr-228 to occupy the same location as PaeM Tyr-243. This may explain the rise of activity observed for this ColM mutant. In PaeM, Ala-238 overlaps with ColM Ser-223 and cannot form a hydrogen bond with Tyr-243. Replacement of Ala-238 by a serine (A238S mutant) decreased PaeM activity 3-fold, supporting the assumption that the serine side chain may lock the active site in a less active conformation. Another putative active site mutant, N249A (see Fig. 2D), displayed wild-type activity, indicating that this asparagine residue did not play any role in the catalytic process or the substrate binding.
TABLE 3.
In vitro and in vivo activities of PaeM mutant proteins
| Protein | Plasmid | Enzymatic activitya | Cytotoxicity against P. aeruginosab |
|---|---|---|---|
| nmol·min−1·mg−1 | μg | ||
| Wild type | pMLD245 | 13 (100%) | 0.04 |
| A238S | pTTB54 | 4.5 (34.5%) | 0.1 |
| D241A | pTTB240 | 0.1 (0.6%) | ND |
| D244A | pTTB243 | 0.6 (4.9%) | ND |
| D241A/D244A | pTTB38 | NDc | ND |
| Y243A | pTTB26 | 0.2 (1.5%) | ND |
| N249A | pTTB60 | 17 (130%) | 0.04 |
| H250A | pTTB30 | 11 (83.2%) | 0.1 |
| R251A | pTTB36 | 0.1 (1.1%) | ND |
| W54K | pTTB67 | 203 | –d |
| E55A | pTTB72 | 206 | – |
| W54K/E55A | pTTB78 | 217 | – |
a The enzymatic activity was measured in vitro in the presence of 2.3 μm of [14C]lipid II and the appropriate amounts of enzyme. Values represent the mean of triplicate determinations, the S.D. being within 15% of the presented values.
b The cytotoxicity of PaeM proteins was measured by spotting various amounts of pure protein samples on a lawn of the susceptible P. aeruginosa DET08 strain. The cytotoxicity was expressed as the minimal amount of protein required to obtain a clear halo indicative of growth inhibition.
c ND, no detectable activity (for in vitro assays, up to 15 μg of protein tested, except for Y243A, 1.5 μg tested; for cytotoxicity assays, up to 3.5 μg of protein tested).
d The cytotoxicity of these variants was not tested.
Isolation of the Catalytic/Killing Domain
Protein dissection experiments showed earlier that it was possible to individually express the C-terminal domain of ColM while maintaining its functionality (15, 31). Interestingly, the specific activity of the isolated catalytic domain of ColM was much higher (50-fold) than that of the full-length protein, suggesting that the presence of the N-terminal and central domains involved in receptor binding and import steps interfered with the activity of the C-terminal domain. The comparison between ColM and PaeM structures may explain this observation. Indeed, the loop at the exit of strand β6 of PaeM, which contains Phe-245, His-250, and Arg-251, flips out of the active site compared with its conformation in ColM. This further creates a 35° swing of helix α9 compared with ColM so as to avoid steric clashes with Phe-245 (Fig. 2C). A similar movement of helix α9 in ColM would be more difficult because it is tightly packed against helices α2 and α6. This observation could rationalize the higher in vitro enzymatic activity of a truncated form of ColM, for which the N-terminal fragment comprised between Ala-33 to Met-122 is deleted (hence lacking helix α2) (15). In this truncated protein helix α9 could more freely adopt the position as observed in the PaeM structure. It was also shown that deletion of the ColM translocation domain (first 32 N-terminal residues) was sufficient to improve the in vitro enzymatic activity of ColM by 50-fold (15). Interestingly, the “TonB” box from this domain (residues 1–6) is stacked onto the C-terminal extremity of helix α9 in the ColM structure. Hence, it would prevent helix α9 from ColM to swing and to adopt the same position as in PaeM without introducing a steric clash. We can imagine that after entry of ColM across the outer membrane into the periplasm, the TonB box will remain bound to the TonB/ExbB/ExbD translocation machinery and will not prevent FkpA-assisted refolding of the catalytic domain in a conformation similar to that of PaeM.
