Abstract
Background:
The T allele of the hepatic lipase (HL) C-514T polymorphism was previously found to be associated with lower plasma HL activity. Here, we examined the association between this polymorphism and plasma HDL-cholesterol concentrations in patients with coronary arteries stenosis.
Methods:
We studied 342 subjects undergoing coronary angiography in two groups of non CAD (n=146) and CAD (n=196). −514C→T polymorphism was determined using polymerase chain reaction and restriction fragment length polymorphism (PCR-RFLP).
Results:
After adjustment for age, smoking and body mass index, HDL-cholesterol concentrations were significantly higher in men with the C/T&T/T genotype than those with the C/C genotype(mean 38.6 and 34.7 respectively P=0.01). The frequency of T allele in non CAD was 0.136 and 0.226 in female and male respectively and 0.170 and 0.223 for female and male in CAD subjects. There was no difference in T allele frequency in CAD and none CAD groups in male and female (P=0.466 and 0.722 respectively).
Conclusion:
−514C→T of LIPC gene have a positive effect on HDL-C concentration especially in male gender. However, no difference was determined in frequency of T allele between CAD and normal arteries subjects.
Keywords: Hepatic lipase gene, HDL-C, Coronary artery stenosis, T allele, Iran
Introduction
Atherosclerotic diseases are a major cause of morbidity and mortality worldwide. Among the classic risk factors known, elevated levels of low density lipoprotein cholesterol (LDL-C), and reduced levels of high density lipoprotein cholesterol (HDL-C) are associated with a significant increase in the incidence of atherosclerotic cardiovascular disease.
HDL are a family of heterogeneous particles that vary in size, density and chemical composition as a result of their rates of synthesis and catabolism and of continuous intravascular remodeling by the action of enzymes and transport proteins (1).
Plasma HDL concentration is modulated by both environmental and genetic factors. Findings from several studies suggest that polymorphisms at the hepatic lipase (HL), apolipoprotein AI/CIII/AIV, and cholesteryl ester transfer protein genes are major sources of genetically determined variation in plasma HDL-C concentrations (2). Notably, allelic variations at the HL gene account for up to 25% of the variability in plasma HDL-C concentrations (2).
Hepatic Lipase (HL), a glycoprotein member of the lipase super family, plays an important role in the metabolism and modeling of both pro-and anti atherogenic lipoproteins. Synthesized and secreted by the liver, HL carries out several metabolic functions, including the hydrolysis of triglycerides, the lipolysis of phospholipids, the modeling of small, dense atherogenic LDL particles, and the catabolism of HDL (3).
The human hepatic lipase gene (LIPC), located on chromosome 15q21, and is 35kb in size, with 9 exons spans >120 kb of DNA and encodes a protein with 449 amino acids (4).
The C→T base pair substitution at Position-514 is associated with up to 30% reduction in promoter activity in vitro (5–7), marked decreased plasma HL activity, and increased concentrations of plasma HDL, HDL2, and large buoyant LDL particles. The association between −514C/T polymorphisms in LIPC gene with the serum HDL-C levels was demonstrated. (8–10).
Thus, there is considerable evidence of associations between promoter polymorphisms of the HL gene and HL activity and plasma lipoprotein concentrations. The C-514T polymorphism of LIPC gene is not associated with CHD susceptibility (11). However, the effects of the LIPC genotype on atherosclerosis have been controversial (12).
With this background, the present study investigated the effects of LIPC promoter −514 C→T polymorphism on coronary atherosclerosis inpatients in Tabriz city in Iran.
Materials and Methods
Subjects
A total of 342 unrelated patients with possible coronary heart stenosis ages 17–80 years (mean 54 years) attending the specialist clinics at the Emam Reza and Shahid madani hospitals in Tabriz, Iran were studied. All subjects underwent coronary angiography. The coronary angiograms were classified as non CAD, stenosis of one vessel (1VD), 2 vessels (2VD) and 3 vessels (3VD).
