Abstract
The spatial and temporal expression patterns of metallothionein (MT) isoforms MT1a and MT2a were investigated in vegetative and reproductive tissues of untreated and copper-treated Arabidopsis by in situ hybridization and by northern blotting. In control plants, MT1a mRNA was localized in leaf trichomes and in the vascular tissue in leaves, roots, flowers, and germinating embryos. In copper-treated plants, MT1a expression was also observed in the leaf mesophyll and in vascular tissue of developing siliques and seeds. In contrast, MT2a was expressed primarily in the trichomes of both untreated and copper-treated plants. In copper-treated plants, MT2a mRNA was also expressed in siliques. Northern-hybridization studies performed on developing seedlings and leaves showed temporal variations of MT1a gene expression but not of MT2a expression. The possible implications of these findings for the cellular roles of MTs in plants are discussed.
MTs are defined as low-Mr, Cys-rich proteins that bind heavy metals. MTs are widely distributed in eukaryotic and prokaryotic organisms (for review, see Kagi, 1991; Robinson et al., 1993). In animals and fungi MTs have been shown to play a role in the detoxification of heavy metals, although their exact function is not completely understood. In plants a correlation has been observed between MT RNA levels and tolerance to heavy metals in different Arabidopsis ecotypes (Murphy and Taiz, 1995a), suggesting a role in metal homeostasis in plants.
In animals and yeast MT expression is regulated by metals (Robinson et al., 1993). In plants the effect of metals on the expression of MTs varies with the plant species, tissue, and MT type. In Mimulus guttatus (de Miranda, 1990), soybean (Kawashima, 1991), and barley (Okumura et al., 1991), MT mRNA levels were decreased by copper treatment, whereas in bean (Foley and Singh, 1994; Foley et al., 1997), wheat germ (Lane et al., 1987), and Nicotiana glutinosa (Choi et al., 1996), MT expression was not affected by metals. In Arabidopsis (Zhou and Goldsbrough, 1994, 1995; Murphy and Taiz, 1995a), wheat (Snowden and Gardner, 1993), pea (Evans et al., 1992), and rice (Hsieh et al., 1995), transcription of MTs was enhanced by certain metals only. As in animals (Robinson et al., 1993), a variety of other stimuli, including ABA, heat shock, cold shock, wounding, viral infection, senescence, salt stress, and Suc starvation, have been shown to influence expression of plant MTs (Buchanan-Wollaston, 1994, 1997; Foley and Singh, 1994; Hsieh et al., 1995; Murphy and Taiz, 1995a; Snowden et al., 1995; Choi et al., 1996; Foley et al., 1997).
In Arabidopsis, three MT gene families have been identified: MT1, MT2, and MT3 (Zhou and Goldsbrough, 1994; Murphy et al., 1997), homologs of which have been identified in other species. The data available regarding the expression of MT genes from a variety of plant species indicate that each MT gene type exhibits characteristic temporal and tissue-specific expression patterns. Expression of most MT1-like sequences has been detected primarily in roots (de Miranda et al., 1990; de Framond, 1991; Evans et al., 1992; Zhou and Goldsbrough, 1994; Hsieh et al., 1995; Hudspeth et al., 1996) and senescent leaves (Kawashima et al., 1991; Buchanan-Wollaston, 1994, 1997; Hsieh et al., 1995; Foley et al., 1997). MT2-type transcripts have been detected primarily in leaves (Snowden and Gardner, 1993; Foley and Singh, 1994; Zhou and Goldsbrough, 1994, 1995; Coupe et al., 1995; Choi et al., 1996) and roots of mature plants (Zhou, 1994; Snowden et al., 1995; Murphy, 1996). MT3-like mRNAs have been detected in leaves (Murphy, 1996; Bundithya and Goldsbrough, 1997), fruits (Ledger and Gardner, 1994), and developing embryos (Dong and Dunstan, 1996).
In Arabidopsis each MT type appears to belong to a small gene family, the members of which appear to exhibit differential gene expression patterns. MT1 consists of three isoforms, MT1a, MT1b, and MT1c. MT1a is constitutively expressed in seedlings and is induced by copper in excised leaves, whereas MT1b seems to be a pseudogene. MT1c is expressed in young and mature roots and in mature leaves and is not affected by copper treatment. The MT2 gene family consists of MT2a and MT2b. They are both constitutively expressed in mature leaves, and only the MT2a gene is copper inducible in seedlings (Zhou and Goldsbrough, 1994; Murphy and Taiz, 1995a; Zhou and Goldsbrough, 1995; Murphy et al., 1997). Immunocytochemical studies have recently shown similar patterns of accumulation of the gene products (Murphy et al., 1997). All of these data suggest that each MT isoform may have specialized functions in different tissues. Some of the functions proposed for plant MTs include a role during development (Kawashima et al., 1992; Ledger and Gardner, 1994; Dong and Dunstan, 1996), in senescence (Buchanan-Wollatson, 1994; Coupe et al., 1995; Hsieh et al., 1995), and in protection against oxidative stress (Choi et al., 1996).
To date, in situ-hybridization studies of metallothionein gene expression have been performed in just two plant species, bean and wheat, and only for MT2 transcripts. In bean MT2 expression was localized specifically in foliar trichomes and veins (Foley and Singh, 1994). In wheat the MT2-like WALI 1 gene was specifically expressed in the apical meristem of roots (Snowden et al., 1995).
Additional information about the tissue-specific expression of MTs is needed to help clarify the biological function(s) of MT genes in plants. We focused our studies on Arabidopsis because its response to copper is well characterized, and because it is the only plant species in which more than one MT gene family has been identified. To further characterize the developmental regulation of MTs in Arabidopsis, we also performed northern-hybridization analysis on developing seedlings and aging leaves. Our results have confirmed that there are differences in the spatial localization of MT1a and MT2a expression in the tissues examined. MT1a was expressed in most vegetative and reproductive organs in vascular tissues and in trichomes, whereas MT2a was predominantly expressed in leaf trichomes. Moreover, developmental studies showed a very distinct pattern of expression for the two MT genes.
MATERIALS AND METHODS
Plant Material
Arabidopsis ecotype Wassilewskija (Ws) was used in all experiments. Plants were grown in a medium consisting of 1.1 g/L Murashige-Skoog basal salt mixture (Sigma) and 1 mm Mes, pH 4.8. For growth periods of less than 8 d, the seedlings were germinated using the vertical mesh transfer system (Murphy and Taiz, 1995a). For longer growth periods, the Aquamist hydroponic system (Pure Food Hydroponics, San Jose, CA) was used. For copper treatment, 40 μm CuCl2 was added to the growth medium 36 h before harvesting.
Preparation of Probes
The coding and flanking untranslated regions of MT1a and MT2a cDNAs were amplified, using sequence-specific primers, by RT-PCR from total RNA prepared from 30 μm CuCl2-treated seedlings of the Ws ecotype as previously described (Murphy and Taiz, 1995a). The MT1a RT-PCR product contained the complete translated region (135 residues) plus 24 nucleotides upstream of the start codon and 182 nucleotides of the 3′ untranslated region. The MT2a PCR product consisted of the complete translated region (236 bp) plus 12 nucleotides upstream of the start codon and 224 nucleotides of the 3′ untranslated region. Both sequences were cloned into the pZero-2 plasmid (Invitrogen, San Diego, CA) in the antisense orientation. The orientation of the inserts was verified by dye-termination dideoxy sequencing utilizing a DNA sequencer (ABI 310, Applied Biosystems) with M13 forward and reverse primers.
