Abstract
Background
Lymphocytes act as a major component of the adaptive immune system, taking very crucial responsibility for immunity. Differences in proportions of T-cell subpopulations in peripheral blood among individuals under same conditions provide evidence of genetic control on these traits, but little is known about the genetic mechanism of them, especially in swine. Identification of the genetic control on these variants may help the genetic improvement of immune capacity through selection.
Results
To identify genomic regions responsible for these immune traits in swine, a genome-wide association study was conducted. A total of 675 pigs of three breeds were involved in the study. At 21 days of age, all individuals were vaccinated with modified live classical swine fever vaccine. Blood samples were collected when the piglets were 20 and 35 days of age, respectively. Seven traits, including the proportions of CD4+, CD8+, CD4+CD8+, CD4+CD8−, CD4−CD8+, CD4−CD8− and the ratio of CD4+ to CD8+ T cells were measured at the two ages. All the samples were genotyped for 62,163 single nucleotide polymorphisms (SNP) using the Illumina porcineSNP60k BeadChip. 40833 SNPs were selected after quality control for association tests between SNPs and each immune trait considered based on a single-locus regression model. To tackle the issue of multiple testing in GWAS, 10,000 permutations were performed to determine the chromosome-wise and genome-wise significance levels of association tests. In total, 61 SNPs with chromosome-wise significance level and 3 SNPs with genome-wise significance level were identified. 27 significant SNPs were located within the immune-related QTL regions reported in previous studies. Furthermore, several significant SNPs fell into the regions harboring known immunity-related genes, 14 of them fell into the regions which harbor some known T cell-related genes.
Conclusions
Our study demonstrated that genome-wide association studies would be a feasible way for revealing the potential genetics variants affecting T-cell subpopulations. Results herein lay a preliminary foundation for further identifying the causal mutations underlying swine immune capacity in follow-up studies.
Keywords: T lymphocyte subpopulations, Genome-wide association study, Swine
Background
Infectious diseases cause many serious economic and welfare problems in current swine industry. Some of porcine diseases belong to zoonoses, and put people at risk to infections. Even though there are many ways, such as vaccination injection, antibiotic treatment and isolation, to deal with issue of diseases, infectious diseases could not be solved completely [1]. On the other hand, genetics and breeding are working on improving immune capacity of animals through artificial selection, which maybe provides a promising strategy. Moreover, swine is increasingly used as a large animal model for several human diseases [2-4]. The porcine immune system is becoming more and more interesting in the field of both basic and applied research.
The immune system plays an essential role in disease resistance of animals. Lymphocytes have been widely recognized as a major component of the adaptive immune system, bearing very crucial responsibility for immunity. Lymphocytes are basically divided into two categories, namely T and B lymphocytes, each responsible for a particular branch of the immune system. T-lymphocytes (T-cells) are mostly responsible for fighting microbes, antigens or foreign substances inside the cells, triggering cell-mediated immunity. The proportions of T-cell subpopulations in peripheral blood vary with health and disease status. CD4 (an antigenic marker of helper T-cell) binds to relatively invariant sites on class II major histocompatibility complex (MHC) molecules outside the peptide-binding groove, which interacts with the T-cell receptor (TCR) [5,6]. The functions of CD4 are to initiate or augment the early phase of T-cell activation. CD4+ T cells are responsible for activating and directing other immune cells. They are essential in determining B cell antibody class switching, activating cytotoxic T cells, and maximizing bactericidal activity of phagocytes such as macrophages. Shedlock and Shen [7] showed that CD4+ cells are required in the priming phase for functional CD8 memory. CD8 antigen is a cell surface glycoprotein found in most cytotoxic T-cells that mediates efficient cell-cell interactions within the immune system. CD8 antigen, together with other T-cell receptors on T-cells, recognizes antigen processed by antigen presenting cells (APCs) in the context of class I MHC molecules [8].
In addition to the individual functions of CD4 and CD8, different combinations of them, i.e., CD4+CD8+, CD4+CD8−, CD4−CD8+ and CD4−CD8−, as well as the ratio of CD4+ to CD8+ also vary with health and disease status, and thus are highly relevant to immune capability of individuals. CD4-CD8+ are MHC class I restricted and mainly recognize replicating viral antigens. CD4+CD8- are MHC class II restricted and respond to nonreplicating protein antigens processed by APCs [9,10]. Differences in ratios of CD4 to CD8 are MHC haplotype-dependent [11].
Immunology is a fast growing research area. Gerner et al. summarized the current knowledge about porcine T lymphocytes and porcine T-lymphocyte subpopulations [12]. Lymphocytes expressing markers CD4 or CD8 alone and CD4 and CD8 together are important in viral clearance by secreting IFN-γ and mediating pathogen specific cytotoxicity [13]. Classical Swine Fever Virus (CSFV)-specific T-cell epitopes, the epitope peptide 290, harbors a CSFV-specific helper T-cell epitope and a CTL epitope, which could elicit both CD4+ and CD8+ T-cell responses [14]. Previous studies have shown that production of CSFV-specific CD8+ CTLs represents an important defense mechanism in the elimination of cells infected by CSFV [15]. Antigen specific lysis of CSFV-infected targets was found to be performed by CD4+ T cells. It has previously been demonstrated that CD8+ T cells lysed pseudorabies virus (PRV)-infected cells [16] and CD4+ producing T cells play important roles in conferring protection against a lethal PRV infection [17]. The immune system of swine differs markedly from that of humans and mice. Swine has a substantial number of CD4-CD8-T lymphocytes in peripheral blood [10,18-20]. Swine is also the only species so far known to hold a substantial number of resting extrathymic CD4+CD8+ T-cells [10,20-22]. Summerfield et al.[23] demonstrated that CD4+CD8+ cells can function as memory T-helper cells which proliferate upon stimulation with recall antigen. The CD4+CD8+ T cells were found to be associated with protection in pigs vaccinated against PRV [24]. The study of the cellular immune response to virus or vaccines in the natural host is of the utmost importance for understanding the interaction between the pathogen and the swine immune system [25].
