Abstract
Purpose
Mutations in the epidermal growth factor receptor (EGFR) have been confirmed as predictors of the efficacy of treatment with EGFR-tyrosine kinase inhibitors (TKIs). We investigated whether polymorphisms of the EGFR gene were associated with clinical outcomes in non-small cell lung cancer (NSCLC) patients treated with EGFR-TKI.
Materials and Methods
A polymorphic dinucleotide repeat in intron 1 [CA simple sequence repeat in intron 1(CA-SSR1)] in intron 1 and single nucleotide polymorphisms (SNP-216) in the promoter region of the EGFR gene were evaluated in 71 NSCLC patients by restriction fragment length polymorphism and DNA sequencing. The relationship between genetic polymorphisms and clinical outcomes of treatment with EGFR-TKIs was evaluated.
Results
SNP-216G/T polymorphisms were associated with the efficacy of EGFR-TKI. The response rate for the SNP-216G/T tended to be higher than that for G/G (62.5% vs. 27.4%, p=0.057). The SNP-216G/T genotype was also associated with longer progression-free survival compared with the GG genotype (16.7 months vs. 5.1 months, p=0.005). However, the length of CA-SSR1 was not associated with the efficacy of EGFR-TKI.
Conclusion
SNP-216G/T polymorphism was a potential predictor of clinical outcomes in NSCLC patients treated with EGFR-TKI.
Keywords: Polymorphism, lung cancer, EGFR tyrosine kinase inhibitor
INTRODUCTION
Small molecule epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs), such as erlotinib and gefitinib, were initially developed for use as a second-line therapy after failure of cytotoxic chemotherapy regimens.1,2 In this setting, erlotinib was shown to increase survival, with the magnitude of benefit similar to that of second-line chemotherapy.1 Both clinical and molecular parameters are helpful in predicting which patients with advanced non-small cell lung cancer (NSCLC) are most likely to benefit from treatment with EGFR-TKIs. The clinical parameters associated with response to EGFR-TKI therapy include adenocarcinoma, female sex, non-smokers and Asian ethnicity.3-5 In addition, specific activating mutations in the TK domain of the EGFR are associated with increased responsiveness to EGFR-TKIs.6-9
However, a previous study found that gefitinib was not effective in EGFR mutation-negative patients, although the patients were expected to demonstrate a good response to treatment with EGFR TKIs, as all patients had adenocarcinoma, were Asian, and were either never smokers or former light smokers.10,11 Therefore, the most exact predictive marker of EGFR-TKIs may be EGFR mutation. Unfortunately, the detection of an EGFR mutation is difficult due to a limited amount of available tissue.12 Thus, another biomarker that can improve the prediction of response to these targeted drugs is needed.
Recently, EGFR amplification has been identified as a predictive marker for response to EGFR-TKI therapy.13,14 The polymorphisms of the gene may regulate protein expression. The CA simple sequence repeat in intron 1 (CA-SSR1) is a highly polymorphic dinucleotide CA repeat in intron 1 of the EGFR gene that is related to transcriptional activity and may predict the outcome of EGFR-TKI therapy in NSCLC patients.15,16 In addition, single nucleotide polymorphisms (SNPs) in the promoter region of the EGFR gene may correlate with increased promoter activity and EGFR expression. One such SNP, SNP-216, is located 216 base pairs upstream from the initiator ATG and exerts a strong influence on EGFR transcription in vivo.17
Therefore, the length of CA-SSR1 and the genotype of SNP-216 may affect EGFR expression and function, which may determine outcomes of EGFR-TKI treatment. In addition, polymorphisms are consistent features that can easily be assessed from blood cells, therefore making them useful markers, especially when the patients do not have available tissue. Moreover, there are allele frequency differences between ethnic groups, which may contribute to different drug responses. Thus, we investigated whether these two genotypes of EGFR can serve as predictive markers for clinical outcomes in Korean NSCLC patients treated with EGFR-TKIs.
