Abstract
The Rho small GTP-binding proteins are versatile, conserved molecular switches in eukaryotic signal transduction. Plants contain a unique subfamily of Rho-GTPases called Rop (Rho-related GTPases from plants). Our previous studies involving injection of antibodies indicated that the pea Rop GTPase Rop1Ps is critical for pollen tube growth. In this study we show that overexpression of an apparent Arabidopsis ortholog of Rop1Ps, Rop1At, induces isotropic cell growth in fission yeast (Schizosaccharomyces pombe) and that green fluorescence protein-tagged Rop1At displays polar localization to the site of growth in yeast. We found that Rop1At and two other Arabidopsis Rops, Rop3At and Rop5At, are all expressed in mature pollen. All three pollen Rops fall into the same subgroup as Rop1Ps and diverge from those Rops that are not expressed in mature pollen, suggesting a coupling of the structural conservation of Rop GTPases to their gene expression in pollen. However, pollen-specific transcript accumulation for Rop1At is much higher than that for Rop3At and Rop5At. Furthermore, Rop1At is specifically expressed in anthers, whereas Rop3At and Rop5At are also expressed in vegetative tissues. In transgenic plants containing the Rop1At promoter:GUS fusion gene, GUS is specifically expressed in mature pollen and pollen tubes. We propose that Rop1At may play a predominant role in the regulation of polarized cell growth in pollen, whereas its close relatives Rop3At and Rop5At may be functionally redundant to Rop1At in pollen.
In angiosperms male gametophyte development can be divided into two major phases: microsporophyte development and pollen development. Microsporophyte development is the division of a diploid sporophytic cell, giving rise to the tapetal initial cell and the microspore mother cell. This diploid microspore mother cell undergoes meiosis to produce haploid microspores. Microspores then enter the phase of pollen development, which begins with an asymmetric mitotic division, resulting in the formation of a pollen grain containing a large, vegetative cell and a small, generative cell enclosed within it. In some species, such as Arabidopsis, the generative cell undergoes a second mitotic division in developing pollen before anthesis to produce a tricellular mature pollen grain. In other species, mature pollen grains are released as bicellular cells, and the second mitotic division occurs during pollen tube growth within the style (Mascarenhas, 1993; McCormick, 1993).
Pollen development involves complex developmental control of gene expression by the haploid genome. It has been estimated that 10% of the 20,000 different genes expressed in pollen grains at anthesis are pollen specific (for review, see Mascarenhas, 1993; McCormick, 1993; Taylor and Helper, 1997). Pollen-specific genes can be divided into two groups: Genes expressed before the first pollen mitosis are referred to as “early” pollen genes and are believed to be involved in pollen development; genes activated after this mitosis are called “late” pollen genes and are presumably involved in pollen maturation and germination (Mascarenhas, 1993). At least 23 late pollen genes have been identified from different plant species (for review, see McCormick, 1993; Twell, 1994; Taylor and Helper, 1997). Several of these late pollen genes encode signaling proteins such as a Ca2+-dependent protein kinase involved in self-incompatibility in Nicotiana alata (Kunz et al., 1996), a Ca2+-dependent calmodulin-independent protein kinase involved in pollen germination in maize (Estruch et al., 1994), a receptor-like kinase, PRK1, essential for normal pollen development in petunia (Lee et al., 1996), and a mitogen-activated protein kinase activated upon pollen hydration in Nicotiana tabacum (Wilson et al., 1997).
We previously reported a small GTP-binding protein, Rop1Ps, that preferentially accumulated in mature pollen of the garden pea (Lin et al., 1996). Rop1Ps belongs to the Rho family of small GTPases, which has become an important group of conserved signaling proteins in eukaryotes. Rho-dependent signaling controls a large variety of key cellular processes in animals and fungi, e.g. actin cytoskeletal reorganization, the establishment of cell polarity, polarized cell growth, membrane trafficking and organization (e.g. exocytosis and endocytosis), focal adhesion, and cell movement (Hall, 1994; Chant and Stowers, 1995; Lamaze et al., 1996; Larochelle et al., 1996; Murphy et al., 1996; Nagata and Hall, 1996; Ridley, 1996).
Plants possess a family of genes encoding proteins closely related to Rop1Ps, including 10 reported genes from Arabidopsis (Yang and Watson, 1993; Delmer et al., 1995; Lin et al., 1996; Winge et al., 1997). Indirect immunofluorescence studies in pea suggest that Rop1Ps is localized to the tip of pollen tubes (Lin et al., 1996). We showed that injected anti-Rop1Ps antibodies inhibited pollen tube elongation in pea, and that this inhibition was independent of cytoplasmic streaming and potentiated by low extracellular Ca2+ and caffeine treatment (Lin and Yang, 1997). These results suggest that Rop1Ps plays a pivotal role in the control of pollen tube growth, probably by interacting with Ca2+ signaling (Lin and Yang, 1997). However, the precise function of these GTPases in pollen needs to be determined using a reverse-genetics approach. Such an approach is most feasible in Arabidopsis due to the recent development of homology-based gene replacement (Kempin et al., 1997) and PCR-mediated identification of T-DNA insertion into genes of known sequences (McKinney et al., 1995; Krysan et al., 1996).
In this paper we report the identification of a novel member of the Arabidopsis Rop gene family, Rop1At, the only Rop gene known to be specifically expressed in the anther. Rop1At appears to have a conserved function in regulating polarized cell growth in fission yeast (Schizosaccharomyces pombe). Analyses of promoter:GUS reporter fusion gene expression show that Rop1At is a late pollen gene. Rop3At and Rop5At, which are most closely related to Rop1At, are also expressed in mature pollen, although at a lower level, whereas other Rop genes divergent from Rop1At are not expressed in mature pollen. These results imply a functional constraint on the structural conservation of the Rop subfamily of GTPases, with the three most closely related members having a potential redundant function in the control of polarized cell growth in pollen.
MATERIALS AND METHODS
Plant Material
Arabidopsis ecotype Columbia plants were grown in growth chambers at 22°C under constant light. Rosette leaves from 4-week-old plants were harvested for genomic DNA isolation. For RNA extractions roots, stems, rosette leaves, open and closed flowers, siliques, and pollen grains were harvested from 4- to 6-week-old plants.
cDNA and Genomic DNA Cloning and Sequencing
The Arabidopsis Columbia cDNA library, λPRL-2 (Tom Newman, Michigan State University, obtained through the Arabidopsis Biological Resource Center, Ohio State University, Columbus), was screened with a 32P-labeled, 167-bp fragment of Rop1Ps cDNA, which corresponds to the most conserved region within the Rho gene family (Yang and Watson, 1993). Plasmids containing positive clones were excised in vivo from lambda phage, and inserts were subcloned into pBluescript II SK (Stratagene) and sequenced using the dideoxynucleotide chain-termination method (Sanger et al., 1977) and Sequenase version 2.0 (United States Biochemical). The cDNA library screen identified two distinct genes, Rop1At and Rop2At. To isolate additional Rop1Ps-related genes, an Arabidopsis genomic library (Voytas et al., 1990) obtained from the Arabidopsis Biological Resource Center was screened with a 369-bp SacII fragment of Rop2At cDNA under moderate hybridization stringency. Positive clones were subcloned and sequenced as described above.