The clone encoding the 33-kD PSII-binding protein O from Arabidopsis (psbO) was a gift from Neil Hoffman (Carnegie Institution of Washington, Stanford, CA). The psbO cDNA was cloned into pGEM4 and contained the complete translated region flanked by 5′ and 3′ untranslated regions of 80 and 135 nucleotides, respectively.
The MT1a and MT2a cDNA sequences were amplified by PCR with M13 sequencing primers and digested with the appropriate restriction enzymes (SacI for sense MT1a, XhoI for antisense MT1a, HindIII for sense MT2, and NotI for antisense MT2) to remove most of the remaining vector sequences. The psbO clone was linearized with BamHI to produce a template of 386 nucleotides. The purified cDNA fragments were then used in the preparation of digoxigenin-labeled riboprobes by in vitro transcription. Sense and antisense strands were transcribed with SP6 or T7 polymerase according to the manufacturer's instructions (Boehringer Mannheim), except for the transcription buffer (5 mm each ribonucleotide triphosphate, 40 mm Tris-HCl, pH 8.0, 26 mm MgCl2, 3 mm spermidine, 0.01% Triton X-100, and 10 mm DTT).
In Situ Hybridization
Plant tissues were fixed in 4% paraformaldehyde and 50 mm Pipes, pH 7.2, and washed twice for 15 min with the same buffer. Roots, leaves, germinating seeds, and siliques were fixed for 2 to 4 h at room temperature. Flowers were fixed overnight at 4°C. Roots were embedded in 0.6% agarose to facilitate further handling. After fixation, specimens were dehydrated in a graded ethyl alcohol series (15%, 30%, 50%, 70%, 85%, 95%, and 100%) and embedded in paraffin. Sections (8 μm thick) from paraffin-embedded material were mounted on glass slides coated with 3-aminopropyltriethoxysilane (Aldrich) in acetone. Paraffin was removed by incubating slides twice in xylene for 10 min. Section pretreatment and hybridization were performed according to the method of Lincoln et al. (1994) with some modifications. Slides were incubated with 2 μg/mL proteinase K (Boehringer Mannheim) for 30 min at 37°C. Hybridization was carried out overnight at 50°C in 50% formamide, 300 mm NaCl, 10 mm Tris-HCl, pH 7.5, 1 mm EDTA, 5% dextran sulfate, 1% blocking reagent, 150 μg/mL tRNA, and about 500 ng/mL riboprobe. Slides were covered with HybriSlip (Research Products International, Mt. Prospect, IL) and sealed with rubber cement. RNase treatment was with 10 μg/mL RNase A (Boehringer Mannheim) for 30 min at 37°C. Detection of hybridized transcripts with antidigoxigenin antisera conjugated with alkaline phosphatase (1:500 dilution; Boehringer Mannheim) was performed according to the method of Coen et al. (1990). Slides were passed through an ethyl alcohol series and xylene before mounting in Eukit (Calibrated Instruments, Inc., Hawthorne, NY). The sections were observed using differential interference contrast optics on an Aristoplan microscope (Leitz, Wetzlar, Germany). Photographs were taken with Kodak Ektachrome 160T film using a Leitz Orthomat E camera.
Image Processing
Photographs were scanned by using a Sprintscan 35 (Polaroid, Inc., Cambridge, MA). For RNA blots, an Arcus II flatbed scanner (AGFA Division, Miles Inc., Ridgefield, NJ) was used. Images were processed using Photoshop, version 4.0. (Adobe, Mountain View, CA), and printed with an NP1600 printer (Codonics, Inc., Middleburg Heights, OH).
RT-PCR of Seedling Tissues
RNA isolation and limiting (22 cycle) quantitative RT-PCR of MT1a and MT2a expression was as described previously (Murphy and Taiz, 1995b). Primary leaves and apical buds, cotyledons, hypocotyls, and roots were excised from 200 seedlings with a razor blade and placed directly into liquid nitrogen before total RNA extraction. Results are summarized from two separate experiments.
RNA Isolation and Northern Blotting
RNA was isolated from liquid-nitrogen-ground plant tissues using Trizol reagent (GIBCO-BRL) following the instructions provided by the manufacturer. Total RNA was fractionated and transferred onto a nylon membrane (Nytran). Filters were hybridized with digoxigenin-labeled riboprobes and washed according to the method of Zhou and Goldsbrough (1994). Detection of transcripts with antidigoxigenin antisera coupled to alkaline phosphatase (Boehringer Mannheim) was carried out following the directions of the manufacturer. Each northern blotting experiment was repeated at least three times, and representative experiments are shown.
RESULTS
Expression of MT1a and MT2a Genes in Seedling Roots
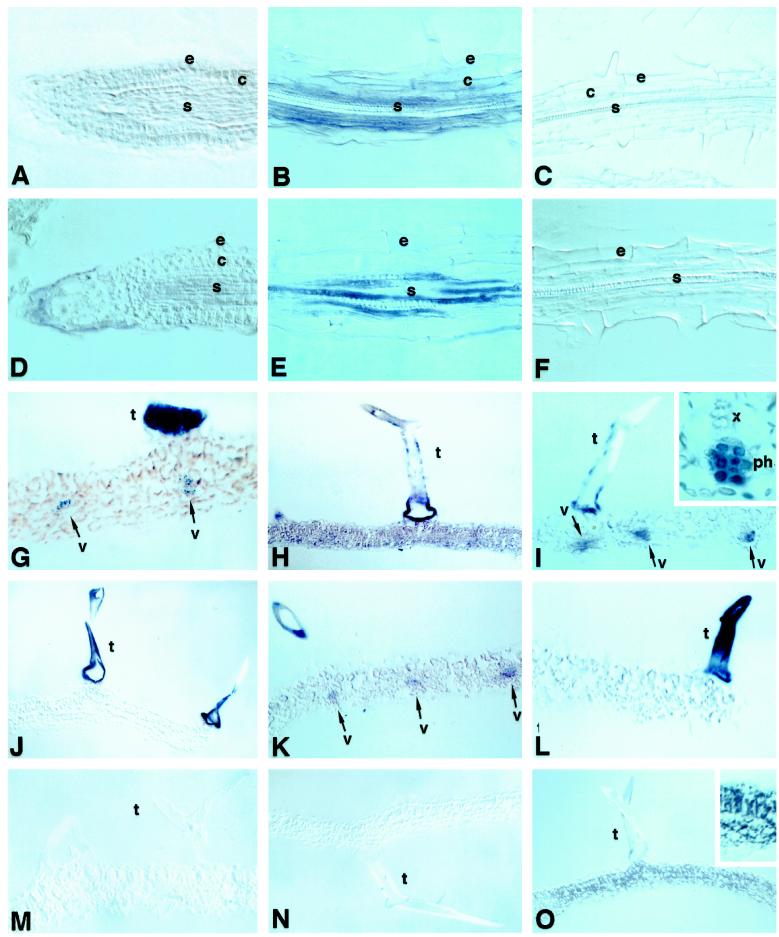
All of the experiments with roots were carried out with 6-d-old seedlings. The results of in situ-hybridization analyses of MT1a and MT2a RNAs using antisense riboprobes are summarized in Figure 1, A to F. In the meristematic region of the root, the hybridization signal obtained for MT1a was low and in many cases difficult to distinguish from the background (Fig. 1A). However, significant amounts of MT1a transcript in the elongation and maturation regions of the roots was detected (Fig. 1B). Accumulation of MT1a transcript in these regions appeared to be higher in cells of the stele and cortex (Fig. 1B). In epidermal cells the hybridization signal obtained with MT1a probes was much less intense. Similar hybridization patterns were observed in roots from copper-treated seedlings (data not shown). No hybridization signals above background levels were detected in sections treated with the MT1a sense probe (Fig. 1C), indicating that the reactions observed with the antisense probe were specific.
