Background: APRIL binds two receptors, BCMA and TACI, so separating signaling outcomes is difficult.
Results: We used an algorithm to design a variant of APRIL that specifically binds BCMA and two variants that selectively bind TACI.
Conclusion: TACI and BCMA signals differ in the context of B cell stimulation.
Significance: These variants will help decipher APRIL signaling in physiology and disease settings.
Keywords: Computational Biology, Innate Immunity, Protein Structure, Protein-Protein Interactions, Receptor Regulation, Signaling, TRAF, APRIL, BCMA, TACI
Abstract
A proliferation-inducing ligand (APRIL), a member of the TNF ligand superfamily with an important role in humoral immunity, is also implicated in several cancers as a prosurvival factor. APRIL binds two different TNF receptors, B cell maturation antigen (BCMA) and transmembrane activator and cylclophilin ligand interactor (TACI), and also interacts independently with heparan sulfate proteoglycans. Because APRIL shares binding of the TNF receptors with B cell activation factor, separating the precise signaling pathways activated by either ligand in a given context has proven quite difficult. In this study, we have used the protein design algorithm FoldX to successfully generate a BCMA-specific variant of APRIL, APRIL-R206E, and two TACI-selective variants, D132F and D132Y. These APRIL variants show selective activity toward their receptors in several in vitro assays. Moreover, we have used these ligands to show that BCMA and TACI have a distinct role in APRIL-induced B cell stimulation. We conclude that these ligands are useful tools for studying APRIL biology in the context of individual receptor activation.
Introduction
A proliferation-inducing ligand (APRIL) is a member of the TNF ligand superfamily originally described as a tumor-promoting factor (1). Physiologically, it plays a prominent role in humoral immunity, in particular by driving antibody class switch toward IgG and IgA (2, 3) and by promoting survival of plasma cells (4). APRIL has also been identified as a prosurvival factor for several B cell malignancies, possibly via the activation of transcription factor NF-κB (reviewed in Ref. 5). APRIL is also thought to promote tumor formation in a number of solid malignancies, either indirectly via infiltrating cells or directly via autocrine stimulation of the tumor itself (1, 6, 7). In line with these results, we recently identified a clear role for APRIL in supporting tumorigenesis in the gastrointestinal tract (8).
APRIL binds two different receptors of the TNF receptor superfamily: B cell maturation antigen (BCMA) and transmembrane activator and cyclophilin ligand interactor (TACI), which are also bound by its homolog B cell-activating factor (BAFF) (9–12). In addition, APRIL binds to heparan sulfate proteoglycans, which appear to play a predominantly structural role by enabling APRIL cross-linking (13, 14), although a distinct signaling role in different contexts cannot be eliminated. In addition to APRIL, TACI can also bind to heparan sulfate proteoglycans, which is suggested to lead to its activation (15). All of these potential binding partners make it difficult to unravel APRIL signaling in a given context and to assess the individual contributions of TACI and BCMA. Therefore, it is not surprising that little is known about the individual signaling pathways activated in response to signals via each of the APRIL receptors or precisely how these are separated in terms of the formation of distinct intracellular complexes and recruitment of signaling adaptors. Much of what is currently known with regard to activation of transcription factors and recruitment of internal adaptors, such as TNF receptor-associated factors (TRAFs), has been carried out using transfection studies (16, 17) or with RNAi-mediated knock-down studies (18), which poses possible problems associated with overexpression or simultaneous removal of multiple interactions, respectively.
In order to generate a tool by which to study APRIL interaction with the individual TNF receptors, we decided to take advantage of computational protein design in order to generate APRIL variants that can bind only one of its receptors. We used the FoldX protein design algorithm (19, 20) to predict amino acid substitutions in APRIL that might switch ligand binding toward one or the other receptor. Previously, FoldX was successfully used to engineer variants of TRAIL that specifically recognize its receptor DR4 or DR5 (21–24). Such a “branch-pruning” approach has the added advantage that it allows selective removal of a single interaction, which contrasts with knock-out or knock-down studies that remove all interactions in which the target protein is involved (25).
In this study, we generated a total of 21 mutant forms of the APRIL protein and tested their ability to bind either BCMA or TACI. Three mutants were of particular interest: APRIL-R206E, which showed clear specificity toward both human and mouse BCMA, and APRIL-D132F and APRIL-D132Y, which showed considerable selectivity for TACI. Following initial ELISAs using immobilized receptors, we further confirmed the binding characteristics in the context of cell-based assays, using either transfected cells in which receptors were overexpressed or endogenously expressing BCMA or TACI cell species. Finally, we used these APRIL variants in a B cell assay to show distinct roles for TACI and BCMA in B cell function.
EXPERIMENTAL PROCEDURES
Computational Design of Selective Variants
X-ray crystal structures of the extracellular domain (ECD)6 of murine APRIL in complex with the ECD of human TACI (Protein Data Bank entry 1xu1) and the ECD of human BCMA (Protein Data Bank 1xu2) have been solved at a resolution of 1.9 and 2.35 Å, respectively (21). Computational design of receptor selective mutants was performed as described previously (22, 23, 26). In short, amino acid residues with Van der Waals clashes, with bad torsion angles, or with a high energy in the crystal structure were repaired by replacing the side chain conformations (rotamers) observed in the x-ray structure by lower energy rotamers; hydrogen bond networks were optimized using the Repair PDB option of the FoldX protein design algorithm (19, 27). Next, a model of human APRIL in complex with human TACI and human BCMA was constructed by FoldX in silico mutagenesis of each of the non-conserved murine residues to its homologous human counterpart. Each residue in the receptor binding interface of human APRIL was subsequently mutated by FoldX to all of the other 19 naturally occurring amino acids using the BuildModel function. The effect on the interaction energy with TACI and BCMA was calculated as the difference in interaction energy (ΔΔG, in kcal/mol) between the interaction energy of the mutant and the wild-type amino acid, using the AnalyseComplex option. In the case of preformed trimers, such as APRIL, the AnalyseComplex option was set as before (22, 23, 26) to consider the monomer subunits comprising APRIL as a single molecule and therefore did not discriminate in terms of binding energy on which particular monomer subunit a residue that interacts with the receptor is located.
Amino acid substitutions were selected that 1) caused a decrease in interaction energy toward one of the receptors, 2) caused an increase in interaction energy toward one receptor while causing either a decrease in interaction energy or showing a neutral effect toward the other receptor, or 3) caused an increase in interaction energy for both receptors but showed a different magnitude in change for one receptor over the other. Because some variants (mainly from the Asp-132 series) showed severe intrachain Van der Waals clashes upon mutation, it was decided to report the average interaction energy corrected for intrachain Van der Waals clashes. This corrected interaction energy was calculated by summing the average interaction energy and the average ΔΔGintraclash energy, where ΔΔGintraclash is the difference in intrachain clash energy (ΔΔGintraclash, in kcal/mol) between the intrachain clash energy of the mutant and the wild-type amino acid. The reported corrected interaction energy was capped at +4 kcal/mol.
Generation of Variants
FLAG-tagged APRIL variants selective for a receptor were generated using a QuikChange site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA) according to the manufacturer's guidelines, using the primers listed in the supplemental material. A pcDNA3.1 construct containing FLAG-tagged wild-type soluble APRIL (amino acids 105–250, numbering according to UniprotKB/Swiss-Prot O75888) was used as the PCR template and was described previously (14). Plasmid DNA of clones was isolated (BIOKÉ, Leiden, The Netherlands), and the presence of the mutation(s) was verified by DNA sequencing. Positive clones were selected and grown for large scale plasmid DNA isolation and used for subsequent transfections.
Cell Culture
Human Jurkat and Raji cells and mouse A20 cells were cultured in RPMI 1640 (Invitrogen). Primary mouse B cells were cultured in RPMI 1640 supplemented with 50 μm 2-mercaptoethanol. 293T cells were cultured in Iscove's modified Dulbecco's medium. All media were supplemented with 8% FCS, 2 mm l-glutamine, 40 μg/ml penicillin, and 40 units/ml streptomycin and maintained at 37 °C with 5% CO2.
Expression of APRIL Variants
To test the expression of the individual FLAG-tagged APRIL variants, 293T cells were grown to 60% confluence in a 6-well plate and transfected with the different variants using calcium phosphate precipitation. Following transfection, the cells were kept in culture for 72 h before supernatant containing the soluble mutants was harvested and stored at −20 °C. Expression of each of the APRIL mutants was then tested by Western blotting, and the relative concentrations were assessed. Briefly, supernatants were resolved on a 15% polyacrylamide gel, transferred to a nitrocellulose membrane, and blocked overnight using Odyssey blocking buffer (LI-COR Biosciences, Cambridge, UK) diluted 1:1 with PBS. The membrane was then incubated with mouse anti-FLAG-M2 (Sigma-Aldrich) at a concentration of 1:5000 diluted in PBS, 0.2% Tween 20; the secondary antibody used was IRD800-coupled anti-mouse IgG1 (Westburg, Leusden, The Netherlands) diluted in PBS, 0.2% Tween 20, and 0.02% SDS. Blots were visualized using a near infrared imaging system (Odyssey, LI-COR Biosciences) according to the manufacturer's instructions, which allows quantification of bands to give a relative estimate of protein concentration.