To address whether the killing domain of PaeM can also be expressed individually and whether it displays the same catalytic activity as the full-length protein, truncated variants of PaeM were generated. Based on the structure, we decided to cut the protein in the linker region connecting the receptor binding and catalytic domains, i.e. within the long loop connecting helices α4 and α6. The resulting PaeM-Δ1–126 and Δ1–133 constructs thus encompassed essentially the C-terminal half of the protein that is well conserved among ColM-like proteins. Contrary to what was observed previously for ColM (15), these truncated PaeM proteins were soluble and could be purified without denaturation/renaturation step. As shown in Table 4, they exhibited a lipid II-degrading activity (∼1 μmol·min−1·mg−1) that was much higher (∼70 fold) than that of the full-length PaeM. It was noteworthy that it was the highest activity ever found for a ColM-like protein-isolated killing domain. First, this finding clearly demonstrated that the catalytic domain of PaeM was functionally independent. It also showed, as observed previously for ColM (15), that the presence of the N-terminal protein domain affected the catalytic activity of the enzyme. Importantly, in contrast to the full-length PaeM protein, the two truncated proteins were shown to exhibit a highly potent cytotoxic effect against E. coli cells under bypass conditions (osmotic shock treatment), allowing them to get access to the periplasmic space of the bacteria (Table 4). To our knowledge this is the first example of a ColM-like protein capable of killing other bacterial species when targeted to the periplasm of these bacteria. The fact that the full-length PaeM was only poorly toxic against E. coli cells in these conditions substantiated the hypothesis that it may require a maturation process once it has reached this cell compartment. As ColM requires FkpA to exert its lethal effect, we can, therefore, speculate that PaeM follows a similar pathway involving specific chaperone/peptidyl-proline isomerase activities from P. aeruginosa.
TABLE 4.
Activity of truncated forms of PaeM
| Protein | Enzymatic activity | Cytotoxicity against E. colia |
|---|---|---|
| nmol·min−1·mg−1 | % of surviving cells | |
| Control (buffer) | 100% | |
| Full-length PaeM | 13 | 53% |
| PaeM-Δ1–126 | 850 | 0.1% |
| PaeM-Δ1–133 | 1010 | 0.1% |
a The cytotoxicity of full-length and truncated forms of PaeM was tested on E. coli BW25113 strain by the bypass procedure. Exponentially growing cells were washed and then incubated in a high osmolarity solution (48% sucrose) before being suddenly diluted in M9-salts solution (1/50, v/v) in the presence or not of 10 μl of pure protein sample (40 and 1.5 μg of protein for the full-length and truncated forms, respectively). After 30 min of incubation in the cold, the number of surviving cells was determined by plating cell suspensions on 2YT-agar medium. The result is expressed as the percentage of survivals as compared with the condition without added PaeM.
These data suggest that in both PaeM and ColM proteins the translocation/reception domains may apply constraints to the killing domain to keep it in an intermediate conformation that is not fully active. In most colicins the three functional domains are clearly separated by peptide linkers, which is not the case in ColM-like bacteriocins. In the structures of PaeM and ColM, the three domains are interlaced altogether, and the overall structure is rather compact. We then searched for specific interactions existing at the interface between the activity and receptor domains of PaeM that could stabilize such a compact structure. Indeed, we speculated that the disruption of these constraints could perturb this conformation of PaeM and increase its activity. As mentioned above, the side chain of the essential Arg-251 residue of the activity domain of PaeM forms a salt bridge with the Glu-55 side chain from the receptor binding domain. Also, the tryptophan residue Trp-54 from the receptor binding domain tightly packs against helix α6 from the catalytic domain (Fig. 2D). To destabilize this interface, we therefore constructed three PaeM mutants carrying the W54K, E55A, and W54K/E55A substitutions. Expression of these proteins yielded inclusion bodies, but they were successfully solubilized and purified (see “Experimental Procedures”). All three mutants exhibited a similar activity toward the lipid II that was 15-fold higher than that observed for the wild-type protein (Table 3) but was still lower than that of the isolated killing domain (Table 4). However, these variants did not inhibit growth of E. coli cells when tested under bypass conditions (100% surviving cells observed after treatment with 1.5 μg of these purified proteins). Even though these PaeM mutants were not capable of killing E. coli cells, this analysis clearly substantiated the fact that weakening the interactions between the killing and reception domains of PaeM improves the in vitro enzymatic activity.
