Abstract
Background
Resistance to contemporary broad-spectrum β-lactam antibiotics mediated by extended-spectrum β-lactamases (ESBLs) is increasing worldwide. Klebsiella pneumoniae, an important cause of nosocomial and community acquired urinary tract infections has rapidly become the most common ESBL producing organism. We examined ESBL production in urinary isolates of K. pneumoniae in relation to the presence of blaSHV, blaTEM and blaCTX-M genes.
Methods:
Antibiotic resistance of 51 clinical isolates of K. pneumoniae was determined to amoxicillin, amikacin, ceftazidime, cefotaxime, cefteriaxon, ceftizoxime, gentamicin, ciprofloxacin and nitrofurantoin by disc diffusion. Minimum inhibitory concentrations were also measured for ceftazidime, cefotaxime, cefteriaxon, ceftizoxime and ciprofloxacin. ESBL production was detected by the double disc synergy test and finally, presence of the blaSHV, blaTEM and blaCTX-M genes were shown using specific primers and PCR.
Results:
Disc diffusion results showed that 96.08 % of the isolates were resistant to amoxicillin followed by 78.43 % resistance to nitrofurantoin, 49.02 % to amikacin and ceftazidime, 41.17 % to ceftriaxone, 37.25% resistance to cefotaxime and ceftizoxime, and 29.42 % to gentamicin and ciprofloxacin. Both resistant and intermediately resistant organisms were resistant in MIC determinations. Twenty two isolates (43.14%) carried blaSHV, 18 (35.29%) had blaTEM and 16 (31.37%) harbored blaCTX-M genes. ESBL production was present in 14 isolates (27.45 %) of which, 3 did not harbor any of the 3 genes. Among the non-ESBL producers, 9 lacked all 3 genes and 2 carried them all.
Conclusion:
No relation was found between gene presence and ESBL expression.
Keywords: Klebsiella pneumoniae, Urinary infection, Extended spectrum β-lactamases, blaSHV, blaTEM, blaCTX-M
Introduction
Klebsiella pneumoniae is an opportunistic pathogen that causes a significant proportion of community and hospital acquired infections including urinary tract, pneumonia, septicemia and soft tissue infections (1). Extended-spectrum β-lactam antibiotics have been widely used for treatment of serious Gram-negative infections since the 1980s. However, bacterial resistance has emerged quickly due to the production of ESBLs (2–6). These enzymes are capable of hydrolyzing extended spectrum β-lactam antibiotics such as ceftazidime, ceftriaxone, cefotaxime and aztreonam. ESBLs are derived from genes for the narrower-spectrum β-lactamases (TEM-1, TEM-2 or SHV-1) by mutations that alter the amino acid configuration around the enzyme active site (4, 5). They are typically encoded by plasmids that can be exchanged between bacterial species (7, 8). It is also reported that the predominant types of ESBLs in K. pneumoniae are SHV type followed by TEM and CTX-M (2, 4, 9, 10).
The prevalence of ESBL producing K. pneumoniae varies around the world and has been reported to be around 1–5% in European countries, 7.5% in North America, 13.3% in Europe, 22.4% in Asia Pacific Rim, 44% in Latin America, 48.5% in Turkey, 51% in China, 71.4% in Mexico and 72% in India (4, 11–13). The prevalence of ESBL producing K. pneumoniae in Iran is reported to range from 19.6 to 75% with SHV as the dominant type (14–18). We studied ESBL production in the urinary isolates of Klebsiella pneumoniae in relation to the presence of SHV, TEM and CTX-M β-lactamase genes among the isolates.
Materials and methods
Fifty one urinary isolates of K. pneumoniae were chosen from a collection of urinary isolates between March and August 2008 from two hospitals in Tehran (Taleghani and Imam Hussein). The isolates were tested for antimicrobial susceptibility by the disc diffusion method according to the NCCLS guidelines (19). The antibiotic discs were obtained from Padtan Teb (Tehran, Iran) and the following antibiotics were used: amoxicillin (30 μg), amikacin (30μg), ceftazidime (30μg), cefotaxime (30μg), cefteriaxon (30μg), ceftizoxime (30μg), gentamicin (10μg), ciprofloxacin (10μg), and nitrofurantoin (300μg). Minimum inhibitory concentrations were also measured for 5 antibiotics using the broth microdilution method and the NCCLS guidelines (20).