Receptor-binding ELISA
The relative binding of the variants to either BCMA or TACI was tested using a binding ELISA. The following proteins were used: human BCMA-Fc, human TACI-Fc (both generated in house), mouse BCMA-Fc (R&D Systems, Abingdon, UK), and mouse TACI-Fc (R&D Systems); in all cases, the Fc portion was from human IgG1. BCMA-Fc and TACI-Fc were coated on 96-well Nunc Maxisorp plates (Thermo Fisher Scientific, Roskilde, Denmark) at concentrations of 1 and 2 μg/ml, respectively, in 0.5 m sodium bicarbonate buffer, pH 9.5, overnight at 4 °C. Plates were then blocked with 5% BSA for 1 h at 37 °C. Soluble APRIL variants in the form of tissue culture conditioned medium were then added to the plate for 2 h at 37 °C. Following APRIL binding, plates were washed three times with PBS, 0.05% Tween 20. Bound APRIL was detected with 1 μg/ml HRP-coupled anti-FLAG M2 and visualized with ABTS (Sigma-Aldrich). The absorbance was read at 405 nm using a UV-visible microplate reader (Bio-Rad).
Surface Plasmon Resonance
In order to further assess the binding properties of the mutants and to measure apparent affinities and kinetics of receptor binding, a surface plasmon resonance-based receptor-binding assay was performed on a Biacore 2000 system (GE Healthcare). Anti-FLAG-M2 monoclonal antibody (Sigma-Aldrich) was diluted at a concentration of 5 μg/ml in 10 mm sodium acetate, pH 4.5, and covalently immobilized (approximately 1500 resonance units) to a CM-5 sensor chip (GE Healthcare), using standard amine coupling chemistry according to the manufacturer's guidelines (amine coupling kit, GE Healthcare). The different FLAG-APRIL variants in the form of tissue culture supernatants were captured onto the chip via the FLAG tag at 10 μl/min for 5 min, giving capture levels ranging from 159 to 207 resonance units. As a reference lane, one of the flow cells was left free of soluble FLAG-APRIL, to control for any background binding. BCMA- or TACI-Fc was then injected for 3 min over all four flow cells at 30 μl/min at increasing concentrations (ranging from 1 to 50 nm) using the single cycle kinetics method at 25 °C. Dissociation was monitored during 5 min. The resulting curves were fitted using a 1:1 Langmuir model using BIAevaluation software version 4.1. For each combination of FLAG-APRIL and BCMA- or TACI-Fc, the apparent ka, kd, and KD were calculated from a global fit of binding curves from at least three separate experiments. Nonspecific binding in the reference cell was subtracted before curve fitting. Regeneration of the anti-FLAG antibody surface was carried out with a 10-min injection at 30 μl/min of a mixture of one-third volume of TBS, pH 11.5, one-third volume of ionic solution, and one-third volume of water. The ionic solution was composed of KSCN (0.46 m), MgCl2 (1.83 m), urea (0.92 m), and guanidine HCl (1.83 m).
Generation of Jurkat-BCMA:Fas and TACI:Fas Reporter Cells and Killing Assay
Jurkat-BCMA:Fas-2309 cl13 reporter cells were generated as described (28). TACI:Fas Jurkat cell reporter cell lines were generated essentially as described previously for EDAR:Fas cells (29). 293T cells were transiently transfected with pMSCVpuro-TACI:Fas and co-transfected with the pHIT60 and VSV-G plasmids, containing the sequences for gag-pol and VSV-G, respectively. pMSCVpuro-TACI:Fas encodes the hemaglutinin signal peptide (amino acid sequence MAIIYLILLFTAVRG), part of the extracellular domain of human TACI (amino acids 2–118), amino acids VD, and the transmembrane and intracellular domains of human Fas (amino acids 169–335). After transfection, 293T cells were incubated for 48 h in RPMI supplemented with 10% FCS. 6 ml of virus-containing 293T cell supernatants supplemented with 8 μg/ml of Polybrene were added to 106 Fas-deficient Jurkat-JOM2 cells (a kind gift of Olivier Micheau, University of Dijon, France) in two additions of 3 ml each for 3 and 16 h, respectively, after which time cells were cultured in 10% RPMI for 72 h and then selected with 0.5 μg/ml puromycin and cloned. Clones were screened for their selective sensitivity to Fc-BAFF but not Fc-EDA1 (30), and one clone (clone 112) was selected for further experimentation. For the killing assay, 3 × 104 Jurkat cells were seeded/well in a 96-well plate and stimulated with doubling dilutions of quantity-matched supernatants for a period of 16 h. Cells were subsequently harvested, spun down, and resuspended in 250 μl of Nicoletti buffer containing 50 μg/ml propidium iodide and stored for at least 24 h at 4 °C (as described in Ref. 31). Analysis of apoptosis was assessed by flow cytometric measurement of propidium iodide-stained nuclei using a FACSCalibur system (BD Biosciences).
Internalization Assay
To determine receptor internalization, 5 × 105 cells were labeled with phycoerythrin (PE)-conjugated anti-human or anti-mouse TACI antibodies (clones 1A1-K21-M22 and 8F10, respectively, BD Biosciences) and incubated with conditioned medium containing matched amounts of APRIL variants for a period of 90 min at 37 °C (the optimal time point was determined in a time course experiment). Following incubation, cells were cooled on ice to halt endocytosis; treated for 1 min with either acid solution (0.154 m NaCl, pH 2, to strip off surface-exposed antibody) or PBS, 1% BSA as a control; and then analyzed by FACS for the presence of the remaining PE label. The efficacy of the acid stripping was tested and optimized previously (32). All APRIL receptor staining on lymphoma cells (mouse and human) were performed after incubation with FcR blocking reagent (Miltenyi Biotech, Leiden, The Netherlands). TACI internalization was also studied using confocal microscopy. Cells were stained with PE-conjugated anti-mouse TACI antibody, incubated with APRIL in conditioned medium (as described above for FACS analysis), resuspended in mounting medium (VectaShield, Brunschwig Chemie, Amsterdam, The Netherlands), and transferred onto a glass slide. Cells were kept on ice before microscopy.
B Cell Assay
B cells were purified from murine splenocytes using magnetic activated cell separation with CD45R/B220 magnetic activated cell separation beads (Miltenyi Biotec, Utrecht, The Netherlands). Purified B cells were then seeded in 96-well round-bottomed microtiter plates at a density of 2 × 105 cells/well and incubated with diluted conditioned media containing the APRIL variants. After 6 days of incubation, viability was assessed using propidium iodide exclusion, and supernatants were assayed for IgA by ELISA. Coated 96-well plates (2 μg/ml anti-mouse Ig; Southern Biotech) were blocked with PBS, 5% BSA and, following washes with PBS, 0.05% Tween 20, incubated at 37 °C for 1 h with the collected supernatants. Bound IgA was detected with HRP-labeled anti-mouse IgA (Southern Biotech) and ABTS (Sigma-Aldrich).
RESULTS
Computational Design of Receptor-selective APRIL Variants
The x-ray crystal structures of murine APRIL in complex with human TACI or human BCMA (21) were used as templates for designing receptor-selective variants of human APRIL. The ECD of murine APRIL shares 85% sequence identity with the human form. In addition, neither sequence contains an insertion or deletion relative to the other (supplemental Fig. 1A). Models of human APRIL in complex with TACI or BCMA were constructed by assuming an identical protein backbone conformation and by in silico mutating all non-conserved murine residues to its homologous human counterpart.
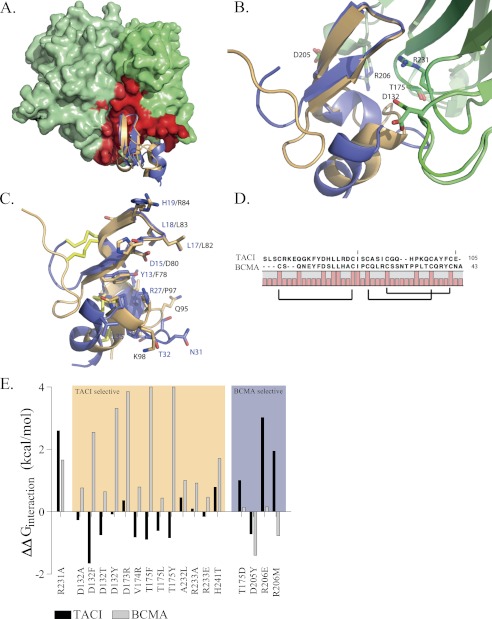
Like most other TNF family ligands, APRIL is expressed as a homotrimer that binds three receptor monomers. In contrast to many other TNF ligands, the main interaction surface of a single receptor binding interface is not located in the cleft between two adjacent APRIL monomers but instead resides mainly on the central surface of an APRIL monomer (Fig. 1A). The same can be observed for BAFF in complex with BAFF-R (33). Inspection of the interface between APRIL and BCMA or TACI reveals that the main chain conformation of APRIL hardly changes upon interaction with the two different receptors and that many side chains only show minor conformational changes. In contrast, the main chain conformation of TACI and BCMA show considerable deviation in the binding interface (Fig. 1B) and relatively few conserved interactions at the amino acid level (Fig. 1, C and D), which is a favorable starting condition for the computational protein design approach.
