Background: Expression patterns of E2F1 target genes differ during cellular senescence.
Results: Rb protein selectively represses specific E2F1 target genes via a TAAC element in senescent cells.
Conclusion: Cellular senescence is influenced by selective repression of E2F1 target transcription by Rb.
Significance: An understanding of how E2F1 target genes that participate in proliferation are regulated is crucial for elucidating the mechanisms of cellular senescence.
Keywords: Aging, Chromatin Remodeling, DNA Transcription, E2F Transcription Factor, Retinoblastoma (Rb), CSIG, Heterochromatin
Abstract
The retinoblastoma (Rb) protein mediates heterochromatin formation at the promoters of E2 transcription factor 1 (E2F1) target genes, such as proliferating cell nuclear antigen and cyclin A2 (CCNA2), and represses these genes during cellular senescence. However, the selectivity of Rb recruitment is still not well understood. Here, we demonstrate that a senescence-associated gene is a direct target of E2F1 and is also repressed by heterochromatin in senescent cells. In contrast, ARF and p27KIP1, which are also E2F1 targets, are not repressed by Rb and heterochromatin formation. By comparing the promoter sequences of these genes, we found a novel TAAC element that is present in the cellular senescence-inhibited gene, proliferating cell nuclear antigen, and CCNA2 promoters but absent from the ARF and p27KIP1 promoters. This TAAC element associates with Rb and is required for Rb recruitment. We further determined that TAAC element-mediated Rb association requires the E2F1 binding site, but not E2F1 protein. These results provide a novel molecular mechanism for the different expression patterns of E2F1 targets and afford new mechanistic insight regarding the selectivity of Rb-mediated heterochromatin formation and gene repression during cellular senescence.
Introduction
Cellular senescence causes irreversible exit from the cell cycle and functions as a general protective mechanism against proliferative stress responses and cancer in vivo (1–3). In fact, senescent cells accumulate in benign tumors. Senescence-associated growth arrest mainly occurs through the stable down-regulation of cell cycle regulatory and DNA replication genes, such as CDC2, cyclin A2 (CCNA2), PCNA,2 and MCM3, many of which are E2F1 targets. Based on the current understanding of the field, the members of the retinoblastoma (Rb) gene family, including pRb, p107, and pRb2/p130, negatively regulate the activities of the E2F family of proteins (4). When cells undergo senescence, Rb mediates the formation of senescence-associated heterochromatin foci (SAHF) by recruiting other heterochromatin proteins, such as HP1γ, to E2F1-responsive promoters, which then stably repress these E2F1 targets (5). Importantly, this mechanism may be the reason why growth factors cannot induce E2F1 targets during cellular senescence (6).
ARF (cyclin-dependent kinase inhibitor 2A, p14ARF) and p27KIP1 are also reported to be E2F1 targets (7, 8), and the expression of these genes is up-regulated during cellular senescence. In fact, overexpression of E2F1 may induce a senescence response through the activation of ARF (9). In addition, one of the most important cyclin-dependent kinase inhibitors, p27KIP1, was reported to accumulate in senescent cells (10). The mechanism by which some E2F1 targets, such as PCNA and CCNA2, are transcriptionally repressed by SAHF, whereas others are not, such as ARF and p27KIP1, is important for understanding how E2F1 targets are transcriptionally regulated during cellular senescence.
In our previous study, we identified a new senescence-associated gene called cellular senescence-inhibited gene (CSIG; RSL1D1) (11). CSIG is abundantly expressed in growing human diploid fibroblast cells, but its expression decreases upon replicative senescence. Overexpression of CSIG significantly prolonged the progression of replicative senescence by down-regulating PTEN and p27KIP1 (12). CSIG was also reported to facilitate nucleostemin transport between the nucleolus and nucleoplasm (13). Therefore, it is important to explore the mechanism by which CSIG is down-regulated in senescent human cells to further characterize its function and understand the regulatory mechanisms of other senescence-associated genes.
In this study, we demonstrate that CSIG is a direct target of E2F1. The transcriptional profiles of CSIG, PCNA, CCNA2, ARF, and p27KIP1 are distinct during senescence. CSIG, PCNA, and CCNA2 are repressed by Rb-mediated heterochromatin, but ARF and p27KIP1 are not repressed. A comparison of the promoter sequences demonstrated the existence of a novel TAAC element in the CSIG, PCNA, and CCNA2 promoters, which was absent in the ARF and p27KIP1 promoters. Rb associates with these proteins through the TAAC element, and it is required for Rb recruitment. We further determined that the association of the TAAC element with Rb is E2F binding site-dependent, but E2F1 protein-independent. These results provide a novel molecular explanation for the different expression patterns of E2F1 targets and afford new mechanistic insight regarding the selectivity of Rb-mediated heterochromatin formation and gene repression during cellular senescence.
EXPERIMENTAL PROCEDURES
Plasmids and Vectors
The following retroviral vectors were used in this study: pWZL-Hygro (oncogenic Ras (H-RasV12) and E2F1); pLPC-Puro (E2F1, E1A, and E1AΔN; E1A and E1AΔN were generously provided by Dr. Scott Lowe (5)); pMSCV-miR30-puro (E2F1, p16INK4A, HMGA1, Rb, p130, and p107); the shRNA sequences shp16INK4A, shHMGA1, and shRb were generously provided by Dr. Narita (14); shRNA sequences targeting p130 and p107 were generously provided by Dr. Scott Lowe (15); the short hairpin RNAs specific for human E2F1 (5′-GACGTGTCAGGACCTTCGT-3′) were cloned into the pMSCV-miR30-puro vector (16) and pLNCX2-Neo (Ras and E2F1). The ARF-promoter-PGL3 plasmid was generously provided by Dr. Hongti Jia. The primer sequences used to construct the promoters were as follows: CSIG promoter, forward, 5′-CTAGCTAGCCATTTCTTCTACAACTTGATTA-3′ and reverse, 5′-CCGCTCGAGCATCTTGTTTCCACCTC-3′; CCNA2 promoter, forward, 5′-CGGGGTACCCATAGAAAGATAACGACG-3′ and reverse, 5′-CCCAAGCTTAAAGAGAAACAGACAAGC-3′; PCNA promoter, forward, 5′-CGGGGTACCATGAACGATTGAGTGATT-3′ and reverse, 5′-CCCAAGCTTGGCTGAGACCTAGAAAGA-3′; and p27KIP1 promoter, forward, 5′-CGGGGTACCCCAGGGATGGCAGAAACT-3′, and reverse, 5′-CCCAAGCTTACACCCCGAAAAGACGAG-3′. The E2F1 siRNA (siE2F1) sequence was as follows: 5′-GACGUGUCAGGACCUUCGU-3′.
Cell Culture and Gene Transfer
Human diploid 2BS fibroblasts (National Institute of Biological Products, Beijing, China) as well as IMR90 and WI38 cells (American Type Culture Collection) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% FBS and antibiotics. Retroviruses were packed using Phoenix cells (Dr. Narita, Cancer Research UK, Cambridge Research Institute), and infections were performed as previously described (17). The infected population was selected with 0.8 μg/ml puromycin (Sigma) for 2–3 days, 25 μg/ml hygromycin B (Roche) for 2–3 days, or 150 μg/ml G418 for 5 days. For coinfection, the cells were sequentially selected with puromycin, hygromycin, and G418. Post-selection day 7 refers to 7 days after the puromycin selection. The wild-type and TAAC-mutated CSIG promoter constructs in Fig. 5 (C and D) were integrated into U2OS cells using a retroviral system (pWZL). Integrated cell populations were selected with hygromycin B (Roche Applied Science) for 10 days.
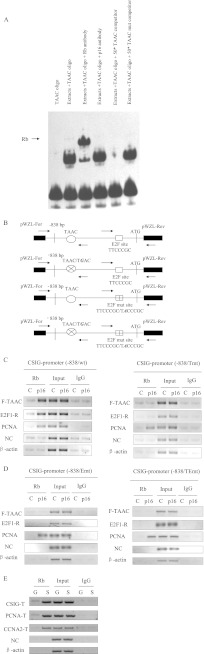
FIGURE 5.