Computer Analysis of the Rop Subfamily
Predicted amino acid sequences for Arabidopsis Rop1Ps-related genes were compared with known members of the Rho family of GTP-binding proteins available from the GenBank database using computer software from DNASTAR, Inc. (Madison, WI). Alignments of these sequences were carried out using the MegAlign program (DNASTAR, Inc., Madison, WI). Phylogenetic analyses of the aligned sequences were conducted using PAUP (Phylogenetic Analysis Using Parsimony) software (version 3.1.1, D.L. Swofford [1993], Smithsonian Institution, Washington, DC).
Reverse Transcription and PCRs
Total RNA was isolated from different Arabidopsis tissues as described previously (Logemann et al., 1987). The first-strand cDNAs were synthesized using murine leukemia virus RT (GIBCO-BRL) in a 50-μL reaction containing 2.5 μm oligo-dT primers (GIBCO-BRL), 5 μg of total RNA, 10 mm DTT, 1 unit/μL RNase inhibitor (GIBCO-BRL), 0.20 mm dNTP mix, and 10 units/μL RT. Reverse-transcription reactions were carried out at 42°C for 60 min and were terminated by heating to 99°C for 5 min and chilling to 4°C for 5 min. Five microliters of the reaction mixture was used as a template for each of the PCRs described below. PCR reactions were carried out in 25 μL of a mixture containing 2 mm MgCl2, 0.25 unit of Taq polymerase (GIBCO-BRL), and 0.5 μm gene-specific primers (see Table I). For Rop1At and Rop2At, 25 cycles of PCR amplification were carried out at 94°C for 30 s (denaturation), at 60°C for 30 s (annealing), and at 72°C for 30 s (synthesis). Five microliters of each PCR product was loaded on a 1.5% agarose gel to visualize the amplified cDNAs.
Table I.
Rop isogene-specific primers for RT-PCR
| Genes | Sense Primers | Antisense Primers | Expected cDNA Length |
|---|---|---|---|
| bp | |||
| Rop1At | 5′-GAAATTAATAAACTTTGAGGGG-3′ (−24 to −3) | 5′-AGAGATTTCCAATCATCATAG-3′ (+615 to +595) | 639 |
| Rop2At | 5′-GCGGCAGAGATGGCGTCAAGG-3′ (−9 to +11) | 5′-CTTATCACAAGAACGCGCAACG-3′ (+592 to 571) | 601 |
| Rop3At | 5′-TACGTAGCTCCATTTCTGGTGGAG-3′ (−41 to −18) | 5′-CCACAATCCAAGATTGACAGT-3′ (+177 to 157) | 218 |
| Rop4At | 5′-CATTATTATCTCTCATCGATTTGG-3′ (−184 to −161) | Same as above | 361a |
| Rop5At | 5′-GTGACATATTTTGGCTCGTCG-3′ (−38 to −18) | Same as above | 215a |
| Rop6At | 5′-CGTCCGTGAGGATGAGTAGT-3′ (−172 to −153) | Same as above | 349 |
Expected genomic DNA lengths for Rop4At and Rop5At are approximately 800 and 335 bp, respectively.
The same procedures were used for Rop3At and Rop4At, except that the annealing temperature was 55°C. The same PCR reaction conditions were used for Rop5At and Rop6At, but the number of PCR cycles was increased to 45, and 25 μL of the PCR reaction was loaded on a 2% low-melting agarose gel. As PCR amplification and loading controls, the same template cDNA was amplified using primers for the constitutive Act2 gene (An et al., 1996b). Act2 PCR amplification was conducted at 94°C for 30 s, at 55°C for 30 s, and at 72°C for 1 min for 25 cycles, and 5 μL of the PCR reaction was loaded onto a 1% agarose gel. RT-PCR reactions were repeated twice using total RNAs extracted with a different isolation procedure (Thompson et al., 1983).
To ensure gene-specific PCR amplifications, at least one of the two PCR primers was designed according to sequences of divergent 5′ untranslated regions (Table I). To confirm the specificity of each pair of primers, two sets of PCRs were performed separately, one containing 1 ng of a specific cDNA or a genomic DNA clone corresponding to the primers (positive control), and the other containing a mixture of an equal amount (1 ng of each) of cDNA or genomic DNAs for each of the other five Rop genes. PCR conditions were identical to those used for RT-PCR described above.
Construction of Rop1At Promoter:GUS Fusion Gene and Plant Transformation
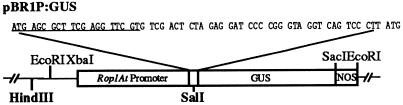
To direct the expression of Rop1At promoter:GUS fusion gene in Arabidopsis plants, a binary vector containing the fusion gene was constructed as follows. A 1.5-kb XbaI/PstI genomic fragment flanking the 5′ end of the Rop1At coding sequence was subcloned into pUC19. This fragment was sequenced by the dideoxynucleotide chain-termination method (Sanger et al., 1977) to confirm that it contained 1.3 kb upstream of the Rop1At translation-initiation codon (see Fig. 4). The 1.5-kb genomic DNA fragment was then subcloned into the EcoRI/PstI sites of pBluescript II SK to allow the use of a HindIII site at the 5′ end of the genomic sequence for further subcloning into a binary vector. To introduce a SalI site 20 bp downstream of the Rop1At ATG codon, the sense T7 sequencing primer and the antisense primer containing a SalI site were used for PCR amplification of the putative Rop1At promoter. The amplified fragment was digested with HindIII and SalI, and then translationally fused with the GUS gene in pBI101.2 vector (Clontech, Palo Alto, CA). This plasmid was designated pBR1P:GUS (Fig. 4).
Figure 4.
Construction of Rop1At promoter:GUS fusion gene. Rop1At genomic clone containing the putative promoter region was cloned into the binary vector as described in the text. Shown is the region joining Rop1At and the GUS gene. The restriction sites used for translational fusion with the GUS gene are shown in bold; underlined sequences are the partial coding region for Rop1At. The last ATG codon shown is the translation-initiation codon for the GUS gene.
pBR1P:GUS was mobilized into Agrobacterium tumefaciens strain LB4404 by the freeze-thaw method (An et al., 1988) and introduced into Arabidopsis (ecotype RLD) by the root-transformation method (Valvekens et al., 1988). Primary transgenic plants were selected on Murashige and Skoog medium (Sigma) containing 50 μg/mL kanamycin. Primary transgenic plants and their progenies were analyzed by histochemical GUS staining.
Histochemical GUS Staining and DNA Staining
Histochemical assays for GUS activity in transgenic Arabidopsis plants were performed as described previously (Jefferson et al., 1987). To examine cell-specific GUS activity, tissues were photographed under a microscope (Zeiss) equipped with Nomarski optics. Nuclei of transgenic pollen grains were briefly costained with DAPI and visualized under an epifluorescence microscope (Coleman and Goff, 1985).
Overexpression of the Rop1At Gene in Fission Yeast (Schizosaccharomyces pombe)
The Rop1At coding region was amplified by PCR using primers covering the translation start and stop codons, respectively. The PCR fragment was first cloned into the EcoRV site of pBluescript II SK and then subcloned into SalI and SmaI sites downstream of the nmt1 promoter in the thiamine-repressible fission yeast expression vector pREP3X (Forsburg, 1993). The fission yeast strain FY254 (h-, can1–1, leu1–32, ade6-M210, and ura4-D18) was transformed by the electroporation method (Prentice, 1992), and transformants were plated on Edinburgh minimal medium/uracil plates containing 5 mm of thiamine, and were then incubated at 30°C. Overexpression of the Rop1At gene was induced by growing yeast cells in liquid medium lacking thiamine for 24 h. As a a control, yeast cells containing the vector alone were treated in the same manner. Yeast morphology was examined under a microscope (Oxioskop, Zeiss) and recorded using a 35-mm camera.