The observation that ColM and PaeM did not discriminate in vitro between lipids II originating from different bacterial species prompted us to test these bacteriocins on Gram-positive species. Indeed, the absence of outer membrane in the latter species eliminated one of the barriers potentially limiting accessibility of these proteins to their target located in the inner membrane. Preliminary experiments showed that neither ColM and PaeM nor their truncated forms, in particular the PaeM-Δ1–133 variant exhibiting the highest lipid II hydrolase activity in vitro, showed antibacterial activity against the tested Gram-positive species (S. aureus, E. faecalis, and E. faecium). The reasons for this lack of activity remain to be determined.
Conclusion
The crystal structure of the ColM-like bacteriocin PaeM of P. aeruginosa has been determined in the presence and absence of Mg2+, a cation that is essential for the enzyme activity. Compared with ColM, this structure reveals significant differences in the spatial positions of the conserved residues identified as essential for the bacteriocin enzyme activity and cytotoxicity. These structural differences could rationalize the higher specific enzymatic activity (by 600-fold) of PaeM relative to ColM and highlight a structural plasticity of the active site of this enzyme family. Although PaeM is much more active than ColM in vitro, it exhibits only poor killing efficiency against E. coli cells when targeted to the periplasm of these bacteria. PaeM may not be in the fully active conformation that is required to meet and degrade lipid II molecules exposed to the periplasmic face of the membrane, or the presence of the translocation/reception domains inhibits the expression of the cytotoxic effect. In contrast, a truncated PaeM construct that only contains the catalytic domain is 70-fold more active than the full-length protein and displays a potent toxic effect when targeted to the periplasm of E. coli cells. A proteolytic cleavage event that releases the catalytic domain from the rest of the molecule has been described for some nuclease colicins but not for ColM (1). Whether proteolytic maturation of PaeM follows its import in P. aeruginosa cells cannot be totally excluded yet, and this should be explored in the future. The analysis of truncated variants of PaeM allowed us to identify a small-size killing domain (14 kDa) whose lipid II-degrading activity is 43,000-fold higher than that of ColM and that is capable of exerting its cytotoxic effect once targeted to the periplasm of E. coli cells. Moreover, in contrast to the killing domain of ColM, that of PaeM showed a superior ability to fold without the assistance of the rest of the protein. This isolated domain thus represents an attractive weapon that could potentially be exploited as a broad spectrum antibacterial agent. Indeed, all types of eubacteria, including both Gram-negative and Gram-positive bacteria, contain peptidoglycan as an essential cell-wall polymer, and any novel agent interfering with the biosynthesis of this polymer could, therefore, represent an alternative way to circumvent the problem of antibiotic bacterial resistance. The peptidoglycan structure varies to some extent in the bacterial world, in particular in the amino acid composition, structure, and cross-linking of the peptide chains. In fact, we recently showed that ColM hydrolyzed lipids II of different structures originating from E. coli, E. faecalis, E. faecium, and S. aureus at a similar rate (23). In this work we demonstrated that this is also the case for PaeM. These two bacteriocins and their homologues could thus be considered as interesting candidates to be exploited in a perspective of development of new antibacterial agents. As preliminary experiments showed that these proteins and their isolated catalytic domains did not inhibit growth of the few Gram-positive species tested, one of the main challenges in the future will be to render these enzymes accessible to their targets in the different pathogenic bacterial species considered.
Acknowledgments
We are indebted to Anthony Doizy and Dominique Liger for technical assistance. We acknowledge SOLEIL for provision of synchrotron radiation facilities and in particular staff members from beamline Proxima-1.