Rb associates with the TAAC element in vitro and in vivo. A, an EMSA was performed using the protein extracts of senescent 2BS cells. Extracts, core protein extracted from the senescent 2BS cells; TAAC oligo, 3′ end-labeled oligonucleotide probes containing the TAAC element on the CSIG promoter 5′-ATAACCGGTACCCGCCCCCT-3′; TAAC mut competitor, unlabeled probes containing the TAAC mutant element 5′-ATGACCGGTACCCGCCCCCT-3′. B, schematic view of the promoter constructs used for integration into U2OS cells and the primer locations (small arrows) for the ChIP assay. pWZL-For, forward sequencing primer for pWZL; pWZL-Rev, reverse sequencing primer for pWZL. C, ChIP analysis of the CSIG promoter constructs in U2OS cells ectopically expressing p16. Lanes C, pcDNA3.1 vector control; F-TAAC, fragment from pWZL-For to the TAAC element; E2F1-R, fragment from E2F binding site to pWZL-Rev; NC, nonsense control fragment on the CSIG promoter; Tmt, TAAC mutant. D, ChIP analysis of the CSIG promoter constructs in U2OS cells ectopically expressing p16. Lanes C, pcDNA3.1 vector control; F-TAAC, fragment from pWZL-For to the TAAC element; E2F1-R, fragment from E2F binding site to pWZL-Rev; NC, nonsense control fragment on the CSIG promoter; Emt, E2F binding site mutant; TEmt, TAAC and E2F binding site mutants. E, growing (G) and Ras-induced senescent (S) 2BS cells were processed for ChIP using Rb antibodies or using nuclear extract (Input) or IgG as a negative control. The primer sets used for CSIG-T, PCNA-T, and CCNA2-T include the CSIG/PCNA/CCNA2 promoter regions containing the TAAC element. NC, nonsense control fragment on the CSIG promoter.
Cell Proliferation and DAPI Staining
2BS cells were plated on coverslips and subsequently labeled with BrdU (100 μg/ml, Sigma) for 6 h. Nuclei incorporating BrdU were visualized by immunolabeling using an anti-BrdU antibody (Pharmingen, 1:400) as previously described (18). DNA was visualized by DAPI (1 μg/ml) after permeabilization with 0.2% Triton X-100 in phosphate-buffered saline.
Electrophoretic Mobility Shift Assay
The EMSAs in Fig. 1J were performed using a human GST-E2F1 fusion protein that was produced in Escherichia coli BL21 transfected with pGEX-4T-E2F1. The EMSA probes were generated by end-labeling 30-bp duplexes using T4 polynucleotide kinase and [γ-32P]ATP. DNA-protein binding reactions were performed by mixing 5 μg of purified recombinant GST-E2F1 fusion protein, 2 μl of 5× binding buffer, 2.5 μg/μl BSA, and 4,000 cpm/μl probes. The competition experiments included a 50-fold molar excess of unlabeled double-stranded oligonucleotide. The binding mixtures were incubated for 20 min at room temperature. DNA-protein complexes were resolved on 6.5% polyacrylamide gels in 1× Tris borate-EDTA buffer at room temperature. The EMSAs in Fig. 5A were 3′-biotinylated using the biotin 3′-end DNA labeling kit (Pierce) according to the manufacturer's instructions and were annealed for 2 h at room temperature. The sequences of the oligonucleotides used are 5′-ATAACCGGTACCCGCCCCCT-3′ for the TAAC oligonucleotides and 5′-ATGACCGGTACCCGCCCCCT-3′ for the TAAC mutant oligonucleotides. The binding reactions were carried out for 20 min at room temperature in the presence of 50 ng/μl poly(dI-dC), 0.05% Nonidet P-40, 5 mm MgCl2, 10 mm EDTA, and 2.5% glycerol in 1× binding buffer (LightShiftTM chemiluminescent EMSA kit; Pierce) using 20 fmol of biotin end-labeled target DNA and 4 μg of nuclear extract from senescent 2BS cells. Unlabeled target DNA (4 pmol), anti-Rb (2 μl; Cell Signaling), or anti-p16 (2 μl; Santa Cruz Biotechnology) was added per 20 μl of binding reaction where indicated. The assays were loaded onto native 6% polyacrylamide gels that had been pre-electrophoresed for 60 min in 0.5× Tris borate-EDTA. Samples were electrophoresed at 100 V before being transferred onto a positively charged nylon membrane (HybondTM-N+) in 0.5× Tris borate-EDTA at 100 V for 30 min. Transferred DNA was cross-linked to the membrane at 120 mJ/cm2 and detected using horseradish peroxidase-conjugated streptavidin (LightShiftTM chemiluminescent EMSA kit) according to the manufacturer's instructions.
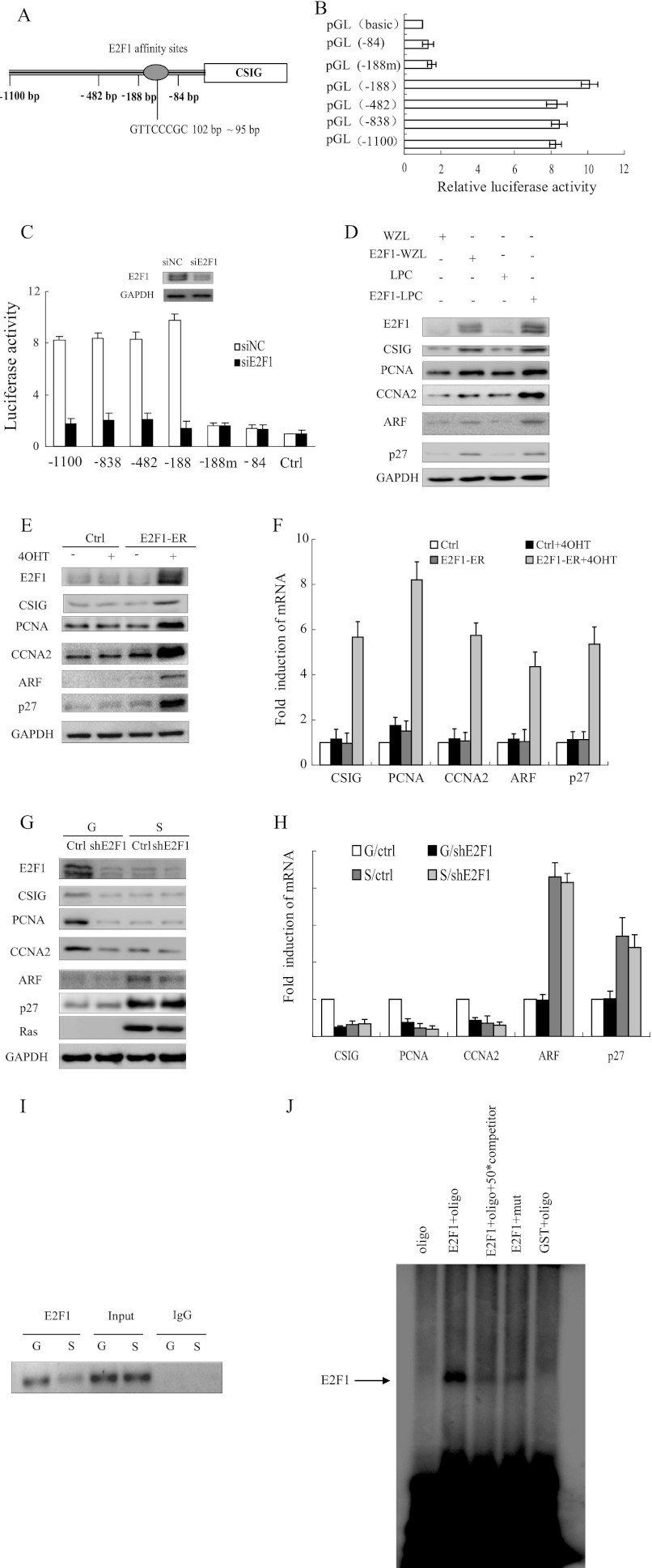
FIGURE 1.
CSIG is one of the target genes of E2F1. A, schematic representation of E2F1 affinity sites. B, different constructs were analyzed for luciferase expression in HeLa cells. The values represent the means and standard errors of three independent experiments. C, deletion mutants of human CSIG promoter constructs were cotransfected with siE2F1 oligonucleotides and small interference nonsense control (siNC) in HeLa cells and were analyzed for luciferase expression. The values represent the means and standard errors from three independent experiments. D, 2BS cells stably expressing retroviral E2F1 expression plasmids were analyzed for the indicated proteins 7 days after retroviral transduction. E and F, the 2BS cells stably expressing the hormone-regulated E2F1-ER fusion construct were treated with 100 nm 4-hydroxytamoxifen (4OHT) for 18 h, and the protein and mRNA levels of the indicated genes were determined by Western blot and quantitative RT-PCR. The values represent the means and standard errors from three independent experiments. G and H, the 2BS cells expressing vector alone (G) or a combination of pWZL-Ras-Hygro (S), and vector (Ctrl) or shRNA against E2F1 (shE2F1) were harvested to determine the protein and mRNA levels of the indicated genes by Western blot and quantitative RT-PCR. The values represent the means and standard errors from three independent experiments. I, growing (G) and Ras-induced senescent (S) 2BS cells were processed for ChIP using an anti-E2F1 antibody (C-20x; Santa Cruz Biotechnology). The primer set used includes the region from −255 to −35 bp of the CSIG promoter. J, EMSA was performed using E2F1 protein expressed in vitro through the GST expression system. oligo, end-labeled oligonucleotide probes containing the E2F1 binding site on the CSIG promoter 5′-ATGACCGGTTCCCGCCCCCT-3′; mut, end-labeled probes containing the mutant E2F1 binding site oligonucleotide 5′-ATGACCGGTACCCGCCCCCT-3′; competitor, unlabeled probes containing the E2F1 binding site; Ctrl, control.