FIGURE 1.
Crystal Structures of APRIL in complex with BCMA and TACI and prediction of the APRIL-selective mutants. A, front view of APRIL (light and dark green) in complex with TACI (orange) or BCMA (blue). The APRIL-TACI and APRIL-BCMA complexes are superimposed. APRIL monomers are depicted using a molecular surface representation, and main chain coordinates of the receptors are depicted schematically. For clarity, only a single receptor unit is depicted, and two ligand monomers are shown. The TACI and BCMA receptor-binding interface of APRIL is mapped in red on the APRIL surface. In contrast to most other TNF family ligands, the receptor-binding interface resides only for a small part in the cleft between two adjacent ligand monomers because most of the receptors interactions are located on the central surface of a single APRIL monomer. B, detailed view of TACI and BCMA in complex with APRIL. Selected APRIL residues involved in an interaction with the receptors are depicted (APRIL structure in complex with BCMA and TACI is depicted in light green or dark green, respectively). C, detailed view of TACI (orange) and BCMA (blue) residues involved in APRIL binding. TACI and BCMA show a root mean square deviation of 1.45 Å upon superposition (calculated over 95 main chain atoms); the main chain coordinates show a larger displacement C-terminally of the β-sheet. The interacting residues of BCMA or TACI are relatively non-conserved. Labels of BCMA residues are colored blue, TACI is shown in black, and cysteine bridges are colored yellow. D, structure-based alignment of the ECD ligand-binding domain of human BCMA and human TACI. Brackets indicate cysteine bridge connectivity. Full bars, conserved residues. E, FoldX interaction energy. Interaction free energy between APRIL variants and BCMA or TACI is calculated as the difference with the interaction energy of wild type APRIL and expressed as ΔΔG in kcal/mol. The FoldX interaction energy is corrected for unfavorable intrachain Van der Waals clashes upon mutation (see “Experimental Procedures”). Variants are grouped as TACI-specific or BCMA-specific. R231A, a previously constructed APRIL variant unable to bind both receptors, was used as control. Structure images were generated using PyMOL (available on the World Wide Web) and based on Protein Data Bank entries 1xu1 and 1xu2 (21).
Due to the 3-fold symmetry of the APRIL-receptor complex, a “design unit” consisting of only two adjacent APRIL monomer subunits and a single receptor monomer subunit was used in the design process, similar to the one used in previous TRAIL design work (22, 23, 26). We used such a design unit for APRIL despite the fact that the receptor-binding interface of APRIL, in contrast to TRAIL, is largely confined to a single monomer subunit. However, the presence of a few direct contacts between the receptor and an adjacent APRIL monomer (i.e. APRIL residues 205, 206, and 208) in addition to long range electrostatic interactions made inclusion of the adjacent APRIL monomer necessary (Fig. 1A). Residues comprising the receptor interface of APRIL were identified, and each of these residues was subsequently mutated into all of the other 19 natural occurring amino acids, and contribution to the interaction energy was calculated by the FoldX protein design algorithm. Evaluation of the calculated interaction energy revealed several mutations that could confer APRIL receptor selectivity toward BCMA or TACI. Subsequently, several combinations of single BCMA or TACI specificity-conferring mutants were combined in single APRIL variants to evaluate the effect on receptor binding by FoldX. The best performing single mutants and combination mutants were selected for experimental characterization (Fig. 1E). For the purpose of clarity, APRIL mutants will be referred to by only the amino acid substitution (e.g. R231A represents APRIL-R231A).
Generation of APRIL Variants
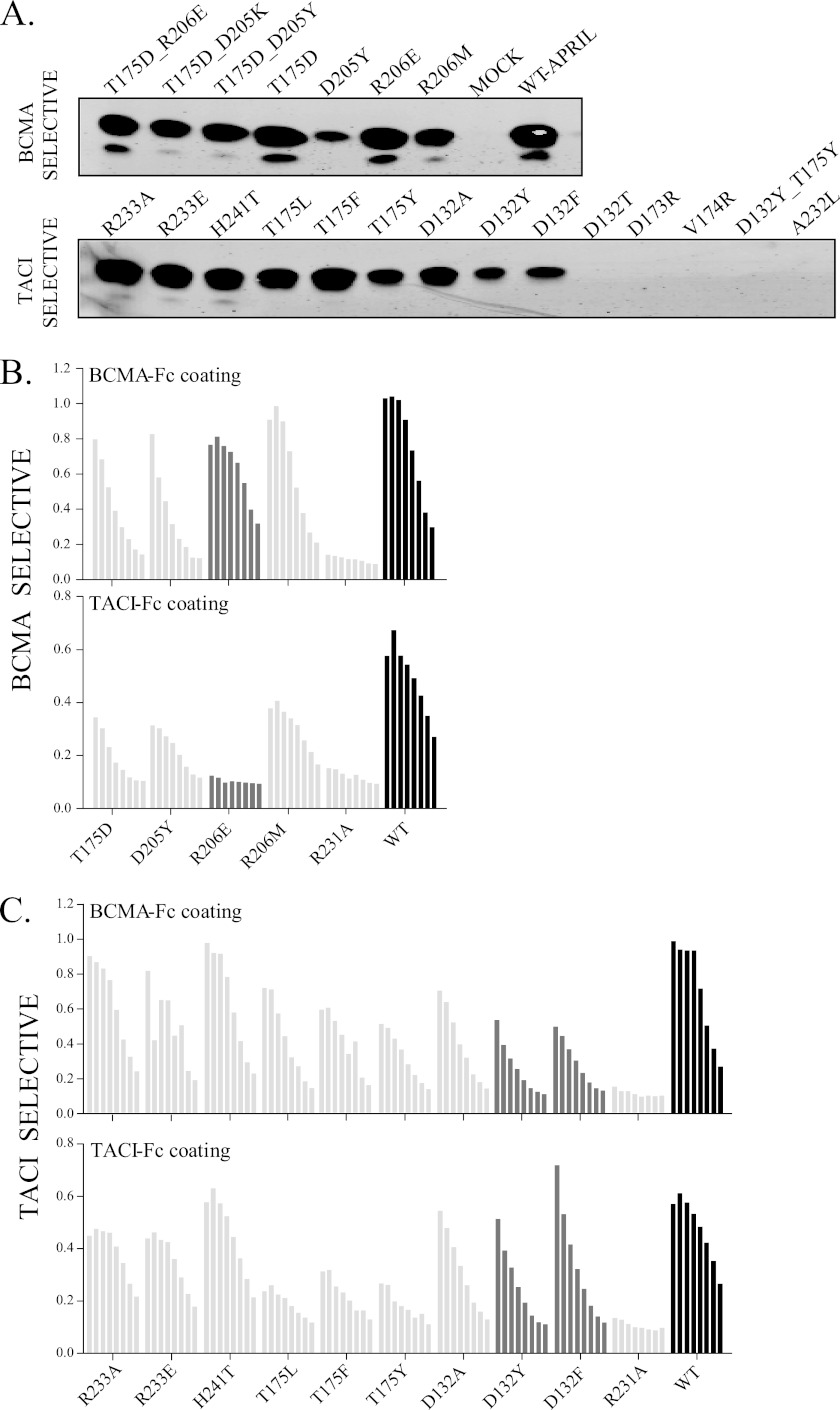
Because human WT APRIL does not express well as a soluble recombinant protein in Escherichia coli and is difficult to purify with high yield from mammalian cell cultures, it was decided to test FLAG-tagged APRIL variants directly from conditioned culture medium of transfected HEK-293T cells. This is a validated approach that has been used previously (14, 32). Protein expression was quantified by Western blot using an anti-FLAG tag antibody (Fig. 2A). Several variants (R233A, R233E, H241T, T175L, T175F, T175D, R206E, and R206M) expressed well, others (D132A, D205Y, D132Y, T175Y, and D132F) displayed reduced expression levels, and some (D132T, D173R, V174R, and A232L) were not secreted at all. Some of these non-secreted mutants were detected in cell lysates, suggesting folding and/or secretion problems, whereas others were not expressed at all, possibly as a result of mRNA instability or another problem (supplemental Fig. 2). Because our goal was to generate a usable soluble APRIL variant, potential causes were not further investigated, and all non-expressers were omitted from any further studies. In addition, some selectivity-conferring mutations were combined into double mutant variants; however, these mutants either failed to express (D132Y/T175Y) or did not show any binding toward both BCMA and TACI (T175D/D205Y/K and T175D/R206E) (data not shown).