Differences in proportions of T-cell subpopulations in peripheral blood among individuals under same conditions provide evidence of genetic control on these traits, but little is known about the genetic mechanism of them, especially in swine. Identification of the genetic control on these variants may help the genetic improvement of immune capacity in animals through selection. So far only two reports focused on QTL mapping for T-cell subpopulations in peripheral blood in pig [26,27]. QTL mapping has been very successful in domestic animals but the identification of quantitative trait mutations (QTMs) is still a challenge although a few prominent successful cases have been reported [28].
Recently, the first high-density 60K porcine SNP array has been developed [29], which offers the prerequisite for genome-wide association study (GWAS), a powerful approach for high-resolution mapping of loci controlling complex traits in domestic animals. Duijvesteijn et al. [30] firstly reported a GWAS for androstenone levels in pigs by 60K SNP array and revealed a cluster of candidate genes on Sus scrofa chromosome (SSC) 6. More recently, Fan et al.[31] preformed a GWAS for body composition and structural soundness traits of pigs and identified several genes by functional clustering analyses. Up to now, GWAS have been becoming a most commonly-used strategy for gene identification for complex traits in animals as well as humans.
In this study, we performed GWAS for T lymphocyte traits in swine by genotyping 675 pigs from 3 breeds (including a Chinese indigenous breed) based on the 60K SNP array, with measuring seven phenotypes of T-cell subpopulations in 562 piglets. A suite of significant SNPs associated with T-cell subpopulations at either the genome-wise or chromosome-wise were identified. These promising loci may be considered as preliminary foundation for further replication studies and eventually unraveling the causal mutations for T-cell subpopulations traits in swine.
Methods
Animal resource
The animal resource used in this study consists of 675 pigs of three breeds (Landrace, Yorkshire and Songliao Black pigs) including 562 piglets and their parents. The structure of experimental population was given in Table 1. All individuals were raised under standard indoor conditions at the experimental farm of the Institute of Animal Sciences, Chinese Academy of Agricultural Sciences, Beijing, China. At 21 days of age, all piglets were vaccinated with 4 doses live Classical Swine Fever Virus (CSF) Vaccine (Rabbit origin, tissue virus ≥ 0.01 mg/dose) (Qilu Animal Health Products Co., Ltd., Shandong, China) through intramuscular injection. Blood samples were collected from the jugular vein of each piglet one day before the vaccination (day 20), and two weeks after the vaccination (day 35), respectively. And blood samples were directly injected into eppendorf tubes containing 60 μl of 20% EDTA in phosphate-buffered saline (PBS). In addition, ear tissues of all individuals were also collected. The whole procedure for collection of the samples (blood and ear tissue) was carried out in strict accordance with the protocol approved by the Animal Welfare Committee of China Agricultural University (Permit number: DK997).
Table 1.
Animal resource for GWAS
| Breed | Sires | Dams | Piglets | Total |
|---|---|---|---|---|
| Landrace |
4 |
13 |
68 |
85 |
| Yorkshire |
16 |
63 |
415 |
494 |
| Songliao Black |
3 |
14 |
79 |
96 |
| Total | 23 | 90 | 5621 | 6752 |
Note 1: The total number of the piglets which were phenotyped 2: The total number of all the animals (including piglets and their parents) which were genotyped.
Measurement of phenotypes
For all individuals, seven different types of phenotypes of T-cell subpopulations, including the proportions of CD4+, CD8+, CD4+CD8+, CD4+CD8−, CD4−CD8+, CD4−CD8− and the ratio of CD4+ to CD8+ T cells, were obtained by the double cytofluorometric analysis. The blood cells were incubated with 10 μl of mouse anti porcine CD4-FITC (Serotec UK) and 10 μl of mouse anti porcine CD8-RPE (Serotec UK) for 30 min, and then washed with 0.1 M PBS (pH 7.2, containing 0.3% bovine serum albumin). The red blood cells were digested by 0.1% ammonium oxalate solution. The stained cells were detected by EPICS Flow Cytometer (Beckman-Coulter Company, USA).
Genotyping
DNA was extracted from ear tissue samples of all pigs, including piglets and their parents and genotyped using the Illumina PorcineSNP60K BeadChip containing 62,163 SNPs. Features of the Illumina PorcineSNP60K BeadChip have been detailed previously [29]. All individuals were genotyped using BeadStudio (Illumina) in terms of a custom cluster file developed from the 675 samples investigated.
Genotype quality control
To assess the technical reliability of the genotyping panel, a randomly selected DNA sample was genotyped twice and over 99% identity of called genotypes (two mismatches) was obtained. This demonstrates the technically robust feature of the 60K SNP BeadChip panel employed herein. All the samples included are with a minimum of 95% call rate.
We performed a 2-step quality control procedure for all genotyped piglets as follows. In first quality control, we discards meaningless SNPs (minor allele frequency (MAF) = 0 or call rate = 0) out of the initial full–set of 62,163 SNPs for each breed. So 8307/6258/4838 SNPs were moved out for Landrace, Yorkshire and Songliao Black respectively. Then BEAGLE software (Version 3.3.2) [32] was adopted to impute missing genotype for all the SNPs with assigned physical positions. In second quality control, out of the imputed SNPs, SNPs with a call rate < 90% (n = 286/276/428 for Landrace, Yorkshire and Songliao Black respectively) were discarded, and SNPs with MAF < 0.03 in the resource population (n = 3704/5887/7868 for Landrace, Yorkshire and Songliao Black respectively) were discarded, and SNPs with Hardy–Weinberg equilibrium (p < 10-6) (n = 171/602/220 for Landrace, Yorkshire and Songliao Black respectively) were discarded. In this way, 49,140/49,695/48,809 SNPs were available for Landrace, Yorkshire and Songliao Black respectively. Finally 40833 common SNPs in three breeds were selected for the subsequent analyses. The distribution of common SNPs across genome after filtering was presented in Additional file 1: Table S1.
Statistical analyses
Parentage test
Considering the probability of potential parentage mistakes in the original pedigree records, we adopted Cervus [33] to estimate the most possible parent-offspring pair with maximum likelihood method using 100 randomly chosen autosomal SNPs with 100% call rate. Eventually, about 10% piglets had incorrect recorded sires or dams. Among these individuals with pedigree errors, 64% of them were reassigned to correct parents based on the information of SNP genotypes, and the corrected pedigree information were used in the subsequent analyses, while the remaining individuals which could not be referred to true parents were treated as offspring with unknown parents.