This work was supported by grants from the Agence Nationale de la Recherche (PEPGLYCOL project, ANR-07-MIME-020) and the Centre National de la Recherche Scientifique (UMR 8619).
The atomic coordinates and structure factors (codes 4G75 and 4G76) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- ColM
- colicin M
- PaeM
- ColM-like protein from P. aeruginosa
- MurNAc
- N-acetylmuramic acid
- GlcNAc
- N-acetylglucosamine
- Ni2+-NTA
- Ni2+-nitrilotriacetic acid.
REFERENCES
- 1. Cascales E., Buchanan S. K., Duché D., Kleanthous C., Lloubès R., Postle K., Riley M., Slatin S., Cavard D. (2007) Colicin biology. Microbiol. Mol. Biol. Rev. 71, 158–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pugsley A. P. (1984) The ins and outs of colicins. Part II. Lethal action, immunity, and ecological implications. Microbiol. Sci. 1, 203–205 [PubMed] [Google Scholar]
- 3. Pugsley A. P. (1984) The ins and outs of colicins. Part I. Production and translocation across membranes. Microbiol. Sci. 1, 168–175 [PubMed] [Google Scholar]
- 4. Schaller K., Höltje J. V., Braun V. (1982) Colicin M is an inhibitor of murein biosynthesis. J. Bacteriol. 152, 994–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barreteau H., El Ghachi M., Barnéoud-Arnoulet A., Sacco E., Touzé T., Duché D., Gérard F., Brooks M., Patin D., Bouhss A., Blanot D., van Tilbeurgh H., Arthur M., Lloubès R., Mengin-Lecreulx D. (2012) Characterization of colicin M and its orthologs targeting bacterial cell wall peptidoglycan biosynthesis. Microb. Drug Resist. 18, 222–229 [DOI] [PubMed] [Google Scholar]
- 6. El Ghachi M., Bouhss A., Barreteau H., Touzé T., Auger G., Blanot D., Mengin-Lecreulx D. (2006) Colicin M exerts its bacteriolytic effect via enzymatic degradation of undecaprenyl phosphate-linked peptidoglycan precursors. J. Biol. Chem. 281, 22761–22772 [DOI] [PubMed] [Google Scholar]
- 7. Braun V., Schaller K., Wolff H. (1973) A common receptor protein for phage T5 and colicin M in the outer membrane of Escherichia coli B. Biochim. Biophys. Acta 323, 87–97 [DOI] [PubMed] [Google Scholar]
- 8. Braun V., Patzer S. I., Hantke K. (2002) Ton-dependent colicins and microcins. Modular design and evolution. Biochimie 84, 365–380 [DOI] [PubMed] [Google Scholar]
- 9. Hullmann J., Patzer S. I., Römer C., Hantke K., Braun V. (2008) Periplasmic chaperone FkpA is essential for imported colicin M toxicity. Mol. Microbiol. 69, 926–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gérard F., Brooks M. A., Barreteau H., Touzé T., Graille M., Bouhss A., Blanot D., van Tilbeurgh H., Mengin-Lecreulx D. (2011) X-ray structure and site-directed mutagenesis analysis of the Escherichia coli colicin M immunity protein. J. Bacteriol. 193, 205–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Usón I., Patzer S. I., Rodríguez D. D., Braun V., Zeth K. (2012) The crystal structure of the dimeric colicin M immunity protein displays a 3D domain swap. J. Struct. Biol. 178, 45–53 [DOI] [PubMed] [Google Scholar]
- 12. Barreteau H., Bouhss A., Fourgeaud M., Mainardi J. L., Touzé T., Gérard F., Blanot D., Arthur M., Mengin-Lecreulx D. (2009) Human- and plant-pathogenic Pseudomonas species produce bacteriocins exhibiting colicin M-like hydrolase activity toward peptidoglycan precursors. J. Bacteriol. 191, 3657–3664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grinter R., Milner J., Walker D. (2012) Ferredoxin containing bacteriocins suggest a novel mechanism of iron uptake in Pectobacterium spp. PLoS One 7, e33033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kulasekara B. R., Kulasekara H. D., Wolfgang M. C., Stevens L., Frank D. W., Lory S. (2006) Acquisition and evolution of the exoU locus in Pseudomonas aeruginosa. J. Bacteriol. 