Figure 1.
Expression of MT1a and MT2a in Arabidopsis roots and leaves. The hybridization signal ranged from purple to dark blue. A, B, G, H, and I, Sections probed with MT1a antisense; C and M, sections probed with MT1a sense; D, E, J, K, and L, sections probed with MT2a antisense; F and N, sections probed with antisense probe with MT2a sense; O, section probed with antisense probe for the chloroplast-specific transcript psbO. All roots sections are from 6-d-old seedlings grown without excess copper. A and D, Longitudinal sections. A and D, Root-tip sections. B, C, E, and F correspond to sections through the maturation region. All leaf sections are from the youngest leaves of 2-week-old untreated plants (G, J, M, and O) or plants treated with excess copper (H, I, K, and L). Inset in I corresponds to a higher magnification of one of the vascular bundles. e, Epidermis; c, cortex; s, stele; t, trichome; v, vascular bundle. Arrows, Locations of hybridization signal.
The level of expression of MT2a in the meristematic region of the root was near the limit of our detection system (Fig. 1D). However, as in the case of MT1a, a strong hybridization signal was detected in the phloem, although the identity of the cells could not be determined (Fig. 1E). No significant MT2a transcript levels were found in other cells of the root. Surprisingly, when roots from plants that had been treated with excess copper were examined, we did not find any difference in the expression patterns of MT2a compared with untreated plants (data not shown). The absence of hybridization signal above background levels obtained with the MT2a sense probe (Fig. 1F) indicates that the signal detected with the MT2a antisense probe was specific.
To determine the relevance of these results to earlier findings (Murphy and Taiz, 1995b; Zhou and Goldsbrough, 1995), which showed copper-induced increases in MT2 expression in Arabidopsis seedlings, MT1a and MT2a mRNA expression in primary leaves and apical buds, cotyledons, hypocotyls, and roots were quantitated by fluorometric assay of RT-PCR products. As shown in Table I, MT2a expression was specifically induced by copper in cotyledons and, to a lesser extent, in hypocotyls but not in the roots. This finding is consistent with the failure to detect MT2a expression in seedling roots by in situ hybridization, even in the presence of copper. Copper increased the level of MT1a expression only in primary leaves and apical buds.
Table I.
RT-PCR quantitation of MT1a and MT2a mRNA expression in excised roots, hypocotyls, cotyledons, and primary leaves from 6.5-d-old Ws seedlings
| Plant Part | MT1a
|
MT2a
|
||
|---|---|---|---|---|
| Control | 40 μm CuCl2 | Control | 40 μm CuCl2 | |
| % | ||||
| Root | 62 ± 29 | 54 ± 24 | 75 ± 32 | 68 ± 22 |
| Hypocotyl | 31 ± 20 | 91 ± 35 | 27 ± 34 | 77 ± 26 |
| Cotyledon | 139 ± 20 | 142 ± 11 | 13 ± 27 | 182 ± 28 |
| Primary leaves | 20 ± 13 | 183 ± 47 | 130 ± 51 | 137 ± 43 |
mRNA expression levels were quantitated fluorometrically after size verification on agarose gels (see Methods) and are expressed as mean percentages ± sd of measured β-tubulin mRNA levels. High sd of primary leaf mRNA is the result of the difficulty in excising small (approximately 7% of total seedling fresh weight) primary leaves at this stage of development.
Localization of MT1a and MT2a mRNAs in Arabidopsis Leaves
The youngest visible leaves of 2-week-old Arabidopsis plants were subjected to RNA in situ-hybridization analysis to determine which cells were responsible for the expression of MT1a and MT2a that had been observed previously by northern hybridization. Leaves from plants of the same age that had been treated with 40 μm CuCl2 for 36 h were also examined. In leaves from control plants, MT1a expression was detected at high levels in trichomes and at lower levels in vascular bundles of minor veins (Fig. 1G). The hybridization pattern of MT1a in plants treated with excess copper was somewhat variable, but in all experiments MT1a was expressed at higher levels in copper-treated plants than in untreated ones (Fig. 1, H and I). Expression of MT1a was high in leaf trichomes of copper-treated plants, but since the hybridization signals were saturated in both control and treated trichomes, it was impossible to determine whether the amount of MT1a transcripts in trichomes of copper-treated plants was higher than in control plants. However, the stimulatory effect of copper on MT1a expression was discernible in other parts of the leaf. In some cases, hybridization signals were evenly distributed in all leaf tissues, including the rest of the epidermis and the mesophyll (Fig. 1H). In other cases, the signal was restricted in the mesophyll to vascular bundles but was much stronger than in control leaves (Fig. 1I). In the vascular bundles, the signal appeared to be localized in the phloem, possibly in the sieve elements (Fig. 1I, inset). No hybridization signal was observed in sections probed with the sense MT1a probe (Fig. 1M).
In the same leaves, MT2a was expressed at very high levels in trichomes of both control (Fig. 1J) and copper-treated (Fig. 1, K and L) plants. In control plants, trichomes were the only leaf cell type in which MT2a mRNA could be detected. In some leaf sections from copper-treated plants, low levels of expression were seen also in vascular bundles (Fig. 1K). In most cases, however, expression of MT2a in leaves from copper-treated plants remained restricted to trichomes exclusively (Fig. 1L). As before, the possibility of higher expression levels of MT2a in trichomes of copper-treated than in control plants cannot be ruled out because of color saturation.
In all cases, the MT2a antisense riboprobe hybridized more strongly with trichomes than the antisense MT1a probe, judging by the much shorter development times needed for the appearance of the MT2a signal (1 h versus more than 9 h for MT1a). This suggests that MT2a may be expressed at higher levels than MT1a in trichomes, although it could also be due to differences in the affinities of the probes for their respective sequences. However, the fact that the MT1a probe gave darker signals than the MT2a probe in other tissues (Fig. 1, H versus K) suggests that MT2a was expressed at higher levels than MT1a in trichomes. Hybridization reactions using the MT2a sense probe did not produce signals above background levels, indicating that the strong trichome staining is specific (Fig. 1N).
As a second, positive control, we also used a probe for a gene clone encoding psbO from Arabidopsis. As shown in Figure 1O, this probe hybridized with only chloroplast-containing mesophyll cells rather than the trichomes, further indicating that the hybridization patterns obtained with our MT antisense probes were specific.
