Abstract
Background
Non- Tuberculous Mycobacteria are environmental opportunistic pathogens that can be found in various terrestrial and aquatic habitats. There are an epidemiological links between species isolated in tap water and those isolated from patients. hsp65 gene has more variability in its sequences, compared to the some more conserved genes in NTM, for identification of mycobacteria to species level. In this study, the prevalence of NTM in Isfahan City water samples was determined using culture, biochemical tests and PCR-RFLP analyses of hsp65 gene.
Methods:
Eighty-five water samples were collected and cultured. The mycobacterial isolates were identified by conventional biochemical tests. A 441 bp fragment of hsp65 genes was amplified and digested by two restriction enzymes, BstEII and HaeII. Digested products were analyzed using polyacrilamid gel electrophoresis (PAGE).
Results:
25.9% of the water samples contained different species of NTM. Dominant isolates were M. fortuitum (26.7%), M. chelonae like organism (13.3%) and M. mucogenicum (13.3%). Nineteen isolates of Mycobacteria were differentiated using hsp65 genes PCR-RFLP. Three isolates could not be identified at the species level because their RFLP patterns were different from other known PCR-RFLP profiles. There were different hsp65 gene PCR-RFLP profiles produced by digestion with BstEII and HaeIII.
Conclusion:
This study showed that PCR-RFLP of hsp65 gene in mycobacteria is more reliable method for identification of NTM at the specie level than conventional phenotypic methods (P<0.05). In comparing of RFLP patterns of this study to other investigation, some minor differences were negligible.
Keywords: NTM, Environment, hsp65 gene, PCR-RFLP
Introduction
Mycobacteria are a heterogeneous group of bacteria in terms of their genotypic features and disease association. These organisms can cause hypersensitivity, pneumonitis, asthma, and bronchitis, infection of skin, wounds and glands. Moreover, these infections are serious threat for cystic fibrosis patients. In recent years, Non- Tuberculous Mycobacteria (NTM) was reported as important agents of infection in immunosuppressive patients (1). NTM can be isolated from different natural sources among them water (1–4). These organisms have been isolated from hard conditions such as low pH and nutrients. Several species of NTM have been identified in different environments including public drinking water, pool, undrinkable tap water, water cooler etc. Therefore, water may act as an important NTM source for transmission to human. In some studies, the presence of NTM in water samples collected from different regions were determined (1, 2, 3, 5).
Traditionally, NTM has been detected in clinical and environmental samples by culture-based techniques; however, these techniques may not be well suited for environmental samples. Indeed, identification of NTM by culture and phenotypic characterization is widely used but it takes 4 to 6 weeks or longer for slow growing species and identification of some species may miss by biochemical methods. The rapid methods used for identification include high performance liquid chromatography (HPLC), DNA probes, restriction fragment length polymorphism (RFLP) using various target regions including heat shock protein 65 KD gene (hsp65), ITS and rpo B (1, 6, 7). Molecular methods linked to PCR are more reliable and faster for identification of NTM (1, 5). hsp65 gene is used widely for identification of NTM to species level because of its variability compare to some other conserved genes such as 16S rRNA (6, 7). Therefore, it is suitable target for identification of NTM to species level than other methods (8).
To identify the presence of NTM species in water samples in Isfahan and demonstrate the usefulness of this protocol for identification and characterization of NTM, we applied culture-based techniques and PCR-RFLP targeting hsp65 for characterization of 22 mycobacteria isolated from water samples in Isfahan, Iran.
Materials and Methods
Collection and preparation of the samples
Eighty five water samples were collected from different sources including mineral water (8.2%), dentistry unit water (10.6%), undrinkable tap water (14.1%), drinkable water supply (14.1%), haemodialysis unit water (8.2%), general pools (8.2%), river water (7.1%), water spout (11.8%), water cooler (11.8%), and water boiler (5.9%) in Isfahan, Iran. To 2-litters’ sterile Erlenmeyer flasks, sodium thio-sulfate as antichlor and 0.04% Cetyl pridinium chloride as antimicrobial agent were added. Sampling of water was done using the grab sampling method (9). At the sampling location, total chlorine content was determined using DPD method (9, 10). Filled containers were tightly capped and delivered to the laboratory and processed the day of collection. Five hundred ml of samples were passed from 0.45 μm filters. The filters were transferred directly onto 7H10 Middle Brook solid media, include 15% OADC (Oleic acid, Albumin, Dextrose, Catalase). The plates were examined once a week for eight weeks. When the colonies appeared, they were subjected to acid-fast staining and acid-fast colonies were transferred to Lowenstein- Jensen (LJ) slant media and incubated in 37°C.