Data for body mass index (BMI), smoking history, blood pressure, diabetes status and lipid profile were collected. Hypertension was considered to be present if systolic blood pressure was >140 mmHg or diastolic blood pressure >90 mmHg and any antihypertensive treatment had been instituted. Diabetes mellitus was diagnosed if fasting plasma glucose was >120 mg/dl. Smoking status was defined as non-smokers, low smokers (<10 cigarettes/day), and ex smoker (> 10 cigarettes/day).
The inclusion criteria were the presence of chest pain whose required angiographic examination. Those of receiving lipid lowering drugs or having a history of coronary bypass surgery, diabetes, hepatic or renal failure (serum creatinine > 2.0 mg/dl) and uncontrolled hypothyroidism, or neoplasias were excluded from the study.
Informed consent was obtained from patients and the study was approved by the Tabriz University of Medical Sciences Ethical Committee.
Blood sampling and biochemical determinations
Blood samples were collected early in the morning after an overnight fast in glass tubes. Glucose, total cholesterol (TC), triglycerides (TG), HDL-C (direct method) and LDL-C (direct method) were determined by standard enzymatic procedures using commercial kits (Pars Azmon Co). Apolipoprotein A1 (Apo A1) and Apolipoprotein B (Apo B) levels were measured by immunoturbidimeteric method (Pars Azmon Co). Creatinine is measured with Jaffe method for exclude renal patients.
Genotyping of hepatic lipase gene polymorphism
DNA was extracted from 1 ml EDTA-containing blood using phenol-chlorophorm method. Amplification of a 711-bp promoter region of the hepatic lipase gene was carried out by polymerase chain reaction (PCR) with 50 ng genomic DNA and 10 pmol of each oligonucleotide primers (F: 5′-GGATCACCTCTCAATGGGTC -3′, and R: 5′-ACCTGGTTTCAGGCTTTGTC-3′) in 25 μl. PCR was performed at 96 °C for 5 min followed by 36 cycles of denaturation at 95 °C for 45 sec, annealing at 60 °C for 1 min, and extension at 72 °C for 1 min.
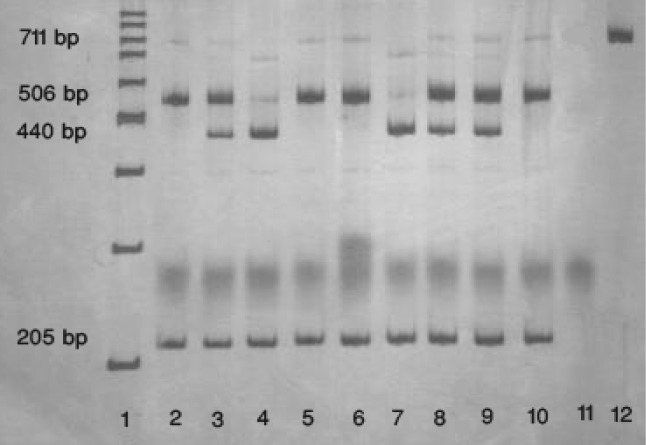
The PCR products (10 μl) were then digested with 5 units of restriction enzyme NlaIII in a total volume of 20 μl as recommended by the manufacture (Fermentas canada). Digested products were separated by electrophoresis on a 6% non-denaturing polyacrylamide gel at 200 V for 2 h. and visualized by silver staining procedure (Fig. 1).
Fig. 1:
Representative polyacrylamide gel electrophoresis picture of −514C/T. Lane 1 is DNA marker, lane2 and 5 and 6 and 10 are related to the CC genotype, lane3 and 8 and 9 are related to the CT genotype, lane 4 and 7 are related to the TT genotype. Lane 11 is negative control (without DNA), Lane 12 is DNA control (without enzyme)
Statistical analysis
Frequency distributions of characteristics of studied subjects were examined according to LIPC genotype. Chi-square tests were used to compare the genotypes. Due to small number of TT genotype (n =17) and absence of any significant differences between CT and TT genotypes regarding HDL-C concentrations, we considered CT and TT genotype as a group.