Expression of MT1 and MT2 in Arabidopsis Flowers
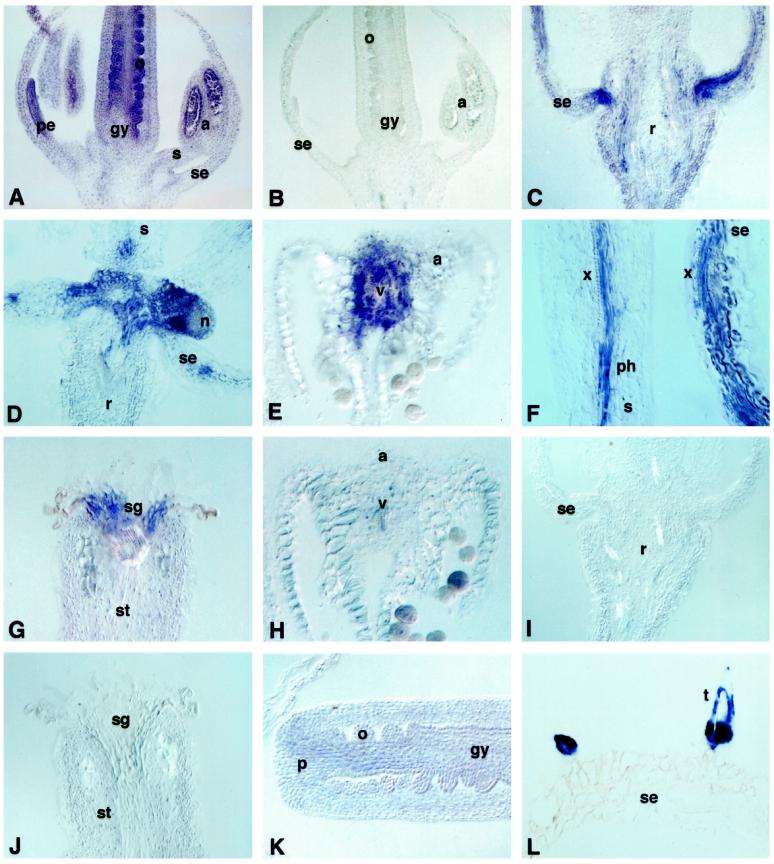
To investigate the expression patterns of MT genes in Arabidopsis flowers, tissue sections from flowers at different stages of development were hybridized with MT1a and MT2a riboprobes. At floral stages 9 to 10 (Smythe et al., 1990), during which the ovule protrusions elongate, the petals are below the level of stamens and the anthers contain microspore mother cells or tetrads, the antisense MT1a probe hybridized to varying degrees with all tissues of the flower (Fig. 2A). MT1a expression was higher in the gynoecium, especially in the developing ovules, in anthers, in cells of the tapetum, and in tetrads. Hybridization with vascular tissues of the flower, especially in the stamens and the receptacle, was also detectable (Fig. 2A). No hybridization signal was observed in sections of flowers treated with the sense MT1a riboprobe (Fig. 2B).
Figure 2.
Localization of MT1a and MT2a in Arabidopsis flowers. A and C to G, Sections hybridized with MT1a antisense riboprobe; B, section probed with MT1a sense; H to L, sections probed with MT2a antisense. A, B, and K, Flowers at stage 8 to 9. C to J and L, Flowers at anthesis. All sections are from plants grown without excess copper. a, Anthers; gy, gynoecium; n, nectary; o, ovule; p, placenta; pe, petal; ph, phloem; r, receptacle; s, stamen; se, sepal; sg, stigma; st, style; t, trichome; v; vascular bundle; x, xylem.
In the next stages of flower development (11–12; after the stigmatic papillae appear, when the petals are level with the long stamens, and the integuments extend toward the apex of the nucellus), MT1a expression was still stronger in all tissues of the gynoecium, especially in the integuments and funiculus of developing ovules, in the placenta, and along some strands of vascular tissue (data not shown). In anthers the strong hybridization signal observed in the pollen sacs in stages 9 to 10 had disappeared. In mature flowers, MT1a was highly expressed in vascular strands of sepals and the receptacle, especially the provascular tissue connecting the receptacle with the sepals (Fig. 2C), in the nectaries (Fig. 2D), in the tissue surrounding the vascular strands of anthers (Fig. 2E), in the vascular tissues of stamens and petals (Fig. 2F), and in the cells of the stigmatic core (Fig. 2G).
In contrast to MT1a, MT2a was expressed at very low levels in the gynoecium and only at the early stages of ovule development (Fig. 2K). At later stages of development, MT2a mRNA levels were below the limits of detection in all floral tissues (Fig. 2, H–J). The only cells expressing MT2a in mature flowers were the trichomes (Fig. 2L).
No effects of copper on the expression of either MT1a or MT2a in flowers was observed (data not shown).
In Situ Hybridization of MT1a and MT2a in Germinating Seeds
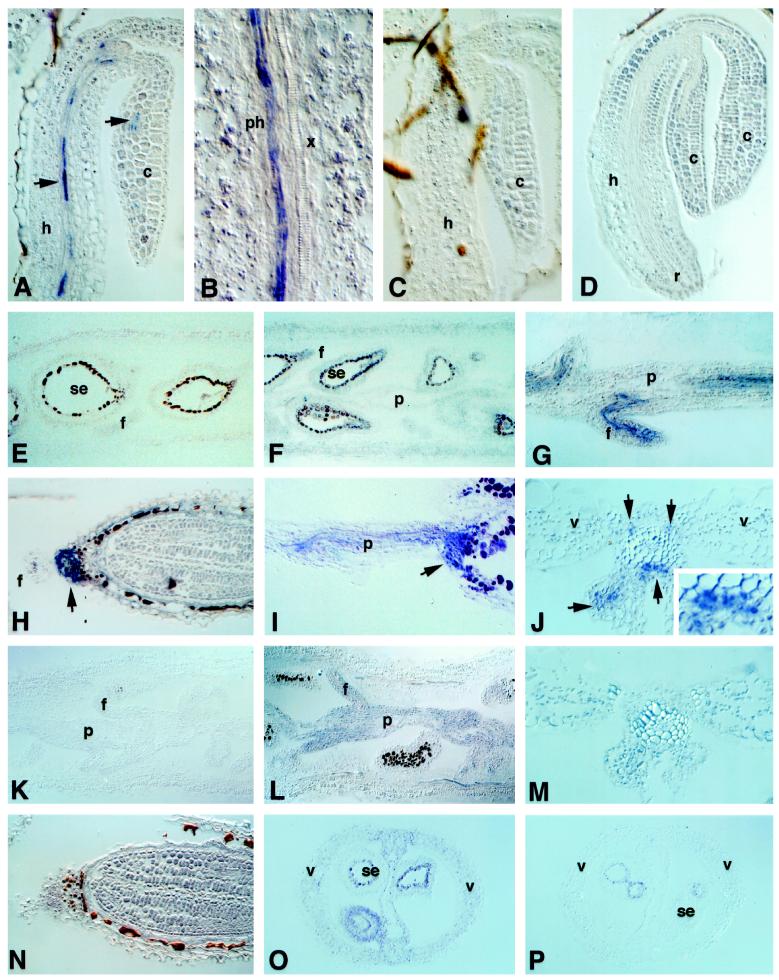
We also studied the localization of MT1a and MT2a gene expression during the early stages of seedling development in the presence or absence of copper. A sample of the results obtained from copper-treated seedlings is shown in Figure 3, A to D. The antisense MT1a riboprobe hybridized with specific cells of the vascular tissue in the hypocotyl and cotyledons (Fig. 3A). In the hypocotyl these cells were tentatively identified as phloem sieve elements (Fig. 3B). No hybridization signal was detected in any other region of the hypocotyl or radical. The hybridization pattern of control plants was essentially the same as in copper-treated seedlings (data not shown). However, the number of seedlings that showed a hybridization signal was 3- to 4-fold higher in the copper-treated seeds than in the untreated controls. From these observations we inferred that MT1a is expressed at low levels in germinating seedlings in phloem tissue and that copper caused a slight induction in the expression of MT1a in the same cell type. The MT1a sense probe control failed to hybridize with any seed tissue (Fig. 3C). In contrast, expression of MT2a was not detected in any tissue in germinating seedlings, either with (Fig. 3D) or without (data not shown) copper treatment.