Expression of the Gene Encoding the Jellyfish GFP:Rop1At Fusion Protein in Fission Yeast
The mGFP4 coding region was amplified from pBIN-mGFP4 (Haseloff et al., 1997) by PCR using a sense primer containing an XbaI site upstream of the ATG codon and an antisense primer containing a BglII site in place of the GFP translation stop codon. The GFP fragment was cloned into XbaI and SmaI sites of pUC19. The Rop1At coding region was amplified by PCR using two primers containing a BglII site upstream of the ATG codon and a SstI site immediately following the UAG codon. This Rop1At fragment was then translationally fused with the mGFP4 gene at BglII/SstI sites in pUC19. The fusion gene was then cloned into pREP3X and introduced into the fission yeast strain FY254 as described above. To observe proper subcellular localization of the fusion protein, yeast cells containing the GFP:Rop1At fusion gene were grown in nonrepressive Edinburgh minimal medium/uracil for 24 h before transfer to a partially repressive medium containing 2 mm thiamine for 5 h. Green fluorescence was observed using an epifluorescent microscope (Oxioskop, Zeiss) and recorded using a 35-mm camera.
RESULTS
Identification of Rop1Ps Homologs in Arabidopsis
To identify Rop1Ps homologs, we screened an Arabidopsis cDNA library using a Rop1Ps probe (Yang and Watson, 1993). Two distinct Rop1Ps-related genes, Rop1At and Rop2At, were identified from this screen. Two additional genes, Rop4At and Rop5At, were isolated from an Arabidopsis genomic DNA library. Another Rop1Ps-related gene, Rop6At, was identified from an Arabidopsis expressed sequence tag database. A cDNA clone for Rop3At was obtained from Dr. Dring N. Crowell (Indiana University-Purdue University at Indianapolis). After this work was completed, a family of Arabidopsis genes related to Rop1Ps, designated Arac, was reported (Winge et al., 1997). Among the genes whose coding regions have been completely sequenced, four are identical to Arac genes: Rop2At (Arac4), Rop3At (Arac1), Rop4At (Arac5), and Rop6At (Arac3). However, Rop1At, which exhibits the highest homology to Rop1Ps, is a novel Rop member.
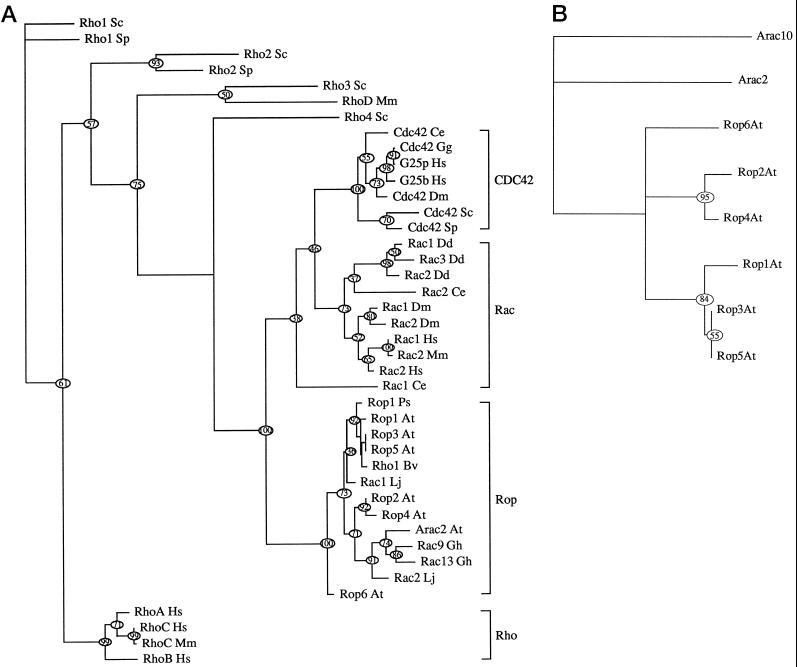
Phylogenetic analysis indicates that the predicted polypeptides encoded by these Rop1Ps-related genes belong to a subfamily of Rho-GTPases designated Rop, which is distinct from all major subfamilies of Rho-GTPases from animals and fungi (Fig. 1A). The Rop subfamily is more closely related to Rac (about 65% identity at the amino acid level) than to CDC42 (55% identity) and Rho (45%–50% identity). Members of the Rop subfamily share 80% or greater amino acid sequence identity. The Arabidopsis Rop subfamily may be further divided into several subgroups based on overall sequence homology and the divergence of C-terminal sequences (Figs. 1B and 2). The C-terminal region of Rho-GTPases typically contains both a C-terminal Cys-X-X-Leu motif required for geranylgeranylation and a proximal polybasic domain involved in subcellular localization (Hancock et al., 1991; Hancock and Marshall, 1993). Members of the Rop1 subgroup, including Rop1At, Rop3At/Arac1, and Rop5At, are most closely related to Rop1Ps and share the Ser-Lys-Ala-Gln-Lys-Ala-Cys-Ser-Ile-Leu sequence with Rop1Ps. Rop2At/Arac5 and Rop4At/Arac4 form a second subgroup with the C-terminal Asn-Lys-Asn-Arg-Cys-Ala/Val-Phe-Leu sequence. Rop6At/Arac3 and Arac2 each diverge from these two subgroups, whereas the most divergent subgroup, represented by Arac10 (Winge et al., 1997), lacks the C-terminal geranylgeranylation motif that is present in almost all known Rho-GTPases.
Figure 1.
Phylogenetic relationship between different Rho-GTPases. Unrooted trees were constructed using PAUP (Swofford, 1993). Amino acid sequences for various plant, animal, and yeast Rho-GTPases were obtained from GenBank using the Blast program. At, Arabidopsis thaliana; Bv, Beta vulgaris; Ce, Caenorhabditis elegans; Dd, Dictyostelium discoideum; Dm, Drosophila melanogaster; Gg, Gallus gallus; Gh, Goosypium hirsutum; Hs, Homo sapiens; Lj, Lotus japonicus; Mm, Mus musculus; Ps, Pisum sativum; Sc, Saccharomyces cerevisiae; Sp, S. pombe. A, Rho family tree showing phylogenetic relationships among major subfamilies from different eukaryotic kingdoms. This tree does not include all known members of Rho-GTPases, since several novel Rho-GTPases that do not fall within any of the major subfamilies are not included. B, Arabidopsis Rop family tree. Members shown include those described in this paper and those whose complete coding sequences are available in the Arabidopsis database.
Figure 2.
Alignment of the C-terminal divergent sequences of Arabidopsis Rop proteins. Predicted amino acid sequences including the C-terminal variable region for Arabidopsis Rop members were aligned using the MegAlign program. Residues identical to Rop1Ps sequences are indicated by dots; dashes represent gaps introduced into the alignment.