MATERIALS AND METHODS
Eligible patients and treatment
In this study, 71 patients with advanced NSCLC were enrolled. Eligible patients had at least one measurable lesion, an Eastern Cooperative Oncology Group (ECOG) performance status (PS) of 0-2, and each patient received gefitinib or erlotinib after receiving prior chemotherapy treatment at Yonsei Cancer Center, Yonsei University College of Medicine, Seoul, Korea, from January 2007 to December 2010. The variables used in the pretreatment analysis were age, sex, clinical stage, ECOG PS, histological type, smoking history, number of prior chemotherapy regimens, and if possible, EGFR mutation status. Histological analysis of tumors was based on the WHO classification for cell types.18 Patients received a daily dose of 150 mg of erlotinib or 250 mg of gefitinib. Erlotinib or gefitinib was continued until disease progression, intolerable toxicity, or patient refusal. Patients were evaluated every eight weeks by computed tomography and clinical responses were defined according to the RECIST 1.1 response evaluation criteria for patients with measurable disease.19
EGFR mutation detection and genotyping
EGFR mutation detection methodologies have been published elsewhere, and we sequenced exons 18-21 of the TK domain of EGFR in tumors.20 For detection of EGFR polymorphisms, genomic DNA was purified from leukocytes after selective lysis of erythrocytes using an automated DNA extractor, according to the manufacturer's instructions (Applied