Phenotypic identification
The mycobacterial isolates were identified by the growth characteristics, including growth at 25, 37 and 42 °C, pigment production, semi quantitative catalase test, Tween 80 hydrolysis, arylsulfatase test (3 and 14 days), heat-stable catalase (pH 7, 68°C), pyrazin amidase (4 and 7 days), urease, nitrate reduction test and colony morphology . The phenotypic identification tests were repeated whenever mycobacteria identification was doubtful. Reference strains of M. smegmatis (PTCC 1307) and M. fortuitum (ATCC 6841) were used as control species in all steps of this study.
Species identification of NTM by PCR-RFLP
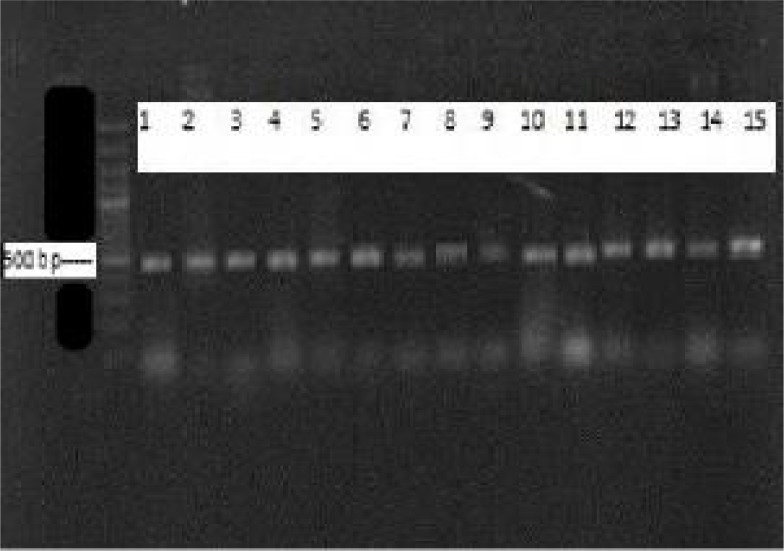
PCR-RFLP of the hsp65 gene was undertaken to identify the species of isolates. Chromosomal DNA was extracted using CTAB (cetyl- Trimethyl Ammonium Bromide) method. PCR of the hsp65 gene was performed using the forward primer for hsp65, Tb11 (5′ACC AAC GAT GGT GTG TCC AT 3’) and the reverse primer, TB12 (5’ CTT GTC GAA CCG CAT ACC CT 3’) (11, 12). In a 25μl PCR mix, 2U of Taq polymerase, 10 mM Tris-HCl (pH 8.3), 1.5mM MgCl2, 200mM dNTP, 20pmol of each primers and 2ng of DNA template were added. PCR was performed using initial denaturation at 95°C for 5min, 45 amplification cycles of 60s at 95°C, 60s at 56°C and 60s at 72°C and a final extention of 72°C for 7min in a Biomerta Gradient thermocycler and an Eppendorf AG 22331. The PCR products were run on 1.5% agarose gel. DNA bands were visualized by ethidium bromide staining and photographed (Fig. 1). The amplified products of hsp65 gene regions were digested with two restriction enzymes of HaeIII and BstEII according to the recommendations of the manufacturers. The digested products were separated on 10% polyacrylamid gel electrophoreses (PAGE) and RFLP patterns were analyzed according to fragments sizes (11–14).
Fig. 1:
Agarose gel electrophoresis of 441 bp hsp 65 PCR products. First Lane: 100 bp DNA marker; lane 1: M. smegmatis (PTCC 1307); lane 2: M. fortutium (ATCC 6841) and lanes 3 to 15: NTM isolates
Results
Sources of samples, percentage of positive samples and NTM species that found in water samples are summarized in Table 1.