All values are expressed as means ± S.D. unless otherwise noted. ANOVA was used to analyze the differences in plasma lipid, ApoA and ApoB levels. When statistically significant effects were demonstrated, Turkey’s post-hoc test was used to identify between-group differences. In order to adjust for sex, age, hypertension, BMI and smoking, we employed a general linear model.
Statistical analyses were carried out using the SPSS statistical package (SPSS, 11.5). Statistical significance was assessed at the 5% level of probability.
Results
A total of 342 subjects were investigated, from which 196 individuals were found to be affected with CAD based on coronary angiography procedure. Among the all subjects, 222 carriers of the CC genotype, 103 of the CT genotype and 17 of the TT genotype of the LIPC promoter gene were identified. This study revealed a T allele frequency of 0.221 and 0.150 in males and females respectively (Table 1). The interaction between LIPC genotypes and CAD groups is represented in Table 2.
Table 1:
Genotype distribution and allele frequency of −514 C >T polymorphism between male and female groups
| Genotype | % Frequency (number) | ||
|---|---|---|---|
| Men | Women | Total | |
| CC | 62.4(151) | 71(71) | 64.9(222) |
| CT | 31(75) | 28(28) | 30.1(103) |
| TT | 6.6(16) | 1(1) | 5(17) |
| T allele | 0.221 | 0.150 | 0.211 |
Table 2:
Characteristics of the subjects according to LIPC genotype and CAD status
| Genotype | non CAD | 1VD | 2VD | 3VD | Total |
|---|---|---|---|---|---|
| Female * | |||||
|
| |||||
| CC | 43 | 11 | 9 | 8 | 71 |
| CT | 16 | 4 | 4 | 4 | 28 |
| TT | 0 | 1 | 0 | 0 | 1 |
| Total | 59 | 16 | 13 | 12 | 100 |
| Male ** | |||||
| CC | 55 | 27 | 35 | 34 | 151 |
| CT | 25 | 21 | 15 | 14 | 75 |
| TT | 7 | 1 | 2 | 6 | 16 |
| Total | 87 | 49 | 52 | 54 | 242 |
P Value is 0.752
P Value is 0.466
CAD; coronary artery disease, 1VD; stenosis of one vessel, 2VD; stenosis of 2 vessels, 3VD; stenosis of 3 vessels.
P< 0.05 is significant
Statistical analysis failed to show any overall relationship between the −514C/T polymorphisms and CAD groups. Also values of the CAD groups were similar among the 3 genotype groups.
The characteristics of the subjects according to the –514C/T polymorphism are shown in Table 3. To investigate the effects LIPC promoter polymorphisms on lipid profile, we compared two subgroups LIPC genotype CC compared with genotype CT/TT.
Table 3:
Characteristics of the subjects according to LIPC genotype status
| Parameter | CC(n=222) | CT&TT(n=120) | P Value |
|---|---|---|---|
| Age[ year] | 53.2±10.7 | 54.7±10.0 | 0.25 |
| BMI [kg/m2] | 27.0±4.0 | 26.7±3.8 | 0.58 |
| Apo A-I [mg/dl] | 123.6±19.0 | 124.1±21.4 | 0.84 |
| Apo B100 [mg/dl] | 103.7±26.5 | 103.3±3 | 0.90 |
| Total Cholesterol [mg/dl] | 172.7±40.9 | 177.2±50.1 | 0.43 |
| Triglyceride [mg/dl] | 196.9±100.9 | 174.1±94.8 | 0.07 |
| HDL-C [mg/dl] | 36.3±9.6 | 39.3±12.1 | 0.02 |
| LDL-C [mg/dl] | 95.1±33.3 | 96.6±35.1 | 0.73 |
P< 0.05 is significant
BMI, Body Mass Index; HDL-C, high density lipoprotein-cholesterol; LDL-C, low density lipoprotein cholesterol, Apo A-I; Apolipoprotein A-I, Apo B100; Apolipoprotein B100
No significant differences were found between subjects with the CC genotype and those with the CT/TT genotype concerning total cholesterol, triglyceride, ApoA1.ApoB, LDL-C, BMI, and age. Finally, a significant association was found between T allele and HDL-C concentration in men group (Table 4).