Mixed model based single locus regression analyses (MMRA)
Similar to our previous study by Jiang et al.[34], we performed association tests for each SNP via regression analysis based on the following linear mixed model:
| (1) |
where y is the vector of phenotypes of all piglets on Day 35; μ is the overall mean; c is the vector of phenotypes of all piglets on Day 20, k is the regression coefficient of the phenotypic observations on day 35 on those on day 20; f is the vector of the fixed effects, including effect of breed and batch of sampling, M is the incidence matrix of f; v is the vector of random litter effects, including effect of different litters, T is the incidence matrix of v; X is the vector of the SNP genotype indicators which take values 0, 1 or 2 corresponding to the three genotypes 11, 12 and 22 (assuming 2 is the allele with a minor frequency), b is the regression coefficient of phenotypes on day 35 on SNP genotypes. a is the vector of the residual polygenic effect with (where A is the additive genetic relationship matrix and is the additive variance), Z is the incidence matrix of a; e is the vector of residual errors with (where is the residual error variance). For each SNP, the estimate of b and the corresponding sampling variances can be obtained via mixed model equations (MME), and a Wald Chi-squared statistic with df = 1 is constructed to examine whether the SNP is associated with the trait surveyed.
We employed Fortran 95 to code the computing program for the method and it is available upon request.
Statistical Inference
For the analyses above, the permutation method was adopted to adjust for multiple testing for the number of SNP loci detected. In our method, the observations of each phenotype were randomly shuffled 10,000 times and the empirical critical value was determined by choosing the 95th percentile of the highest statistic over the 10,000 permutation replicates at both genome-wise and chromosome-wise. We declared a significant SNP at a 0.05 significance level if its raw value of the Wald Chi-squared statistic was larger than the empirical critical value.
Population stratification assessment
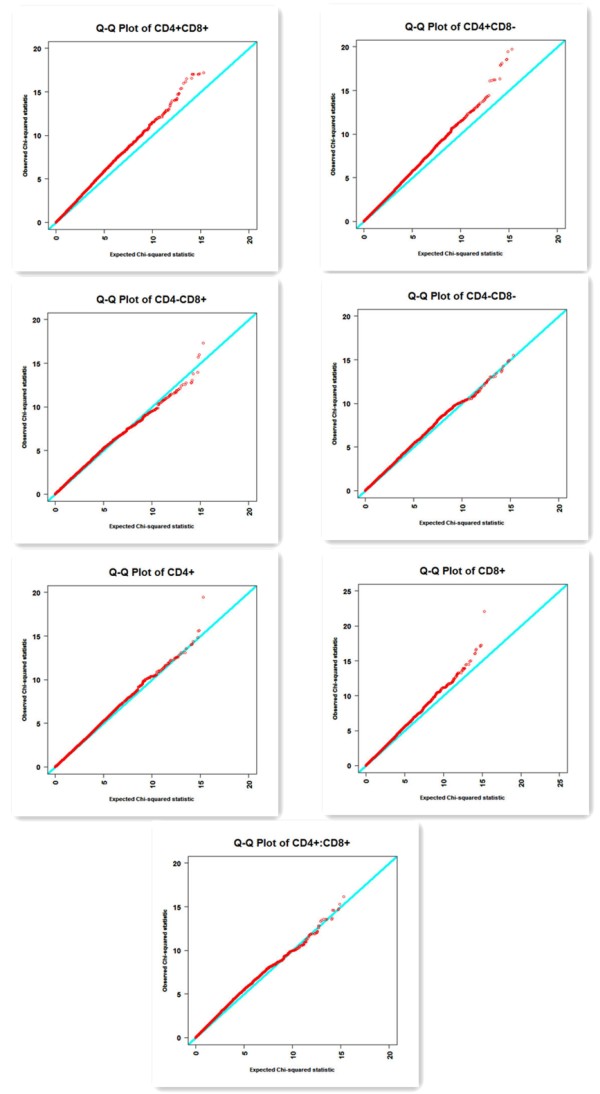
Confounding due to population stratification has been considered as a major plague to the validity of genetic association studies [35]. To view if the population stratification exists in our experimental population, we examined the distribution of the test statistics obtained from the numerous association tests performed and assessed their deviation from the expected distribution of no SNP being associated with the trait of interest by utilizing a quantile-quantile (Q-Q) plot, which is a routine and most frequently used tool for scrutinizing the population stratification in GWAS.
Linkage disequilibrium block analyses
Linkage disequilibrium (LD) block analyses were performed for the chromosomal regions with multiple significant SNPs clustered around genome-wise significant SNPs. The LD blocks were defined using Haploview (Version 4.2) [36], and the LD blocks were defined by the criteria of Gabriel et al. [37] to further pinpoint potential candidate genes affecting T-cell subpopulations.
Results
Alterations of proportions of T-cell subpopulations in peripheral blood after challenge
The descriptive statistics of T-cell subpopulations or their ratio in peripheral blood on day 20 (the day before vaccinating) and day 35 (the day two weeks after vaccinating) are shown in Table 2.
Table 2.
Descriptive statistics analysis and estimates of variance components of immune traits in piglets
| Trait | Test Day | Mean | Standard Deviation | CV(%) | Variance component of genetic effect | Variance component of litter effect |
|---|---|---|---|---|---|---|
| CD4-CD8- T % |
20 |
36.2a |
25.98 |
71.76 |
32.65 |
33.28 |
| |
35 |
35.7 a |
10.77 |
30.18 |
|
|
| CD4+CD8+ T % |
20 |
8.11 a |
3.70 |
45.63 |
3.43 |
3.54 |
| |
35 |
11.23b |
5.06 |
45.07 |
|
|
| CD4+CD8- T % |
20 |
19.87 a |
7.76 |
39.08 |
2.90 |
8.26 |
| |
35 |
16.38b |
7.28 |
44.42 |
|
|
| CD4-CD8+ T % |
20 |
36.8 a |
13.57 |
36.88 |
6.88 |
35.45 |
| |
35 |
36.77 a |
11.53 |
31.37 |
|
|
| CD4+ T % |
20 |
27.92 a |
8.64 |
30.94 |
20.14 |
10.35 |
| |
35 |
27.64 a |
8.83 |
31.96 |
|
|
| CD8+ T % |
20 |
44.93 a |
14.14 |
31.47 |
2.44 |
48.44 |
| |
35 |
47.96b |
12.57 |
26.22 |
|
|
| CD4+ / CD8+ |
20 |
0.72 a |
0.39 |
54.2 |
0.74E-02 |
0.81E-02 |
| 35 | 0.63b | 0.29 | 46.71 |
Note: In the third column, different letters in superscript denote significant difference between ages 20 and 35 for the same trait.