Figure 3.
Expression of MT1a and MT2a in germinating seedlings and developing siliques. Sections were probed with: A, B, and E to J, MT1a antisense; C, MT1a sense; D and K to O, MT2a antisense; P, MT2a sense. A to D, F, G, I, J, L, M, and O were treated with excess copper; E, H, K, and N are untreated plants. c, Cotyledons; h, hypocotyl; f, funiculus; p, placenta; ph, phloem; se, seed; v, vascular tissue; x, xylem. Arrows indicate the location of the transcripts. Blue or purple stain corresponds to hybridization signals. The seed coat stains brown because of its normal pigmentation.
Expression of MT1 and MT2 in Siliques and Developing Seeds
In sections of siliques from control plants, we did not detect hybridization signals above background levels with either MT1a (Fig. 3E) or MT2a (Fig. 3K) antisense probes. In siliques from copper-treated plants, low levels of MT1a expression were detected in most tissues of the silique (Fig. 3F). Higher levels of MT1a expression were detected in vascular strands of the placenta, central septum, and funiculus (Fig. 3, G, I, and J). Cross-sections through the central septum showed MT1a hybridization with cells surrounding the xylem tissue, which appeared to be derived from the phloem (Fig. 3J). In developing seeds we found a strong hybridization signal with MT1a antisense probe in cells in the region where the funiculus attaches to the chalazal end of the seed (Fig. 3, H and I). The signal had a ring shape and surrounded the vascular bundle of the funiculus, in which tracheary elements were visible in the middle (Fig. 3H). In the longitudinal view the signal was localized to tissue that appeared as a continuation of the vascular strand of the funiculus, which is funnel-shaped inside the seed (Fig. 3I). This hybridization pattern was not apparent in very immature seeds, only in seeds in which the embryo occupied at least one-half of the seed volume. The same results were obtained in developing seeds from both untreated and copper-treated plants. In younger, copper-treated developing seeds, low levels of MT1a mRNA were observed in the integuments (Fig. 3F).
MT2a mRNA was not detected in siliques of untreated plants (Fig. 3K). In siliques from copper-treated plants, MT2a was localized at low levels in most tissues (Fig. 3, L, M, and O). MT2a expression was higher in the placenta and funiculus (Fig. 3L), but was evenly distributed in all cells rather than being predominant in vascular strands, as it was with MT1a expression. No significant levels of MT2a expression were observed in tissues of developing seeds at more mature stages, either untreated (Fig. 3N) or copper-treated (data not shown). Younger seeds from copper-treated plants showed low levels of MT2a expression in the integuments (Fig. 3O). No hybridization signals above background were obtained in siliques and developing seed tissues with MT2a sense control probe (Fig. 3P).
Northern-Hybridization Analysis of MT1a and MT2a mRNA in Developing Seedlings
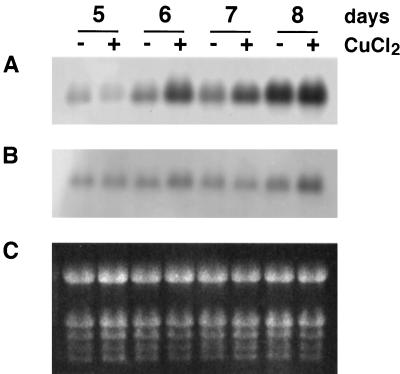
To determine whether the expression of MT1a and/or MT2a was regulated during seedling development, and whether the effects of excess copper would vary with the developmental stage of the plants, total RNA was isolated from Arabidopsis seedlings grown for 5 to 8 d on vertical mesh transfer plates and then analyzed by northern blotting using the same riboprobes that were used for the in situ-hybridization studies. The results show that the amount of MT1a mRNA in the seedlings increased more than 10-fold from d 5 to 8 (Fig. 4A). The effect of copper on MT1a expression depended on the age of the seedlings. In the youngest (5 d old) seedlings, excess copper did not affect MT1a expression. In 6- and 7-d-old seedlings, excess copper caused an increase in MT1a RNA accumulation. In the 8-d-old seedlings, copper had either no effect or caused only a slight increase in MT1a expression.
Figure 4.
Northern analysis of MT1a and MT2a expression in developing Arabidopsis seedlings. Total RNA (10 mg) was extracted from seedlings germinated for 5, 6, 7, or 8 d in the absence (−) or presence (+) of 40 μm copper. RNA was separated in a formaldehyde agarose gel, blotted, and hybridized with digoxigenin-labeled RNA probes. A, Blot probed with MT1a antisense. B, Blot probed with MT2a antisense. C, Gel stained with ethidium bromide showing rRNA.
In contrast to MT1a, the expression of MT2a in the absence of copper did not vary significantly during the stages of seedling development examined (Fig. 4B). Copper caused a slight (approximately 2-fold) increase in MT2a transcript level, but the effect was consistently detected only in 8-d-old seedlings (Fig. 4B). In all cases, the time required for the appearance of hybridization signals on the membranes probed with MT2a was about 3 to 5 times longer than with MT1a. Neither MT1a nor MT2a sense probes hybridized with any other RNA band on the filters (data not shown).
Expression of MT1a and MT2a in Senescing Leaves
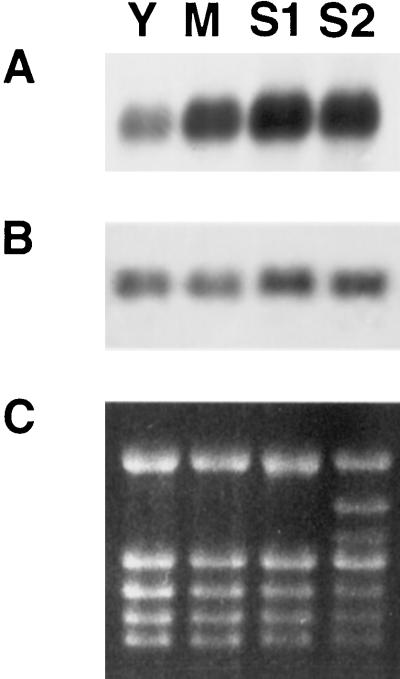
We also investigated the developmental regulation of MT1a and MT2a expression in senescing leaves. Leaves of different ages (young, mature, and early- and late-senescent) were collected and their total RNA analyzed by northern blotting. As shown in Figure 5, the expression of MT1a increased by >2-fold from the young to the mature leaf stage and then increased further from the mature to the early-senescent stage (Fig. 5A).