Chromatin Immunoprecipitation Assay
ChIPs were performed as previously described (19) using anti-E2F1 (C-20x; Santa Cruz Biotechnology), anti-H3K9Me3 (Upstate), anti-HP1γ (Upstate), anti-Rb (4H1; Cell Signaling), anti-p107 (C-18; Santa Cruz Biotechnology), and anti-p130 (C-20; Santa Cruz Biotechnology) antibodies. DNA released from the precipitated complexes was amplified by PCR using sequence-specific primers. The primer sets that were used amplified the promoter regions of CSIG, CCNA2, PCNA, ARF, p27KIP1, and β-actin. The primer sequences for CCNA2, PCNA, and β-actin that were used in the ChIP assays were a kind gift from Dr. Scott Lowe (5). The ChIP primer sequences were as follows: CCNA2 promoter, forward, 5′-CGCTTTCATTGGTCCATTTC-3′ and reverse, 5′-CCGGCCAAAGAATAGTCGTA-3′; PCNA promoter, forward, 5′-GCATGGACACGATTGGCCCT-3′ and reverse, 5′-CTCGAACATGGTGGCGGAGT-3′; β-actin promoter, forward, 5′-AAATGCTGCACTGTGCGGCGAA-3′ and reverse, 5′-TGCTCGCGGGCGGACGCGGTCTCGG-3′; CSIG promoter, forward, 5′-GGCCAGGCTGGTCTGGAAC-3′ and reverse, 5′-GTCCTACCTACTCGGGAAGCT-3′ (PCR of the CSIG promoter between −255 bp and −35 bp generated a 220-bp product); ARF promoter, forward, 5′-GAAGAATGGAAGACTTTCGACGAGG-3′ and reverse 5′-ACCTCCAAGATCTCGGAACGG-3′ (PCR of the ARF promoter between −419 bp and −53 bp generated a 366-bp product); p27KIP1 promoter, forward, 5′-CGGCCGTTTGGCTAGTTTGTTTGT-3′ and reverse 5′-GGAGGCTGACGAAGAAGAAGATGA-3′ (PCR of the p27 promoter between −615 bp and −326 bp generated a 294-bp product); ChIP primers for the CSIG luciferase reporter and its TAAC mutant, F-TAAC, forward, 5′-TACATCGTGACCTGGGAAGC-3′ and reverse, 5′-CGGGAGCCACCCGGAGCCA-3′; E2F1-R, forward, 5′-CACTGCCCCGGAGAGCGA-3′ and reverse, 5′-CGACATTCAACAGACCTTGC-3′; and NC, forward, 5′-TTTGAATACAGCCCAACAC-3′ and reverse, 5′-TGGATAACTGACAAATGGAA-3′. CSIG-T forward, 5′-CGGACTGGTCTCCAACTC-3′ and reverse, 5′-CGGGAGCCACCCGGAGCCA-3′ (PCR of the CSIG promoter between −693 bp and −469 bp generated a 224-bp product); PCNA-T, forward, 5′-CCAAGTGTTTACGGAATGA-3′ and reverse, 5′-GCGCTCGTAGGTGTCACAAGAT-3′ (PCR for the presence of the PCNA promoter DNA between −716 bp and −525 bp, generating a 191-bp product); and CCNA2-T, forward, 5′-GGGGCTCCCAGATTTCGT-3′ and reverse, 5′-ACTCCACGGGCTGCTGCTAC-3′ (PCR for the presence of the CCNA2 promoter DNA between −420 bp and −76 bp, generating a 344-bp product).
Western Blot
Western blot analysis was performed on 20 μg of whole cell lysate using the chemiluminescent HRP substrate (Millipore). Blots were probed with the following antibodies: anti-CSIG (used as previously described (12), 1:5000), anti-p16 (C-20, Santa Cruz Biotechnology, 1:500), anti-Ras (F235, Santa Cruz Biotechnology, 1:1000), anti-Rb (4H1, Cell Signaling, 1:1000), anti-E2F1 (KH95, Santa Cruz Biotechnology, 1:200), anti-CCNA2 (C-19, Santa Cruz Biotechnology, 1:1000), anti-PCNA (I88, Bioworld, 1:1000), anti-ARF (H132, Santa Cruz Biotechnology, 1:1000), anti-p27KIP1 (MBL, 1:1000), anti-HMGA1 (Biosynthesis, 1:500), p107 (C-18, Santa Cruz Biotechnology 1:1000), and p130 (C-20, Santa Cruz Biotechnology, 1:1000).
Real Time PCR
Gene-specific primers were designed using Primer 5 (sequences are available from the authors upon request). Real time PCR was performed in triplicate using the SYBR Green PCR Master Mix (Applied Biosystems) on an ABI Prism 7300 sequence detector (Applied Biosystems). The β-actin gene served as an endogenous control for normalization.
Luciferase Assay
The cells were plated in 24-well culture plates in triplicate for each condition at an initial concentration of 5 × 104 cells/well. The cells were cotransfected with 0.8 μg of the gene promoter reporter constructs (or 0.2 μg of reporter construct and 0.6 μg of protein overexpression plasmid or shRNA) and 8 ng (or 2 ng, respectively) of the Renilla luciferase reporter plasmid pRL-CMV vector, which served as an internal control. Luciferase activity was assessed using a dual luciferase reporter assay system (Promega) according to the manufacturer's instructions. The enzyme activity was normalized for the efficiency of transfection on the basis of Renilla luciferase activity levels and is reported as relative light units. All of the reporter assays were performed in triplicate in at least two individual experiments, and standard errors are denoted by bars in the figures.
RESULTS
CSIG Is a Target Gene of E2F1
As previously reported, CSIG mRNA is significantly decreased in senescent cells compared with actively growing cells (11), which suggests that CSIG may be transcriptionally repressed during cellular senescence. To test this hypothesis, we first constructed a series of plasmids in which various lengths of the CSIG promoter (−1100, −838, −482, −188, and −84 bp upstream from the ATG start codon of the CSIG promoter) were placed upstream of the pGL3-basic vector. The activity of the firefly reporter remained high in all cases except when the region between −188 and −84 bp was excised (Fig. 1B). This result indicates that the major transcriptional regulatory elements of CSIG are located in the region between −188 and −84 bp from the ATG start codon. Analysis of the sequence from −188 to −84 bp revealed a putative E2F1 binding site from −102 to −95 bp (Fig. 1A). Moreover, the activity of the CSIG promoter-luciferase reporter containing the wild-type E2F1 binding site was almost 5-fold higher than the E2F1 binding site mutant (−188m) (Fig. 1B). Furthermore, when we introduced E2F1 siRNA oligonucleotides and the CSIG promoter luciferase reporter into HeLa cells, the activities of the different fragments of the CSIG promoter (−1100, −838, −482, and −188 bp) were down-regulated. However, this decrease did not occur with the promoter reporters in which the E2F1 binding site was mutated (−188m) or deleted (−84 bp) (Fig. 1C). These experiments suggest that the CSIG luciferase reporter is induced by E2F1.
We next assessed whether E2F1 could also stimulate endogenous CSIG expression by using a 2BS cell line stably expressing retroviral E2F1 expression plasmids. CSIG was up-regulated when E2F1 was overexpressed, as were other E2F1 targets, including PCNA and CCNA2 (Fig. 1D). This result was confirmed by the activation of a 2BS cell line stably expressing the E2F1-ER fusion construct upon the addition of 4-hydroxytamoxifen (Fig. 1, E and F). Furthermore, CSIG was down-regulated after the knockdown of E2F1 (Fig. 1, G and H). These results suggest that E2F1 stimulates endogenous CSIG expression.
We next sought to determine whether E2F1 binds to the CSIG promoter in vivo and in vitro by using ChIP and EMSAs (Fig. 1, I and J). The results of the ChIP assays suggested that E2F1 binds to the endogenous CSIG promoter in growing cells but not senescent cells in vivo. Using an EMSA, we found a high affinity E2F1 binding site located −102 bp upstream of the ATG codon. Importantly, a point mutation of this binding site blocked E2F1 binding (Fig. 1J). These results demonstrate that E2F1 can bind to the CSIG promoter in vivo and in vitro at the E2F1 binding site.