Biosystems, Foster City, CA, USA). Genotyping of the promoter region of EGFR-216 was performed using polymerase chain reaction (PCR), and polymorphism assignment was determined by restriction enzyme digestion using previously described methods.17 CA-SSR1 was amplified by PCR and sequenced using a previously reported method.15
Statistical methods
The association between the presence of CA-SSR1 or SNP-216 and other categorical clinical variables was assessed using the χ2 test or Fisher's exact test. Progression-free survival (PFS) was defined as the time from the start date of TKI treatment until the date of tumor progression or death. Overall survival (OS) was measured from the start date of TKI therapy to the date of death or final follow-up. Survival was estimated using a Kaplan-Meier curve and compared using the log-rank test. A p-value of less than 0.05 was considered statistically significant.
RESULTS
Patient characteristics
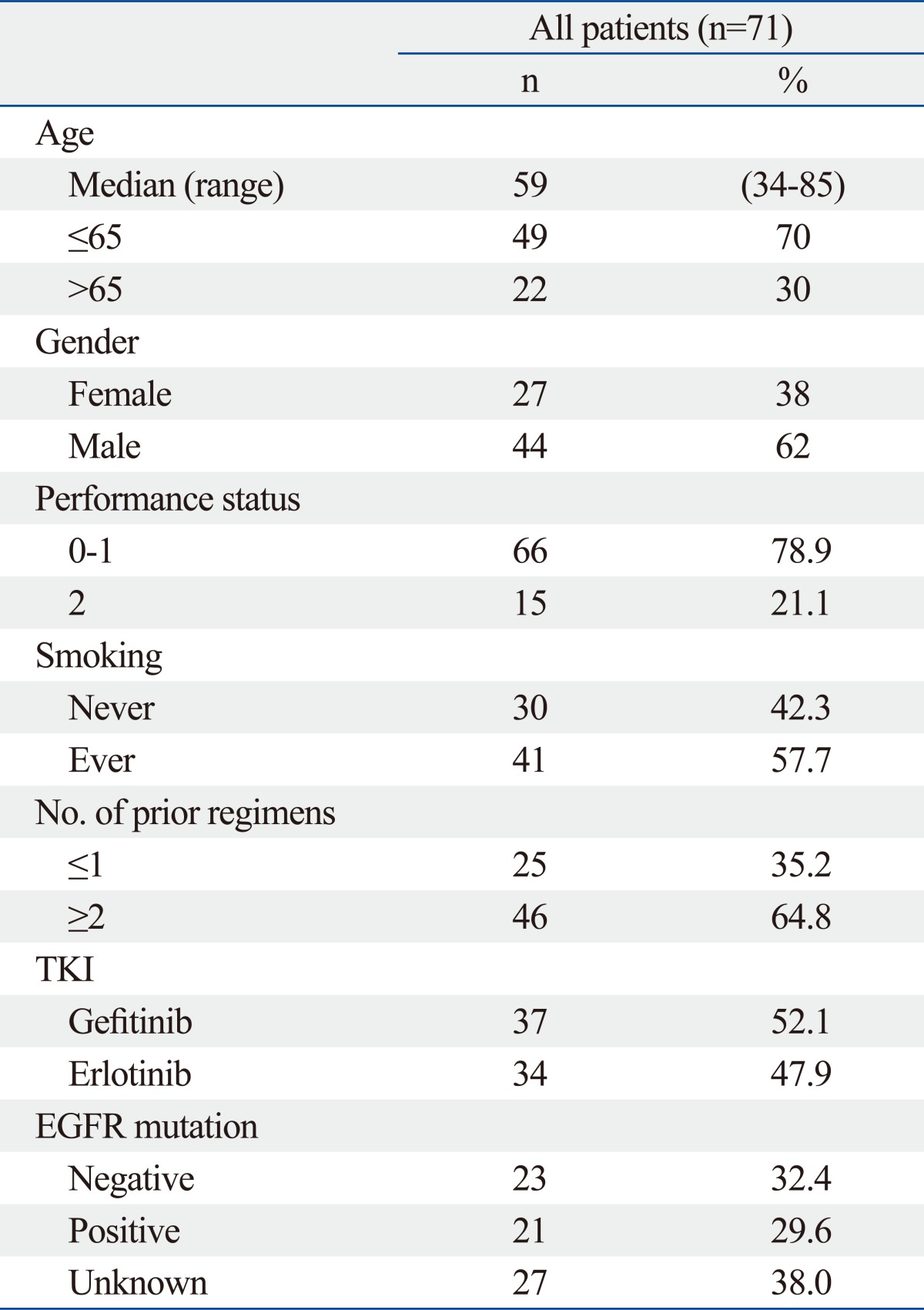
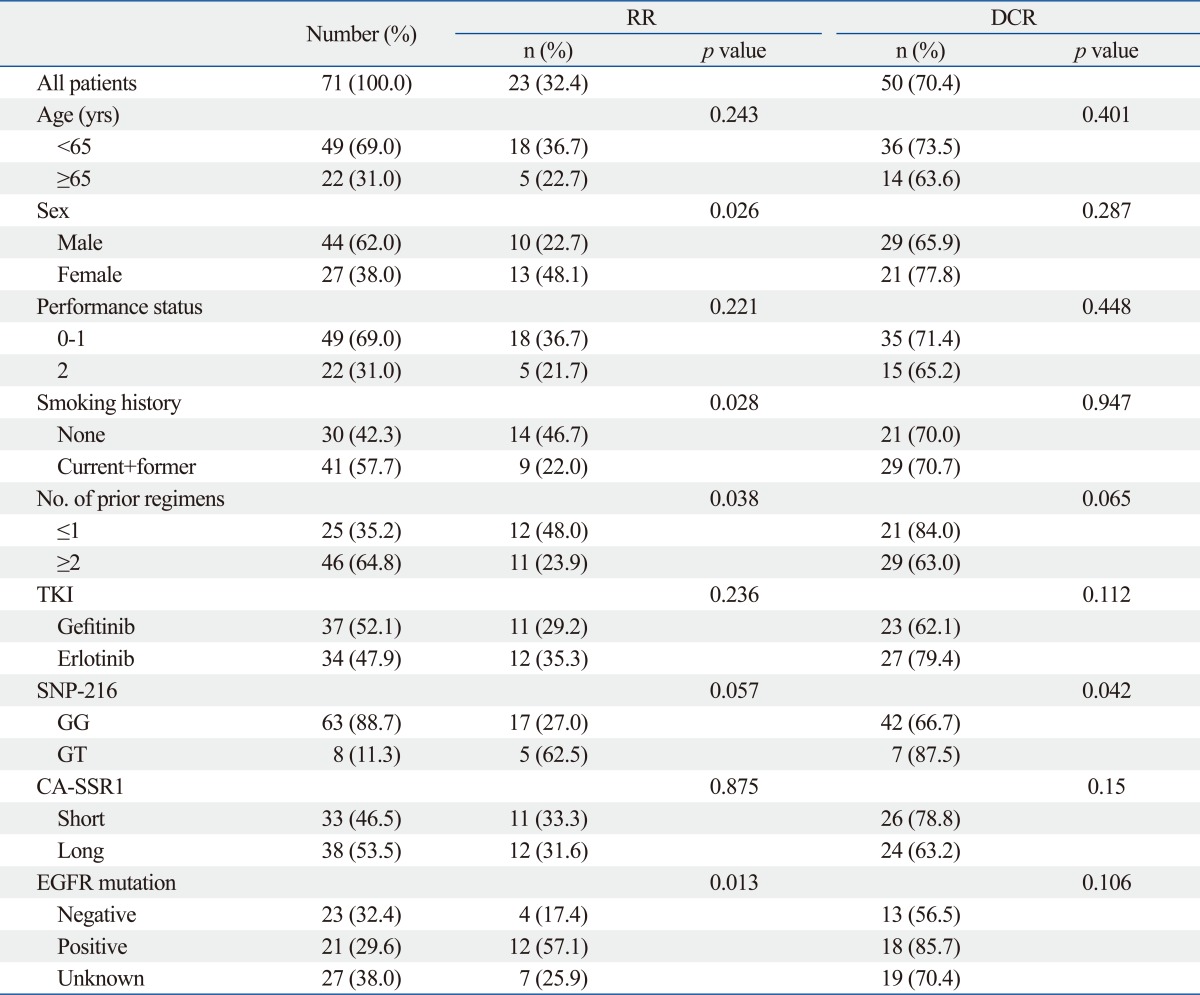
The demographic characteristics of the 71 subjects are shown in Table 1. The median age of the patients was 59 years (range, 34-85). Females and never smokers accounted for 38% and 42.3% of patients, respectively. EGFR mutation status was analyzed in 44 of 71 patients (62%), and 21 of the 44 patients (44.7%) were positive. Overall, 37 patients (52.1%) were treated with gefitinib.
Table 1.
Patient Baseline Characteristics
TKI, tyrosine kinase inhibitor; EGFR, epidermal growth factor receptor.