Differential Expression of Different Rop Subgroups in Pollen and Vegetative Tissues
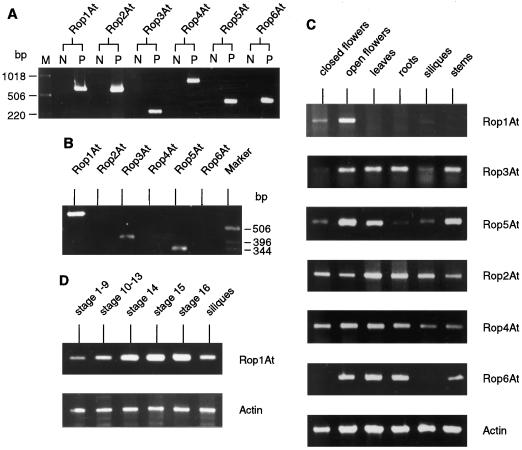
To further address which of the Arabidopsis Rop genes may be a functional homolog of Rop1Ps, we investigated their expression in mature pollen. Since it is difficult to collect large quantities of Arabidopsis pollen for RNA gel-blot hybridization analysis, gene-specific RT-PCR was used to analyze Rop transcript accumulation in pollen. Gene-specific primers were designed according to sequences of the 5′- and 3′-untranslated regions for each gene (Table I). The specificity of primers was confirmed by their ability to amplify only the corresponding Rop gene, not any other Rop genes (Fig. 3A). cDNAs derived from mature pollen were amplified using different pairs of gene-specific primers. RT-PCR results showed that members of the Rop1 subgroup, including Rop1At, Rop3At, and Rop5At, were all expressed in pollen, whereas the other Rop members examined were not (Fig. 3B). Among these three members, Rop1At transcripts were the most abundant.
Figure 3.
RT-PCR analyses of Rop gene expression in various Arabidopsis tissues. A, Demonstration of Rop isogene-specific PCR amplification. Two sets of PCR reactions for each Rop isogene were performed. Lane M, DNA marker; lanes N, negative control, which involves the same primers and template DNAs containing a mixture of equal amounts of cDNA or genomic DNA for each of the other five Rop genes; lanes P, positive control, which involves a specific cDNA (for Rop1At, Rop2At, Rop3At, and Rop6At) or genomic DNA (for Rop4At and Rop5At) as templates and corresponding gene-specific primers. Expected cDNA lengths of amplified fragments are shown in Table I. The PCR reaction conditions are described in text. B, Accumulation of various Arabidopsis Rop transcripts in mature pollen. Total pollen RNA was isolated and the cDNA derived was amplified for 40 cycles using Rop gene-specific primers as described in the text. C, Organ distribution of various Arabidopsis Rop transcripts. RT-PCR was performed using Rop gene-specific primers described in A and total RNAs from different tissues as indicated. Act2 RT-PCR was included as a constitutive control. The number of PCR cycles was: 25 for Rop1At, Rop2At, Rop3At Rop4At, and act2, and 45 for Rop5At and Rop6At. D, Analyses of Rop1At mRNA accumulation during floral development. Total RNAs isolated from Arabidopsis floral buds and flowers at different stages were used for RT-PCR using the reaction conditions described in B. Flower stages were estimated as described previously (Smyth et al., 1990). Stages 1 to 9, Initiation and formation of floral primordia and organ differentiation; stages 10 to 13, organs fully developed, anthesis; stage 14, anthers extended above stigma, pollination; stage 15, stigma extends above anthers; stage 16, petals and sepals wither; siliques, developing siliques before seed maturation.
To assess whether any of the three pollen Rop genes are pollen specific, the expression pattern for Rop members in different tissues was analyzed using RT-PCR as described above. As shown in Figure 3C, Rop1At transcripts were most abundant in open flowers, easily detected in closed flowers, but barely detected in young siliques. No Rop1At PCR products were found in vegetative organs (including rosette leaves, roots, and stems), even after extended PCR cycles. In contrast, all five of the other Rop genes investigated were expressed in various parts of Arabidopsis plants. The organ distribution was similar for Rop3At and Rop5At transcripts. However, the overall level of Rop5At transcripts was much lower than that for Rop3At, since the number of PCR cycles had to be increased from 25 to 45 to detect Rop5At RT-PCR products. Furthermore, Rop5At transcripts were barely detectable in roots, whereas Rop3At transcripts were abundant.
Rop2At and Rop4At transcripts showed similar distribution patterns: both appeared to be constitutively expressed in all organs. Winge et al. (1997) reported similar expression patterns for Rop2At and Rop4At. The expression of Rop6At was very weak and was different from that of other Rop genes. The number of PCR cycles had to be extended to 45 to detect Rop6At transcripts. Rop6At transcripts were only found in open flowers, leaves, roots, and stems, not in closed flowers and siliques. As an internal control, a parallel reaction was performed using primers for the Arabidopsis Act2 gene that is known to be constitutively expressed (An et al., 1996b). Levels of the PCR products for Act2 were constant for different tissues, demonstrating that the observed differences in Rop PCR products were indicative of relative Rop transcript levels.
The expression pattern for Rop1At described above suggests that Rop1At gene expression may be subject to the developmental regulation associated with pollen development and pollination. To investigate the developmental regulation of Rop1At, its transcript levels at various stages of flower development were analyzed using RT-PCR. Flower stages were determined as described previously (Smyth et al., 1990). As shown in Figure 3D, Rop1At transcripts appear in young floral buds prior to pollen development (flower stages 1–9). Levels of Rop1At transcripts greatly increased just prior to anthesis, peaked at anthesis, and gradually declined after fertilization. These results suggest that Rop1At expression is activated before pollen development is initiated and reaches a maximum during pollen maturation.
Rop1At Is a Late Pollen Gene
To determine the precise spatial and temporal expression pattern for Rop1At, we expressed Rop1At promoter:GUS fusion gene in Arabidopsis plants. A 1.3-kb genomic DNA fragment, including 20-bp 5′-coding sequences, was translationally fused with the GUS gene in pBI101.2 (Fig. 4) and was introduced into Arabidopsis plants.
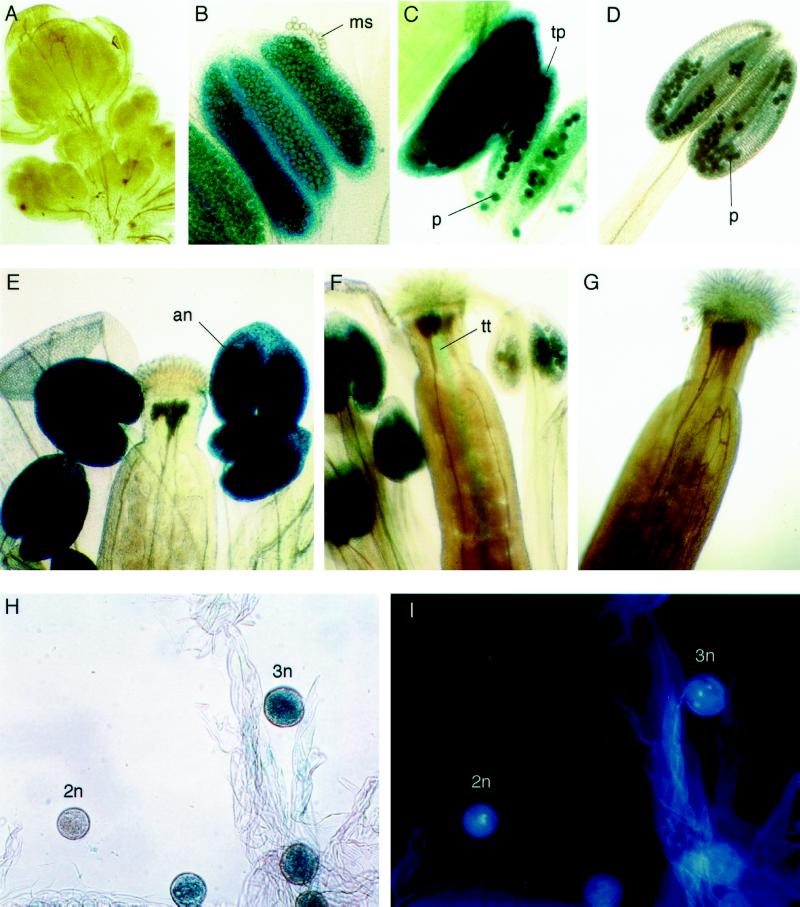
Twelve primary transgenic plants were randomly chosen for histochemical staining of GUS activity (Jefferson et al., 1987) and all showed identical staining patterns, which were further confirmed in the next generation. As shown in Figure 5, GUS activity was specifically detected in anthers and in other parts of the plant. Very low levels of GUS activity were first detected in anthers undergoing microspore development. The GUS activity increases dramatically when all floral organs are differentiated and reaches a maximum at anthesis. Further analysis revealed that the strong GUS activity in the anther of open flowers was the result of GUS expression in mature pollen grains.