Compared with the measurements on day 20, the proportion of CD4+CD8- T cells and the ratio of CD4+ to CD8+ in blood on day 35 decreased, while the proportion of CD4+CD8+ T cells and CD8+ T cells in blood at day 35 increased significantly. The proportion of CD4+, CD4-CD8- and CD4-CD8+ T cells on day 35 did not changed significantly.
Significant SNPs
Both genome-wise significant SNPs and chromosome-wise significant SNPs for the seven traits are presented in Table 3. The profiles of p values (in terms of –log10p value) of all tested SNPs for the seven phenotypes of T-cell subpopulations are shown in Additional file 2: FigureS1. In total, 64 significant (P < 0.05) SNPs reached the chromosome-wise level. Among these identified SNPs, three reached the genome-wise significance level. In addition, some of these SNPs were identified to be associated with more than one trait, e.g., ALGA0006170 on SSC1 (associated with CD8+ and CD4-CD8- T cells), H3GA0041012 on SSC14 (associated with the ratio of CD4+ to CD8+, CD4+CD8- and CD4-CD8+ T cells), ALGA0122248 on SSC16 (associated with CD4+ and the ratio of CD4+ to CD8+ T cells), INRA0055474 on SSC18 (associated with CD4+ and CD4+CD8- T cells), ALGA0098112 on SSC18 (associated with CD4+CD8- and the ratio of CD4+ to CD8+ T cells) and MARC0089391 on SSC18 (associated with CD4+CD8- and the ratio of CD4+ to CD8+ T cells). Several regions harbour more than one significant SNP, e.g., 10 significant SNP in the 0.8 Mb region (form 6.6 Mb to 7.4 Mb) on SSC12 and 6 significant SNP in the 1.4 Mb region (form 116.7 Mb to 118.1 Mb) on SSC13.
Table 3.
Significant SNPs for T-cell subpopulations
|
Trait |
No. SNPs |
SNP Name |
SSC. |
Position |
Significance levelb |
Nearest gene |
Distance (bp) |
|---|---|---|---|---|---|---|---|
| (bp)a | |||||||
| CD4-CD8- T% |
3 |
ALGA0006170 |
SSC1 |
147676962 |
A |
SELS |
101263 |
| |
|
ALGA0027442 |
SSC4 |
106255554 |
A |
LOC100515138 |
15207 |
| |
|
MARC0035862 |
SSC17 |
26683704 |
A |
FLRT3 |
999166 |
| CD4+CD8+ T% |
15 |
ASGA0000475 |
SSC1 |
5393161 |
A |
QKI |
197563 |
| |
|
MARC0090836 |
SSC5 |
18263549 |
A |
LOC100523435 |
8399 |
| |
|
H3GA0016197 |
SSC5 |
30846602 |
A |
LOC100514286 |
within |
| |
|
DRGA0005876 |
SSC5 |
52631994 |
A |
LOC100153016 |
7905 |
| |
|
ASGA0093882 |
SSC8 |
27821389 |
A |
LOC100515221 |
68339 |
| |
|
MARC0103793 |
SSC9 |
117134341 |
A |
NCF2 |
within |
| |
|
ALGA0061180 |
SSC11 |
18408471 |
A |
LOC100518860 |
within |
| |
|
ALGA0062506 |
SSC11 |
55628913 |
A |
RNF219 |
152990 |
| |
|
DRGA0012994 |
SSC13 |
116721928 |
A |
LOC100518275 |
Within |
| |
|
ASGA0059251 |
SSC13 |
116794774 |
A |
LOC100518275 |
within |
| |
|
H3GA0037561 |
SSC13 |
117065279 |
A |
LOC100518275 |
within |
| |
|
MARC0024545 |
SSC13 |
117511306 |
A |
LOC100739759 |
within |
| |
|
H3GA0037568 |
SSC13 |
117963270 |
A |
CLDN11 |
within |
| |
|
ALGA0072642 |
SSC13 |
118134975 |
A |
SLC7A14 |
22223 |
| |
|
ASGA0077977 |
SSC17 |
59215829 |
A |
CBLN4 |
within |
| CD4+CD8- T% |
31 |
SIRI0000967 |
SSC1 |
65386385 |
A |
MAP3K7 |
93369 |
| |
|
ALGA0109882 |
SSC1 |
65402776 |
A |
MAP3K7 |
76978 |
| |
|
ALGA0003935 |
SSC1 |
67256904 |
A |
LOC100152346 |
within |
| |
|
MARC0008049 |
SSC6 |
74003688 |
A |
LOC100738715 |
within |
| |
|
ALGA0105115 |
SSC9 |
127202082 |
A |
CD46 |
12474 |
| |
|
MARC0076632 |
SSC9 |
127394064 |
B |
LOC100514786 |
within |
| |
|
ASGA0046812 |
SSC10 |
16205420 |
A |
LOC100513811 |
within |
| |
|
ALGA0057450 |
SSC10 |
16220722 |
A |
LOC100513811 |
within |
| |
|
ALGA0061535 |
SSC11 |
23576083 |
A |
TRNAE-UUC |
42913 |
| |
|
M1GA0015162 |
SSC11 |
61231196 |
A |
LOC100154696 |
within |
| |
|
ASGA0051093 |
SSC11 |
61450492 |
A |
LOC100154696 |
112456 |
| |
|
ALGA0108362 |
SSC12 |
6596691 |
A |
LOC100518214 |
225784 |
| |
|
MARC0009109 |
SSC12 |
6637944 |
A |
SOX9 |
229926 |
| |
|
ALGA0106073 |
SSC12 |
6663787 |
A |
SOX9 |
204083 |
| |
|
ASGA0052986 |
SSC12 |
6791202 |
A |
SOX9 |
76668 |
| |
|
ASGA0052974 |
SSC12 |
6826155 |
A |
SOX9 |
41715 |
| |
|
ALGA0064738 |
SSC12 |
6891741 |
A |
SOX9 |
20500 |
| |
|
H3GA0033370 |
SSC12 |
6976387 |
B |
LOC100518393 |
21514 |
| |
|
ASGA0053002 |
SSC12 |
7105246 |
A |
LOC100519105 |
55408 |
| |
|
ASGA0104370 |
SSC12 |
7185885 |
A |
LOC100519105 |
136047 |
| |
|
ALGA0064767 |
SSC12 |
7400177 |
A |
LOC100519105 |
350339 |
| |
|
ALGA0078815 |
SSC14 |
76804400 |
A |
LOC100521406 |
85124 |
| |
|
H3GA0041012 |
SSC14 |
77946383 |
A |
PSAP |
within |
| |
|
ASGA0065687 |
SSC14 |
107127514 |
A |
HTR7 |
260753 |
| |
|
ALGA0087956 |
SSC15 |
123078022 |
A |
LOC100516232 |
22744 |
| |
|
INRA0055474 |
SSC18 |
21991584 |
A |
LOC100622672 |
28674 |
| |
|
ALGA0097499 |
SSC18 |
22003396 |
A |
LOC100513190 |
3075 |
| |
|
ASGA0079256 |
SSC18 |
22055859 |
A |
LOC100525548 |
1899 |
| |
|