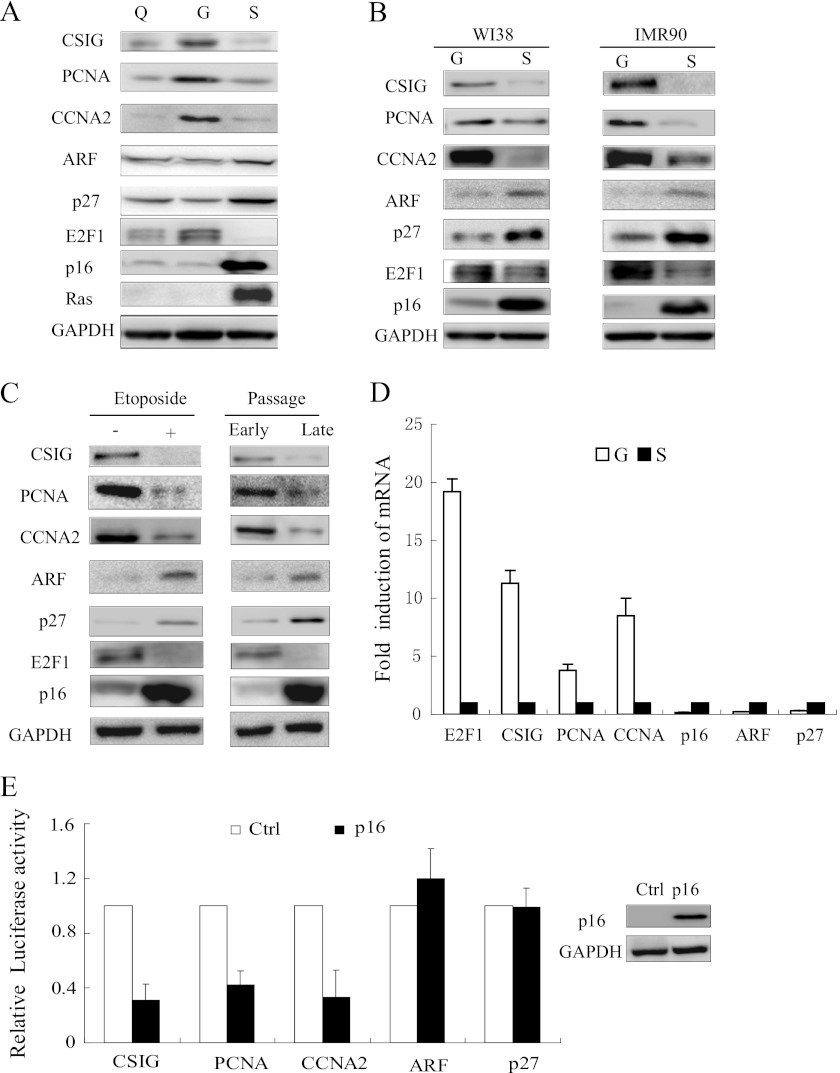
CSIG, but Not ARF and p27KIP1, Is Transcriptionally Repressed during Cellular Senescence
Because E2F1 is a well known transcription factor, we sought to characterize the expression patterns of E2F1 targets during cellular senescence by examining CSIG, PCNA, CCNA2, ARF, and p27KIP1 in growing, quiescent, and Ras-induced senescent 2BS cells. We found that the protein levels of CSIG, PCNA, and CCNA2, but not ARF or p27KIP1, decreased in Ras-induced senescent cells relative to growing cells (Fig. 2A). The same result was confirmed in two other fibroblast cell lines, WI38 and IMR90 (Fig. 2B). Moreover, the same trend in the protein levels of E2F1 targets was observed in replicative senescent 2BS cells and etoposide-induced 2BS senescent cells with DNA damage (Fig. 2C). In addition, the mRNA levels of CSIG, PCNA, and CCNA2, but not those of ARF and p27KIP1, decreased in senescent cells (Fig. 2D). To further determine the transcriptional activities of these genes, we cotransfected p16INK4 constructs together with CSIG, PCNA, CCNA2, ARF, or p27KIP1 promoter-luciferase reporters into U2OS cells, in which the responsiveness to p16INK4 overexpression is quite similar to that of senescent fibroblast cells (20). In fact, p16INK4 overexpression-induced cellular senescence has been widely reported (5, 21, 22). The p16INK4 protein accumulates in senescent cells and engages the Rb/E2F1 pathway by inhibiting cyclin D-dependent kinases. As shown in Fig. 2E, when p16 was overexpressed in U2OS cells, the activities of the CSIG, PCNA, and CCNA2 promoters were down-regulated. In contrast, the activities of the ARF and p27KIP1 promoters were not affected by p16 overexpression. These data indicate that the transcriptional programs that regulate CSIG, PCNA, CCNA2, ARF, and p27KIP1 are different during cellular senescence.
FIGURE 2.
CSIG, but not ARF and p27KIP1, is transcriptionally repressed during cellular senescence. A and C, Western blot of the 2BS cells lysates for CSIG, PCNA, CCNA2, ARF, p27KIP1, p16, E2F1, and Ras. GAPDH served as a loading control. The following conditions are shown: Q, quiescent (by low serum); G, growing; S, Ras-induced senescence (pWZL-Ras-Hygro). Etoposide +, DNA damage-induced senescent cells. Early passage, 21 PD 2BS cells. Late passage, 65 PD 2BS cells. B, Western blot of WI38 or IMR90 cell lysates for CSIG, PCNA, E2F1, and p16. GAPDH served as a loading control. G indicates growing, and S indicates Ras-induced senescent cells. D, the mRNA levels of the indicated genes were determined by quantitative RT-PCR in growing (G) and Ras-induced senescent (S) 2BS cells. The values represent the means and standard errors from three independent experiments. E, different gene promoters were analyzed for luciferase expression in U2OS cells transfected with p16-pcDNA. The values represent the means and standard errors of three independent experiments.
Heterochromatin Accumulates at the CSIG Promoter, but Not the ARF and p27KIP1 Promoters
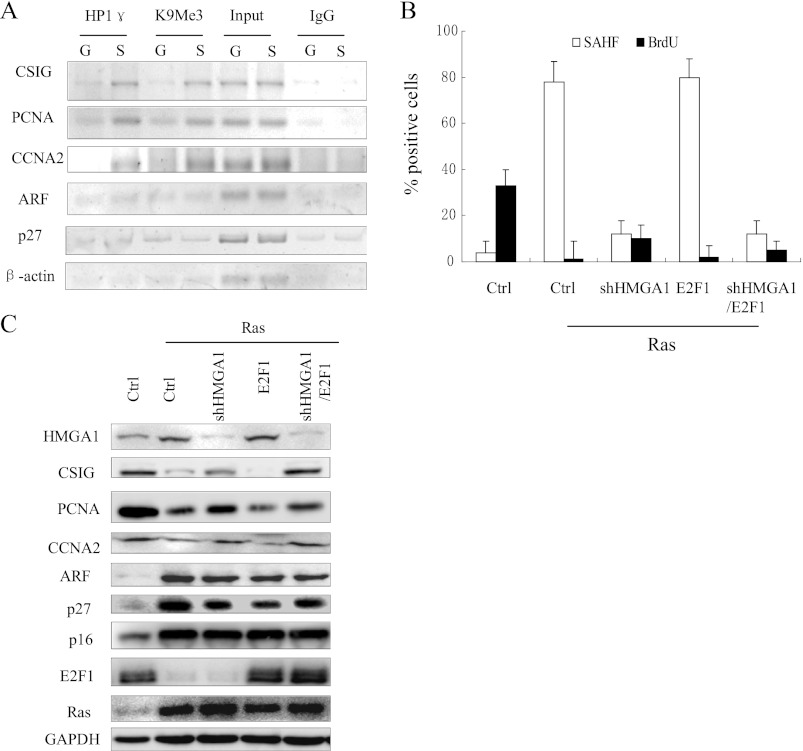
One central question that remains to be elucidated is why E2F1 target genes are differentially transcribed during cellular senescence. Because it is known that SAHF are involved in the stable repression of several E2F1 targets, including PCNA, CCNA2, and MCM3 (5), we next sought to determine whether the chromatin remodeling was different at the CSIG, ARF, and p27KIP1 promoters. We examined the association of the histone H3 trimethylated lysine 9 (H3K9Me3) and HP1γ with the promoters of CSIG, ARF, and p27KIP1 using a ChIP assay (Fig. 3A). As expected, the quantities of both H3K9Me3 and HP1γ bound to the CSIG promoter were higher in senescent cells compared with growing 2BS cells, which was consistent with the PCNA and CCNA2 data. In contrast, H3K9Me3 and HP1γ did not accumulate at the ARF and p27KIP1 promoters during cellular senescence. Furthermore, when we knocked down high mobility group AT-hook 1 (HMGA1), one of the essential structural components of SAHF, using shRNA in 2BS cells transfected with Ras, SAHF accumulation was almost completely prevented (Fig. 3B). These results are consistent with previous reports. Similarly, knockdown of HMGA1 activated CSIG, but did not up-regulate ARF and p27KIP1 (Fig. 3C). These results demonstrate that heterochromatin formation is responsible for the stable repression of CSIG, but not ARF and p27KIP1 in senescent cells.