Figure 5.
Northern hybridization of MT1a and MT2a in aging Arabidopsis leaves. Total RNA (8 μg) was isolated from young (Y), mature (M), mid-senescent (S1), and almost completely senescent (S2) leaves. RNA was processed as in Figure 4. Filters were hybridized with MT1a antisense (A) or MT2a antisense (B) riboprobes. C, Ethidium bromide-stained gel.
In contrast to MT1a, the level MT2a mRNA did not vary significantly during the four stages of leaf maturation and senescence examined (Fig. 5B).
DISCUSSION
We have investigated the spatial distribution of MT1a and MT2a mRNAs by in situ hybridization in vegetative and reproductive tissues of Arabidopsis in the presence and absence of excess copper. Our in situ hybridization results indicate that the two MT genes have contrasting expression patterns, suggesting that they play distinct roles in metal ion homeostasis and development.
The riboprobes used in the in situ and northern hybridization experiments were synthesized using clones of MT1a and MT2a that contained the complete translated regions and most of the 3′ untranslated regions. The sequence identities between MT1a and MT1c and between MT2a and MT2b in these regions were 67% and 60%, respectively. These values are sufficiently low to expect isoform specificity of the probes under the high-stringency conditions used. In the case of MT2a, we were able to confirm by northern blotting that the antisense probe did not hybridize with sense transcripts prepared from the MT2b clone (data not shown). We were unable to obtain a cDNA clone of MT1c for parallel tests of the specificity of the MT1a probe. Although a recently deposited expressed sequence tag sequence has been identified, which appears to correspond to MT1c (expressed sequence tag clone no. 222N3T7, accession no. N38326), the abundance of this transcript is probably much lower than that of MT1a (P. Goldsbrough, personal communication). Thus, the two probes used in this study appear to be relatively isoform specific.
In young control roots, both MT1a and MT2a genes were expressed in the maturation zone of the root. However, compared with MT1a, the level of expression of MT2a was low, and its detection was not always possible.
Little or no MT1a or MT2a expression was detected in control root tips by in situ hybridization. This result is consistent with the finding that the pea MT1-like PsMTa gene promoter directed GUS expression in Arabidopsis to all tissues of the root except the apex (Fordham-Skelton et al., 1997). However, the wheat MT2-like WALI1 gene was expressed predominantly in the root apical meristem (Snowden et al., 1995), and other expression studies of cotton using GUS fused to the MT1 promoter showed the highest stain at the root tip (Hudspeth et al., 1996). These discrepancies may reflect the complexity of the expression regulation of the different MT genes, as well as the possible pitfalls associated with various methods of detection. For example, localization data obtained by reporter genes may not always reflect in vivo gene-expression pattern (Taylor, 1997).
Copper treatment failed to cause a significant increase in the expression of either MT1a or MT2a in growing tips and maturation zones of 6-d-old roots, as measured by in situ hybridization. This finding was unexpected, since previous northern blotting and RT-PCR studies with Arabidopsis seedlings had demonstrated an increase in the total MT2 transcript level in the presence of copper (Zhou and Goldsbrough, 1994; Murphy and Taiz, 1995a). However, these studies were based on total mRNA extracted from whole seedlings. When we repeated our RT-PCR measurements of MT1 and MT2 mRNA using excised regions of the seedling, we were able to confirm that the copper-induced increase in MT2 mRNA was restricted to the cotyledons and, to a lesser extent, the hypocotyl (Table I). This finding indicates that the previous correlation we observed between seedling copper tolerance (as measured by root growth) and MT2 mRNA (Murphy and Taiz, 1995a) probably reflected MT2 expression in the cotyledons rather than the root, suggesting that the cotyledon plays a key role in the copper tolerance of the seedling as a whole. Since MT2a expression is low in seedling roots, it does not appear to play a role in copper homeostasis in young roots. However, it is important to point out that the response of mature roots to copper appears to be different from that of seedling roots (Zhou, 1994; Snowden et al., 1995; Murphy et al., 1997).
Both MT1a and MT2a were expressed at very high levels in young leaf trichomes of both untreated and copper-treated plants. MT2a expression in trichomes appeared to be higher than that of MT1a. Perhaps because of the high levels of expression, which saturated the signal, it was not possible to detect any copper stimulation relative to the controls. In control plants low levels of MT1a expression in young leaves were also detected in minor veins, and copper stimulated the expression of MT1a in the vascular bundles, primarily in the phloem. In some cases, low levels were also detected in the mesophyll cells. In contrast, MT2a in leaves appeared to be expressed almost exclusively in the trichomes. Occasionally, very low levels of MT2a transcript were observed in the mesophyll of copper-treated plants, primarily in the minor veins. Overall, these results are in good agreement with previous studies showing copper stimulation of MT1, but not MT2, in Arabidopsis leaves (Zhou and Goldsbrough, 1994; Murphy et al., 1997). In bean, MT2 was also found to be specifically expressed in trichomes and to be absent from the mesophyll (Foley and Singh, 1994).
The expression patterns of MT1a and MT2a in leaves suggest possible functions for these proteins. The high levels of expression of MT1a and MT2a in trichomes are particularly striking and may indicate that trichomes play an important role in metal detoxification in leaves. In Indian mustard, for example, Cd accumulates preferentially in trichomes (Salt, 1995), and nickel accumulation has been demonstrated in the trichomes of Alyssum lesbiacum (Krämer et al., 1997). By analogy to the salt-secreting trichomes of halophytes, leaf trichomes may provide a pathway for secreting excess heavy metals outside the mesophyll. The volume of trichome cells is enormous compared with that of a typical mesophyll cell (Fig. 1H), allowing them to serve as large reservoirs for sequestering potentially toxic metal ions. Eventually, as the trichomes senesce, the metal ions would be deposited harmlessly onto the leaf surface.
Alternatively, the high expression levels of MT1a and MT2a in trichomes may reflect a high requirement for copper. Trichome cells are active in sulfur (Gotor et al., 1997), flavonoid (Charrier et al., 1996), and anthocyanin (Lloyd et al., 1994) metabolism. A number of genes involved in defense mechanisms, including polyphenol oxidase (Shahar et al., 1992; Thipyapong et al., 1997), peroxidase (Mohan et al., 1993), Phe ammonia-lyase (Prasad et al., 1995), and chalcone synthase (Sistrunk et al., 1994), are expressed in trichomes. In addition, trichomes have been shown to develop lignified cell walls (Wyatt et al., 1993). Many of the enzymes in the above pathways require copper for activity. Copper is a cofactor in at least two enzymes involved in lignin biosynthesis: polyphenol oxidase and diamine oxidase. MT1a could be involved in lignification processes in all of these tissues, whereas MT2a might act only in trichomes. The expression of MTs has been shown to increase after wounding (Snowden et al., 1995; Choi et al., 1996). Since rapid lignification is associated with wound healing, the increase in MTs following wounding might reflect an increase in copper demand. The function of MTs in these cells might be to facilitate the transfer of free copper ions to copper-requiring enzymes. Further studies are needed to determine whether copper accumulates in the trichomes of Arabidopsis.