Figure 5.
Histochemical localization of GUS expression in transgenic Arabidopsis plants carrying the Rop1At promoter:GUS fusion gene. Various parts of transgenic T2 plants were stained with 5-bromo-4-chloro-3-indolyl-β-glucuronide cyclohexylamine salt, as described in the text. Typical staining patterns are shown. Anthers were costained with DAPI to determine the developmental stages of pollen. an, Anther; ms, microspore; p, pollen; tp, tapetum; tt, transmitting tissue; 2n, pollen at bicellular stage; 3n, pollen at tricellular stage. A, Early stages of floral buds; B, anthers at the stage of microspore development; C, anthers just prior to anthesis; D, anthers just after anthesis; E, stigma and anthers at anthesis; F, stigma and anthers after anthesis; G, stigma from stage-16 flowers; H, pollen at various developmental stages; I, pollen from H costained with DAPI.
To define the stages of pollen development at which GUS activity was expressed, pollen was costained with 5-bromo-4-chloro-3-indolyl-β-glucuronide cyclohexylamine salt and DAPI (Coleman and Goff, 1985). No GUS activity was detected in microspores prior to the binuclear stage, and very weak GUS activity was detected in binuclear microspores (48 h of staining was required to detect any GUS activity). GUS expression started to increase in trinuclear pollen and reached a maximum in mature pollen (at anthesis), being detectable only after 6 h of staining. GUS was also found in transmitting tissue of carpels during pollination but not before or after pollination. Staining of in vitro-germinated transgenic pollen tubes suggests that the staining observed in the transmitting tissue resulted from GUS expression in pollen tubes (data not shown).
A weak GUS activity was also detected in the tapetum at the early stages of microspore development, which would be consistent with the accumulation of low levels of Rop1At transcripts during early flower development. This activity was only detectable following an extended staining (48 h). The tapetal GUS expression remained throughout pollen development until the degeneration of tapetal cells (see Fig. 5).
The GUS expression pattern described above is in accordance with the accumulation of Rop1At transcripts during flower development, indicating that the 1.3-kb genomic fragment truly represents the Rop1At promoter. The 1.3-kb fragment was sequenced to determine whether Rop1At promoter sequences contain the cis-acting elements required for the expression of pollen genes. It contained a region located 370 bp upstream of ATG (GTAATTGTGA) with a strong homology (9 of 10 bp matches to the 56/59 box) to a pollen-specific enhancer sequence shared by the LAT56 and LAT59 promoters (Twell et al., 1991). The GTGA motif within this box is essential for high levels of pollen-specific expression (Twell et al., 1991; Twell, 1994). At least two additional GTGA motifs, located 573 and 461 bp upstream of ATG, are present in the Rop1At promoter. Similar GTGA motifs are found in the promoters of several other pollen genes, e.g. Zmg13 from maize, chiA from petunia, and GBAN215–6 and GBAN215–12 from Chinese cabbage (Hamilton et al., 1989; van Tunen et al., 1990; Kim et al., 1997). The Rop1At transcript accumulation and GUS fusion gene expression patterns, together with the presence of pollen-specific cis-elements, demonstrate that Rop1At is a late pollen gene.
Rop1At May Function in Fission Yeast to Regulate Polarized Cell Growth
On the basis of its tip localization and involvement in pollen tube growth, we speculated that Rop1Ps and its homologs might be a molecular switch in the signal transduction pathway leading to polarized cell growth (Lin and Yang, 1997). The function of certain Rho-type GTPases is conserved across kingdoms, e.g. human CDC42 is able to complement the temperature-sensitive yeast cdc42 mutants defective in polarity control. Therefore, we wanted to determine whether Rop1At is able to control polarized cell growth in fission yeast, which, like pollen tubes, elongates by tip growth. First, we overexpressed Rop1At in fission yeast under the control of the thiamine-repressible nmt1 promoter. As shown in Figure 6, Rop1At overexpression induced dramatic morphological changes. The majority of cells become shorter and fatter and rounded or pear-shaped, in contrast to the elongated, rod-shaped wild-type cells. When cells containing the Rop1At gene were grown in a repressive medium, or when cells containing the pREP3X plasmid alone were grown in a nonrepressive medium, the cell morphology was normal. The Rop1At-induced morphological changes were similar to those caused by the overexpression of constitutively active mutants of the fission yeast CDC42 gene, which is implicated in the control of polarized cell growth (Miller and Johnson, 1994). A Rop1Ps-related gene (designated here as Rop6At) was isolated during the screening of an Arabidopsis cDNA library for cDNA clones that induced nonpolarized growth in fission yeast (Xia et al., 1996).
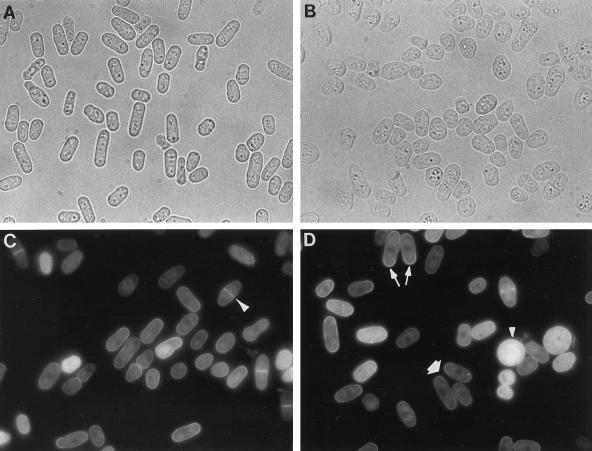
Figure 6.
Overexpression of Rop1At and polar localization of GFP:Rop1At fusion in fission yeast. The Rop1At or GFP:Rop1At fusion genes were cloned into pREP3X under the control of a thiamine-repressible promoter and introduced into fission yeast, as described in the text. A, Yeast cells with pREP3X-Rop1At grown in a repressive medium containing 5 mm thiamine. Cells have a normal morphology. B, Yeast cells containing pREP3X-Rop1At grown in a nonrepressive medium lacking thiamine. Greater than 90% were abnormal in shape. C and D, Yeast cells containing pREP3X-GFP:Rop1At grown in a partially repressive medium containing 2 mm thiamine and examined under an epifluorescence microscope. Typical GFP localization patterns are indicated: long arrow, unipolar localization; thick arrow, bipolar localization; long arrowhead, localization to the septum; short arrowhead, nonpolar localization in GFP-Rop1At-overexpressing cells.