Genotyping of EGFR
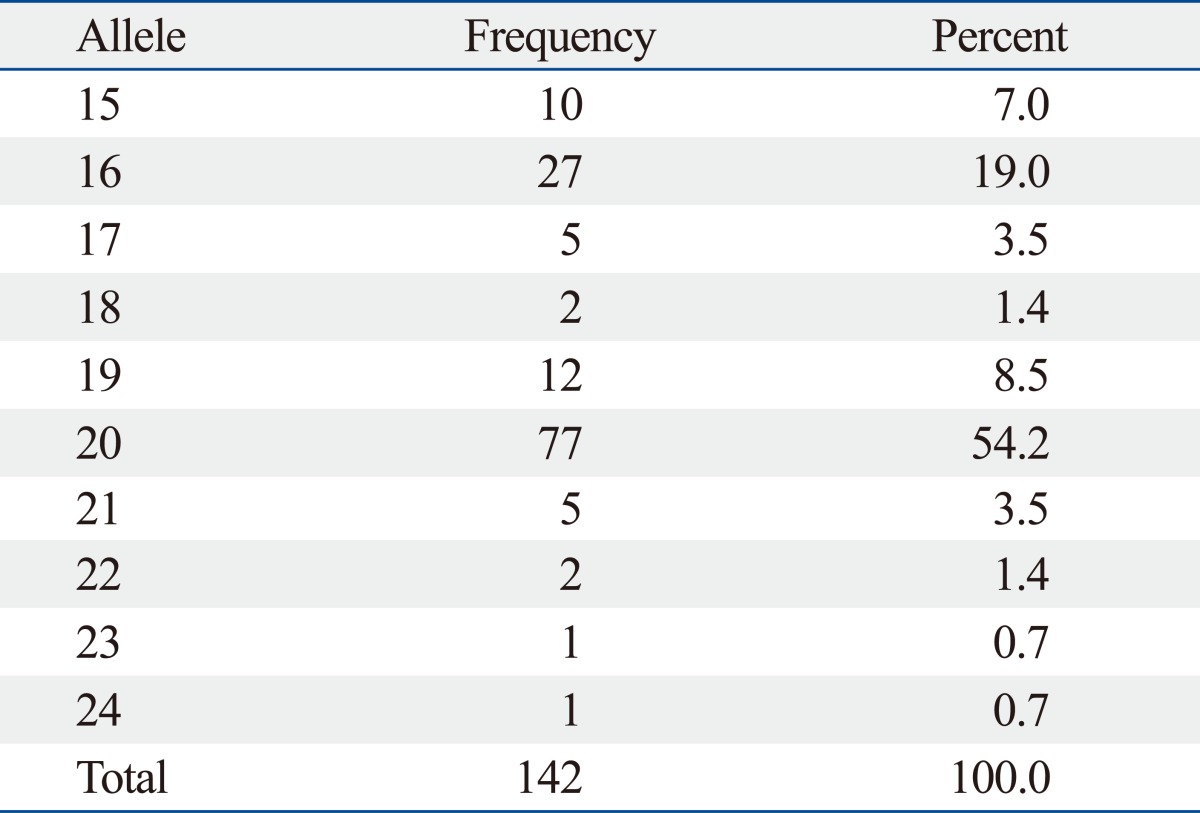
In 142 chromosomes, the median number of CA repeats was 20 (range, 15-24) (Table 2). The most frequent CA-SSR1 genotype was 20/20 repeats found in 18 patients (25.3%); allele 16/16 was found in only two patients (2.8%). The median of the sum of CA repeat numbers in both alleles was 37 (range, 31-44), and we classified patients as "longer" for those with greater than 37 repeats and as "shorter" for those with 37 repeats or fewer, according to the median of the sum of repeat numbers in both alleles based on a previous study.15 For SNP-216 polymorphism, 63 patients (88.7%) exhibited the GG genotype; GT was present in only 8 patients (11.3%); and TT was present in none.
Table 2.
Allele Distribution of CA-SSR1 Repeat Length
CA-SSR1, CA simple sequence repeat in intron 1.
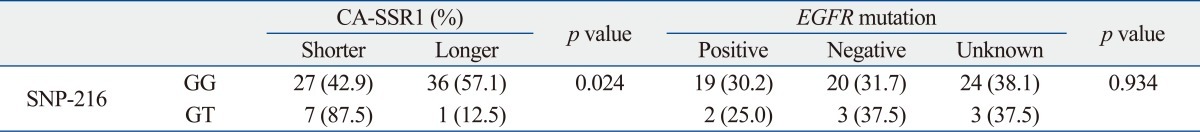
There were no differences in clinical characteristics according to CA-SSR1 and SNP-216 genotypes (data not shown). EGFR mutation positivity was not associated with CA repeats and SNP-216. However, seven of eight patients who had the GT genotype of SNP-216 exhibited a shorter CA-SSR1 (Table 3).
Table 3.
The Relationship between SNP-216 and Repeat Length of CA-SSR1 or EGFR Mutation
SNP, single nucleotide polymorphism; CA-SSR1, CA simple sequence repeat in intron 1; EGFR, epidermal growth factor receptor.