ALGA0098112 |
SSC18 |
38463920 |
A |
BMPER |
113622 |
| |
|
MARC0089391 |
SSC18 |
38488630 |
A |
BMPER |
88912 |
| |
|
MARC0024065 |
SSC18 |
50965324 |
A |
LOC100622432 |
within |
| CD4-CD8+ T% |
3 |
ASGA0022812 |
SSC4 |
126976845 |
A |
LOC100737939 |
107579 |
| |
|
H3GA0041012 |
SSC14 |
77946383 |
A |
PSAP |
within |
| |
|
ALGA0103427 |
SSC15 |
127742340 |
A |
IKZF2 |
within |
| CD4+ T% |
6 |
ASGA0023262 |
SSC4 |
130826499 |
A |
LOC100151966 |
174827 |
| |
|
ALGA0030335 |
SSC5 |
7606208 |
A |
LOC100512654 |
6682 |
| |
|
ALGA0122248 |
SSC16 |
73417692 |
A |
LOC100518939 |
70693 |
| |
|
INRA0055474 |
SSC18 |
21991584 |
A |
LOC100513190 |
within |
| |
|
ASGA0079256 |
SSC18 |
22055859 |
A |
LOC100525548 |
1899 |
| |
|
ASGA0080319 |
SSC18 |
49513089 |
A |
LOC100516656 |
43287 |
| CD8+ T% |
1 |
ALGA0006170 |
SSC1 |
147676962 |
B |
SELS |
101263 |
| CD4+:CD8+ |
5 |
H3GA0041012 |
SSC14 |
77946383 |
A |
PSAP |
within |
| |
|
ALGA0117513 |
SSC15 |
123215784 |
A |
LOC100518938 |
11678 |
| |
|
ALGA0122248 |
SSC16 |
73417692 |
A |
LOC100738933 |
403285 |
| |
|
ALGA0098112 |
SSC18 |
38463920 |
A |
BMPER |
113622 |
| MARC0089391 | SSC18 | 38488630 | A | BMPER | 88912 |
a: Derived from the recent porcine genome sequence assembly (Sscrofa9.2) ( http://www.ncbi.nlm.nih.gov/projects/mapview/map_search.cgi?taxid=9823&build=previous).
b: A means chromosome-wise significance level, B means genome-wise significance level.
Population stratification assessment
The Q-Q plots for the test statistics of MMRA are shown in Figure 1. From these plots, it is apparent that there is no clear overall systematic bias in most of traits. However, for the traits of CD4+CD8-, CD4+CD8+ and CD8+ T cells, the effects of population stratification caused an overall slightly systematic bias, and the points where the observed statistics of the significant SNPs are higher than the expected χ2 statistics at the significance level defined by the permutation tests.
Figure 1 .
Quantile-quantile (Q-Q) plots of test statistics in GWA for T-cell subpopulations. Under the null hypothesis of no association at any SNP locus, the points would be expected to follow the slope lines. Deviations from the slope lines correspond to loci that deviate from the null hypotheses.
Discussion
T cells are the major cell populations mediating the adaptive arm of the immune system. Several studies on T cell subpopulations showed that variations in CD4 and CD8 T cell levels and the ratio of CD4 to CD8 are significantly heritable [38-40]. Heritability estimates were around 65% for the ratio of CD4 to CD8, 50% for CD4+ counts, and 56% for CD8+ counts [40]. Therefore, as a category of immune-related traits with high heritability, T-cell subpopulations can be potentially implemented to selection for disease resistance and susceptibility in swine breeding. The present results clearly show that a number of loci contribute to the variation of T-cell subpopulations in peripheral blood in pig. These findings would enhance our understanding of genetic control of the variations of T-cell subpopulations.
In this study, we treated breed as a fixed effect to avoid potential confounding between effects of SNP and breed. Hence, to check whether significant SNPs actually were segregating in the different breeds and showing the same effects in the different breeds, we tested all the significant SNPs based on the improved model considering the interaction of SNP by breed. Finally, no SNP has a significant interaction effect was detected. And the main purpose of our study is to detect common SNPs influencing the T-cell subpopulations in swine, so we did not put the interaction effect in our association model in GWAS.
In this study, we carried out GWAS to explore potential causal genes for the T-cell subpopulations in swine. To our knowledge, this is the first study aiming to reveal the genetic mechanism of those immune traits in swine based on a high density SNP chip panel. Our results revealed 64 significant SNPs associated with the seven traits. Among these identified SNPs, 27 fall into previously reported immune-related QTL regions [27,41,42], including 10 significant SNPs in the regions which have been reported to harbor QTL for lymphocyte previously [27,42-44]. In particular, the significant SNP (ASGA0077977) with effect on the CD4+CD8+ on SSC17 is located within the reported QTL for CD4+CD8+ region [27]; The significant SNP (ASGA0000475) for CD4+CD8+ on SSC1 is located within the reported QTL for CD4+/CD8+ [27]; The significant SNP (ALGA0027442) for CD4-CD8- on SSC4 is located within the reported QTL for CD4+ [27]; The significant SNP for CD4+ on SSC5 (ALGA0030335) fell into the region which has been reported to harbor QTL for CD4+ in our previous studies [27].