Table 1:
Rapid and slow growing mycobacteria isolated from different water sources in Isfahan
| Water sources | NTM positive | Species of NTM |
|---|---|---|
| Water supply | (41.7%) | M. gordonae, M. mucogenicum, M. furtuitum ss. fortuitum, M. chitae, M. neoaurum |
| Undrinkable tap water | (50%) | M. chelonae, M. furtuitum, M. mucogenicum, M. chelonae like organism, 2 Unidentified species |
| Waterspout | (20%) | M. mucogenicum, M. furtuitum ss. fortuitum |
| Cold water dispenser | (50%) | M. chelonae like organism(2), M. furtuitum 3th variant(2), 1 Unidentified species |
| Dentistry unit | (22.2%) | M. mucogenicum, M. fortuitum 3th variant |
| Pool and baths | (28.6%) | M. chelonae like organism, M. duvalii |
| Haemodialysis center | 0 | - |
| Mineral | 0 | - |
| River | 0 | - |
| Hot water dispenser | 0 | - |
Twenty-two mycobacteria were isolated and identified by growth characteristics and conventional biochemical tests. Dominant isolates were M. fortuitum- (26.7%), M. chelonae like organism (13.3%) and M. mucogenicum (13.3%).
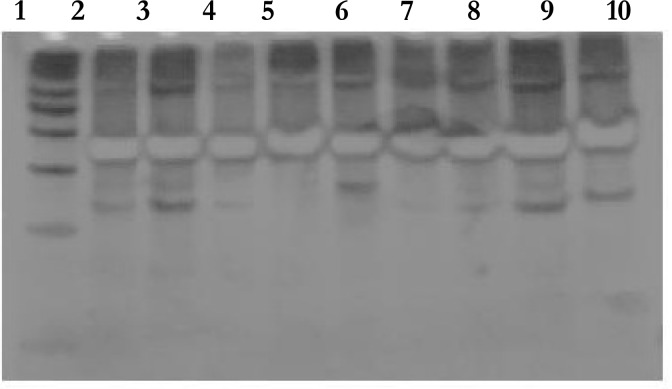
PCR-RFLP of the hsp65 gene was undertaken to identify the species of isolates. A 441 bp fragment of hsp65 genes was amplified (Fig. 1). After restriction digest of the PCR products with BstEII and HaeIII, the patters of digested fragments were analyzed by polyacrilamid gel electrophoresis (PAGE). Nineteen isolates of Mycobacteria were differentiated using hsp65 genes PCR-RFLP (Table 2). Three isolates could not be identified at the species level because their patterns were different with the other known PCR-RFLP profiles. The patters of digested hsp65 PCR products are shown in Fig. 2 .
Table 2:
Fragment sizes of mycobacterial 441 bp hsp 65 PCR products after digestion by HaeIII and BstEII
| BstEII Digestion | HaeIII Digestion | Mycobacterial isolates |
|---|---|---|
| No digestion | 140/130/65/55 | M. duvalii |
| 305–325/130–140 | 145/70/60/45/40 | M. chelonae |
| 140/65/55 | M. mucogenicum | |
| 140/70/60 | M. mucogenicum | |
| 145/100/70/60 | Unidentified | |
| 140/70/60 | M. mucogenicum | |
| 140/80/70/60 | M. mucogenicum | |
| 305–325/115–125 | 155/140/45/40 | Unidentified |
| 140/70/60/45/40 | M. chelonae like organism | |
| 140/80/65/45 | M. chitae | |
| 140/70 | M. chelonae like organism | |
| 170/140/70 | M. neoaurum | |
| 140/80 | M. chelonae like organism | |
| 140/75 | M. chelonae like organism | |
| 230–245/130–140/80–85 | 150/60 | Unidentified |
| 230–245/117–125/90–100 | 135/115 | M. gordonae (type III) |
| 230–245/117–125/80–85 | 135/130 | M. furtuitum |
| 155/135/70 | M. furtuitum ss. furtuitum | |
| 155/135 | M.furtuitum ss. furtuitum | |
| 150/135/75/45 | M.furtuitum 3th variant | |
| 150/135/65 | M.furtuitum 3th variant | |
| 150/135/65 | M.furtuitum 3th variant |
Fig. 2:
Differential identification of NTM by digestion of hsp65 441-bp PCR amplicons by HaeIII, 1:50 bp ladder; 2: Unidentified, 3:M. chelonae like organism, 4: M. chitae, 5: M. duvalii, 6:M. mucogenicum, 7:M. chelonae like organism, 8:M. chelonae like organism, 9: M. neoaurum, 10: M. chelonae like organism
Discussion
NTM are widely distributed in the environment; soil, water and other natural reservoirs. The majorities of NTM are less harmful and not virulent particularly in humans with a normal immune system but some species cause serious infections especially in immunocompromised individuals. However, in recent years, NTM have emerged as a major cause of opportunistic infections. Therefore, it is necessary to perform risk analysis and identify the species of NTM that are present in the environment for understanding their incidence and making aware clinician. Phenotypic characterization including pigment production, growth rate and biochemical test algorithms has been used for the identification of Mycobacterium species. However, a number of advantages for the molecular techniques over conventional testing for identifying mycobacteria species are described (14–20).