The double disc synergy test (DDST) was used to detect ESBL production (21). Briefly, the organisms were swabbed on to Mueller-Hinton agar plates (as performed for disc diffusion), an amoxiclav disc (20 + 10 μg) was placed in the center of the plate and ceftizoxime (30μg), cefotaxime (30μg), cefteriaxon (30μg) discs were placed 15 mm away from the central disc. The plates were incubated at 37 °C for up to 24 h. An increase of ≥ 5 mm inhibition zone for antibiotics around the amoxyclav disc compared to the cephalosporin discs alone was considered ESBL production. ESBL production was further confirmed by the phenotypic confirmatory test (PCT) using ceftazidime and ceftazidime/clavlanic acid (22).
Genomic DNA was prepared from overnight cultures grown on Lauria-Bertani broth [LB] (Hi-Media, Mumbai, India) by inoculating a single colony into 100 μl of double distilled water and boiling for 10 min. An equal volume of chloroform: isoamyl alcohol (24:1 v/v) was added, mixed by end over end rotation and the mixture was centrifuged at 8000 × g for 15 min. The upper layer was then used in the PCR reaction mixtures (21).
PCR amplification was carried out with specific primers for blaSHV (forward: 5′-TGG TTA TGC GTT ATA TTC GCC-3′, and reverse: 5′-GGT TAG CGT TGC CAG TGCT-3′) amplifying an 800 bp fragment (Fazapajouh, Tehran), blaTEM (forward: 5′-ATA AAA TTC TTG AAG ACG AA-3′, reverse: 5′-GAC AGT TAC CAA TGC TTA ATC-3′) amplifying an 850 bp fragment and blaCTX-M (forward; 5′-TGG TTA AAA ATC ACT ACT GCG-3′, reverse: 5′-ATT ACA AAC CGT CGG TGA C-3′) amplifying an 891 bp product (Takapou Zist, Tehran) (22–24). PCR was carried out in 25 μl volume reaction mixtures containing 10 pM of each primer, 200 μM dNTP, 1.5 mM MgCl2, 1.5 μl of crude template DNA and 1 U Taq polymerase in the reaction buffer provided by the manufacturer (CinnaGen, Tehran, Iran). The following thermocycler program was carried out for PCR experiments: 4 min denaturation at 94 °C followed by 32 cycles of 1 min at 94 °C, 1 min at the annealing temperature (55 °C for blaSHV and blaCTX-M and 58 °C for blaTEM) and 1 min at 72 °C with a final extension period of 10 min at 72 °C.
Results
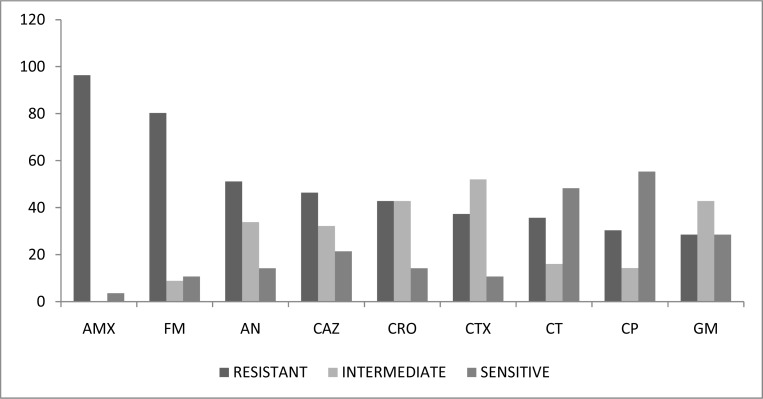
The antibiotic susceptibility profile measured by disc diffusion is shown in figure 1. As observed the highest rate of resistance was found for amoxicillin (96.08 %) and the lowest for gentamicin and ciprofloxacin (29.42 %). Among the broad spectrum β-lactam antibiotics, the level of resistance ranged between 37.25 to 41.17 %. The MIC results showed 88.23% resistance to cefotaxime, 83.35% to ceftriaxone, 82.35% to ceftazidime, 50.98% to ceftizoxime and 47.06% to ciprofloxacin. These values were much higher compared to the resistance patterns obtained by disc diffusion. However, when the numbers of resistant and intermediately resistant organisms were compiled, there was total agreement between the two phenotypic tests.