FIGURE 2.
Production and receptor binding properties of the APRIL mutants. A, protein expression of APRIL variants in conditioned medium by transient transfection of 293T cells. Supernatants (10 μl of each) were analyzed by anti-FLAG immunoblotting. Top, mutants predicted to be selective for BCMA. Bottom, mutants predicted to be selective for TACI. All mutants were checked more than three times following independent rounds of transfection to assess their expression. B and C, receptor-binding ELISA to compare binding of the predicted BCMA- and TACI-specific APRIL variants to human BCMA-Fc and TACI-Fc. Bars (from left to right), doubling dilutions of the conditioned media starting from undiluted media. Relevant APRIL variants are shown with dark gray bars, WT APRIL with black bars, and other variants with light gray bars. This is representative of three separate experiments performed with independent APRIL-containing cell conditioned media. R231A APRIL variant does not bind any of the APRIL receptors (negative binding control).
Determination of Receptor Binding by ELISA
APRIL variants were tested using a receptor-binding ELISA as an initial screening assay to examine the real in vitro TACI and BCMA receptor binding and to determine receptor selectivity. Because we were interested in the relative changes in affinity of the APRIL variants for the TACI and BCMA receptors, we did not correct at this point for the different expression levels of the selected mutants. Thus, in the following experiments, although binding to the target receptor could seem weaker than for the WT variant, this is not necessarily the case (see below). Variants were grouped according to their predicted binding properties (i.e. being either TACI- or BCMA-specific; Fig. 1E). R231A, previously shown by us to be a mutation that leads to loss of both TACI and BCMA binding, but not binding to heparan sulfate proteoglycans, was included as a negative control (14, 32). Binding to human BCMA-Fc was retained by all variants predicted to selectively bind BCMA (Fig. 2B). Although T175D was well expressed, it showed relatively lower binding to BCMA when compared with WT APRIL; D205Y also showed decreased binding to BCMA. However, R206M and R206E retained a binding profile toward BCMA comparable with WT APRIL.
In contrast, when tested for binding toward human TACI-Fc, all of these single variants showed significantly reduced binding compared with WT APRIL. Therefore, all variants predicted to selectively bind BCMA indeed showed enhanced selectivity toward BCMA. However, binding of R206E to TACI-Fc was completely lost, indicating not only enhanced BCMA selectivity but complete specificity. All APRIL mutants designed for being TACI-selective retained binding to both BCMA and TACI yet showed a preferential binding for TACI at the expense of BCMA (Fig. 2C). Mutants with the best TACI to BCMA binding ratio were D132Y and D132F.
Although both the TACI and BCMA variants were not specifically designed to bind murine receptors, the ligands were also tested for their binding to the homologous mouse receptors, which share ∼70% sequence identity with their human counterparts (supplemental Fig. 1B). Although one variant, H241T, showed an improved selectivity toward the mouse TACI, R206E, D132F, and D132Y showed similar binding toward mouse BCMA-Fc and mouse TACI-Fc as that observed with human receptors (mBCMA-Fc and mTACI-Fc, supplemental Fig. 3).
The binding of APRIL to TACI and BCMA was quantified by surface plasmon resonance (supplemental Fig. 4 and Tables 1 and 2). WT APRIL bound TACI- and BCMA-Fc, both human and mouse, with affinities comparable with previously published values (Tables 1 and 2) (9, 11, 12, 34). The BCMA-specific mutant R206E bound both human and mouse BCMA with affinities comparable with WT APRIL, whereas its binding to TACI was obviously reduced. Comparison of the affinities of R206E for BCMA and TACI shows that this variant is 25-fold more selective for BCMA than for TACI. Unfortunately, the TACI-selective variants D132Y and D132F could not be produced in sufficient quantities to obtain reliable surface plasmon resonance readings. Taken together, these initial screening assays highlight one variant, R206E, with specificity for BCMA and two variants, D132F and D132Y, with selectivity toward TACI. All of these mutants bind similarly to human and mouse receptors.
TABLE 1.
Affinities for human and mouse BCMA as measured by BIAcore
Values represent the average of a global fit using three curves from separate experiments. Ru, resonance units.
| Protein | ka | kd | KD |
|---|---|---|---|
| m−1 s−1 | s−1 | m | |
| Affinities for human BCMA-Fc | |||
| WT | 5.3 ± 0.1 × 105 | 2.0 ± 0.1 × 10−4 | 3.8 ± 0.1 × 10−10 |
| R206E | 3.3 ± 0.0 × 105 | 1.5 ± 0.0 × 10−4 | 4.6 ± 0.0 × 10−10 |
| Affinities for mouse BCMA-Fc | |||
| WT | 8.9 ± 0.0 × 105 | 1.3 ± 0.0 × 10−4 | 1.4 ± 0.1 × 10−10 |
| R206E | 16 ± 0.0 × 105 | 1.5 ± 0.0 × 10−4 | 0.95 ± 0.3 × 10−10 |
TABLE 2.
Affinities for human and mouse TACI as measured by BIAcore
| Protein | ka | kd | KD |
|---|---|---|---|
| m−1 s−1 | s−1 | m | |
| Affinities for human TACI-Fc | |||
| WT | 1.1 ± 0.1 × 105 | 4.1 ± 0.1 × 10−4 | 39 ± 3 × 10−10 |
| R206E | 20 ± 29 × 105 | 100 ± 90 × 10−4 | 110 ± 120 × 10−10 |
| R206M | 4.9 ± 0.0 × 105 | 1.6 ± 0.0 × 10−4 | 3.2 ± 0.0 × 10−10 |
| Affinities for mouse TACI-Fc | |||
| WT | 1.7 ± 0.1 × 105 | 7.5 ± 0.1 × 10−4 | 44 ± 1 × 10−10 |
| R206E | RU too low, no fit | ||
R206E Shows Specificity for BCMA, whereas D132F and D132Y Show Selectivity toward TACI
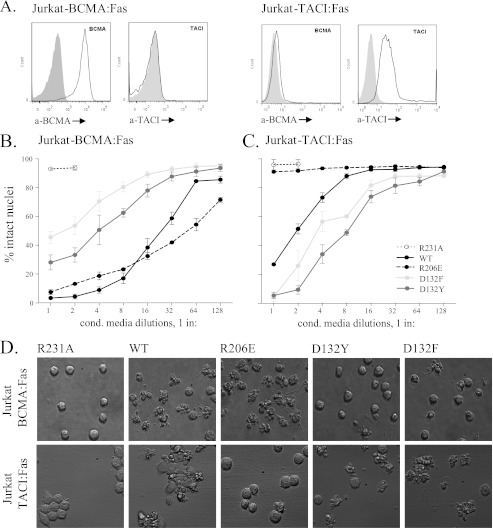
In order to study receptor selectivity of APRIL variants in a standardized cell-based assay, BCMA:Fas- and TACI:Fas-expressing Jurkat cells were used as a reporter system (28). In this assay, binding of APRIL to chimeric receptors triggers the proapoptotic Fas signaling pathway, leading to cell death. Reporter cells indeed expressed their respective chimeric receptors on the surface, as shown by FACS staining (Fig. 3A). Both WT APRIL and R206E efficiently killed BCMA:Fas-expressing cells (Fig. 3B), but only WT APRIL, and not R206E, killed TACI:Fas reporter cells (Fig. 3C). Conversely, D132F and D132Y showed reduced activity on BCMA:Fas Jurkat cells (Fig. 3B) but enhanced activity on TACI:Fas Jurkat cells when compared with WT APRIL (Fig. 3C). Cell death was also evident at the morphological level, with numerous apoptotic blebs forming as early as 1 h post-treatment initiation (Fig. 3D). Thus, the receptor specificity of R206E and selectivity of D132F and D132Y were confirmed in a cell-based assay.
FIGURE 3.
R206E shows specificity for BCMA, whereas D132Y and D132F show selectivity for TACI. Binding activity of the ligands was tested on TACI:Fas- and BCMA:Fas-expressing reporter cells. The ligand binding is directly associated with induced cell death. A, staining for human BCMA and TACI on Jurkat BCMA:Fas (left) and Jurkat JOM2 TACI:Fas cells (right). B and C, measurement of cell death produced after a 16-h treatment with doubling dilutions of the APRIL variants on BCMA:Fas (B) and TACI:Fas (C) reporter cells. D, microscopic pictures (×40) of Jurkat-BCMA-Fas (top) and Jurkat-TACI-Fas (bottom) cells after a 1-h stimulation with the indicated APRIL variants. Conditioned media were matched for APRIL amounts before incubation.