FIGURE 3.
Heterochromatin accumulates at the CSIG promoter but not at the ARF and p27KIP1 promoters. A, growing and Ras-senescent 2BS cells were processed for ChIP using anti-HP1γ and anti-H3K9Me3 antibodies, nuclear extract (Input), or IgG as a negative control. B, the 2BS cells expressing vector alone (Ctrl), Ras with the indicated combination of vectors, shRNA against HMGA1 (shHMGA1), or the hormone-regulated E2F1-ER fusion construct were assessed for BrdU incorporation and SAHF formation after treatment with 100 nm 4-hydroxytamoxifen for 18 h. The values represent the means and standard errors for three independent experiments. C, the cell populations described in B were analyzed for the indicated proteins after treatment with 100 nm 4-hydroxytamoxifen for 18 h. Ctrl, control.
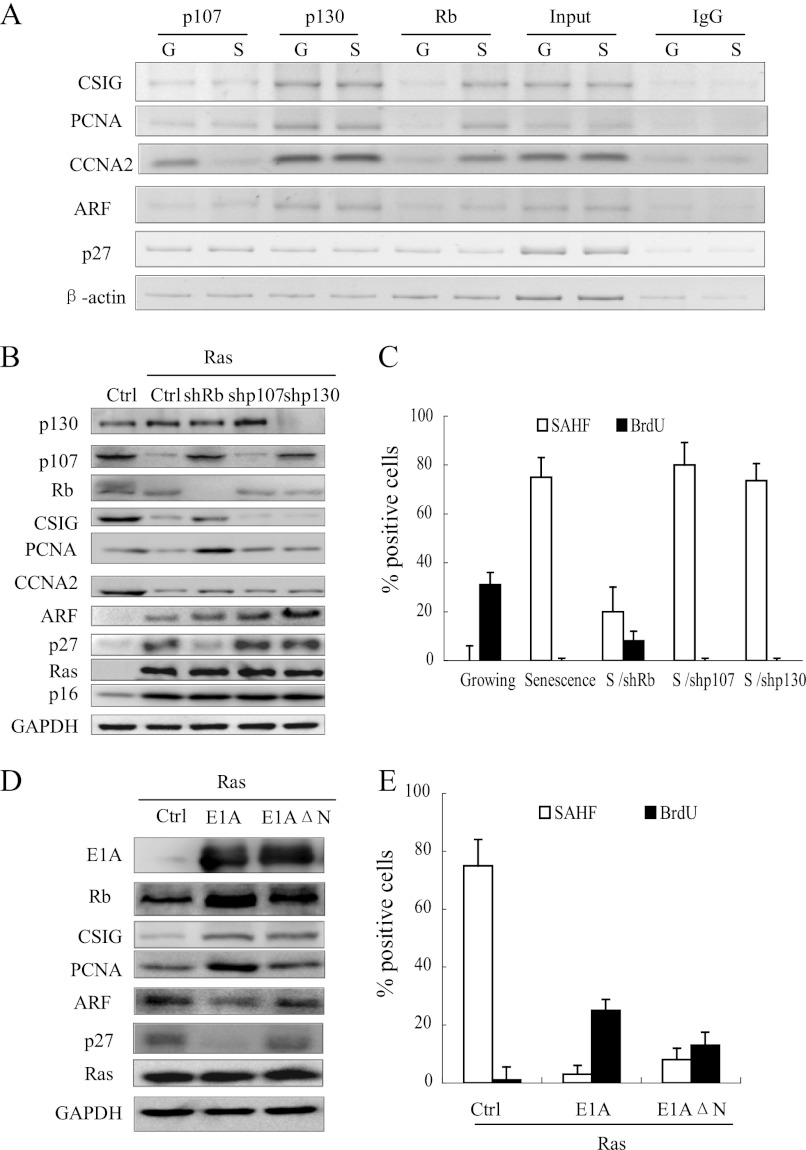
Rb Represses the Transcription of CSIG but Does Not Affect the Transcription of ARF and p27KIP1 during Senescence
It has been reported that Rb is required for SAHF formation at the promoters of E2F1 targets, such as PCNA and CCNA2 (5). Therefore, we investigated whether Rb is responsible for the differential chromatin remodeling between CSIG, ARF, and p27KIP1 by examining the association of Rb with the promoters of these genes using a ChIP assay. As shown in Fig. 4A, Rb was absent from these gene promoters in growing 2BS cells. In contrast, Rb was detected only on the CSIG promoter and not on the ARF and p27KIP1 promoters in senescent cells. To further investigate the role of Rb, we utilized 2BS cells infected with retroviral shRNA against Rb, p107, or p130. shRNA targeting of Rb, but not p107 or p130, reduced SAHF formation and induced CSIG in Ras-induced senescent cells. However, shRb did not induce the up-regulation of ARF or p27KIP1 expression (Fig. 4, B and C). Moreover, considering that E1A can block Rb function (5), we examined the effect of E1A on the expression of CSIG/PCNA/CCNA2/ARF/p27KIP1 in response to Ras (Fig. 4, D and E). As expected, full-length E1A abrogated Ras-induced SAHF formation and up-regulated CSIG, but not ARF or p27KIP1. To verify that these effects are derived from E1A countering Rb, we chose E1A-ΔN, which cannot bind to p300 and p400 but still possesses the binding domain that interacts with the Rb LXCXE motif (5). In this setting, although E1A-ΔN was not as effective as its full-length in preventing cell cycle arrest (Fig. 4E), it can still prevent SAHF formation and repression of E2F1 targets (Fig. 4, D and E). These data demonstrate that the selectivity of Rb binding is responsible for the differential chromatin remodeling of the promoters for these genes during cellular senescence.
FIGURE 4.
Rb represses the transcription of CSIG but does not affect the transcription of ARF or p27KIP1 during senescence. A, growing (G) and Ras-senescent (S) 2BS cells were processed for ChIP using an antibody against Rb, p107, or p130 or using nuclear extract (input) or IgG as a negative control. DNA fragments were amplified by PCR from the promoter regions of CSIG, PCNA, CCNA2, ARF, p27, and β-actin. B, the 2BS cells expressing vector alone (Ctrl) or H-Ras (pWZL-Ras-Hygro) with the indicated combination of vector, shRNA against Rb (shRb), p107 (shp107), and p130 (shp130) were analyzed for the indicated proteins. C, the cell populations described in B were assessed for BrdU incorporation and SAHF formation. The values represent the means and standard errors for the three independent experiments. D, the 2BS cells expressing a combination of Ras and the vector (Ctrl), E1A, or E1AΔN were analyzed for the indicated proteins. E, the cell populations described in D were assessed for BrdU incorporation and SAHF formation. The values represent the means and standard errors from three independent experiments. Ctrl, control.
The TAAC Element Is Present in the Promoters of CSIG, PCNA, and CCNA2 but Is Not in the ARF and p27KIP1 Promoters
Our data showed that Rb is responsible for the differential expression of the E2F1 targets examined. However, it was unclear what events regulate the specificity of the Rb association. To address this question, we characterized the promoter sequences of the E2F1 target genes (supplemental Table S1) using the online tool TFSCAN. We found that apart from some general transcription factors, such as E2F1, SP1, and c-Ets (Table 1), C-Myb was predicted to be involved in CSIG/PCNA/CCNA2 transcriptional regulation, but not in ARF/p27KIP1 regulation. Moreover, it has been reported that Myb is involved in Rb binding activity (23, 24). Based on our TFSCAN results and previous reports (25, 26), the core binding element for C-Myb is TAAC. Therefore, we further characterized the role of the TAAC element in CSIG transcriptional regulation.
TABLE 1.
Predicted distribution of representative transcription factors on the CSIG, PCNA, CCNA2, ARF, and p27KIP1 promoter from TFSCAN
| Representative TFs | Down-regulated E2F target genes in senescent cells |
Up-regulated E2F target genes in senescent cells |
|||
|---|---|---|---|---|---|
| CSIG | PCNA | CCNA2 | ARF | p27KIP1 | |
| Sp1 | + | + | + | + | + |
| c-Ets-2 | + | + | + | + | + |
| E2F | + | + | + | + | + |
| GATA-1 | + | − | − | + | + |
| MAZ | + | − | − | + | + |
| CREB | + | − | − | + | − |
| TBP | + | − | − | − | + |
| LF-A1 | + | − | + | + | + |
| H4TF-2 | + | + | + | + | + |
| c-Myb | + | + | + | − | − |
Rb Associates with the TAAC Element in Vitro and in Vivo
To assess the role of the TAAC element, we first investigated whether Rb binds to the TAAC element in vitro using EMSA. As shown in Fig. 5A, using extracts from senescent 2BS cells, we detected Rb complexes bound to a biotin-labeled CSIG promoter fragment containing the TAAC element. Wild-type cold probes, but not TAAC mutant cold probes, were able to compete for Rb complex binding. These results suggest that Rb can bind to the TAAC element in vitro. We next assessed binding in a ChIP assay, which showed that Rb binds to the TAAC element of endogenous CSIG/PCNA/CCNA2 promoters in senescent cells but not growing cells in vivo (Fig. 5E). To further investigate whether Rb can bind to the TAAC element in vivo, we generated two U2OS cell lines containing an integrated CSIG promoter or its TAAC mutant and performed a ChIP assay using these cell lines. Rb bound the TAAC element and the E2F1 binding site of the integrated wild-type CSIG promoter, but not the TAAC element mutant (Fig. 5C). These data demonstrate that the TAAC element associates with Rb and is required for Rb binding to the CSIG promoter in vivo during cellular senescence.