Several significant SNPs were found associated with more than one trait in this study. Specifically, SNP H3GA0041012 on SSC14 was associated with CD4+/CD8+, CD4+CD8- and CD4-CD8+ T cells. SNPs ALGA022248 on SSC16 and INRA0055474, ASGA0079256 and ALGA0098112 on SSC18 were associated with two traits, respectively. The traits of T-cell subpopulations are correlated, so a change in one trait may result in a change also in other traits, which should be the reason that these SNP significantly associated with more than one trait.
Several regions harboured more than one significant SNP. Six SNPs which have significantly associated with the proportion of CD4+CD8+ T cells fell in the region of 116.7 Mb to 118.1 Mb on SSC13. Ten SNPs which fell in the region of 6.6-7.4 Mb on SSC12 were significantly associated with the proportion of CD4+CD8- T cells. In this region, QTLs for lymphocyte were mapped previously [42]. Additionally, the LD block analysis for SSC12 showed that a LD block exists between ALGA0064738 and ALGA006476, where the genome-wise significant SNP H3GA0033370 was included (Figure 2).
Figure 2 .
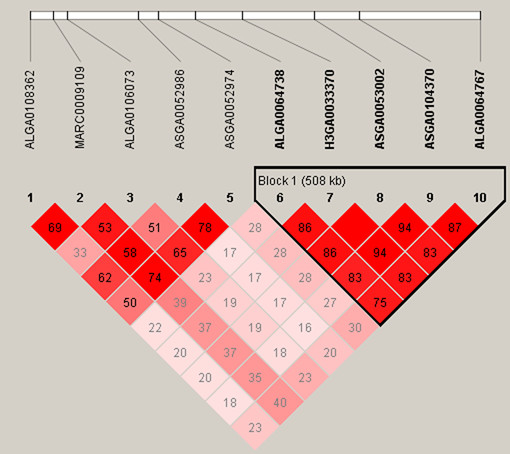
Linkage disequilibrium (LD) pattern for significant SNPs on SSC12. LD blocks are marked with triangles. Values in boxes are LD (r²) between SNP pairs and the boxes are coloured according to the standard Haploview colour scheme: LOD > 2 and D' = 1, red; LOD > 2 and D' < 1, shades of pink/red; LOD < 2 and D' = 1, blue; LOD < 2 and D' < 1, white (LOD is the log of the likelihood odds ratio, a measure of confidence in the value of D'.
Several significant SNPs fell into the regions which harbour a number of known immunity-related genes. On SSC12, ten SNPs (one of them with genome-wise significance level) for the proportion of CD4+CD8- T cells fell in the 0.8 Mb region, which harbours SOX9 ((sex determining region Y)-box 9) gene. This gene has important role in chondrogenesis, sex determination, pigmentation, organ maintenance and cancer. To our knowledge, differential Sox9 expression is critical for the establishment and maintenance of a regular thymic microenvironment, where specialized stromal cells promote thymocytes development and selection to functionally mature T cells. And the LD block analysis for this region indicated that SOX9 gene was in LD with the most significant SNP of CD4+CD8- T%, so it has a great potential value of investigation its function on T-cell subpopulations in further research. SNPs ALGA0105115 and MARC0076632 with effects on the CD4+CD8- T cells were found in the region which harbours CD34 (CD34 molecule) gene. SNPs ASGA0046812 and ALGA0057450 with effects on the CD4+CD8- T cells were found in the region which harbours AKT3 (v-akt murine thymoma viral oncogene homolog 3 (protein kinase B, gamma)) gene. The AKT gene family has been implicated in signal transmission leading to activation, differentiation as well as cellular survival of T-lymphocytes. The candidate genes discussed there need further research to confirm the genetic mechanism on the traits in this study.
Conclusions
Summary, our study revealed 64 SNPs associated with T-cell subpopulation in peripheral blood in pigs at chromosome-wise significance level (including 3 SNPs at genome-wise significance level) and 27 significance SNPs were located within the immune-related QTL regions reported in previous studies. Furthermore, 14 significant SNPs fell into the regions harboring known immunity-related genes. Findings herein lay a preliminary foundation for further identifying the causal mutations affecting swine immune capacity in follow-up studies.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
XL and WXF are the major executive persons of all jobs of this study, including collection of the phenotypes, SNP genotyping, statistical analysis, and drafting this manuscript. YRL and YL assisted in phenotype collecting and SNP genotyping. XDD and JPZ assisted in the statistical analysis. JFL and QZ planed and supervised the whole study. All authors read and approved the final manuscript.
Supplementary Material
Distributions of SNPs after quality control on each chromosome.
Manhattan plots of GWA for T-cell subpopulations. Different chromosomes are represented by different colors. Chromosome 19 stands for the X chromosome of swine.
Contributor Information
Xin Lu, Email: xinlu1981@gmail.com.
Wei-Xuan Fu, Email: weixuan.fu@gmail.com.
Yan-Ru Luo, Email: luoyanru-1204@163.com.
Xiang-Dong Ding, Email: xding@cau.edu.cn.
Jia-Peng Zhou, Email: zhoujp111@gmail.com.
Yang Liu, Email: yangliucau@gmail.com.
Jian-Feng Liu, Email: liujf@cau.edu.cn.
Qin Zhang, Email: qzhang@cau.edu.cn.
Acknowledgements
The authors appreciate the financial support provided by the National Natural Science Foundations of China (30972092), the National High Technology Research and Development Program of China (863 Program 2011AA100302), the National Major Special Project of China on New Varieties Cultivation for Transgenic Organisms (2009ZX08009-146B), the Natural Science Foundations of Beijing (6102016), New-Century Training Programme Foundation for the Talents by the State Education Commission of China (NETC-10-0783) and Scientific Research Foundation for the Returned Overseas Chinese Scholars of State Education Ministry.