Fig. 1:
Antibiotic susceptibility of 51 urinary isolates of Klebsiella pneumoniae measured by disc diffusion. AMX, amoxicillin, FM, nitrofurantoin, AN, amikacin, CAZ, ceftazidime, CRO, cefriaxone, CTX, cefotaxime, CT, ceftizoxime, CP, ciprofloxacin, GM, gentamicin
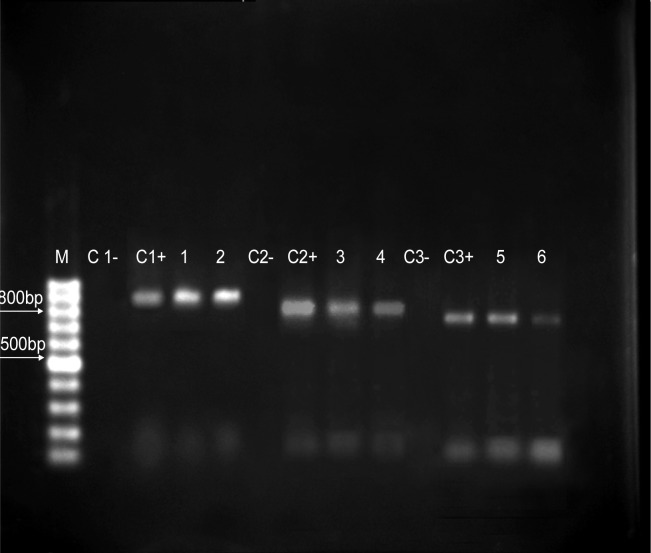
Figure 2 shows the PCR amplification products of β-lactamase genes. The results showed that 22 isolates (43.14%) carried the blaSHV gene, 18 (35.29%) had blaTEM and 16 (31.37%) harbored the blaCTX-M genes. The DDST results detected ESBL production in 14 isolates (27.45%) of which, 4 carried the blaSHV gene, 3 had blaCTX-M, 3 harbored blaTEM, 1 carried blaSHV-TEM and surprisingly 3 did not have any of the 3 genes tested. Of the 37 Organisms which did not produce ESBL, 9 lacked all 3 genes, 7 carried blaSHV, 4 blaCTX-M, 3 blaTEM, 4 blaSHV-TEM, 4 blaSHV-CTX-M, 3 blaTEM-CTX-M and finally, 2 carried all 3. These results suggest that other types of ESBL may be responsible for the ESBL phenotype.
Fig. 2:
PCR amplification of β-lactamase genes in urinary isolates of K. pneumoniae. M, 100 bp DNA ladder, C-, negative control, C+, positive control, lanes 1–2, blaCTX-M (891 bp), lanes 3–4, blaTEM (850 bp), lanes 5–6, blaSHV (800 bp) amplification products
Discussion
Klebsiella pneumoniae has rapidly become the most common ESBL producing organism, making its eradication difficult from high risk wards such as intensive care units. In our study, 27.45% of the urinary isolates of K. pneumoniae were ESBL producers. Despite the fact that a small number of isolates were tested, ESBL production among our isolates was higher than a study performed in Semnan, Iran where ESBL production in urinary isolates was reported to be 19.6% (17). On the other hand, our results showed a lower rate of ESBL production compared to most of the other studies carried out in Iran and other parts of the Middle or Far East (11–18). The prevalence rates of ESBL production are different in different countries or different parts of each country and are very much dependent upon antibiotic policies in every region. The conventional disc diffusion method does not usually allow for routine differentiation of ESBL producing strains. Several methods have been devised to differentiate between ESBL and non-ESBL producers among which, the double disc diffusion test has been reported to be the most discriminating (25–26). Also, molecular detection methods such as PCR have been used for molecular differentiation of β-lactamase producing isolates. However, most of the studies performed, aim for the detection of the more commonly occurring β-lactamase genes such as the TEM and SHV family and it is not practical to screen for a large number of β-lactamase genes. In a study carried out in Taiwan, a correlation was reported between phenotypic resistance to cefotaxime, ceftazidime and ceftriaxone in ESBL producing K. pneumoniae isolates and presence of blaSHV, blaCTX-M and blaVEB-1 like genes but not blaTEM (22). Our results showed no relation between presence of TEM, SHV and CTX-M β-lactamase genes with β-lactamase production or ESBL phenotype. Presence of ESBLs can be masked by the expression of chromosomal or plasmid mediated AmpC β-lactamases. Also, ESBL producing strains with AmpC β-lactamases can cause a false negative in ESBL detection (25–27). More importantly, expression of β-lactamase genes depend upon the environmental conditions such as the presence of antibiotics and gene presence shown by PCR does not necessarily indicate its expression.