188, 4037–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Barreteau H., Bouhss A., Gérard F., Duché D., Boussaid B., Blanot D., Lloubès R., Mengin-Lecreulx D., Touzé T. (2010) Deciphering the catalytic domain of colicin M, a peptidoglycan lipid II-degrading enzyme. J. Biol. Chem. 285, 12378–12389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zeth K., Römer C., Patzer S. I., Braun V. (2008) Crystal structure of colicin M, a novel phosphatase specifically imported by Escherichia coli. J. Biol. Chem. 283, 25324–25331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Amrein K. E., Takacs B., Stieger M., Molnos J., Flint N. A., Burn P. (1995) Purification and characterization of recombinant human p50csk protein-tyrosine kinase from an Escherichia coli expression system overproducing the bacterial chaperones GroES and GroEL. Proc. Natl. Acad. Sci. U.S.A. 92, 1048–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Miller J. H. (1972) Experiments in Molecular Genetics. pp. 431–435, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 19. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, New York [Google Scholar]
- 20. Dagert M., Ehrlich S. D. (1979) Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene 6, 23–28 [DOI] [PubMed] [Google Scholar]
- 21. Laemmli U. K., Favre M. (1973) Maturation of the head of bacteriophage T4. I. DNA packaging events. J. Mol. Biol. 80, 575–599 [DOI] [PubMed] [Google Scholar]
- 22. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 23. Patin D., Barreteau H., Auger G., Magnet S., Crouvoisier M., Bouhss A., Touzé T., Arthur M., Mengin-Lecreulx D., Blanot D. (2012) Colicin M hydrolyses branched lipids II from Gram-positive bacteria. Biochimie 94, 985–990 [DOI] [PubMed] [Google Scholar]
- 24. Kabsch W. (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 26, 795–800 [Google Scholar]
- 25. Schneider T. R., Sheldrick G. M. (2002) Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 [DOI] [PubMed] [Google Scholar]
- 26. Bricogne G., Vonrhein C., Flensburg C., Schiltz M., Paciorek W. (2003) Generation, representation, and flow of phase information in structure determination. Recent developments in and around SHARP 2.0. Acta Crystallogr. D Biol. Crystallogr. 59, 2023–2030 [DOI] [PubMed] [Google Scholar]
- 27. Morris R. J., Zwart P. H., Cohen S., Fernandez F. J., Kakaris M., Kirillova O., Vonrhein C., Perrakis A., Lamzin V. S. (2004) Breaking good resolutions with ARP/wARP. J. Synchrotron Radiat. 11, 56–59 [DOI] [PubMed] [Google Scholar]
- 28. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) PHENIX. Building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 29. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 30. Vollmer W., Blanot D., de Pedro M. A. (2008) Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32, 149–167 [DOI] [PubMed] [Google Scholar]
- 31. Barnéoud-Arnoulet A., Barreteau H., Touzé T., Mengin-Lecreulx D., Lloubès R., Duché D. (2010) Toxicity of the colicin M catalytic domain exported to the periplasm is FkpA independent. J. Bacteriol. 192, 5212–5219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Helbig S., Braun V. (2011) Mapping functional domains of colicin M. J. Bacteriol. 193, 815–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pilsl H., Glaser C., Gross P., Killmann H., Olschläger T., Braun V. (1993) Domains of colicin M involved in uptake and activity. Mol. Gen. Genet. 240, 103–112 [DOI] [PubMed] [Google Scholar]
- 34. Helbig S., Patzer S. I., Schiene-Fischer C., Zeth K., Braun V. (2011) Activation of colicin M by the FkpA prolyl cis-trans isomerase/chaperone. J. Biol. Chem. 286, 6280–6290 [DOI] [PMC free article] [PubMed] [Google Scholar]