Genotypes and response to EGFR-TKI therapy
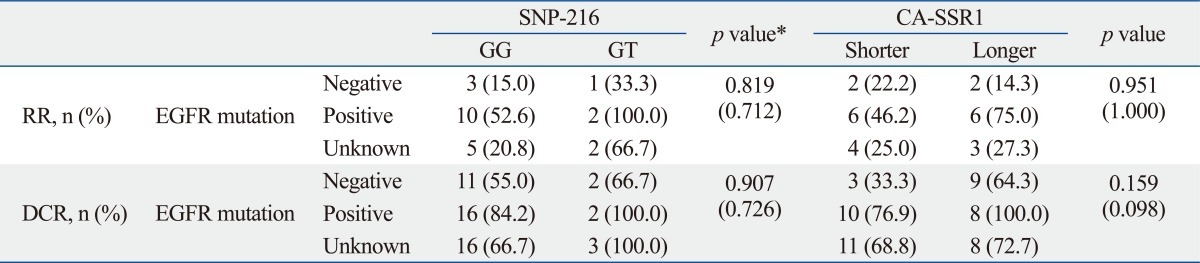
Responses were evaluated in all 71 patients. A partial response was noted in 23 patients, generating an RR of 32.4%, and an additional 27 patients (37.6%) demonstrated the best response of stable disease. No significant association was found between response and the length of CA repeats. Also, there was no difference in the relationship between SNP-216 or CA-SSRI polymorphism and the efficacy of EGFR-TKIs according to EGFR mutation status (Supplementary Table 1). Patients with the GT genotype of SNP-216 tend to show higher response rates than patients with the GG genotype (62.5% vs. 27%, p=0.057). In addition, patients with the GT genotype had a higher disease control rate than those with GG (87.5% vs. 66.7%, p=0.042). Better responses were also achieved in those that never smoked, had received TKI therapy as a second line therapy, were positive for the EGFR mutation, and were female (Table 4).
Supplementary Table 1.
The Relationship between SNP-216 or CA-SSR1 Polymorphism and the Efficacy of EGFR-TKIs according to EGFR Mutation Status
TKI, tyrosine kinase inhibitor; SNP, single nucleotide polymorphism; CA-SSR1, CA simple sequence repeat in intron 1; EGFR, epidermal growth factor receptor; RR, response rate; DCR, disease-control rate.
*p values were obtained by comparing between EGFR mutation and SNP-216 or CA- SSR1 polymorphism using the chi-square or Fisher's exact test (expected cell value <5). Data in the '( )' represent the p values by comparing the EGFR mutation negative and positive group.
Table 4.
Correlation of Baseline Characteristics and Genetic Polymorphism with Response to EGFR-TKIs
TKI, tyrosine kinase inhibitor; SNP, single nucleotide polymorphism; CA-SSR1, CA simple sequence repeat in intron 1; EGFR, epidermal growth factor receptor; RR, response rate; DCR, disease-control rate.