References
- Visscher AH, Janss LL, Niewold TA, de Greef KH. Disease incidence and immunological traits for the selection of healthy pigs. A review. Vet Q. 2002;24(1):29–34. doi: 10.1080/01652176.2002.9695121. [DOI] [PubMed] [Google Scholar]
- Alving K. Airways vasodilatation in the immediate allergic reaction. Involvement of inflammatory mediators and sensory nerves. Acta Physiol Scand Suppl. 1991;597:1–64. [PubMed] [Google Scholar]
- Kenmochi T, Mullen Y, Miyamoto M, Stein E. Swine as an Allotransplantation Model. Vet Immunol Immunop. 1994;43(1–3):177–183. doi: 10.1016/0165-2427(94)90134-1. [DOI] [PubMed] [Google Scholar]
- Misfeldt ML, Grimm DR. Sinclair Miniature Swine - an Animal-Model of Human-Melanoma. Vet Immunol Immunop. 1994;43(1–3):167–175. doi: 10.1016/0165-2427(94)90133-3. [DOI] [PubMed] [Google Scholar]
- Doyle C, Strominger JL. Interaction between CD4 and Class II MHC Molecules Mediates Cell Adhesion (Reprinted from Nature, vol 330, pg 256–259, 1987) J Immunol. 2010;184(11):5935–5938. [PubMed] [Google Scholar]
- Regueiro JR, Rodríguez-Gallego C, Arnaiz-Villena A. Human T-lymphocyte activation deficiencies. R.G. Landes; CRC Press distributor, Austin, TX Boca Raton, FL; 1994. [Google Scholar]
- Shedlock DJ, Shen H. Requirement for CD4 T cell help in generating functional CD8 T cell memory. Science. 2003;300(5617):337–339. doi: 10.1126/science.1082305. [DOI] [PubMed] [Google Scholar]
- Swain SL, Dutton RW, Schwab R, Yamamoto J. Xenogeneic Human Anti-Mouse T-Cell Responses Are Due to the Activity of the Same Functional T-Cell Subsets Responsible for Allospecific and Major Histocompatibility Complex-Restricted Responses. J Exp Med. 1983;157(2):720–729. doi: 10.1084/jem.157.2.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty PC, Topham DJ, Tripp RA. Establishment and persistence of virus-specific CD4(+) and CD8(+) T cell memory. Immunol Rev. 1996;150:23–44. doi: 10.1111/j.1600-065X.1996.tb00694.x. [DOI] [PubMed] [Google Scholar]
- Ober BT, Summerfield A, Mattlinger C, Wiesmuller KH, Jung G, Pfaff E, Saalmuller A, Rziha HJ. Vaccine-induced, pseudorabies virus-specific, extrathymic CD4(+)CD8(+) memory T-helper cells in swine. J Virol. 1998;72(6):4866–4873. doi: 10.1128/jvi.72.6.4866-4873.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damoiseaux JG, Cautain B, Bernard I, Mas M, van Breda Vriesman PJ, Druet P, Fournie G, Saoudi A. A dominant role for the thymus and MHC genes in determining the peripheral CD4/CD8 T cell ratio in the rat. J Immunol. 1999;163(6):2983–2989. [PubMed] [Google Scholar]
- Gerner W, Käser T, Saalmüller A. Porcine T lymphocytes and NK cells–an update. Dev Comp Immunol. 2009;33(3):310–320. doi: 10.1016/j.dci.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Dwivedi V, Manickam C, Binjawadagi B, Linhares D, Murtaugh M, Renukaradhya G. Evaluation of immune responses to porcine reproductive and respiratory syndrome virus in pigs during early stage of infection under farm conditions. Virol J. 2012;16(9):45. doi: 10.1186/1743-422X-9-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armengol E, Wiesmüller K, Wienhold D, Büttner M, Pfaff E, Jung G, Saalmüller A. Identification of T-cell epitopes in the structural and non-structural proteins of classical swine fever virus. J Gen Virol. 2002;83(Pt 3):551–560. doi: 10.1099/0022-1317-83-3-551. [DOI] [PubMed] [Google Scholar]
- Rau H, Revets H, Balmelli C, McCullough K, Summerfield A. Immunological properties of recombinant classical swine fever virus NS3 protein in vitro and in vivo. Vet Res. 2006;37(1):155–168. doi: 10.1051/vetres:2005049. [DOI] [PubMed] [Google Scholar]
- Kimman T, DeBruin T, Voermans J, Bianchi A. Cell-mediated immunity to pseudorabies virus: cytolytic effector cells with characteristics of lymphokine-activated killer cells lyse virus-infected and glycoprotein gB- and gC-transfected L14 cells. J Gen Virol. 1996;77(Pt 5):987–990. doi: 10.1099/0022-1317-77-5-987. [DOI] [PubMed] [Google Scholar]
- Bianchi A, Moonen-Leusen H, Van Milligen F, Savelkoul H, Zwart R, Kimman T. A mouse model to study immunity against pseudorabies virus infection: significance of CD4+ and CD8+ cells in protective immunity. Vaccine. 1998;16(16):1550–1558. doi: 10.1016/S0264-410X(98)00044-9. [DOI] [PubMed] [Google Scholar]
- Carr MM, Howard CJ, Sopp P, Manser JM, Parsons KR. Expression on Porcine Gamma-Delta Lymphocytes of a Phylogenetically Conserved Surface-Antigen Previously Restricted in Expression to Ruminant Gamma-Delta T-Lymphocytes. Immunology. 1994;81(1):36–40. [PMC free article] [PubMed] [Google Scholar]
- Hirt W, Saalmuller A, Reddehase MJ. Distinct Gamma-Delta-T-Cell Receptors Define 2 Subsets of Circulating Porcine Cd2-Cd4-Cd8- Lymphocytes-T. Eur J Immunol. 1990;20(2):265–269. doi: 10.1002/eji.1830200206. [DOI] [PubMed] [Google Scholar]
- Saalmuller A, Hirt W, Reddehase MJ. Porcine Gamma-Sigma-Lymphocyte-T Subsets Differing in Their Propensity to Home to Lymphoid-Tissue. Eur J Immunol. 1990;20(10):2343–2346. doi: 10.1002/eji.1830201026. [DOI] [PubMed] [Google Scholar]
- Pescovitz MD, Lunney JK, Sachs DH. Murine Anti-Swine T4 and T8 Monoclonal-Antibodies - Distribution and Effects on Proliferative and Cyto-Toxic T-Cells. J Immunol. 1985;134(1):37–44. [PubMed] [Google Scholar]
- Saalmuller A, Reddehase MJ, Buhring HJ, Jonjic S, Koszinowski UH. Simultaneous Expression of Cd4 and Cd8 Antigens by a Substantial Proportion of Resting Porcine Lymphocytes-T. Eur J Immunol. 1987;17(9):1297–1301. doi: 10.1002/eji.1830170912. [DOI] [PubMed] [Google Scholar]