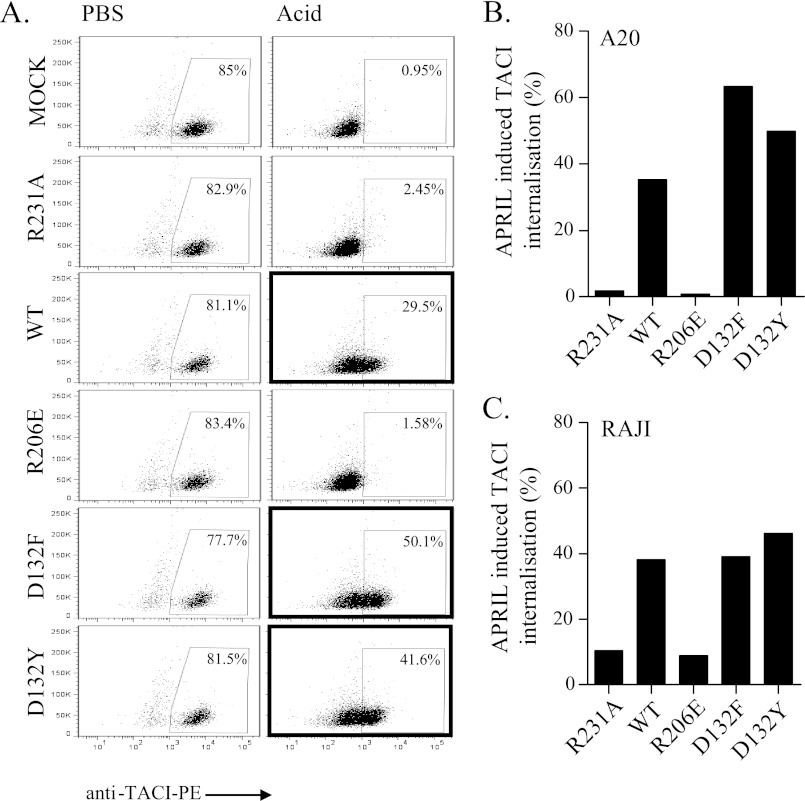
D132F and D132Y, but Not R206E, Triggered TACI Internalization on Endogenously Expressed Receptors
In order to test whether the R206E variant would also be unable to stimulate endogenous WT TACI, we used a receptor internalization assay (32). We chose the mouse A20 cell line that has been shown to express high amounts of TACI (10, 32). Treatment with WT APRIL for 90 min at 37 °C triggered TACI internalization, as shown by the high PE signal retained after acid treatment, which marks the antibody that was internalized, together with the receptor (Fig. 4A, panel 3, marked box). Visualization of stimulated cells by confocal microscopy confirmed in a more direct way that TACI was internalized (supplemental Fig. 5). The two TACI-selective ligands, D132F and D132Y, efficiently triggered TACI internalization at levels even higher than those achieved with WT APRIL (Fig. 4B, panels 5 and 6, marked boxes), in accordance with the ELISA binding and Jurkat killing assays. In contrast, R206E failed to trigger TACI internalization and was comparable with the “receptor-dead” R231A variant or with the mock control, where acid treatment completely quenched the extracellular PE signal due to lack of TACI internalization (Fig. 4, A and B). Similar results were obtained on the human lymphoma cell line Raji, for which we recently showed expression of both TACI and BCMA (32) (Fig. 4C). These results point to the inability of R206E to stimulate endogenous TACI.
FIGURE 4.
D132F and D132Y, but not R206E, triggered TACI internalization on endogenously expressed receptors. Cells were stained with a PE-coupled anti-TACI antibody and incubated with the indicated ligands for 1 h at 37 °C to allow receptor internalization. Subsequently, cells were placed on ice to halt membrane movements and then treated with either PBS (control) or acid solution (pH 2) to strip off labeled receptors that were not internalized. A, example of FACS profile of the TACI internalization for A20 cells. The high PE signal that remained after acid treatment (marked boxes) reflects TACI being internalized and protected inside cells. B, quantification of A expressed as percentage of APRIL-induced TACI internalization. C, quantification of TACI internalization for human Raji cells.
Distinct Effects of APRIL Variants on B Splenocyte Survival and IgA Production
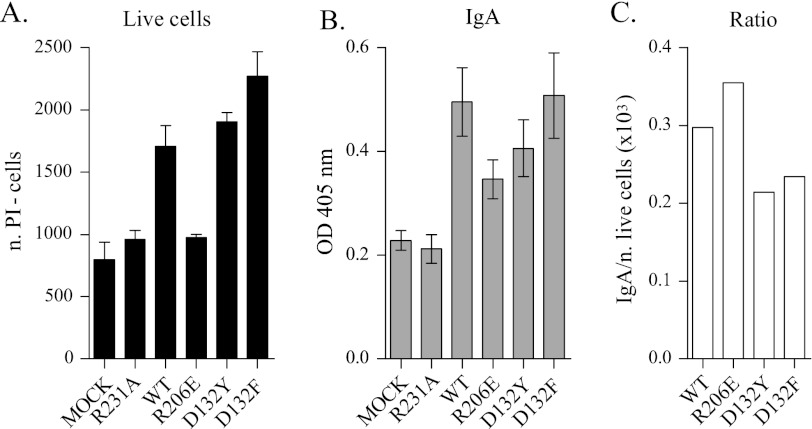
APRIL variants were tested for their activity on freshly isolated mouse B220+ splenocytes, which are known to respond to APRIL by increasing survival and IgA production (14, 32). WT APRIL increased the number of live murine B cells remaining after 6 days of culture by a factor of 2 (Fig. 5A) and also doubled the levels of IgA compared with the control (Fig. 5B). TACI-selective APRIL variants were slightly more potent than WT at increasing cell survival (Fig. 5A), although IgA production was not found to be proportionally increased (Fig. 5, B and C). In contrast, the BCMA-specific variant R206E failed to increase live cell numbers yet partially increased IgA levels in cell supernatants (Fig. 5, A and B). This suggests that APRIL variants might prove useful at dissecting TACI- or BCMA-dependent B cell responses.
FIGURE 5.
Differential effects of APRIL variants on B splenocytes survival and IgA production. Primary mouse splenocytes were positively selected for B220 and stimulated for 6 days with the indicated ligands in conditioned medium diluted 1:1 in normal medium. After 6 days, propidium iodide-negative cells (live cells) were counted (A), and supernatants were screened for soluble IgA levels (B). C, graphs representing the ratio between IgA and number of live B cells stimulated. Due to the different concentrations of ligands produced in conditioned medium, the concentrations of all of the variants were adjusted to that of the lowest expresser, D132F. Error bars, S.E. among triplicates.
DISCUSSION
APRIL is a well studied protein as a consequence of its role in several pathological settings, such as tumor growth and autoimmunity (reviewed in Ref. 5). The high expression of APRIL in many B-cell malignancies prompted a therapeutic study in our laboratory aimed at blocking APRIL signaling for therapeutic purposes (32). APRIL binds two receptors (BCMA and TACI), but the knowledge of which receptor mediates APRIL signaling in different contexts is limited. In order to address this, we set out to develop receptor-selective variants.
Receptor-selective TNF family ligands have been previously generated by employing rational design- or directed evolution-based methods. Rational design approaches have been previously used to design receptor-selective TNF-α variants (35) and TRAIL variants (22, 23, 26, 36). Directed evolution methods employing phage display were also used to generate receptor-selective TRAIL variants (37), a receptor-selective TNFα antagonist (38), and a LIGHT/LTβ variant that does not bind DcR3 (39). In addition, selective receptor activation can also be achieved by the use of receptor-specific agonistic antibodies (40, 41) or other binding scaffolds (42). Although these approaches can be equally useful, factors such as stoichiometry of receptor activation (40) and prolonged activation due to a longer half-life must be taken into account.
We decided to use a rational design strategy employing a protein design algorithm to engineer receptor-selective APRIL ligands that are structurally similar to the WT ligand. Our previous work on TRAIL variants showed that this approach was feasible, although we had to construct a model of human APRIL in complex with human TACI and BCMA (22, 26).
Using the FoldX protein design algorithm, we generated a panel of amino acid substitutions that were predicted to direct binding of APRIL toward one or the other of its receptors. Although the majority of the variants exhibited good expression, some showed a reduced expression (D205Y, T175Y, D132Y, D132F, and D132A), and others were not expressed (D132T, D173R, V174R, and A232L). Analysis of these non-expressing variants suggests that they had folding or secretion problems, indicating that some residues in APRIL could play a role in those processes aside from being involved in receptor binding. In the case of the BCMA-selective variants, the four designed variants showed preferred binding to BCMA compared with APRIL, and one, R206E, showed clear specificity toward both human and mouse BCMA. R206E retained an affinity toward BCMA comparable with that of WT APRIL and also demonstrated comparable activity in cellular assays. R206E showed no binding to mouse TACI-Fc and much decreased binding to human TACI-Fc, as assessed by surface plasmon resonance, as well as an inability to bind and activate TACI at the cell surface. The design of variants specific toward TACI was less successful because only two variants (D132F and D132Y) were found to have significantly reduced binding to and activation of BCMA while retaining an acceptable level of TACI binding.