We expected that Rop1At would be localized to polar sites if its function is to regulate polarized growth in fission yeast. We investigated the subcellular localization of Rop1At in living yeast cells using jellyfish GFP. The GFP-Rop1At fusion gene is expressed in fission yeast under the control of the thiamine-repressible nmt1 promoter. Like the overexpression of the wild-type Rop1At gene, the fusion gene also induced nonpolarized phenotypes under nonrepressive conditions (data not shown). The severity of the phenotype was correlated with the level of fusion protein expression, as indicated by the intensity of the green fluorescence. Under partially repressive conditions (2 mm thiamine), most cells contained low fluorescence and were relatively normal in shape. Several fluorescence patterns were observed in these cells.
Overall, the fusion protein appeared to be localized to the plasma membrane, which is consistent with the presence of a polybasic domain at the C terminus of Rop1At (the polybasic domain has been shown to mediate targeting of isoprenylated proteins to the plasma membrane; Hancock et al., 1991; Adamson et al., 1992). Fluorescence was concentrated in the septum in dividing cells. Soon after cell division was completed, however, fluorescence shifted to old ends, where tip growth is re-initiated (unipolar growth). When cell elongation was shifted to a bipolar pattern, GFP was concentrated at both ends of the cell. Under partially repressive conditions, certain cells contained strong cytoplasmic fluorescence and were completely rounded and somewhat enlarged. In these cells GFP became uniformly distributed on the plasma membrane. Such cells most likely contained a high copy number of plasmids. In nonrepressive conditions most cells were rounded and did not show polarized localization of the fusion protein. The Rop1At-induced isotropic growth, together with its polar localization in fission yeast, suggest that Rop1At has a conserved function in regulating polarized cell growth.
DISCUSSION
Although the Rho-GTPases are conserved in eukaryotic cells as key regulators of actin cytoskeletal organization, emerging evidence from fungi and mammals suggests that members of the Rho family have also diverged considerably in both structure and function as various eukaryotic phyla evolve. The current data suggest that the plant-specific Rop subfamily of Rho-GTPases has a conserved function in the regulation of polarized cell growth. However, phylogenetically distinct subgroups of the Arabidopsis Rop subfamily exhibit different developmental expression patterns. One of these subgroups is of particular interest, in that all of its members are expressed in mature pollen implicating them in pollen tip growth.
Plants Have Evolved a Distinct Subfamily of Rho-GTPases
The Rho family of small GTP-binding proteins characterized in fungi and animals include three major subfamilies: CDC42, Rac, and Rho (Chardin, 1993; Hall, 1994; Nobes and Hall, 1995). In mammalian cells each of the three subfamilies controls a specific actin-dependent process (Ridley and Hall, 1992; Ridley et al., 1992; Luo et al., 1994; Chant and Stowers, 1995; Nobes and Hall, 1995). The distinct function for each subfamily is reflected in their amino acid sequence and in their ability to interact with specific downstream effector proteins (Chardin, 1993; Nagata and Hall, 1996; Ridley, 1996). For example, members within a subfamily share 80% or greater amino acid sequence identity, whereas sequence identity between subfamilies ranges from 45% to 70% (Chardin, 1993).
We propose that plants possess a distinct subfamily of Rho-GTPases called Rop. Although Rop is most similar to Rac, phylogenetic analysis suggest that Rop evolved prior to the divergence of Rac and CDC42. Rop members share many unique motifs or residues that presumably determine functional specificity of these GTPases. Within the conserved effector domain (residues 29–49) exist several Rop-specific residues (T30, T33, F43, and V48). A Rho family-specific insert region consists of 12 amino acid residues (Thr-Arg-Arg-Glu-Leu-Ala-Lys-Met-Lys-Ala-Glu-Pro) in all fungal and animal Rho-GTPases (Chardin, 1993) and functions as an effector domain (Nisimoto et al., 1997). Although the Rop subfamily also contains a corresponding region of 10 residues (residues 128–137), its consensus amino acid sequence (Phe-Phe-Val-Asp-His-Pro-Gly-Ala-Val-Pro) is quite different. The fact that the Rop subfamily is absent from S. cerevisiae and has not yet been found in animals indicates that it is unique to plants.
Each eukaryotic kingdom has evolved a specific set of Rho-GTPases (Fig. 1A). The Rho subfamily is found both in fungi and animals and is most likely to exist in plants (Lin and Yang, 1997). The ancestor of the CDC42/Rac/Rop group may have diverged from the Rho subfamily and split into CDC42 and the plant-specific Rop. CDC42, which controls cell polarity and cortical actin formation in both fungi and animals, has not been found in plants. Rac, which appears to have split from CDC42 and become animal specific, regulates actin-dependent cell movement. Therefore, it is logical to speculate that Rop has retained certain conserved functions of the Rho family (e.g. cell polarity control) and has also evolved to perform specific functions that are unique to plant cells. This hypothesis is clearly supported by our studies showing that Rop plays an essential role in pollen tube growth in pea (Lin et al., 1996; Lin and Yang, 1997), that Rop1At exhibits polarized localization and induces isotropic growth when overexpressed in fission yeast (Fig. 6), and that different subgroups of Rop GTPases exhibit distinct patterns of developmentally regulated gene expression in the male gametophyte and various vegetative tissues (Figs. 3 and 5).
A Specific Rop Subgroup Is Conserved in Protein Structure and Developmental Gene Expression in the Male Gametophyte
Our studies show that the Rop subfamily can be further divided into several subgroups on the basis of primary structure and gene-expression patterns. Like Rop1Ps, all members of the Rop1 subgroup, Rop1At, Rop3At, and Rop5At, are expressed in mature pollen, whereas members of the second subgroup, Rop2At and Rop4At, are constitutively expressed in vegetative tissues but not in pollen. Furthermore, two other divergent Rop GTPases, Rop6At (this study) and Arac2 (Winge et al., 1997), exhibit differential expression patterns. Thus, the Rop subfamily may be divided into the reproductive class (the Rop1 subgroup expressed in pollen) and the vegetative class (Rop2At, Rop4At, Rop6At, Arac2), as do actins and the actin-binding proteins called profilins; Staiger et al., 1993; An et al., 1996a, 1996b; Christensen et al., 1996; Huang et al., 1996a, 1996b, 1997; McDowell et al., 1996). It is interesting that all three types of conserved proteins implicated in the organization of the actin cytoskeleton (actin, profilin, and Rho) exhibit a tight linkage between their structural conservation and developmental gene regulation in pollen. An important question is whether such a correlation reflects a possible functional conservation of these proteins in the regulation of certain pollen-specific arrays of the actin cytoskeleton. The ability to systematically knock out specific genes in Arabidopsis should allow this question to be addressed.
Rop1At May Have a Distinct Role in the Control of Polarized Growth in Pollen
We have demonstrated that Rop1At displays a unique expression pattern associated with the development and the function of pollen. Although Rop3At and Rop5At are also expressed in pollen, their transcript levels are only a small fraction of Rop1At transcripts in pollen. This suggests that Rop1At has a predominant role in pollen development and function, whereas Rop3At and Rop5At may be functionally redundant to Rop1At.