In this study, 85 water samples were collected from different sources (Table 1). The prevalence of NTM was determined using culture, biochemical tests and PCR-RFLP analyses on hsp65 gene. 25.9% of water samples contained one to three different species of NTM. Twenty-two NTM were isolated and 19 species identified by conventional methods. Dominant isolates were M. fortuitum- (26.7%), M. chelonae like organism and M. mucogenicum (13.3%). The results analyses did not shown any correlation between the mycobacteria presence in water samples and the range of the total chlorine concentrations (P>0.05) but the presences of NTM were correlated with the water sample temperature (P <0.05%).
Covert and Rodgers isolated different species of NTM from 54% of ice samples and 35% of public drinking water (5). Shin and Lee showed that half of tap water samples in hospital environment are positive for mycobacteria (21). Argueta and Yoder reported that 25 of 121 food samples (20.6%) were positive for NTM (18). The results of this study showed that the incidence of NTM was similar to other geographical environments.
In this study, a 441 bp fragment of NTM hsp65 gene was amplified and digested by BstEII and HaeIII and their patterns were analyzed on polyacrilamid gel. As shown in Fig. 2 and 3, there were different PCR-RFLP profiles. Nineteen isolates (86.4%) of NTM were identified to the species level. Three isolates presented profiles that were different from the known RFLP profiles and could not be identified that is agree with other studies (18–21). Turenne and Tschetter showed that hsp65 PRA was useful for identification of some species such as M. gastri and M. kansasii that cannot be identified by other methods (17). Wong and Yip, reported that PCR-RFLP targeting hsp65 gene region could identify 74.5% of NTM (19). Telenti and Marchesi identified 10 NTM isolates to the species level using PCR-RFLP in which 439 bp PCR products were digested with BstEII and HaeIII (20). However, in this study, some isolates shown patterns that are different with the results of other studies (8, 11–15). Absence of standardization for all of NTM species may cause some confusing in pattern analysis especially in new species. In other hand, interpretation of bands is ambiguous for highly polymorphic species. Interpretation of the patterns sometimes leads to miss identification, because of the sequence base banding patterns, which does not conform the electrophoretic banding pattern exactly (8).
In this studym, 2 positive controls were used: M. smegmatis (Hae III: 154/129/63/45/41 and BstEII: 245/140/85) and M. fortuitum (HaeIII: 137/127/67/56 and BstEII: 229/129/82). In comparing of patterns of this study to other investigation, some minor different was negligible. For example HaeIII enzyme for M. mucogenicum produces segments size 139/65/58 bp (8) and in this study NTM 6, NTM 13 and NTM 18 (with same segments of BstEII enzyme products, 300–325/130–140), shown partly different in HaeIII products: 140/65/55, 140/70/60 and 140/80/70/60, but they identified as M. mucogenicum. Of course patterns of bands that reported in different researches sometimes varied by 10–12 bp different (5, 8, 14). In other example for NTM identified as M. chelonae in referents paper (13), HaeIII products segments were shown, 140/85 and 140/65 but in this study segments for NTM 140/80 and they were detected as M. chelonae. In addition, the bands lower than 50 bp difficultly were distinguished; however, bigger segments mostly were sufficient for primitive identification of species.