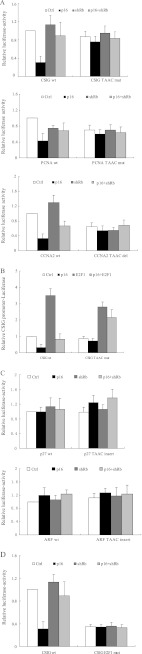
The Selective Repression of CSIG, PCNA, and CCNA2 by Rb Is TAAC Element-dependent
To further investigate the role of the TAAC element in the repression of CSIG, PCNA, and CCNA2 by Rb, we utilized U2OS cells that were cotransfected with p16 and shRb constructs together with the CSIG promoter-luciferase reporter. When p16 was overexpressed, the activity of wild-type CSIG promoter was down-regulated. The shRb construct rescued the decrease observed with the wild-type CSIG promoter, but not with the TAAC element mutant (Fig. 6A). Similar results were observed for the PCNA and CCNA2 promoters. Moreover, E2F1 did not induce the wild-type CSIG promoter when p16 was overexpressed. In contrast, the TAAC mutant was up-regulated by E2F1, even when p16 was introduced (Fig. 6B). These data suggest that the TAAC element is required for the repression of CSIG, PCNA, and CCNA2 by Rb during cellular senescence. Because the loss of the TAAC element in the CSIG, PCNA, and CCNA2 promoters inhibited Rb-mediated repression, we wondered whether the introduction of the TAAC element into the p27KIP1/ARF promoter would enable Rb binding or Rb-mediated repression during senescence. Using U2OS cells, we found that the p27KIP1 and ARF promoters containing the TAAC element were still not repressed by p16 overexpression or affected by the knockdown of Rb (Fig. 6C). These results suggest that introducing the TAAC element alone is not sufficient for Rb recruitment or gene repression.
FIGURE 6.
The selective repression of CSIG, PCNA, and CCNA2 by Rb is TAAC element-dependent. A, U2OS cells cotransfected with vectors alone (Ctrl) or p16 and shRNA against Rb (shRb) with CSIG, PCNA, or CCNA2 promoter luciferase reporter constructs or their TAAC element mutants or deletions. CSIG TAAC mut, TAAC to TGAC mutant of the CSIG promoter; PCNA TAAC mut, TAAC to TGAC mutant of the PCNA promoter; CCNA2 TAAC del, the fragment of the CCNA2 promoter with a TAAC element deletion. The values represent the means and standard errors for three independent experiments. B, U2OS cells cotransfected with vectors alone (Ctrl) or with p16 and E2F1 overexpression vectors with CSIG promoter luciferase reporter constructs or the TAAC element mutant were analyzed for luciferase expression. The values represent the means and standard errors for three independent experiments. C, U2OS cells cotransfected with vectors alone (Ctrl) or p16 and shRNA against Rb (shRb) with ARF and p27KIP1 promoter luciferase reporter constructs or their TAAC insertion mutants. Insertion of the TAAC element into p27 and ARF promoters was constructed by introducing a TAAC element into the −244-bp position of the p27 promoter and the −478-bp position of the ARF promoter. The values represent the means and standard errors for three independent experiments. D, U2OS cells cotransfected with vectors alone (Ctrl) or p16 and shRNA against Rb (shRb) together with wild-type CSIG promoter luciferase reporter constructs or their E2F binding site mutant. The values represent the means and standard errors for three independent experiments. Ctrl, control.
An E2F1 Binding Site Is Required for the Recruitment of Rb to the TAAC Element on the CSIG Promoter
We next investigated whether TAAC element-mediated Rb binding activity is E2F1 binding site-dependent using a ChIP assay in U2OS cells transfected with the CSIG promoter or its E2F binding site mutant. We found that when the E2F1 binding site was mutated, Rb did not bind to the TAAC element or the CSIG promoter (Fig. 5D). Moreover, when we overexpressed p16 in U2OS cells, the activity of wild-type CSIG promoter was down-regulated, and shRb rescued this down-regulation (Fig. 6D). In contrast, p16 overexpression or the expression of shRb did not affect activity of the CSIG promoter containing a mutated E2F1 binding site. Therefore, these data demonstrate that the E2F1 binding site is required for recruiting Rb to the TAAC element on the CSIG promoter.
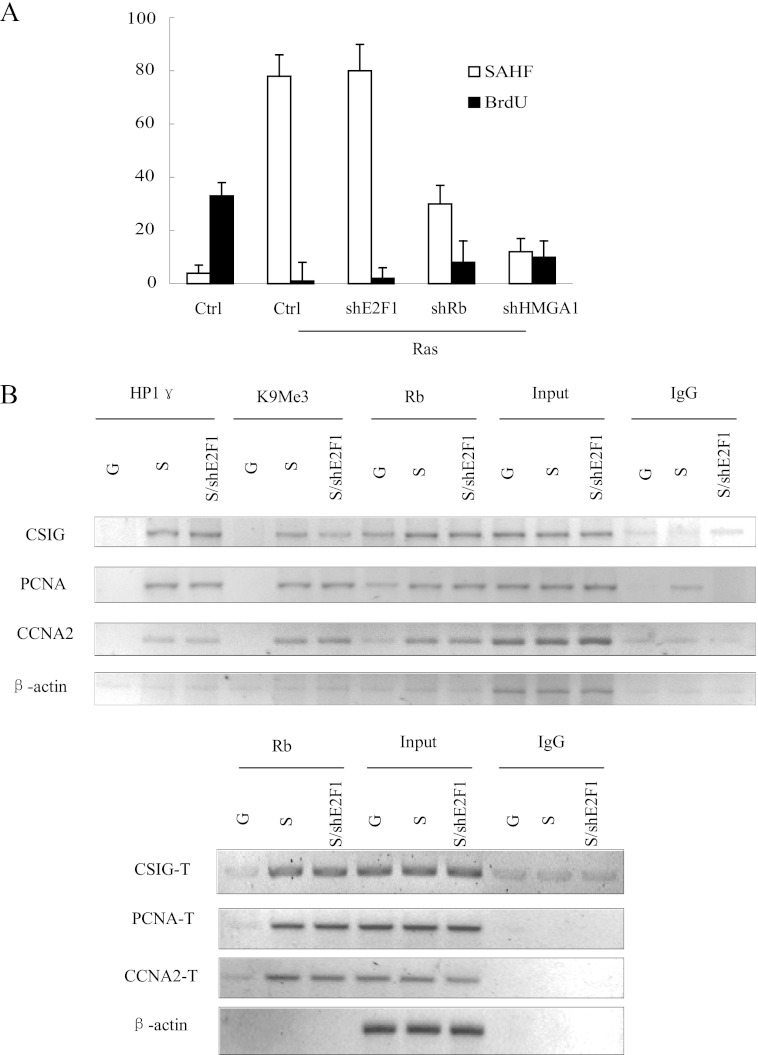
The E2F1 Protein Is Dispensable for Rb Binding to Both the E2F1 Binding Site and the TAAC Element
Despite the observation that the E2F1 binding site is required for the recruitment of Rb to the TAAC element on the CSIG promoter, it is difficult to explain why Rb accumulation on the promoters of E2F1 targets coincides with a decrease in E2F1 expression in senescent cells (9, 20). It is possible that E2F1 protein is dispensable for Rb-mediated heterochromatin formation. Therefore, we assessed this notion in 2BS cells cotransfected with E2F1 shRNA and the Ras overexpression plasmid. We found that knockdown of E2F1 did not affect SAHF formation during cellular senescence (Fig. 7A). Furthermore, using a ChIP assay, we found that shE2F1 did not affect Rb binding to either the E2F1 binding site or the TAAC element in the CSIG, PCNA, and CCNA2 promoters (Fig. 7B). These data suggest that the E2F1 protein is dispensable for Rb-mediated heterochromatin formation on the CSIG, PCNA, and CCNA2 promoters.