Further insight into potential roles for Rop1At was gained by investigating its temporal and spatial expression patterns and by functional analyses in fission yeast. Analyses of GUS fusion gene expression in transgenic plants indicate that Rop1At is specifically expressed in pollen. Rop1At transcription is activated after the first postmeiotic cell division, reaches a maximum when pollen is mature, and remains active during pollen tube growth. This expression, which is typical of a late pollen gene, is consistent with a role for Rop1At in pollen germination and tube growth. Nonetheless, the functional analyses in fission yeast suggest that Rop1At has a conserved function in the regulation of polarized cell growth. Polar localization of GFP-Rop1At fusion protein to the site of cell growth in yeast is analogous to that of Rop proteins in pea and Arabidopsis pollen tubes suggested by our immunofluorescence studies (Lin et al., 1996; Y. Lin and Z. Yang, unpublished results). These results led us to conclude that Rop1At and Rop1Ps may be orthologs in regard to their potential roles in the regulation of polarized cell growth in Arabidopsis and pea pollen tubes.
ACKNOWLEDGMENTS
We thank Dr. Dring N. Crowell for providing the Rop3At cDNA clone, Dr. Susan Forsburg for the yeast expression vector pREP3X and strain FY254, Dr. Jim Haseloff for pBIN-mGFP4, and the Ohio State-NSF Arabidopsis Biological Resources Center for Arabidopsis cDNA and genomic libraries. We also thank Yakang Lin for his assistance with microscopy work.
Abbreviations:
- DAPI
4′,6-diamidino-phenylindole
- GFP
green fluorescent protein
- RT
reverse transcriptase
Footnotes
This work was supported by grants from the National Science Foundation (no. MCB-9724047) and the U.S. Department of Agriculture (no. 96-35304-3861) to Z.Y. and by a National Institutes of Health grant (no. GM 45570) to K.R.D.
LITERATURE CITED
- Adamson P, Paterson HF, Hall A. Intracellular localization of the p21rho proteins. J Cell Biol. 1992;119:617–627. doi: 10.1083/jcb.119.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An G, Eber PR, Mitra A, Ha SB. Binary vectors. In: Gelvin S, Schilperoort RA, editors. Plant Molecular Biology Manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1988. pp. 1–19. [Google Scholar]
- An Y-Q, Huang S, McDowell JM, McKinney EC, Meagher RB. Conserved expression of the Arabidopsis ACT1 and ACT3 actin subclass in organ primordia and mature pollen. Plant Cell. 1996a;8:15–30. doi: 10.1105/tpc.8.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An Y-Q, McDowell JM, Huang S, McKinney EC, Chamliss S, Meagher RB. Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J. 1996b;10:107–121. doi: 10.1046/j.1365-313x.1996.10010107.x. [DOI] [PubMed] [Google Scholar]
- Chant J, Stowers L. GTPase cascades choreographing cellular behavior: movement, morphogenesis, and more. Cell. 1995;81:1–4. doi: 10.1016/0092-8674(95)90363-1. [DOI] [PubMed] [Google Scholar]
- Chardin P. ras homologs: a comparison of primary structures. In: Lacal JC, McCormick F, editors. The Ras Superfamily of GTPases. Boca Raton, FL: CRC Press; 1993. pp. 203–229. [Google Scholar]
- Christensen HEM, Ramachandran S, Tan C-T, Surana U, Dong C-H, Chua N-H. Arabidopsis profilins are functionally similar to yeast profilins: identification of a vascular bundle-specific profilin and a pollen-specific profilin. Plant J. 1996;10:269–279. doi: 10.1046/j.1365-313x.1996.10020269.x. [DOI] [PubMed] [Google Scholar]
- Coleman AW, Goff LJ. Applications of fluorochromes to pollen biology. I. Mithramycin and 4′,6-diamidino-2-phenylindole (DAPI) as vital stains and for quantitation of nuclear DNA. Stain Technol. 1985;60:145–154. doi: 10.3109/10520298509113905. [DOI] [PubMed] [Google Scholar]
- Delmer DP, Pear JR, Andrawis A, Stalker DM. Genes for small GTP-binding proteins analogous to mammalian Rac are preferentially expressed in developing cotton fibers. Mol Gen Genet. 1995;248:43–51. doi: 10.1007/BF02456612. [DOI] [PubMed] [Google Scholar]
- Estruch J, Kadwell S, Merlin E, Crossland L. Cloning and characterization of a maize pollen-specific calcium-dependent calmodulin-independent protein kinase. Proc Natl Acad Sci USA. 1994;91:8837–8841. doi: 10.1073/pnas.91.19.8837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg SL. Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res. 1993;21:2955–2956. doi: 10.1093/nar/21.12.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. Small GTP-binding proteins and the regulation of the actin cytoskeleton. Annu Rev Cell Biol. 1994;10:31–54. doi: 10.1146/annurev.cb.10.110194.000335. [DOI] [PubMed] [Google Scholar]
- Hamilton DA, Bashe DM, Stinson JR, Mascarenhas JP. Characterization of a pollen-specific genomic clone from maize. Sex Plant Reprod. 1989;2:208–212. doi: 10.1105/tpc.1.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock JF, Cadwallader K, Paterson H, Marshall CJ. A CAAX or CAAL motif and a second signal are sufficient for plasma membrane targeting of ras proteins. EMBO J. 1991;10:4033–4039. doi: 10.1002/j.1460-2075.1991.tb04979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock JF, Marshall CJ. Posttranslational processing of ras and ras-related proteins. In: Lacal JC, McCormick F, editors. The Ras Superfamily of GTPases. Boca Raton, FL: CRC Press; 1993. pp. 65–84. [Google Scholar]
- Haseloff J, Siemering KR, Prasher D, Hodge S. Removal of a cryptic intron and subcellular localisation of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA. 1997;94:2122–2127. doi: 10.1073/pnas.94.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, An Y-Q, McDowell JM, McKinney EC, Meagher RB. The Arabidopsis thaliana ACT4/ACT12 actin gene subclass is strongly expressed throughout pollen development. Plant J. 1996a;10:189–202. doi: 10.1046/j.1365-313x.1996.10020189.x. [DOI] [PubMed] [Google Scholar]
- Huang S, An Y-Q, McDowell JM, McKinney EC, Meagher RB. The Arabidopsis ACT11 actin gene is strongly expressed in tissues of the emerging inflorescence, pollen, and developing ovules. Plant Mol Biol. 1997;33:125–139. doi: 10.1023/a:1005741514764. [DOI] [PubMed] [Google Scholar]
- Huang S, McDowell JM, Weise MJ, Meagher RB. The Arabidopsis profilin gene family. Plant Physiol. 1996b;111:115–126. doi: 10.1104/pp.111.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Burgess SM, Hirsh D. GUS fusion:β glucuronidase as a sensitive and versatile gene fusion marker. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempin SA, Liljegren S, Block LM, Rounsley SD, Yanofsky MF. Targeted disruption in Arabidopsis. Nature. 1997;389:802. doi: 10.1038/39770. [DOI] [PubMed] [Google Scholar]