Table 4:
HDL-C concentration in subjects according to LIPC genotype status and gender
| HDL-C(mg/dl) | −514C/T | n | Mean± SD | P Value |
|---|---|---|---|---|
| Male | CC | 151 | 34.8±9.1 | 0.01 |
| CT&TT | 91 | 38.61±10.8 | ||
| Female | CC | 71 | 39.7±10.0 | 0.54 |
| CT&TT | 29 | 41.5±15.6 | ||
| Total | CC | 222 | 36.3±9.6 | 0.02 |
| CT&TT | 120 | 39.3±12.2 |
P< 0.05 is significant
Discussion
The present study investigated the effects of −514 C→T, LIPC polymorphism on lipid profile in subjects with coronary arteries stenosis. This polymorphism was detected in about 36% of patients with angiographically established coronary artery disease and in about 33% of non CAD control subjects (Table 1).
This study revealed a T allele frequency of 0.226, 0.136 and 0.223, 0.170 for non CAD (males and females) and CAD (males and females) respectively. These finding are in agreement with previously reported T allele frequency in Caucasians (13, 14).
It has been suggested that HL may modulate the development of cardiovascular disease through its catalytic activity and independently of ligand-binding function pathways (15). In addition, HL has been identified to be present in the vessel wall where it modulates atherogenic risk in apoE-deficient and lecithin-cholesterol acyl transferase transgenic mice (16). These multiple functions of HL, which facilitate not only plasma lipid metabolism but also cellular lipid uptake, can be anticipated to have a major and complex impact on atherogenesis. Therefore, the C-514T HL polymorphism may confer atherosclerosis susceptibility by affecting HL synthesis and activity. However, no significant difference was founded concerning T allele frequency in non CAD and CAD groups in this study (P=0.752 and P= 0.466 for women and men respectively). There is ample evidence to demonstrate that HL affects both HDL cholesterol and triglyceride-rich lipoproteins (17, 18). It can be postulated that TT genotype with low HL activity could changed production of preβ HDL and delivery of HDL -C to the liver (19). We didn’t find any statistical significance in other heart risk factors in CC and CT&TT genotype in our study groups, therefore we suggest substitution C→T in −514 of LIPC don’t change the heart risk factors (Table 3). Very few data are available concerning the relation between the LIPC −514C→T polymorphism and plasma HDL-C concentrations among patients with coronary artery stenosis.
In this study, serum HDL-C concentration was found to be higher among men subjects with T allele (Table 4). These findings are in consistent with literature documenting elevated plasma HDL-cholesterol concentration in association with the T allele among the general white population (20–23).
It has been widely accepted that such an association is likely to be mediated by a reduction in HL activity induced by the- 514 T allele. In vitro studies reported upto30% reduction in promoter activity attributed to the C→T base substitution (24, 25). The role of HL in the metabolism of HDL is well recognized (9). The association of T allele with HDL-C levels in a specific Gender may be due to differences in sex hormones which may differentially modulate lipoprotein metabolism or HL activity between males and females. The gender difference in HL activity has led some to hypothesize that HL activity is a major determinant of the more atherogenic. Lipoprotein profile in men compared with women. Moreover As HL activity is regulated by sex- steroid hormones which are presented in higher level in men than in women, the −514C→T of LIPC gene may cause further reduction of HL activity in men, leading to denser HDL-C concentration (Table 4).
In conclusion, these data demonstrate two findings. First, there was a significant association of T allele with HDL-C concentration in men groups and second there was no difference at T allele frequency in non CAD and CAD groups.
Acknowledgments
We thank all members of Shahrekord Cellular and Molecular Research Center. This study was supported by Biotechnology Research Center of Tabriz University. The authors have declared no conflict of interest.
Abbreviations
- BMI
body mass index;
- CAD
coronary artery disease;
- HDL
high-density lipoprotein;
- HDL-C
HDL cholesterol;
- LDL
low-density lipoprotein;
- LDL-C
LDL cholesterol;
- LIPC
human hepatic lipase gene.