Genotypes and survival for EGFR-TKI therapy
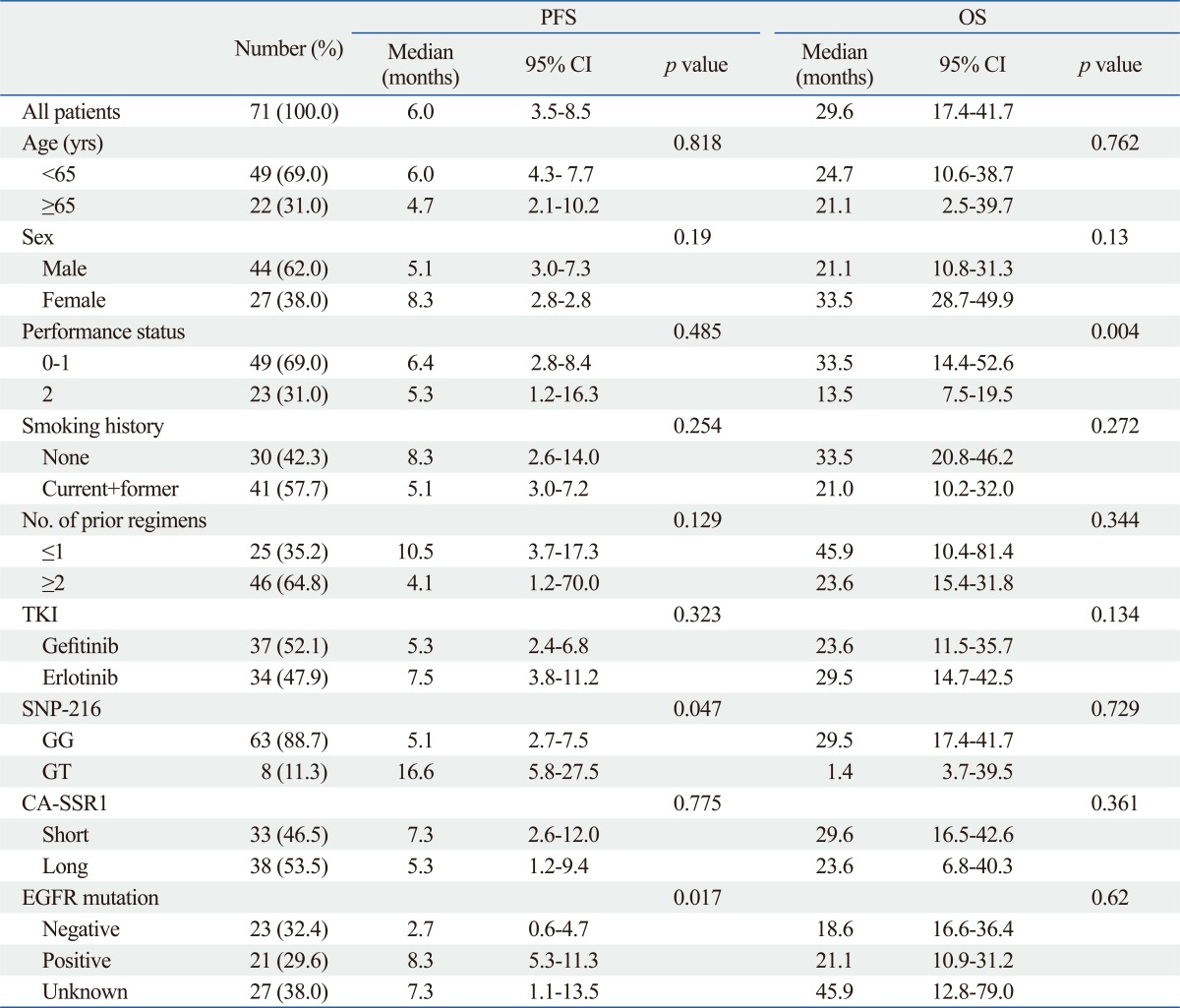
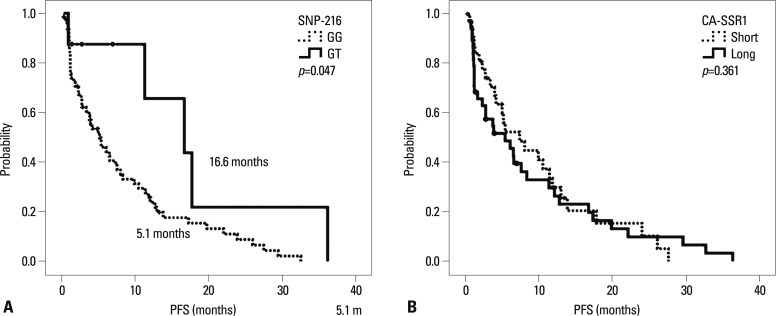
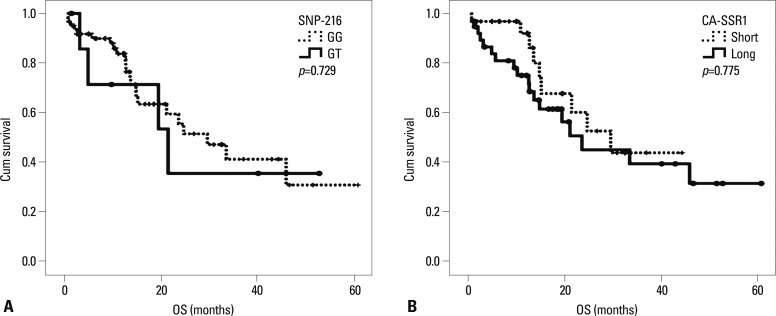
The median follow-up duration for all 71 patients was 12.7 months (range, 1.1-60.8 months). The median PFS was 6.0 months (95% CI, 3.5-8.5 months), and the median OS was 29.6 months (95% CI, 17.4-41.7 months). Patients with the GT genotype of SNP-216 had significantly longer PFS than those with the GG genotype (16.6 months vs. 5.1 months, p=0.047). In addition, the patients with an EGFR mutation reported longer survival than those without (8.3 months vs. 2.7 months, p=0.017). Notably, among the patients for whom EGFR mutation status was unknown, the patients with SNP-216G/T exhibited a longer survival than those with SNP-216-G/G (median PFS 17.7 months vs. 5.3 months, p=0.068). However, there was no difference in PFS according to the lengths of the CA repeat (Table 5) (Fig. 1). The patients with good performance status exhibited longer overall survival than those with poor performance status, and there was no difference in overall survival according to other clinical characteristics and genotypes (Fig. 2).
Table 5.
Correlation of Baseline Characteristics and Genetic Polymorphism with Survival
TKI, tyrosine kinase inhibitor; SNP, single nucleotide polymorphism; CA-SSR1, CA simple sequence repeat in intron 1; EGFR, epidermal growth factor receptor; PFS, progression-free survival; OS, overall survival; CI, confidential interval.
Fig. 1.
Kaplan-Meier curve of progression-free survival according to genotype. (A) SNP-216. (B) CA-SSR1 length. SNP, single nucleotide polymorphism; CA-SSR1, CA simple sequence repeat in intron 1; PFS, progression-free survival.
Fig. 2.
Kaplan-Meier curve of overall survival according to genotype. (A) SNP-216. (B) CA-SSR1 length. SNP, single nucleotide polymorphism; CA-SSR1, CA simple sequence repeat in intron 1; OS, overall survival.