In conclusion, this study showed that PCR-RFLP of hsp65 gene in mycobacteria is more reliable method for identification of NTM at the specie level than conventional phenotypic methods (P<0.05).
Ethical considerations
Ethical issues (Including plagiarism, Informed Consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc) have been completely observed by the authors.
Acknowledgments
The study was supported by Grant No. 186056 from Isfahan University of Medical Sciences, Isfahan, Iran. We thank Tahmineh Narimani and Mohamad Kazemi for assisting in technical support. The authors declare that there is no conflict of interests.
List of abbreviations
- DPD method
n, n- diethyl- p- phenylene diamine (DPD) colorimetric method
- hsp65 gene
Heat Shock Protein 65 gene
- NTM
Non- tuberculous Mycobacteria
- PCR-RFLP
Polymerase Chain Reaction- Restriction Fragment Length Polymorphism
- PRA
PCR-RFLP analyses
References
- 1.Koning B, Amer I, Sollich V, Koning W. Intra and interpatient variability of the hsp65 and 16S–23S intergenic gene region in Mycohaterium abscessus strains from patients with cystic fibrosis. J Clin Microbiol. 2005;43(7):3500–3503. doi: 10.1128/JCM.43.7.3500-3503.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Groote MAD, Huitt G. Infections due to rapidly growing Mycobacteria. Clin Infect Dis. 2006;42:1756–63. doi: 10.1086/504381. [DOI] [PubMed] [Google Scholar]
- 3.Kankya C, Muwonge A, Djønne B, Munyeme M, Opuda-Asibo J, Skjerve E, Oloya J, Edvardsen V, Johansen T. Isolation of non-tuberculous mycobacteria from pastoral ecosystems of Uganda: Public Health significance. BMC Public Health. 2011;11(320):1–9. doi: 10.1186/1471-2458-11-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mazurek Gerald H, Jereb John, Vernon Andrew, LoBue Phillip, Goldberg Stefan, Castro Kenneth. Updated Guidelines for Using Interferon Gamma Release Assays to Detect Mycobacterium tuberculosis Infection, United States. CDC; Atlanta: 2010. pp. 1–25. No 59, RR- 5. [PubMed] [Google Scholar]
- 5.Covert TC, Rodgers MR, Reyes AL, Stelma GN. Occurrence of nontoberculous mycobacteria in environmental sample. Appl Environ Microbiol. 1999;68(6):3159–3161. doi: 10.1128/aem.65.6.2492-2496.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nasr Isfahani B, Tavakoli A, Salehi M, Tazhibi M. Detection of rifampin resistance patterns in Mycobacterium tuberculosis strains isolated in Iran by polymerase chain reaction-single-strand conformation polymorphism and direct sequencing methods. Mem Inst Oswaldo Cruz. 2006;101(6):597–602. doi: 10.1590/s0074-02762006000600004. [DOI] [PubMed] [Google Scholar]
- 7.Harmsen D, Dostal S, Roth A, Niemann S, Rpthganger Ridom: Comprehensive and public sequence database for identification of mycobacterium species. BMC Infect Dis. 2003;3(26):1–10. doi: 10.1186/1471-2334-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hafner B, Haag H, Geiss Hk, Noite O. Different molecular methods for the identification of rarely isolated nontuberculous mycobacteria and description of new hsp65 restriction fragment length polymorphism patterns. Mol Cell Probes. 2004;18(1):59–65. doi: 10.1016/j.mcp.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 9.Lenore SC, Arnold EG, Andrew DE. Standard methods for the examination of water and wastewater. 20th ed. American Public Health Association, American Water Works Association, Water Environment Federation, United Book Press Inc; Baltimore, Maryland: 1998. pp. 1–37. Part 1000. [Google Scholar]