Ethical considerations
Ethical issues (Including plagiarism, Informed Consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc) have been completely observed by the authors.
References
- 1.National Cholesterol Education Program (NCEP): Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (AdultTreatment Panel III) Third Report of the National Cholesterol Education Program (NCEP) final report. Circulation. 2002;106:3143–421. [PubMed] [Google Scholar]
- 2.Sviridov D, Nestel PJ. Genetic factors affecting HDL levels, structure, metabolism and function. Curr Opin Lipidol. 2007;18(2):157–63. doi: 10.1097/MOL.0b013e32803dbdd6. [DOI] [PubMed] [Google Scholar]
- 3.Bensadoun A, Berryman DE. Genetics and molecular biology of hepatic lipase. Curr Opin Lipidol. 1996;7:77–81. doi: 10.1097/00041433-199604000-00005. [DOI] [PubMed] [Google Scholar]
- 4.Carr MC, Brunzell JD, Deeb SS. Ethnic differences in hepatic lipase and HDL in Japanese, black, and white Americans: role of central obesity and LIPC polymorphisms. J Lipid Res. 2004;45:466–73. doi: 10.1194/jlr.M300295-JLR200. [DOI] [PubMed] [Google Scholar]
- 5.Deeb SS, Zambon SS, Carr MC, Ayyobi AF, Brunzell JD. Hepatic lipase and dyslipidemia: interactions among genetic variants, obesity, gender, and diet. J Lipid Res. 2003;44:1279–86. doi: 10.1194/jlr.R200017-JLR200. [DOI] [PubMed] [Google Scholar]
- 6.Botma GJ, Verhoeven AJ, Jansen H. Hepatic lipase promoter activity is reduced by the C-480T and G-216A substitutions present in the common LIPC gene variant, and is increased by upstream stimulatory factor. Atherosclerosis. 2001;154:625–32. doi: 10.1016/s0021-9150(00)00478-0. [DOI] [PubMed] [Google Scholar]
- 7.Deeb SS, Peng R. The C-514T polymorphism in the human hepatic lipase gene promoter diminishes its activity. J Lipid Res. 2000;41:155–8. [PubMed] [Google Scholar]
- 8.Kashani Farid MA, Azizi F, Hedayati M, Daneshpour MS, Shamshiri AR, Siassi F. Association between CETP Taq1B and LIPC −514C/T pol-ymorphisms with the serum lipid levels in a group of Tehran’s population: a cross sectional study. Lipids in Health and Disease. 2010;9:96–103. doi: 10.1186/1476-511X-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang C, Ridaura RL, Rimm EB, Rifai N, Hunter DJ, Hu FB. Interactions between the −514C/T polymorphism of the hepatic lipase gene and life style factors in relation to HDL concentrations among US diabetic men. Am J Clin Nutr. 2005;81:1429–35. doi: 10.1093/ajcn/81.6.1429. [DOI] [PubMed] [Google Scholar]
- 10.Zambon A, Deeb SS, Hokanson JE, Brown BG, Brunzell JD. Common variants in the promoter of the hepatic lipase gene are associated with lower levels of hepatic lipase activity, buoyant LDL, and higher HDL2 cholesterol. Arterioscler Thromb Vasc Biol. 1998;18:1723–9. doi: 10.1161/01.atv.18.11.1723. [DOI] [PubMed] [Google Scholar]
- 11.Wang HR, Jiang M, Qiu JP. Quantitative Assessment of the Effect of Hepatic Lipase Gene Polymorphism on the Risk of Coronary Heart Disease. Archives of Medical Research. 2010;41:383–390. doi: 10.1016/j.arcmed.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 12.Jansen H, Verhoeven AJ, Sijbrands EJ. Hepatic lipase: a pro- or anti-atherogenic protein? J Lipid Res. 2002;43:1352–1362. doi: 10.1194/jlr.r200008-jlr200. [DOI] [PubMed] [Google Scholar]