DISCUSSION
We hypothesized that genetic polymorphisms of EGFR might be associated with the efficacy of EGFR-TKI treatment in advanced NSCLC. We found that the incidence of patients with the G/T genotype of SNP-216, a minor form of the SNP, was very low (11.8%), and these patients had a better response to and longer survival after EGFR-TKI therapy. However, the CA-SSR1 was longer than that seen in a Western population15 and was not associated with response to EGFR-TKI.
The EGFR gene comprises a highly polymorphic sequence in intron 1 with variable numbers of the CA dinucleotide simple repeat, ranging from 9 to 22. Previous studies reported that patients with shorter CA-SSR1 demonstrated better responses and longer survival than those with longer repeats.15,16,21 However, in the current study, there was no difference in the efficacy of EGFR-TKI according to the length of CA-SSR1. The length of CA-SSR1 polymorphism differs by ethnicity and tends to be longer in Asians than in Europeans or African-Americans.22 In the current study, the lack of an association between the length of CA-SSR1 and the efficacy of EGFR-TKI therapy may be due to the small number of patients with shorter CA-SSR1 in our cohort.
In addition to CA-SSR1, there are two kinds of SNPs in the promoter region of EGFR that might be associated with promoter activity and mRNA expression. One of the SNPs is located 191 base pairs upstream from the initiator ATG and may be correlated with increased protein expression; however, the minor forms, -191C/A and A/A, are extremely rare among Asians. Liu, et al.17 reported that no patients in an Asian cohort had SNP-191C/A or A/A, and Han, et al.23 reported there was only one in a group of 71 NSCLC patients studied. Therefore, we did not analyze the sequence of the -191 region.
The other SNP, 216 base pairs upstream from the initiator, is in a region that encodes an important binding site for the transcription factor Sp1, which is necessary for activation of the EGFR gene. The incidence of SNP-216 G/T was low (11.3%) in the current study, and these patients tend to show higher responses (p=0.057) and a significantly higher disease control rate (p=0.042) than the others. This result was similar to a previous report.24 In addition, the patients with SNP-216 G/T showed a longer PFS (p=0.047) than those with the GG genotype, especially among the patients for whom the EGFR mutation status was unknown. This result suggests that the SNP-216 genotype could be used as a predictive marker for response to EGFR-TKI therapy, especially among patients that do not have sufficient tumor tissue for EGFR mutation test. However, the OS was not different according to SNP-216, possibly due to additional treatment administered after EGFR-TKI therapy. Of the 71 total patients in the current study, 25 (35.2%) were treated with EGFR-TKI as a second line chemotherapy and 46 (64.8%) received further treatment after EGFR-TKI therapy.
In conclusion, this study showed that SNP-216 in the promoter region of the EGFR gene might be a predictive maker of EGFR TKI treatment effectiveness. Though the most important predictive marker for EGFR TKI effectiveness is EGFR mutation, some patients do not have sufficient tissue available for EGFR mutation test. Our findings are important to patients in whom EGFR mutation status is unknown due to lack of sufficient tumor tissue.
ACKNOWLEDGEMENTS
This study was supported by a faculty research grant from Yonsei University College of Medicine (6-2010-0061).
Footnotes
Previously presented: Material from this manuscript was presented as a poster presentation at the AACR-IASLC Joint Conference on Molecular Origins of Lung Cancer on January 10, 2012.
The authors have no financial conflicts of interest.