- 10.Danial LH. Current Technology of Chlorine Analysis for Water and Wastewater, Technical Information Series. Hach Company Inc; U.S.A.: 2002. pp. 2–11. Booklet 17. [Google Scholar]
- 11.Chang CT, Wang LY, Liao CY, Hvang SP. Identification of nontuberculous mycobacteria existing in tap water by PCR restriction fragment length polymorphism. Appl Environ Microbiol. 2002;68(6):3159–3161. doi: 10.1128/AEM.68.6.3159-3161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Telenti A, Marcehesi F, Balz Marianne. Rapid Identification of Mycobacteria to the Species Level by Polymerase Chain Reaction and Restriction Enzyme Analysis. J Clin Microbiol. 1993;31:175–178. doi: 10.1128/jcm.31.2.175-178.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vincent A, Steingrube, Jeremy L, Gibson PCR Amplification and Restriction Endonuclease Analysis of a 65-Kilodalton Heat Shock Protein Gene Sequence for Taxonomic Separation of Rapidly Growing Mycobacteria. J Clin Microbiol. 2003;33:149–153. doi: 10.1128/jcm.33.1.149-153.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chimara E, Ferrazoli L, Ueky S, Martins M, Durham AM, Arbeit RD, Leão SC. Reliable identification of mycobacterial species by PCR-restriction enzyme analysis (PRA)-hsp65 in a reference laboratory and elaboration of a sequence-based extended algorithm of PRA-hsp65 patterns. BMC Microbiol. 2008;8(48):1–12. doi: 10.1186/1471-2180-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin C, Xue C, Ting ZC, Huan DB, Zhong ZJ. Identification of Mycobacterium marinum 65 KD heat shock protein gene by polymerase chain reaction restriction analysis from lesion of swimming pool granuloma. Chin Med J. 2006;119(1):43–48. [PubMed] [Google Scholar]
- 16.Marchi A, Juttel I, Kawacubo E, Dalmarco E, Blatt S, Cordova C. Evaluation of methods for detection and identification of Mycobacteria species in patients suspected of having pulmonary tuberculosis. Braz J Microbiol. 2008;39:613–618. doi: 10.1590/S1517-83822008000400003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turenne TC, Tschetter L, Wolfe J, Kabani A. Necessity of quality controlled 16S rRNA gene sequence databases identifying nontuberculous mycobacterium species. J Clin Microbiol. 2001;39:3637–3648. doi: 10.1128/JCM.39.10.3637-3648.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ricardo de Souza Moraes P, Chimara E, Alice da Silva Telles M, Yoko Misuka Ueki S, Atsuko Totumi Cunha E, Robin Honer M, Cardoso Leão S. Identification of nontuberculosis mycobacteria from the central public health laboratory from Mato Grosso Do Sul and analysis of clinical relevance. Braz J Microbiol. 2008;39:268–272. doi: 10.1590/S1517-838220080002000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong DA, Yip PC, Tse DL, Tung VW, Cheung DT, Kam KM. Routine use of a simple low-cost genotypic assay for the identification of mycobacteria in a high throughput laboratory. Diagn Microbiol Infect Dis. 2003;47:421–426. doi: 10.1016/s0732-8893(03)00133-0. [DOI] [PubMed] [Google Scholar]
- 20.Chimara E, Ferrazoli L, Yoko Misuka Ueky S, Conceição Martins M, Mitchel Durham A, Arbeit RD, Cardoso Leão S. Reliable identification of mycobacterial species by PCR-restriction enzyme analysis (PRA)-hsp65 in a reference laboratory and elaboration of a sequence-based extended algorithm of PRA-hsp65 patterns. BMC Microbiol. 2008;8(48):1–12. doi: 10.1186/1471-2180-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin JH, Lee EJ, Lee HR, Ryu SM, Kim RH, Chang CL, Kim YJ, Lee JN. Prevalence of non tuberculosis mycobacteria in a hospital environment. J Hosp Infect. 2007;65:143–148. doi: 10.1016/j.jhin.2006.10.004. [DOI] [PubMed] [Google Scholar]