- 13.Nazu A, Nishimura Y, Terada Y, Mabuchi H. Effects of hepatic lipase gene promoter nucleotide variations on serum HDL cholesterol concentration in the general Japanese population. J Hum Genet. 2001;46:172–177. doi: 10.1007/s100380170084. [DOI] [PubMed] [Google Scholar]
- 14.Allen A, Belton C, Patterson C, Horan P, McGlinchey P, Spence M, et al. Family–based association studies of lipid gene polymorphisms in coronary artery disease. Am J Cardiol. 2005;96(1):52–5. doi: 10.1016/j.amjcard.2005.02.043. [DOI] [PubMed] [Google Scholar]
- 15.Ji J, Herbison CE, Mamotte CD, Burke V, Taylor RR, van Bockxmeer FM. Hepatic lipase gene −514 C/T polymorphism and premature coronary heart disease. J Cardiovasc Risk. 2002;9(2):105–13. doi: 10.1177/174182670200900206. [DOI] [PubMed] [Google Scholar]
- 16.Nong Z, Gonzalez-Navarro H, Amar M, Freeman L, Knapper C, Neufeld EB, et al. Hepatic lipase expression in macrophages contributes to atherosclerosis in apoE-deficient and LCAT-transgenic mice. J Clin Invest. 2003;112:367–78. doi: 10.1172/JCI16484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thuren T. Hepatic lipase and HDL metabolism. Curr Opin Lipidol. 2000;11(3):277–83. doi: 10.1097/00041433-200006000-00008. [DOI] [PubMed] [Google Scholar]
- 18.Zambon A, Bertocco S, Vitturi N, Polentarutti V, Vianello D, Crepaldi G. Relevance of hepatic lipase to the metabolism of triacylglycerol-rich lipoproteins. Biochem Soc Trans. 2003;31(5):1070–4. doi: 10.1042/bst0311070. [DOI] [PubMed] [Google Scholar]
- 19.Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang C, Lopez-Ridaura R, Rimm EB. Genetic variation in the hepatic lipase gene and the risk of coronary heart disease among US diabetic men: potential interaction with obesity. Diabetologia. 2006;49:1552–1559. doi: 10.1007/s00125-006-0235-2. [DOI] [PubMed] [Google Scholar]
- 21.Guerra R, Wang J, Grundy SM, Cohen JC. A hepatic lipase (LIPC)allele Associated with high plasma concentrations of high density lipoprotein cholesterol. Proc Nat Acad Sci USA. 1997;94:4532–7. doi: 10.1073/pnas.94.9.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Acker BA, Botma GJ, Zwinderman AH. High HDL cholesterol does not protect against coronary artery disease when associated with combined cholesteryl ester transfer protein and hepatic lipase gene variants. Atherosclerosis. 2008;200:161–167. doi: 10.1016/j.atherosclerosis.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 23.Jansen H, Chu G, Ehnholm C, Dallongeville J, Nicaud V, Talmud PJ. The T allele of the hepatic lipase promotervariant C-480T is associated With increased fasting lipids and HDL and increased preprandial and postprandial LpCIII:B: European Atherosclerosis Research Study (EARS)II. Arterioscler Thromb Vasc Biol. 1999;19:303–8. doi: 10.1161/01.atv.19.2.303. [DOI] [PubMed] [Google Scholar]
- 24.Isaacs A, Sayed-Tabatabaei FA, Njajou OT, et al. The −514C to T Hepatic Lipase promoter region polymorphism and plasma lipids: a meta-analysis. J Clin Endocrinol Metab. 2004;89:3858–3863. doi: 10.1210/jc.2004-0188. [DOI] [PubMed] [Google Scholar]
- 25.Molly CC, John DB. Ethnic differences in hepatic lipase and HDL in Japanese, black, and white Americans: role of central obesity and LIPC polymorphisms. J Lipid Res. 2004;45:466–473. doi: 10.1194/jlr.M300295-JLR200. [DOI] [PubMed] [Google Scholar]