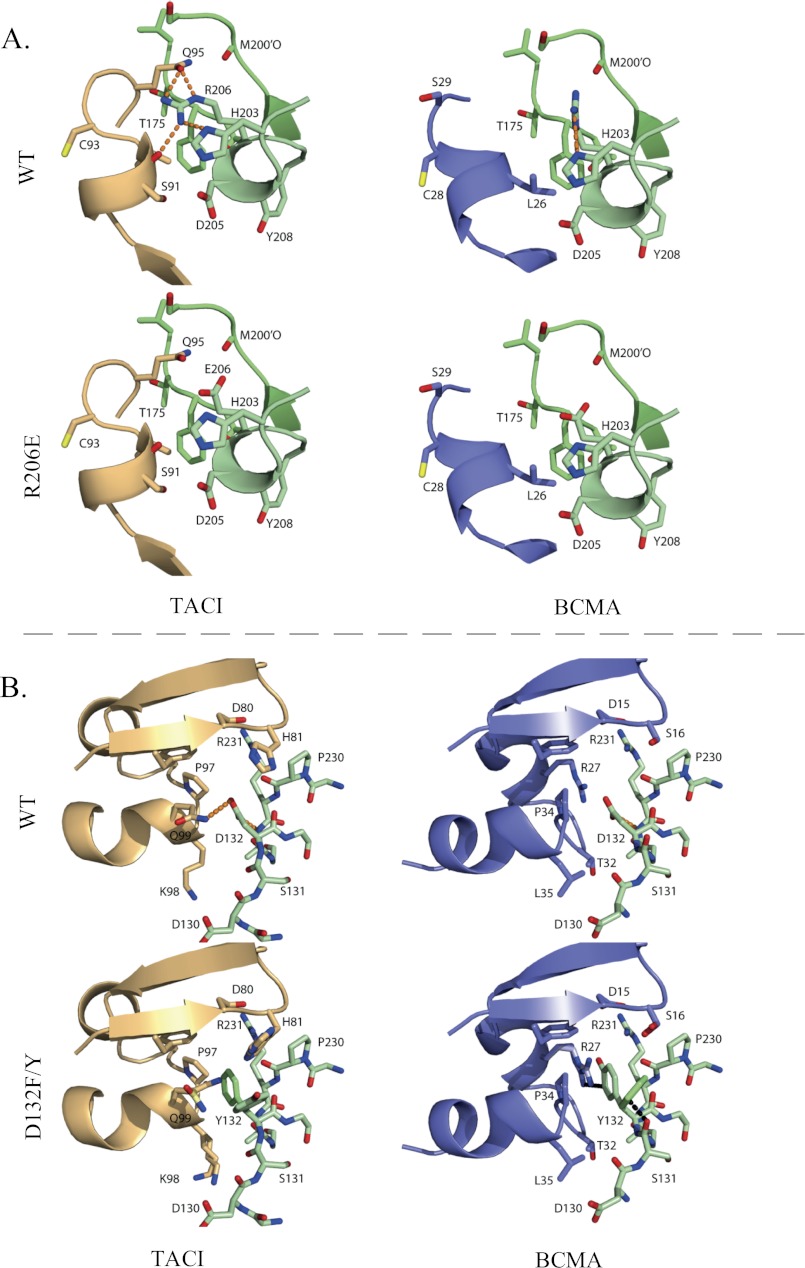
Inspection of the humanized APRIL-BCMA and APRIL-TACI structures does not explain the failure at positions 233, 241, and 175. Explanations could be that the human APRIL may have some backbone changes compared with the murine structure used as a template to model the human sequence, or there could be conformational changes in APRIL upon mutation that eliminate the discriminatory power of the mutations. Evaluation of the FoldX energy terms and structural analysis of R206E indicate that the high specificity for BCMA is mainly achieved by reducing the binding affinity toward the TACI receptor; the binding affinity toward BCMA remains largely unchanged. This is due to the removal of hydrogen bonds with the side chain of Gln-95Taci and the backbone oxygen of Ser-91Taci in the APRIL-TACI complex upon mutation (Fig. 6A). In contrast, the backbone atoms of the corresponding stretch of amino acids in BCMA are further away from Arg-206, such that formation of a side chain/backbone hydrogen bond is not possible.
FIGURE 6.
A, structural consequences of the R206E substitution. In the WT APRIL-TACI crystal structure, Arg-206 makes hydrogen bonds with TACI, whereas in the WT APRIL-BCMA crystal structure, Arg-206 is not involved in the interaction with BCMA. The Glu-206 substitution in TACI and BCMA is not involved in hydrogen bond interactions. B, structural consequences of the D132F and D132Y substitution. In the WT APRIL-BCMA structure, Asp-132 (D132) is involved in a favorable electrostatic interaction with Arg-27, whereas in the WT APRIL-TACI complex, Asp-132 accepts a (weak) hydrogen bond from Gln-99 (Q99) of TACI. The loss of this hydrogen bond due to the Phe-132 and Tyr-132 substitution is compensated in TACI by favorable Van der Waals interactions, whereas in BCMA, the Phe or Tyr either clashes with Arg-27 (R27) or with the main chain oxygen of Ser-131 (S131) of APRIL. TACI is depicted in orange, BCMA in blue, and APRIL in green. The D132F and D132Y structures are superimposed; residues that differ are indicated in lighter shades of blue, green, and orange. Hydrogen bonds are shown as an orange dotted line, and Van der Waals clashes are shown as a black dotted line.
For D132F and D132Y, structural analysis reveals that in the APRIL-BCMA complex, Asp-132 of APRIL is involved in an electrostatic interaction with Arg-27 of BCMA, whereas in the APRIL-TACI complex, Asp-132 is not involved in any specific interactions with the receptor, except for a (weak) hydrogen bond with Gln-99Taci (Fig. 6B). Substituting Asp-132 with Phe or Tyr does not have unfavorable consequences for the interaction with TACI; loss of the hydrogen bond with Gln-99Taci is compensated by a more favorable Van der Waals interaction energy and (de)solvation energy. In contrast, upon substitution of Asp-132 to Phe or Tyr, the favorable electrostatic interaction with Arg-27Bcma is destroyed, and both Phe and Tyr cannot find an energetically favorable conformation. Thus, the loss of an electrostatic interaction and Van der Waals clashes with either receptor residues or intrachain residues upon binding to BCMA, in combination with favorable interactions upon binding to TACI, largely explains the TACI-selective behavior of D132F and D132Y.
The three described APRIL variants were effective at activating both overexpressed and endogenously occurring APRIL receptors. In line with previous studies based on receptor knock-out experiments (28), we observed that only the TACI-selective APRIL ligands, and not the BCMA-specific R206E (Fig. 5A), were able to increase proliferation and/or survival of murine B cells. After 6 days, both D132F and D132Y produced an even higher number of live cells than WT APRIL. This also fits our results, where the two TACI variants were shown to be more active than WT APRIL on the Jurkat TACI:Fas killing (Fig. 3C) and on endogenous TACI internalization (Fig. 4, B and C).
IgA screening of supernatants revealed that different mechanisms could be involved in IgA production (Fig. 5B). BCMA-specific stimulation produced a lower but evident IgA titer without increasing the number of live cells. Of note, stimulation of B cells with undiluted R206E (calculated to be 30 times the concentration used in Fig. 5, A–B) still did not present a difference in live cell numbers compared with the negative controls (supplemental Fig. 6) yet stimulated small amounts of IgA. This might be due to the survival of a small population of preexisting IgA-producing cells (28), perhaps long lived plasma cells shown to depend on BCMA (4). On the other hand, the TACI-selective variants produced higher IgA levels than R206E, despite their lower affinity for BCMA. This might be explained by previous findings where TACI and BAFF-R, but not BCMA, were shown to be involved in class switch recombination, suggesting the formation of newly formed IgA-producing cells (2, 43).
Combined, our data indicate that TACI and BCMA serve different roles during B cell stimulation. Further work should focus on the molecular signals that define this difference. R206E should prove useful in future research to decipher the signal transduction elicited by APRIL through BCMA, and D132F/Y in signal transduction mediated via TACI. We feel that such a “branch-pruning” approach for examining receptor contribution could be more informative than using cells from receptor knock-out mice (or in RNAi-mediated knockdown studies in vitro) because it selectively removes only a single interaction (23). This could be important in terms of downstream signals, which may rely on the presence of one or the other receptor at various stages of development. BCMA and TACI are expressed in a temporal fashion during B cell development (44), potentially leading to different populations in the different KO mice. In addition, the concept of TRAF (the internal regulators) sharing has been reported for the TNF family, which may also be a limitation of the KO approach (45, 46).