- Summerfield A, Rziha HJ, Saalmuller A. Functional characterization of porcine CD4(+)CD8(+) extrathymic T lymphocytes. Cell Immunol. 1996;168(2):291–296. doi: 10.1006/cimm.1996.0078. [DOI] [PubMed] [Google Scholar]
- De Bruin T, Van Rooij E, De Visser Y, Bianchi A. Cytolytic function for pseudorabies virus-stimulated porcine CD4+ CD8dull+ lymphocytes. Viral Immunol. 2000;13(4):511–520. doi: 10.1089/vim.2000.13.511. [DOI] [PubMed] [Google Scholar]
- Xu Y, Cui L, Tian C, Zhang G, Huo G, Tang L, Li Y. Immunogenicity of recombinant classic swine fever virus CD8(+) T lymphocyte epitope and porcine parvovirus VP2 antigen coexpressed by Lactobacillus casei in swine via oral vaccination. Clin Vaccine Immunol. 2011;18(11):1979–1986. doi: 10.1128/CVI.05204-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattrang E, Almqvist M, Johansson A, Fossum C, Wallgren P, Pielberg G, Andersson L, Edfors-Lilja I. Confirmation of QTL on porcine chromosomes 1 and 8 influencing leukocyte numbers, haematological parameters and leukocyte function. Anim Genet. 2005;36(4):337–345. doi: 10.1111/j.1365-2052.2005.01315.x. [DOI] [PubMed] [Google Scholar]
- Lu X, Liu J-F, Gong Y-F, Wang Z-P, Liu Y, Zhang Q. Mapping quantitative trait loci for T lymphocyte subpopulations in peripheral blood in swine. BMC Genet. 2011;12:79. doi: 10.1186/1471-2156-12-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson L. Genome-wide association analysis in domestic animals: a powerful approach for genetic dissection of trait loci. Genetica. 2009;136(2):341–349. doi: 10.1007/s10709-008-9312-4. [DOI] [PubMed] [Google Scholar]
- Ramos AM, Crooijmans RPMA, Affara NA, Amaral AJ, Archibald AL, Beever JE, Bendixen C, Churcher C, Clark R, Dehais P. et al. Design of a High Density SNP Genotyping Assay in the Pig Using SNPs Identified and Characterized by Next Generation Sequencing Technology. PLoS One. 2009;4(8):e6524. doi: 10.1371/journal.pone.0006524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duijvesteijn N, Knol EF, Merks JWM, Crooijmans RPMA, Groenen MAM, Bovenhuis H, Harlizius B. A genome-wide association study on androstenone levels in pigs reveals a cluster of candidate genes on chromosome 6. BMC Genet. 2010;11:42. doi: 10.1186/1471-2156-11-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan B, Onteru S, Du Z, Garrick D, Stalder K, Rothschild M. Genome-wide association study identifies Loci for body composition and structural soundnesstraits in pigs. PLoS One. 2011;6(2):e14726. doi: 10.1371/journal.pone.0014726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A, Crooijmans R, Affara N, Amaral A, Archibald A, Beever J, Bendixen C, Churcher C, Clark R, Dehais P. et al. Design of a high density SNP genotyping assay in the pig using SNPs identified and characterized by next generation sequencing technology. PLoS One. 2009;4(8):e6524. doi: 10.1371/journal.pone.0006524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84(2):210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowski ST, Taper ML, Marshall TC. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol. 2007;16(5):1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x. [DOI] [PubMed] [Google Scholar]
- Jiang L, Liu J, Sun D, Ma P, Ding X, Yu Y, Zhang Q. Genome wide association studies for milk production traits in Chinese Holstein population. PLoS One. 2010;5(10):e13661. doi: 10.1371/journal.pone.0013661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson TA, Manolio TA. How to interpret a genome-wide association study. Jama-J Am Med Assoc. 2008;299(11):1335–1344. doi: 10.1001/jama.299.11.1335. [DOI] [PubMed] [Google Scholar]
- Barrett J, Fry B, Maller J, Daly M. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Amadori A, Zamarchi R, Chieco-Bianchi L. CD4: CD8 ratio and HIV infection: the “tap-and-drain” hypothesis. Immunol Today. 1996;17:414–417. doi: 10.1016/0167-5699(96)10049-9. [DOI] [PubMed] [Google Scholar]
- Evans DM, Frazer IH, Martin NG. Genetic and environmental causes of variation in basal levels of blood cells. Twin Res. 1999;2:250–257. doi: 10.1375/136905299320565735. [DOI] [PubMed] [Google Scholar]
- Hall MA, Ahmadi KA, Norman P, Snieder H, Macgregor A, Vaughan RW, Spector TD, Lanchbury JS. Genetic influence on peripheral blood T lymphocyte levels. Genes Immun. 2000;1:423–427. doi: 10.1038/sj.gene.6363702. [DOI] [PubMed] [Google Scholar]
- Reiner G, Fischer R, Hepp S, Berge T, Kohler F, Willems H. Quantitative trait loci for white blood cell numbers in swine. Anim Genet. 2008;39(2):163–168. doi: 10.1111/j.1365-2052.2008.01700.x. [DOI] [PubMed] [Google Scholar]
- Edfors-Lilja I, Wattrang E, Marklund L, Moller M, Andersson-Eklund L, Andersson L, Fossum C. Mapping quantitative trait loci for immune capacity in the pig. J Immunology. 1998;160:829–835. [PubMed] [Google Scholar]
- Edfors-Lilja I, Wattrang E, Andersson L, Fossum C. Mapping quantitative trait loci for stress induced alterations in porcine leukocyte numbers and functions. Anim Genet. 2000;31:186–193. doi: 10.1046/j.1365-2052.2000.00628.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distributions of SNPs after quality control on each chromosome.
Manhattan plots of GWA for T-cell subpopulations. Different chromosomes are represented by different colors. Chromosome 19 stands for the X chromosome of swine.