The localization of MT1a expression in vascular bundles, especially the phloem, or to tissues that function in the transport of nutrients to the seed (placenta and funiculus), suggests that MT1a may play a role in metal-ion transport and/or vascular development. During leaf senescence micronutrients are remobilized and transported via the phloem to the growing regions of the plant. The elevated expression of MT1a observed by northern blotting in senescing leaves could be related to the remobilization of micronutrients, specifically copper and zinc. Other MTs have been shown to be transcriptionally activated during senescence (Buchanan-Wollaston, 1994; Ledger and Gardner, 1994; Coupe et al., 1995; Hsieh et al., 1995; Clendennen and May, 1997; Foley et al., 1997; Reid and Ross, 1997). Recently, two different MT-like cDNA clones were identified from senescing Arabidopsis leaves (Thomas and Villers, 1996). One of the MT clones followed a pattern of expression similar to the one we observed for MT1a, increasing at mid-senescence and decreasing thereafter.
The stigmatic core is another possible example of the role of MTs in micronutrient remobilization. The cells of the stigmatic core play an important role in pollen germination and growth, supplying the growing pollen tube with water and nutrient reserves. After the passage of the pollen tubes, these stigmatic cells senesce and die. MT1a could be involved in the remobilization of copper and other metals from those cells to the growing pollen tube. In general, MT1a was most highly expressed in either tissues with a high demand of copper or tissues involved in the transport or mobilization of nutrients.
MT1a, but not MT2a, was strongly expressed in flowers, particularly in the gynoecium and the anthers. Given the strong expression of MT1a in various parts of the flower, it may be significant that copper deficiencies typically affect flower formation and maturation much more than vegetative growth, and the ovary and anthers are particularly high in copper content (Märschner, 1995). Thus, MT1a may play an important role in flower development by facilitating the transfer and exchange of copper in those tissues with the highest copper requirement.
Finally, northern-hybridization analysis indicated that MT1a, but not MT2, expression is developmentally regulated in seedlings and leaves. The increasing expression in seedlings could be the reflection of active vascular differentiation in these tissues and/or an increasing demand of copper as the plant grows.
In conclusion, we have confirmed and extended previous observations that MT1 and MT2 are differentially expressed in plant tissues, although there are areas of overlapping expression as well. Of particular interest were the high expression levels of both MTs in leaf trichomes. Further investigations into the role of leaf trichomes in metal homeostasis are thus warranted.
Abbreviations:
- MT
metallothionein
- RT
reverse transcriptase
Footnotes
This study was supported by a grant from the U.S. Department of Agriculture (no. 94-37100-0755) to L.T. and by a Research Scientist Training Postdoctoral Fellowship from the Ministry of Education and Sciences of Spain to M.G.-H.
LITERATURE CITED
- Buchanan-Wollaston V. Isolation of cDNA clones for genes that are expressed during leaf senescence in Brassica napus. Plant Physiol. 1994;105:839–846. doi: 10.1104/pp.105.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston V. cloning of senescence related genes by subtractive hybridization Plant Mol Biol. 1997;33:821–834. doi: 10.1023/a:1005774212410. [DOI] [PubMed] [Google Scholar]
- Bundithya W, Goldsbrough PB. Activity and metal regulation of Arabidopsis metallothionein gene promoters (abstract no. 1292) Plant Physiol. 1997;114:S-251. [Google Scholar]
- Charrier B, Leroux C, Kondorosi A, Ratet P. The expression pattern of alfalfa flavanone 3-hydroxylase promoter-gus fusion in Nicotiana benthamiana correlates with the presence of flavonoids detected in situ. Plant Mol Biol. 1996;30:1153–1168. doi: 10.1007/BF00019549. [DOI] [PubMed] [Google Scholar]
- Choi D, Kim HM, Yun HK, Park J-A, Kim WT, Bok SH. Molecular Cloning of a metallothionein-like gene from Nicotiana glutinosa L. and its induction by wounding and tobacco mosaic virus infection. Plant Physiol. 1996;112:353–359. doi: 10.1104/pp.112.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clendennen S, May GD. Differential gene expression in ripening banana fruit. Plant Physiol. 1997;115:463–469. doi: 10.1104/pp.115.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen ES, Romero JM, Sandra D, Elliott R, Carpenter R. Floricaula: a homeotic gene required for flower development in Antirrhinum majus. Cell. 1990;63:1311–1322. doi: 10.1016/0092-8674(90)90426-f. [DOI] [PubMed] [Google Scholar]
- Coupe SA, Taylor JE, Roberts JA. Characterization of an mRNA encoding a metallothionein-like protein that accumulates during ethylene-promoted abscission of Sambucus nigra L. Planta. 1995;197:442–447. doi: 10.1007/BF00196665. [DOI] [PubMed] [Google Scholar]
- de Framond AJ. A metallothionein-like gene from maize (Zea mays): cloning and characterization. FEBS Lett. 1991;290:103–106. doi: 10.1016/0014-5793(91)81236-2. [DOI] [PubMed] [Google Scholar]
- de Miranda JR, Thomas MA, Thurman DA, Tomsett AB. Metallothionein genes from the flowering plant Mimulus guttatus. FEBS Lett. 1990;260:277–280. doi: 10.1016/0014-5793(90)80122-y. [DOI] [PubMed] [Google Scholar]
- Dong J-Z, Dunstan DI. Expression of abundant mRNAs during somatic embryogenesis of white spruce [Picea glauca (Moench) Voss] Planta. 1996;199:459–466. doi: 10.1007/BF00195740. [DOI] [PubMed] [Google Scholar]
- Evans IM, Gatehouse LN, Gatehouse JA, Robinson NJ, Croy RRD. A gene from pea (Pisum sativum L.) with homology to metallothionein genes. FEBS Lett. 1990;262:29–32. doi: 10.1016/0014-5793(90)80145-9. [DOI] [PubMed] [Google Scholar]
- Evans IM, Gatehouse JA, Lindsay WP, Shi J, Tommey AM, Robinson NJ. Expression of the pea metallothionein-like gene PsMTA in Escherichia coli and Arabidopsis thaliana and analysis of trace metal ion accumulation: implications for PsMTA function. Plant Mol Biol. 1992;20:1019–1028. doi: 10.1007/BF00028889. [DOI] [PubMed] [Google Scholar]
- Foley RC, Liang ZM, Singh KB. Analysis of type 1 metallothionein cDNAs in Vicia faba. Plant Mol Biol. 1997;33:583–591. doi: 10.1023/a:1005790927581. [DOI] [PubMed] [Google Scholar]
- Foley RC, Singh KB. Isolation of a Vicia faba metallothionein-like gene: expression in foliar trichomes. Plant Mol Biol. 1994;26:435–444. doi: 10.1007/BF00039552. [DOI] [PubMed] [Google Scholar]
- Fordham-Skelton AP, Lilley C, Urwin PE, Robinson NJ. GUS expression in Arabidopsis directed by 5′ regions of the pea metallothionein-like gene PsMTA. Plant Mol Biol. 1997;34:659–668. doi: 10.1023/a:1005836632678. [DOI] [PubMed] [Google Scholar]