FIGURE 7.
E2F1 protein is dispensable for Rb binding to both the E2F1 binding site and the TAAC element. A, the 2BS cells expressing vector alone (Ctrl) or a combination of H-Ras and vector control, shRNA against E2F1 (shE2F1), Rb (shRb), or HMGA1 (shHMGA1) were assessed for BrdU incorporation and SAHF formation. The values represent the means and standard errors for three independent experiments. B, the 2BS cells expressing vectors alone (G) or a combination of H-Ras and vector control (S) shRNA against E2F1 (S/shE2F1) were processed for ChIP using anti-HP1γ, anti-H3K9Me3, and anti-Rb antibodies or using nuclear extract (input) or IgG as a negative control. The primer sets used for CSIG-T, PCNA-T, and CCNA2-T include the CSIG/PCNA/CCNA2 promoter regions containing the TAAC element. Ctrl, control.
DISCUSSION
The expression of E2F1 targets varies in senescent cells, and our results suggest that distinct transcriptional programs regulate these genes. ARF and p27KIP1 were not repressed at the transcriptional level by Rb or heterochromatin, which are responsible for the silencing of CSIG, PCNA, and CCNA2 during cellular senescence. Thus, E2F1 targets might be selectively regulated based on their functions. CSIG, PCNA, and CCNA2, which promote cell cycle progression, are repressed permanently in senescent cells, whereas ARF up-regulation is responsible for increasing p53 levels in senescent cells (9). p27KIP1 functions as a cyclin-dependent kinase inhibitor and also contributes to the senescent phenotype (27). However, various mechanisms may influence ARF and p27KIP1 up-regulation during senescence. For instance, ARF can be restored by the removal of the Polycomb genes BMI1 and CBX7 (28, 29), and p27KIP1 can be degraded by the E3 ligase Skp2 (30).
Rb has a nonredundant role in senescent cells by mediating heterochromatin formation at E2F1 targets. Our results demonstrate that Rb binding and the subsequent heterochromatin formation are selective, and Rb does not bind to the promoters of ARF and p27KIP1. This is consistent with the fact that not all E2F1 targets are up-regulated by the knockdown of Rb in senescent cells (15). The mechanism by which Rb binds to its targets has not yet been fully elucidated. Various epigenetic signals and specific sequences on the promoters of E2F1 targets may be responsible for the selectivity of Rb binding and heterochromatin formation. Importantly, our study has identified one potential candidate, the TAAC element. This element is required for Rb recruitment to the CSIG/PCNA/CCNA2 promoters, and a mutation in the element of the CSIG/PCNA/CCNA2 promoters causes a loss of response to Rb knockdown and p16 overexpression in U2OS cells. However, transcriptional activity is a collective process that is dependent on the interaction between the element and cotranscription factors, which results in the recruitment of RNA polymerase II. Thus, the TAAC element itself is not sufficient to repress E2F1 target genes, and the downstream effects of the presence of this element are context-dependent. In this study, we demonstrate that the TAAC element plays a role in the expression of the CSIG, PCNA, and CCNA2 genes. Currently, we cannot yet extend this conclusion to other genes, and therefore additional functions of the TAAC element must be investigated. It has also been reported that other factors are required for Rb binding activity. This is consistent with the fact that introducing the TAAC element alone into the promoters of ARF or p27KIP1 does not confer Rb-mediated repression of gene expression (Fig. 6C). Rb repressor activity and growth suppression also require interaction with BRG1 or BRM, which are central components of SWI/SNF chromatin-remodeling complexes. HPC2 is also required for the repression of CCNA2 and CDC2 by Rb (20). Therefore, elements and factors that assist Rb in specifically repressing genes remain to be identified.
Our data demonstrate that the E2F1 binding site, but not the E2F1 protein, is required for the TAAC element-mediated recruitment of Rb. This result is consistent with previous results demonstrating that Rb cannot directly bind DNA without the help of E2F family proteins (20, 32). In fact, in U2OS cells, upon p16 induction, Rb can be observed on the CCNA2 promoter together with a substantial amount of E2F4 at the same binding site, whereas E2F1 is absent (20). In mouse embryonic fibroblasts lacking E2F4/5, cell cycle regulatory genes are not repressed upon accumulation of hypophosphorylated Rb (33–36). Collectively, E2F4/5, but not E2F1, might facilitate Rb binding to E2F1 target promoters at the E2F1 binding site. Alternatively, although E2F1 was repressed in senescent cells, E2F2 and E2F3 may compensate for E2F1 function (37). In fact, we found that ARF and p27KIP1 are not affected by the decrease in E2F1 when cells undergo senescence (Fig. 1G).
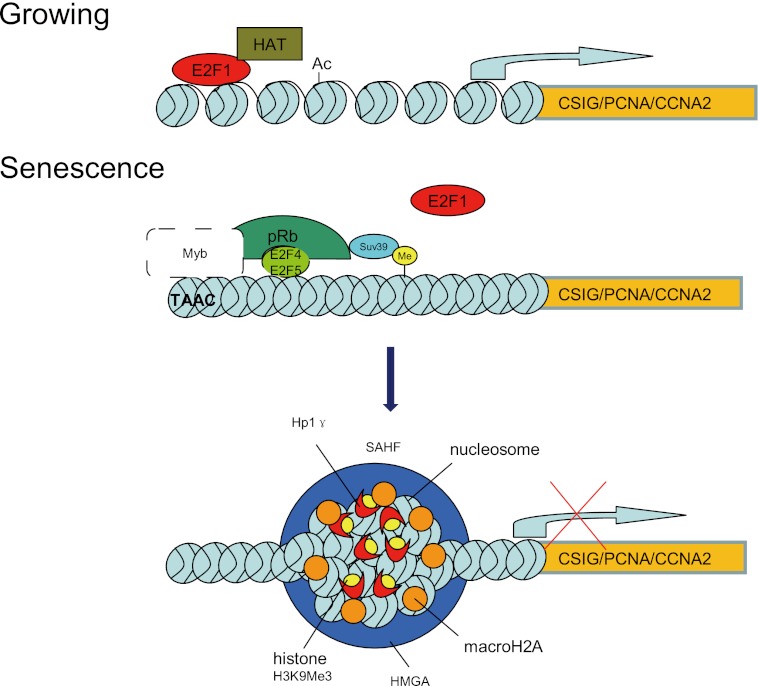
An important unaddressed question is how the TAAC element mediates Rb recruitment. Interestingly, the TAAC element can specifically recruit Myb family proteins (25, 26) according to TFSCAN. Among these, B-Myb recognizes an N-terminal p107 region that overlaps with the larger cyclin-binding domain. In contrast, the E2F transcription factors bind the p107 C-terminal pocket domain (31). Moreover, Rb-dependent repression is mediated in part by the multisubunit protein complex Drosophila RBF, E2F, and Myb (dREAM), which contains homologs of the Caenorhabditis elegans synthetic multivulva class B (synMuvB) gene products (24). These reports suggest that the TAAC element might aid Rb binding to the promoters through Myb (Fig. 8).
FIGURE 8.
Model of CSIG, PCNA, and CCNA2 transcriptional regulation in growing and senescent cells. In growing cells, E2F1 transactivates its target genes, including CSIG. When cells undergo senescence, Rb binds to the CSIG/PCNA/CCNA2 promoters. Both the TAAC element and the E2F1 binding site are required for this association. Subsequently, heterochromatin forms at the CSIG/PCNA/CCNA2 promoters and irreversibly represses CSIG/PCNA/CCNA2 transcription.
In summary, our results provide a novel molecular explanation for differential transcriptional regulation of E2F1 targets and afford new mechanistic insights into the selectivity of Rb-mediated heterochromatin formation and gene repression during cellular senescence.
Acknowledgments
We thank Dr. M. Narita for introducing the retrovirus system and Dr. Scott Lowe, Dr. C. D. Lopez, Dr. J. Campisi, Dr. Jiandong Chen, and Dr. Hongti Jia for plasmids. We also thank members in Dr. M. Narita laboratory for stimulating discussions and helpful advice.
This work was supported by National Basic Research Programs of China Grants 2012CB911200 and 2013CB530801; National Natural Science Foundation of China Grants 30973146, 31000609, and 31071206; and Ministry of Education of the People's Republic of China Grant 20090001120044.

This article contains supplemental Table S1.
- PCNA
- proliferating cell nuclear antigen
- Rb
- retinoblastoma
- CSIG
- cellular senescence-inhibited gene
- SAHF
- senescence-associated heterochromatin foci
- sh
- short hairpin.