References
- 1.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, Tan EH, Hirsh V, Thongprasert S, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005;353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 2.Fukuoka M, Yano S, Giaccone G, Tamura T, Nakagawa K, Douillard JY, et al. Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced non-small-cell lung cancer (The IDEAL 1 Trial) [corrected] J Clin Oncol. 2003;21:2237–2246. doi: 10.1200/JCO.2003.10.038. [DOI] [PubMed] [Google Scholar]
- 3.Pérez-Soler R, Chachoua A, Hammond LA, Rowinsky EK, Huberman M, Karp D, et al. Determinants of tumor response and survival with erlotinib in patients with non--small-cell lung cancer. J Clin Oncol. 2004;22:3238–3247. doi: 10.1200/JCO.2004.11.057. [DOI] [PubMed] [Google Scholar]
- 4.Miller VA, Kris MG, Shah N, Patel J, Azzoli C, Gomez J, et al. Bronchioloalveolar pathologic subtype and smoking history predict sensitivity to gefitinib in advanced non-small-cell lung cancer. J Clin Oncol. 2004;22:1103–1109. doi: 10.1200/JCO.2004.08.158. [DOI] [PubMed] [Google Scholar]
- 5.West HL, Franklin WA, McCoy J, Gumerlock PH, Vance R, Lau DH, et al. Gefitinib therapy in advanced bronchioloalveolar carcinoma: Southwest Oncology Group Study S0126. J Clin Oncol. 2006;24:1807–1813. doi: 10.1200/JCO.2005.04.9890. [DOI] [PubMed] [Google Scholar]
- 6.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, et al. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 8.Tokumo M, Toyooka S, Kiura K, Shigematsu H, Tomii K, Aoe M, et al. The relationship between epidermal growth factor receptor mutations and clinicopathologic features in non-small cell lung cancers. Clin Cancer Res. 2005;11:1167–1173. [PubMed] [Google Scholar]
- 9.Yang CH, Yu CJ, Shih JY, Chang YC, Hu FC, Tsai MC, et al. Specific EGFR mutations predict treatment outcome of stage IIIB/IV patients with chemotherapy-naive non-small-cell lung cancer receiving first-line gefitinib monotherapy. J Clin Oncol. 2008;26:2745–2753. doi: 10.1200/JCO.2007.15.6695. [DOI] [PubMed] [Google Scholar]
- 10.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 11.Fukuoka M, Wu YL, Thongprasert S, Sunpaweravong P, Leong SS, Sriuranpong V, et al. Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS) J Clin Oncol. 2011;29:2866–2874. doi: 10.1200/JCO.2010.33.4235. [DOI] [PubMed] [Google Scholar]
- 12.Hlinkova K, Babál P, Berzinec P, Majer I, Ilencikova D. Rapid and efficient detection of EGFR mutations in problematic cytologic specimens by high-resolution melting analysis. Mol Diagn Ther. 2011;15:21–29. doi: 10.1007/BF03257190. [DOI] [PubMed] [Google Scholar]
- 13.Tsao MS, Sakurada A, Cutz JC, Zhu CQ, Kamel-Reid S, Squire J, et al. Erlotinib in lung cancer - molecular and clinical predictors of outcome. N Engl J Med. 2005;353:133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 14.Cappuzzo F, Hirsch FR, Rossi E, Bartolini S, Ceresoli GL, Bemis L, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst. 2005;97:643–655. doi: 10.1093/jnci/dji112. [DOI] [PubMed] [Google Scholar]
- 15.Nomura M, Shigematsu H, Li L, Suzuki M, Takahashi T, Estess P, et al. Polymorphisms, mutations, and amplification of the EGFR gene in non-small cell lung cancers. PLoS Med. 2007;4:e125. doi: 10.1371/journal.pmed.0040125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nie Q, Yang XN, An SJ, Zhang XC, Yang JJ, Zhong WZ, et al. CYP1A1*2A polymorphism as a predictor of clinical outcome in advanced lung cancer patients treated with EGFR-TKI and its combined effects with EGFR intron 1 (CA)n polymorphism. Eur J Cancer. 2011;47:1962–1970. doi: 10.1016/j.ejca.2011.04.018. [DOI] [PubMed] [Google Scholar]
- 17.Liu W, Innocenti F, Wu MH, Desai AA, Dolan ME, Cook EH, Jr, et al. A functional common polymorphism in a Sp1 recognition site of the epidermal growth factor receptor gene promoter. Cancer Res. 2005;65:46–53. [PubMed] [Google Scholar]
- 18.Travis WD, Colby TV, Corrin B, Shimosato Y, Brambilla E. Histological typing of lung and pleural tumours. 3rd ed. Berlin: Springer-Verlag; 1999. [Google Scholar]
- 19.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, et al. New Guidelines to Evaluate the Response to Treatment in Solid Tumors. Oxford University Press; 2000. pp. 205–216. [DOI] [PubMed] [Google Scholar]
- 20.Shigematsu H, Lin L, Takahashi T, Nomura M, Suzuki M, Wistuba II, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 21.Amador ML, Oppenheimer D, Perea S, Maitra A, Cusatis G, Iacobuzio-Donahue C, et al. An epidermal growth factor receptor intron 1 polymorphism mediates response to epidermal growth factor receptor inhibitors. Cancer Res. 2004;64:9139–9143. doi: 10.1158/0008-5472.CAN-04-1036. [DOI] [PubMed] [Google Scholar]
- 22.Liu W, Innocenti F, Chen P, Das S, Cook EH, Jr, Ratain MJ. Interethnic difference in the allelic distribution of human epidermal growth factor receptor intron 1 polymorphism. Clin Cancer Res. 2003;9:1009–1012. [PubMed] [Google Scholar]
- 23.Han SW, Jeon YK, Lee KH, Keam B, Hwang PG, Oh DY, et al. Intron 1 CA dinucleotide repeat polymorphism and mutations of epidermal growth factor receptor and gefitinib responsiveness in non-small-cell lung cancer. Pharmacogenet Genomics. 2007;17:313–319. doi: 10.1097/FPC.0b013e328011abc0. [DOI] [PubMed] [Google Scholar]
- 24.Puyo S, Le Morvan V, Robert J. Impact of EGFR gene polymorphisms on anticancer drug cytotoxicity in vitro. Mol Diagn Ther. 2008;12:225–234. doi: 10.1007/BF03256288. [DOI] [PubMed] [Google Scholar]