- Gotor C, Cejudo FJ, Barroso C, Vega JM. Tissue-specific expression of ATCYS-3A, a gene encoding the cytosoloc isoform of O-acetylserine(thiol)lyase in Arabidopsis. Plant J. 1997;11:347–352. doi: 10.1046/j.1365-313x.1997.11020347.x. [DOI] [PubMed] [Google Scholar]
- Hsieh H-M, Liu W-K, Huang PC. A novel stress-inducible metallothionein-like gene from rice. Plant Mol Biol. 1995;28:381–389. doi: 10.1007/BF00020388. [DOI] [PubMed] [Google Scholar]
- Hudspeth RL, Hobbs SL, Anderson DM, Rajasekaran K, Grula JW. Characterization and expression of metallothionein-like genes in cotton. Plant Mol Biol. 1996;31:701–705. doi: 10.1007/BF00042243. [DOI] [PubMed] [Google Scholar]
- Kagi JH. Overview of metallothionein. Methods Enzymol. 1991;205:613–626. doi: 10.1016/0076-6879(91)05145-l. [DOI] [PubMed] [Google Scholar]
- Kawashima I, Inokuchi Y, Chino M, Kimura M, Shimizu N. Isolation of a gene for a metallothionein-like protein from soybean. Plant Cell Physiol. 1991;32:913–916. [Google Scholar]
- Kawashima I, Kennedy TD, Chino M, Lane BG. Wheat Ec metallothionein genes. Eur J Biochem. 1992;209:971–976. doi: 10.1111/j.1432-1033.1992.tb17370.x. [DOI] [PubMed] [Google Scholar]
- Krämer U, Grime GW, Smith JAC, Hawes CR, Baker AJM. Micro-PIXE as a technique for studying nickel localization in leaves of the hyperaccumulator plant Alyssum lesbiacum. Nucl Instr Meth Phys Res (B) 1997;130:346–350. [Google Scholar]
- Lane B, Kajoika R, Kennedy T. The wheat-germ Ec protein is a zinc-containing metallothionein. Biochem Cell Biol. 1987;65:1001–1005. [Google Scholar]
- Ledger SE, Gardner RC. Cloning and characterization of five cDNAs for genes differentially expressed during fruit development of kiwifruit (Actinia deliciosa var. deliciosa) Plant Mol Biol. 1994;25:877–886. doi: 10.1007/BF00028882. [DOI] [PubMed] [Google Scholar]
- Lincoln C, Long J, Yamaguchi J, Serikawa K, Hake S. A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell. 1994;6:1859–1876. doi: 10.1105/tpc.6.12.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd AM, Schena M, Walbot V, Davis RW. Epidermal cell fate determination in Arabidopsis: patterns defined by a steroid-inducible regulator. Science. 1994;266:436–439. doi: 10.1126/science.7939683. [DOI] [PubMed] [Google Scholar]
- Märschner H. Mineral Nutrition of Higher Plants. San Diego, CA: Academic Press; 1995. [Google Scholar]
- Mohan R, Vijayan P, Kolattukudy PE. Developmental and tissue-specific expression of a tomato anionic peroxidase Tap1 gene by minimal promoter with wound and pathogen induction by an additional 5′-flanking region. Plant Mol Biol. 1993;22:475–490. doi: 10.1007/BF00015977. [DOI] [PubMed] [Google Scholar]
- Murphy A. Metal ion homeostasis in Arabidopsis thaliana. PhD thesis. Santa Cruz: University of California; 1996. [Google Scholar]
- Murphy A, Taiz L. Comparison of metallothionein gene expression and nonprotein thiols in 10 Arabidopsis ecotypes. Correlation with copper tolerance. Plant Physiol. 1995a;109:1–10. doi: 10.1104/pp.109.3.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy A, Taiz L. A new vertical mesh transfer technique for metal tolerance studied in Arabidopsis. Ecotypic variation and copper-sensitive mutants. Plant Physiol. 1995b;108:29–38. doi: 10.1104/pp.108.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy A, Zhou J, Goldsbrough PB, Taiz L. Purification and immunological identification of metallothioneins 1 and 2 from Arabidopsis thaliana. Plant Physiol. 1997;113:1293–1301. doi: 10.1104/pp.113.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura N, Nishizawa N-K, Umehara Y, Mori S. An iron deficiency-specific cDNA from barley roots having two homologous cysteine-rich MT domains. Plant Mol Biol. 1991;17:531–533. doi: 10.1007/BF00040651. [DOI] [PubMed] [Google Scholar]
- Prasad SVN, Thungapathra M, Mohindra V, Upadhyaya KC. Expression patterns of an Arabidopsis phenylalanine ammonia-lyase promoter in transgenic tobacco. J Genet. 1995;74:111–126. [Google Scholar]
- Reid SJ, Ross GS. Up-regulation of two cDNA clones encoding metallothionein-like proteins in apple fruit during cool storage. Physiol Plant. 1997;100:183–189. [Google Scholar]
- Robinson NJ, Tommey AM, Kuske C, Jackson P. Plant metallothioneins. Biochem J. 1993;295:1–10. doi: 10.1042/bj2950001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salt DE. Mechanisms of cadmium mobility and accumulation in Indian mustard. Plant Physiol. 1995;109:1427–1433. doi: 10.1104/pp.109.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahar T, Henning N, Gutfinger T, Hareven D, Lifschitz E. The tomato 66.3 kD polyphenoloxidase gene:molecular identification and developmental expression. Plant Cell. 1992;4:135–147. doi: 10.1105/tpc.4.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sistrunk ML, Antosiewicz DM, Purgganan MM, Braam J. Arabidopsis TCH3 encodes a novel Ca2+ binding protein and shows environmentally induced and tissue-specific regulation. Plant Cell. 1994;6:1553–1565. doi: 10.1105/tpc.6.11.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden KC, Gardner RC. Five genes by aluminum in wheat (Triticum aestivum L.) roots. Plant Physiol. 1993;103:855–861. doi: 10.1104/pp.103.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden KC, Richards KD, Gardner RC. Aluminum-induced genes: induction by toxic metals, low calcium, and wounding and pattern of expression in root tips. Plant Physiol. 1995;107:341–348. doi: 10.1104/pp.107.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BC. Promoter fusion analysis: an insufficient measure of gene expression. Plant J. 1997;9:273–275. [Google Scholar]
- Thipyapong P, Joel DM, Steffens JC. Differential expression and turnover of the tomato polyphenol oxidase gene family during vegetative and reproductive development. Plant Physiol. 1997;113:707–718. doi: 10.1104/pp.113.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas H, Villers L. Gene expression in leaves of Arabidopsis thaliana induced to senesce by nutrient deprivation. J Exp Bot. 1996;47:1845–1852. [Google Scholar]
- Wyatt RE, Ainley WM, Nagao RT, Conner TW, Key JL. Expression of the Arabidopsis Ataux2–11 auxin-responsive gene in transgenic plants. Plant Mol Biol. 1993;22:731–749. doi: 10.1007/BF00027361. [DOI] [PubMed] [Google Scholar]
- Zhou J (1994) Structure and function of metallothionein genes in Arabidopsis. PhD thesis. Purdue University. West Lafayette, IN
- Zhou J, Goldsbrough PB. Functional homologs of fungal metallothionein genes from Arabidopsis. Plant Cell. 1994;6:875–884. doi: 10.1105/tpc.6.6.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Goldsbrough J. Structure, organization and expression of the metallothionein gene family in Arabidopsis. Mol Gen Genet. 1995;248:318–328. doi: 10.1007/BF02191599. [DOI] [PubMed] [Google Scholar]