REFERENCES
- 1. Prieur A., Peeper D. S. (2008) Cellular senescence in vivo. A barrier to tumorigenesis. Curr. Opin. Cell Biol. 20, 150–155 [DOI] [PubMed] [Google Scholar]
- 2. Campisi J., d'Adda di Fagagna F. (2007) Cellular senescence. When bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 8, 729–740 [DOI] [PubMed] [Google Scholar]
- 3. Courtois-Cox S., Jones S. L., Cichowski K. (2008) Many roads lead to oncogene-induced senescence. Oncogene 27, 2801–2809 [DOI] [PubMed] [Google Scholar]
- 4. Burkhart D. L., Sage J. (2008) Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat. Rev. Cancer 8, 671–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Narita M., Nnez S., Heard E., Narita M., Lin A. W., Hearn S. A., Spector D. L., Hannon G. J., Lowe S. W. (2003) Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113, 703–716 [DOI] [PubMed] [Google Scholar]
- 6. Dimri G. P., Hara E., Campisi J. (1994) Regulation of two E2F-related genes in presenescent and senescent human fibroblasts. J. Biol. Chem. 269, 16180–16186 [PubMed] [Google Scholar]
- 7. Elliott M. J., Dong Y. B., Yang H., McMasters K. M. (2001) E2F-1 up-regulates c-Myc and p14(ARF) and induces apoptosis in colon cancer cells. Clin. Cancer Res. 7, 3590–3597 [PubMed] [Google Scholar]
- 8. Wang C., Hou X., Mohapatra S., Ma Y., Cress W. D., Pledger W. J., Chen J. (2005) Activation of p27Kip1 expression by E2F1. A negative feedback mechanism. J. Biol. Chem. 280, 12339–12343 [DOI] [PubMed] [Google Scholar]
- 9. Dimri G. P., Itahana K., Acosta M., Campisi J. (2000) Regulation of a senescence checkpoint response by the E2F1 transcription factor and p14(ARF) tumor suppressor. Mol. Cell Biol. 20, 273–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Haddad M. M., Xu W., Schwahn D. J., Liao F., Medrano E. E. (1999) Activation of a cAMP pathway and induction of melanogenesis correlate with association of p16(INK4) and p27(KIP1) to CDKs, loss of E2F-binding activity, and premature senescence of human melanocytes. Exp. Cell Res. 253, 561–572 [DOI] [PubMed] [Google Scholar]
- 11. Guo S., Zhang Z., Tong T. (2004) Cloning and characterization of cellular senescence-associated genes in human fibroblasts by suppression subtractive hybridization. Exp. Cell Res. 298, 465–472 [DOI] [PubMed] [Google Scholar]
- 12. Ma L., Chang N., Guo S., Li Q., Zhang Z., Wang W., Tong T. (2008) CSIG inhibits PTEN translation in replicative senescence. Mol. Cell Biol. 28, 6290–6301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tsai R. Y., McKay R. D. (2005) A multistep, GTP-driven mechanism controlling the dynamic cycling of nucleostemin. J. Cell Biol. 168, 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Narita M., Narita M., Krizhanovsky V., Nuñez S., Chicas A., Hearn S. A., Myers M. P., Lowe S. W. (2006) A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell 126, 503–514 [DOI] [PubMed] [Google Scholar]
- 15. Chicas A., Wang X., Zhang C., McCurrach M., Zhao Z., Mert O., Dickins R. A., Narita M., Zhang M., Lowe S. W. (2010) Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence. Cancer Cell 17, 376–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chaussepied M., Ginsberg D. (2004) Transcriptional regulation of AKT activation by E2F. Mol. Cell 16, 831–837 [DOI] [PubMed] [Google Scholar]
- 17. Serrano M., Lin A. W., McCurrach M. E., Beach D., Lowe S. W. (1997) Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88, 593–602 [DOI] [PubMed] [Google Scholar]
- 18. Humbert P. O., Verona R., Trimarchi J. M., Rogers C., Dandapani S., Lees J. A. (2000) E2f3 is critical for normal cellular proliferation. Genes Dev. 14, 690–703 [PMC free article] [PubMed] [Google Scholar]
- 19. Nahle Z., Polakoff J., Davuluri R. V., McCurrach M. E., Jacobson M. D., Narita M., Zhang M. Q., Lazebnik Y., Bar-Sagi D., Lowe S. W. (2002) Direct coupling of the cell cycle and cell death machinery by E2F. Nat. Cell Biol. 4, 859–864 [DOI] [PubMed] [Google Scholar]
- 20. Dahiya A., Wong S., Gonzalo S., Gavin M., Dean D. C. (2001) Linking the Rb and polycomb pathways. Mol. Cell 8, 557–569 [DOI] [PubMed] [Google Scholar]
- 21. McConnell B. B., Starborg M., Brookes S., Peters G. (1998) Inhibitors of cyclin-dependent kinases induce features of replicative senescence in early passage human diploid fibroblasts. Curr. Biol. 8, 351–354 [DOI] [PubMed] [Google Scholar]
- 22. Zhu J., Woods D., McMahon M., Bishop J. M. (1998) Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev. 12, 2997–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Litovchick L., Florens L. A., Swanson S. K., Washburn M. P., DeCaprio J. A. (2011) DYRK1A protein kinase promotes quiescence and senescence through DREAM complex assembly. Genes Dev. 25, 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Litovchick L., Sadasivam S., Florens L., Zhu X., Swanson S. K., Velmurugan S., Chen R., Washburn M. P., Liu X. S., DeCaprio J. A. (2007) Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol. Cell 26, 539–551 [DOI] [PubMed] [Google Scholar]
- 25. Mizuguchi G., Nakagoshi H., Nagase T., Nomura N., Date T., Ueno Y., Ishii S. (1990) DNA-Binding activity and transcriptional activator function of the human B-MYB protein compared with C-MYB B-2145–2011. J. Biol. Chem. 265, 9280–9284 [PubMed] [Google Scholar]
- 26. Saikumar P., Murali R., Reddy E. P. (1990) Role of tryptophan repeats and flanking amino acids in MYB DNA interactions. F-6233–2011. Proc. Natl. Acad. Sci. U.S.A. 87, 8452–8456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Alexander K., Hinds P. W. (2001) Requirement for p27(KIP1) in retinoblastoma protein-mediated senescence. Mol. Cell Biol. 21, 3616–3631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Itahana K., Zou Y., Itahana Y., Martinez J. L., Beausejour C., Jacobs J. J., Van Lohuizen M., Band V., Campisi J., Dimri G. P. (2003) Control of the replicative life span of human fibroblasts by p16 and the polycomb protein Bmi-1. Mol. Cell Biol. 23, 389–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Q., Wang X., Lu Z., Zhang B., Guan Z., Liu Z., Zhong Q., Gu L., Zhou J., Zhu B., Ji J., Deng D. (2010) Polycomb CBX7 directly controls trimethylation of histone H3 at lysine 9 at the p16 locus. PLoS One 5, e13732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhu L. (2010) Skp2 knockout reduces cell proliferation and mouse body size. And prevents cancer? Cell Res. 20, 605–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Joaquin M., Bessa M., Saville M. K., Watson R. J. (2002) B-Myb overcomes a p107-mediated cell proliferation block by interacting with an N-terminal domain of p107. Oncogene 21, 7923–7932 [DOI] [PubMed] [Google Scholar]
- 32. Zhang H. S., Postigo A. A., Dean D. C. (1999) Active transcriptional repression by the Rb-E2F complex mediates G1 arrest triggered by p16INK4a, TGFβ, and contact inhibition. Cell 97, 53–61 [DOI] [PubMed] [Google Scholar]
- 33. Gaubatz S., Lindeman G. J., Ishida S., Jakoi L., Nevins J. R., Livingston D. M., Rempel R. E. (2000) E2F4 and E2F5 play an essential role in pocket protein-mediated G1 control. Mol. Cell 6, 729–735 [DOI] [PubMed] [Google Scholar]
- 34. Peeper D. S., Upton T. M., Ladha M. H., Neuman E., Zalvide J., Bernards R., DeCaprio J. A., Ewen M. E. (1997) Ras signalling linked to the cell-cycle machinery by the retinoblastoma protein. Nature 386, 177–181 [DOI] [PubMed] [Google Scholar]
- 35. Leone G., DeGregori J., Sears R., Jakoi L., Nevins J. R. (1997) Myc and Ras collaborate in inducing accumulation of active cyclin E/Cdk2 and E2F. Nature 387, 422–426 [DOI] [PubMed] [Google Scholar]
- 36. Mittnacht S., Paterson H., Olson M. F., Marshall C. J. (1997) Ras signalling is required for inactivation of the tumour suppressor pRb cell-cycle control protein. Curr. Biol. 7, 219–221 [DOI] [PubMed] [Google Scholar]
- 37. Iaquinta P. J., Aslanian A., Lees J. A. (2005) Regulation of the Arf/p53 tumor surveillance network by E2F. Cold Spring Harb. Symp. Quant. Biol. 70, 309–316 [DOI] [PubMed] [Google Scholar]