In summary, our current data show that APRIL variants have been generated that can selectively activate either TACI or BCMA. These variants open up the possibility for future in vitro and in vivo studies that will provide a better understanding of this intricate subfamily of TNF receptors and ligands and also help to better understand the role of APRIL, both in physiological processes and in various disease settings.
Acknowledgments
We thank Olivier Micheau (University of Dijon, France) for the gift of Fas-deficient Jurkat JOM2 cells, Michael Hahne (Institut de Génetique Moléculaire de Montpellier, France) for the gift of the BCMA-Fc- and TACI-Fc-producing cell lines and Laure Willen (Department of Biochemistry, University of Lausanne) for excellent technical assistance.
This work was supported by Dutch Cancer Society Grant UvA 2007-3750, the Swiss National Science Foundation (to P. S.), and the European Union (PROSPECTS (Grant Agreement HEALTH-F4-2008-201648) and TRIDENT (Grant Agreement FP6-LIFESCIHEALTH-37686); both to L. S.).

This article contains supplemental Figs. 1–6.
- ECD
- extracellular domain
- ABTS
- 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid
- PE
- phycoerythrin.
REFERENCES
- 1. Hahne M., Kataoka T., Schröter M., Hofmann K., Irmler M., Bodmer J. L., Schneider P., Bornand T., Holler N., French L. E., Sordat B., Rimoldi D., Tschopp J. (1998) APRIL, a new ligand of the tumor necrosis factor family, stimulates tumor cell growth. J. Exp. Med. 188, 1185–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Castigli E., Wilson S. A., Scott S., Dedeoglu F., Xu S., Lam K. P., Bram R. J., Jabara H., Geha R. S. (2005) TACI and BAFF-R mediate isotype switching in B cells. J. Exp. Med. 201, 35–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Castigli E., Scott S., Dedeoglu F., Bryce P., Jabara H., Bhan A. K., Mizoguchi E., Geha R. S. (2004) Impaired IgA class switching in APRIL-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 101, 3903–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. O'Connor B. P., Raman V. S., Erickson L. D., Cook W. J., Weaver L. K., Ahonen C., Lin L. L., Mantchev G. T., Bram R. J., Noelle R. J. (2004) BCMA is essential for the survival of long lived bone marrow plasma cells. J. Exp. Med. 199, 91–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kimberley F. C., Medema J. P., Hahne M. (2009) APRIL in B-cell malignancies and autoimmunity. Results Probl. Cell Differ. 49, 161–182 [DOI] [PubMed] [Google Scholar]
- 6. Deshayes F., Laprée G., Portier A., Richard Y., Pencalet P., Mahieu-Caputo D., Horellou P., Tsapis A. (2004) Abnormal production of the TNF homologue APRIL increases the proliferation of human malignant glioblastoma cell lines via a specific receptor. Oncogene 23, 3005–3012 [DOI] [PubMed] [Google Scholar]
- 7. Mhawech-Fauceglia P., Kaya G., Sauter G., McKee T., Donze O., Schwaller J., Huard B. (2006) The source of APRIL up-regulation in human solid tumor lesions. J. Leukoc. Biol. 80, 697–704 [DOI] [PubMed] [Google Scholar]
- 8. Lascano V., Zabalegui L. F., Cameron K., Guadagnoli M., Jansen M., Burggraaf M., Versloot M., Rodermond H., van der Loos C., Carvalho-Pinto C. E., Kalthoff H., Medema J. P., Hahne M. (2012) The TNF family member APRIL promotes colorectal tumorigenesis. Cell Death. Differ., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marsters S. A., Yan M., Pitti R. M., Haas P. E., Dixit V. M., Ashkenazi A. (2000) Interaction of the TNF homologues BLyS and APRIL with the TNF receptor homologues BCMA and TACI. Curr. Biol. 10, 785–788 [DOI] [PubMed] [Google Scholar]
- 10. Rennert P., Schneider P., Cachero T. G., Thompson J., Trabach L., Hertig S., Holler N., Qian F., Mullen C., Strauch K., Browning J. L., Ambrose C., Tschopp J. (2000) A soluble form of B cell maturation antigen, a receptor for the tumor necrosis factor family member APRIL, inhibits tumor cell growth. J. Exp. Med. 192, 1677–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu Y., Bressette D., Carrell J. A., Kaufman T., Feng P., Taylor K., Gan Y., Cho Y. H., Garcia A. D., Gollatz E., Dimke D., LaFleur D., Migone T. S., Nardelli B., Wei P., Ruben S. M., Ullrich S. J., Olsen H. S., Kanakaraj P., Moore P. A., Baker K. P. (2000) Tumor necrosis factor (TNF) receptor superfamily member TACI is a high affinity receptor for TNF family members APRIL and BLyS. J. Biol. Chem. 275, 35478–35485 [DOI] [PubMed] [Google Scholar]
- 12. Yu G., Boone T., Delaney J., Hawkins N., Kelley M., Ramakrishnan M., McCabe S., Qiu W. R., Kornuc M., Xia X. Z., Guo J., Stolina M., Boyle W. J., Sarosi I., Hsu H., Senaldi G., Theill L. E. (2000) APRIL and TALL-I and receptors BCMA and TACI. System for regulating humoral immunity. Nat. Immunol. 1, 252–256 [DOI] [PubMed] [Google Scholar]
- 13. Ingold K., Zumsteg A., Tardivel A., Huard B., Steiner Q. G., Cachero T. G., Qiang F., Gorelik L., Kalled S. L., Acha-Orbea H., Rennert P. D., Tschopp J., Schneider P. (2005) Identification of proteoglycans as the APRIL-specific binding partners. J. Exp. Med. 201, 1375–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kimberley F. C., van Bostelen L., Cameron K., Hardenberg G., Marquart J. A., Hahne M., Medema J. P. (2009) The proteoglycan (heparan sulfate proteoglycan) binding domain of APRIL serves as a platform for ligand multimerization and cross-linking. FASEB J. 23, 1584–1595 [DOI] [PubMed] [Google Scholar]
- 15. Bischof D., Elsawa S. F., Mantchev G., Yoon J., Michels G. E., Nilson A., Sutor S. L., Platt J. L., Ansell S. M., von Bulow G., Bram R. J. (2006) Selective activation of TACI by syndecan-2. Blood 107, 3235–3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hatzoglou A., Roussel J., Bourgeade M. F., Rogier E., Madry C., Inoue J., Devergne O., Tsapis A. (2000) TNF receptor family member BCMA (B cell maturation) associates with TNF receptor-associated factor (TRAF) 1, TRAF2, and TRAF3 and activates NF-κB, Elk-1, c-Jun N-terminal kinase, and p38 mitogen-activated protein kinase. J. Immunol. 165, 1322–1330 [DOI] [PubMed] [Google Scholar]
- 17. Xia X. Z., Treanor J., Senaldi G., Khare S. D., Boone T., Kelley M., Theill L. E., Colombero A., Solovyev I., Lee F., McCabe S., Elliott R., Miner K., Hawkins N., Guo J., Stolina M., Yu G., Wang J., Delaney J., Meng S. Y., Boyle W. J., Hsu H. (2000) TACI is a TRAF-interacting receptor for TALL-1, a tumor necrosis factor family member involved in B cell regulation. J. Exp. Med. 192, 137–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kanno Y., Sakurai D., Hase H., Kojima H., Kobata T. (2010) TACI induces cIAP1-mediated ubiquitination of NIK by TRAF2 and TANK to limit non-canonical NF-κB signaling. J. Recept. Signal. Transduct. Res. 30, 121–132 [DOI] [PubMed] [Google Scholar]
- 19. Guerois R., Nielsen J. E., Serrano L. (2002) Predicting changes in the stability of proteins and protein complexes. A study of more than 1000 mutations. J. Mol. Biol. 320, 369–387 [DOI] [PubMed] [Google Scholar]
- 20. Schymkowitz J. W., Rousseau F., Martins I. C., Ferkinghoff-Borg J., Stricher F., Serrano L. (2005) Prediction of water and metal binding sites and their affinities by using the Fold-X force field. Proc. Natl. Acad. Sci. U.S.A. 102, 10147–10152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hymowitz S. G., Patel D. R., Wallweber H. J., Runyon S., Yan M., Yin J., Shriver S. K., Gordon N. C., Pan B., Skelton N. J., Kelley R. F., Starovasnik M. A. (2005) Structures of APRIL-receptor complexes. Like BCMA, TACI employs only a single cysteine-rich domain for high affinity ligand binding. J. Biol. Chem. 280, 7218–7227 [DOI] [PubMed] [Google Scholar]