- Kim H, Park B, Jin Y, Chung T. Promoter sequences of two homologous pectin esterase genes from Chinese cabbage (Brassica campestris L. ssp. pekinensis) and pollen-specific expression of the GUS gene driven by a promoter in tobacco plants. Molecules and Cells. 1997;7:21–27. [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Tax F, Sussman M. Identification of transferred DNA insertions within Arabidopsis genes involved in signal transduction and ion transport. Proc Natl Acad Sci USA. 1996;93:8145–8150. doi: 10.1073/pnas.93.15.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz C, Chang A, Faure J-D, Clarke AE, Polya GM, Anderson MA. Phosphorylation of style S-RNases by Ca2+-dependent protein kinases from pollen tubes. Sex Plant Reprod. 1996;9:25–34. [Google Scholar]
- Lamaze C, Chuang T-H, Terlecky LJ, Bokoch GM, Schmid SL. Regulation of receptor-mediated endocytosis by Rho and Rac. Nature. 1996;382:177–179. doi: 10.1038/382177a0. [DOI] [PubMed] [Google Scholar]
- Larochelle DA, Vithalani KP, De Lozanne A. A novel member of the rho family of small GTP-binding proteins is specifically required for cytokinesis. J Cell Biol. 1996;133:1321–1329. doi: 10.1083/jcb.133.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-S, Karunanandaa B, McCubbin A, Gilroy S, Kao T-h. PRK1, a receptor-like kinase of Petunia inflata, is essential for postmeiotic development of pollen. Plant J. 1996;9:613–624. [Google Scholar]
- Lin Y, Wang Y, Zhu J, Yang Z. Localization of a rho GTPase implies a role in tip growth and movement of the generative cell in pollen tubes. Plant Cell. 1996;8:293–303. doi: 10.1105/tpc.8.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Yang Z. Inhibition of pollen tube elongation by microinjected anti-Rop1Ps antibodies suggests a crucial role for Rho-type GTPases in the control of tip growth. Plant Cell. 1997;9:1647–1659. doi: 10.1105/tpc.9.9.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann J, Schell J, Willmitzer L. Improved method for the isolation of RNA from plant tissues. Anal Biochem. 1987;163:16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- Luo L, Liao YJ, Jan LY, Jan YN. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 1994;8:1787–1802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- Mascarenhas JP. Molecular mechanisms of pollen tube growth and differentiation. Plant Cell. 1993;5:1303–1314. doi: 10.1105/tpc.5.10.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick S. Male gametophyte development. Plant Cell. 1993;5:1265–1275. doi: 10.1105/tpc.5.10.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell JM, Huang S, McKinney EC, An Y-Q, Meagher RB. Structure and evolution of the actin gene family in Arabidopsis thaliana. Genetics. 1996;142:587–602. doi: 10.1093/genetics/142.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney EC, Ali N, Traut A, Feldmann KA, Belostotsky DA, McDowell JM, Meagher RB. Sequence-based identification of T-DNA insertion mutations in Arabidopsis: actin mutants act2–1 and act4–1. Plant J. 1995;8:613–622. doi: 10.1046/j.1365-313x.1995.8040613.x. [DOI] [PubMed] [Google Scholar]
- Miller PJ, Johnson DI. Cdc42p GTPase is involved in controlling polarized cell growth in Schizosaccharomyces pombe. Mol Cell Biol. 1994;14:1075–1083. doi: 10.1128/mcb.14.2.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy C, Saffrich R, Grummt M, Gournier H, Rybin V, Rubino M, Auvinen P, Lütcke A, Parton RG, Zerial M. Endosome dynamics regulated by a Rho protein. Nature. 1996;384:427–432. doi: 10.1038/384427a0. [DOI] [PubMed] [Google Scholar]
- Nagata K-i, Hall A. The Rho-GTPase regulates protein kinase activity. BioEssays. 1996;7:529–531. [Google Scholar]
- Nisimoto Y, Freeman JLR, Motalebi SA, Hirshberg M, Lambeth JD. Rac binding to p67phox. J Biol Chem. 1997;272:18834–18841. doi: 10.1074/jbc.272.30.18834. [DOI] [PubMed] [Google Scholar]
- Nobes CD, Hall A. Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fiber, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- Prentice HL. High efficiency transformation of Schizosaccharomyces pombe by electroporation. Nucleic Acids Res. 1992;20:621. doi: 10.1093/nar/20.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ. Rho: theme and variations. Curr Biol. 1996;6:1256–1264. doi: 10.1016/s0960-9822(02)70711-2. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating substrates. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger CJ, Goodbody KC, Hussey PJ, Valenta R, Drøbak BK, Lloyd CW. The profilin multigene family of maize: differential expression of three isoforms. Plant J. 1993;4:631–641. doi: 10.1046/j.1365-313x.1993.04040631.x. [DOI] [PubMed] [Google Scholar]
- Taylor LP, Helper PK. Pollen germination and tube growth. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:469–491. doi: 10.1146/annurev.arplant.48.1.461. [DOI] [PubMed] [Google Scholar]
- Thompson WF, Everett M, Polans NO, Jorgensen RA, Palmer JD. Phytochrome control of RNA levels in developing pea and mung bean leaves. Planta. 1983;158:487–500. doi: 10.1007/BF00397240. [DOI] [PubMed] [Google Scholar]
- Twell D. The diversity and regulation of gene expression in the pathway of male gametophyte development. In: Scott RJ, Stead AD, editors. Molecular and Cellular Aspects of Plant Reproduction. Cambridge, UK: University Press; 1994. pp. 83–135. [Google Scholar]
- Twell D, Yamaguchi J, Wing RA, Ushiba J, McCormick S. Promoter analysis of genes that are coordinately expressed during pollen development reveals pollen-specific enhancer sequences and shared regulatory elements. Genes Dev. 1991;5:496–507. doi: 10.1101/gad.5.3.496. [DOI] [PubMed] [Google Scholar]
- Valvekens D, Van Montagu M, can Lijsebettens M (1988) Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA 85: 5536–5540 [DOI] [PMC free article] [PubMed]
- van Tunen AJ, Mur LA, Brouns GS, Rienstra J-D, Koes RE, Mol JNM. Pollen and anther-specific promoters from petunia: tandem promoter regulation of the chiA gene. Plant Cell. 1990;2:393–401. doi: 10.1105/tpc.2.5.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytas DF, Konieczny A, Cummings MP, Ausubel FM. The structure, distribution and evolution of the Tal retrotransposable element family of Arabidopsis thaliana. Genetics. 1990;126:713–721. doi: 10.1093/genetics/126.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C, Voronin V, Touraev A, Vicente O, Heberle-Bors E. A developmentally regulated MAP kinase activated by hydration in tobacco pollen. Plant Cell. 1997;9:2093–2100. doi: 10.1105/tpc.9.11.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winge P, Brembu T, Bones AM. Cloning and characterization of rac-like cDNAs from Arabidopsis thaliana. Plant Mol Biol. 1997;35:483–495. doi: 10.1023/a:1005804508902. [DOI] [PubMed] [Google Scholar]
- Xia G, Ramachandran S, Hong Y, Chan YS, Simanis V, Chua NH. Identification of plant cytoskeletal, cell cycle-related and polarity-related proteins using Schizosaccharomyces pombe. Plant J. 1996;10:761–769. doi: 10.1046/j.1365-313x.1996.10040761.x. [DOI] [PubMed] [Google Scholar]
- Yang Z, Watson JC. Molecular cloning and characterization of rho, a ras-related small GTP-binding protein from the garden pea. Proc Natl Acad Sci USA. 1993;90:8732–8736. doi: 10.1073/pnas.90.18.8732. [DOI] [PMC free article] [PubMed] [Google Scholar]