- 22. Reis C. R., van der Sloot A. M., Natoni A., Szegezdi E., Setroikromo R., Meijer M., Sjollema K., Stricher F., Cool R. H., Samali A., Serrano L., Quax W. J. (2010) Rapid and efficient cancer cell killing mediated by high affinity death receptor homotrimerizing TRAIL variants. Cell Death Dis. 1, e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van der Sloot A. M., Tur V., Szegezdi E., Mullally M. M., Cool R. H., Samali A., Serrano L., Quax W. J. (2006) Designed tumor necrosis factor-related apoptosis-inducing ligand variants initiating apoptosis exclusively via the DR5 receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 8634–8639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. van der Sloot A. M., Quax W. J. (2011) Computational design of TNF ligand-based protein therapeutics. Adv. Exp. Med. Biol. 691, 521–534 [DOI] [PubMed] [Google Scholar]
- 25. Van der Sloot A. M., Kiel C., Serrano L., Stricher F. (2009) Protein design in biological networks. From manipulating the input to modifying the output. Protein Eng. Des. Sel. 22, 537–542 [DOI] [PubMed] [Google Scholar]
- 26. Tur V., van der Sloot A. M., Reis C. R., Szegezdi E., Cool R. H., Samali A., Serrano L., Quax W. J. (2008) DR4-selective tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) variants obtained by structure-based design. J. Biol. Chem. 283, 20560–20568 [DOI] [PubMed] [Google Scholar]
- 27. Schymkowitz J., Borg J., Stricher F., Nys R., Rousseau F., Serrano L. (2005) The FoldX web server. An online force field. Nucleic Acids Res. 33, W382–W388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bossen C., Cachero T. G., Tardivel A., Ingold K., Willen L., Dobles M., Scott M. L., Maquelin A., Belnoue E., Siegrist C. A., Chevrier S., Acha-Orbea H., Leung H., Mackay F., Tschopp J., Schneider P. (2008) TACI, unlike BAFF-R, is solely activated by oligomeric BAFF and APRIL to support survival of activated B cells and plasmablasts. Blood 111, 1004–1012 [DOI] [PubMed] [Google Scholar]
- 29. Swee L. K., Ingold-Salamin K., Tardivel A., Willen L., Gaide O., Favre M., Demotz S., Mikkola M., Schneider P. (2009) Biological activity of ectodysplasin A is conditioned by its collagen and heparan sulfate proteoglycan-binding domains. J. Biol. Chem. 284, 27567–27576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bossen C., Ingold K., Tardivel A., Bodmer J. L., Gaide O., Hertig S., Ambrose C., Tschopp J., Schneider P. (2006) Interactions of tumor necrosis factor (TNF) and TNF receptor family members in the mouse and human. J. Biol. Chem. 281, 13964–13971 [DOI] [PubMed] [Google Scholar]
- 31. Mullauer F. B., Kessler J. H., Medema J. P. (2009) Betulinic acid induces cytochrome c release and apoptosis in a Bax/Bak-independent, permeability transition pore-dependent fashion. Apoptosis 14, 191–202 [DOI] [PubMed] [Google Scholar]
- 32. Guadagnoli M., Kimberley F. C., Phan U., Cameron K., Vink P. M., Rodermond H., Eldering E., Kater A. P., van Eenennaam H., Medema J. P. (2011) Development and characterization of APRIL antagonistic monoclonal antibodies for treatment of B-cell lymphomas. Blood 117, 6856–6865 [DOI] [PubMed] [Google Scholar]
- 33. Kim H. M., Yu K. S., Lee M. E., Shin D. R., Kim Y. S., Paik S. G., Yoo O. J., Lee H., Lee J. O. (2003) Crystal structure of the BAFF-BAFF-R complex and its implications for receptor activation. Nat. Struct. Biol. 10, 342–348 [DOI] [PubMed] [Google Scholar]
- 34. Day E. S., Cachero T. G., Qian F., Sun Y., Wen D., Pelletier M., Hsu Y. M., Whitty A. (2005) Selectivity of BAFF/BLyS and APRIL for binding to the TNF family receptors BAFFR/BR3 and BCMA. Biochemistry 44, 1919–1931 [DOI] [PubMed] [Google Scholar]
- 35. Ameloot P., Fiers W., De Bleser P., Ware C. F., Vandenabeele P., Brouckaert P. (2001) Identification of tumor necrosis factor (TNF) amino acids crucial for binding to the murine p75 TNF receptor and construction of receptor-selective mutants. J. Biol. Chem. 276, 37426–37430 [DOI] [PubMed] [Google Scholar]
- 36. Szegezdi E., van der Sloot A. M., Mahalingam D., O'Leary L., Cool R. H., Munoz I. G., Montoya G., Quax W. J., de J. S., Samali A., Serrano L. (2012) Kinetics in signal transduction pathways involving promiscuous oligomerizing receptors can be determined by receptor specificity. Apoptosis induction by TRAIL. Mol. Cell Proteomics 11, M111.013730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kelley R. F., Totpal K., Lindstrom S. H., Mathieu M., Billeci K., Deforge L., Pai R., Hymowitz S. G., Ashkenazi A. (2005) Receptor-selective mutants of apoptosis-inducing ligand 2/tumor necrosis factor-related apoptosis-inducing ligand reveal a greater contribution of death receptor (DR) 5 than DR4 to apoptosis signaling. J. Biol. Chem. 280, 2205–2212 [DOI] [PubMed] [Google Scholar]
- 38. Shibata H., Yoshioka Y., Ohkawa A., Minowa K., Mukai Y., Abe Y., Taniai M., Nomura T., Kayamuro H., Nabeshi H., Sugita T., Imai S., Nagano K., Yoshikawa T., Fujita T., Nakagawa S., Yamamoto A., Ohta T., Hayakawa T., Mayumi T., Vandenabeele P., Aggarwal B. B., Nakamura T., Yamagata Y., Tsunoda S., Kamada H., Tsutsumi Y. (2008) Creation and X-ray structure analysis of the tumor necrosis factor receptor-1-selective mutant of a tumor necrosis factor-α antagonist. J. Biol. Chem. 283, 998–1007 [DOI] [PubMed] [Google Scholar]
- 39. Morishige T., Yoshioka Y., Inakura H., Tanabe A., Yao X., Tsunoda S., Tsutsumi Y., Mukai Y., Okada N., Nakagawa S. (2010) Creation of a LIGHT mutant with the capacity to evade the decoy receptor for cancer therapy. Biomaterials 31, 3357–3363 [DOI] [PubMed] [Google Scholar]
- 40. Thilenius A. R., Braun K., Russell J. H. (1997) Agonist antibody and Fas ligand mediate different sensitivity to death in the signaling pathways of Fas and cytoplasmic mutants. Eur. J. Immunol. 27, 1108–1114 [DOI] [PubMed] [Google Scholar]
- 41. Motoki K., Mori E., Matsumoto A., Thomas M., Tomura T., Humphreys R., Albert V., Muto M., Yoshida H., Aoki M., Tamada T., Kuroki R., Yoshida H., Ishida I., Ware C. F., Kataoka S. (2005) Enhanced apoptosis and tumor regression induced by a direct agonist antibody to tumor necrosis factor-related apoptosis-inducing ligand receptor 2. Clin. Cancer Res. 11, 3126–3135 [DOI] [PubMed] [Google Scholar]
- 42. Allen J. E., Ferrini R., Dicker D. T., Batzer G., Chen E., Oltean D. I., Lin B., Renshaw M. W., Kretz-Rommel A., El-Deiry W. S. (2012) Targeting TRAIL death receptor 4 with trivalent DR4 atrimer complexes. Mol. Cancer Ther., in press [DOI] [PubMed] [Google Scholar]
- 43. He B., Santamaria R., Xu W., Cols M., Chen K., Puga I., Shan M., Xiong H., Bussel J. B., Chiu A., Puel A., Reichenbach J., Marodi L., Döffinger R., Vasconcelos J., Issekutz A., Krause J., Davies G., Li X., Grimbacher B., Plebani A., Meffre E., Picard C., Cunningham-Rundles C., Casanova J. L., Cerutti A. (2010) The transmembrane activator TACI triggers immunoglobulin class switching by activating B cells through the adaptor MyD88. Nat. Immunol. 11, 836–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mackay F., Schneider P., Rennert P., Browning J. (2003) BAFF and APRIL. A tutorial on B cell survival. Annu. Rev. Immunol. 21, 231–264 [DOI] [PubMed] [Google Scholar]
- 45. Cancro M. P. (2008) Living in context with the survival factor BAFF. Immunity 28, 300–301 [DOI] [PubMed] [Google Scholar]
- 46. Gardam S., Sierro F., Basten A., Mackay F., Brink R. (2008) TRAF2 and TRAF3 signal adapters act cooperatively to control the maturation and survival signals delivered to B cells by the BAFF receptor. Immunity 28, 391–401 [DOI] [PubMed] [Google Scholar]