Ethical considerations
Ethical issues (including plagiarism, informed Consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc) have been completely observed by the authors.
Acknowledgments
The authors wish to thank Shahid Beheshti Research Council for providing a special grant to finance this research.
References
- 1.Podschun R, Ullman U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods and pathogenicity factors. Clin Microbiol Rev. 1998;11:589–603. doi: 10.1128/cmr.11.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford PA. Extended-spectrum β-lactamases in the 21st century: Characterization, epidmiology and detection of this important resistance threat. Am Soc Microbiol. 2001;14:933–51. doi: 10.1128/CMR.14.4.933-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paterson DL, Bonomo RA. Extended-spectrum β-lactamases: a clinical update. Clin Microbiol Rev. 2005;18:657–86. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paterson DL, Hujer KM, Hujer AM, Yeiser B, Bonomo MD, Rice LB, Bonomo RA, the International Klebsiella Study Group Extended-spectrum β-lactamases in Klebsiella pneumoniae bloodstream isolates from seven countries: dominance and widespread prevalence of SHV and CTX-M type β-lactamases. Antimicrob Agents Chemother. 2003;47:3554–60. doi: 10.1128/AAC.47.11.3554-3560.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mulvey MR, Bryce E, Boyd D, Ofner-agostini M, Christianson S, Simor AE, Paton S, the Canadian Hospital Epidemiology Committee, Canadian Nosocomial Infection Surveillance Program, Health Canada Ambler Class A extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella spp. in Canadian hospitals. Antimicrob Agents Chemother. 2004;48(4):1204–14. doi: 10.1128/AAC.48.4.1204-1214.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacoby GA, Han P. Detection of extended-spectrum β-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli. J Clin Microbiol. 1996;34(4):908–11. doi: 10.1128/jcm.34.4.908-911.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brun-Buisson C, Legrand P, Philippon A, Montravers F, Ansquer M, Duval J. Transferable enzymatic resistance to third-generation cephalosporins during nosocomial outbreak of multiresistant Klebsiella pneumoniae. Lancet ii. 1987:302–6. doi: 10.1016/s0140-6736(87)90891-9. [DOI] [PubMed] [Google Scholar]
- 8.Knothe H, Shah P, Kremery V, Antal M, Mitsuhashi S. Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infect. 1983;11:315–7. doi: 10.1007/BF01641355. [DOI] [PubMed] [Google Scholar]
- 9.Sirot D. Extended spectrum plasmid mediated β-lactamases. J Antimicrob Chemother. 1995;36(suppl A):19–34. doi: 10.1093/jac/36.suppl_a.19. [DOI] [PubMed] [Google Scholar]
- 10.Jemima SA, Verghese S. Multiplex PCR for bla(CTX-M) and bla(SHV) in the extended spectrum β-lactamase (ESBL) producing Gram-negative isolates. Ind J Med Res. 2008;128(3):313–7. [PubMed] [Google Scholar]
- 11.Stewart CD, Rasheed JK, Hubert SK, Biddle JW, Raney PM, Anderson GJ, Williams PP, Brittain KL, Oliver A, McGowan JE, Jr, Tenover FC. Characterization of clinical isolates of Klebsiella pneumoniae from 19 laboratories using the National Committee for Clinical Laboratory Standards Extended-Spectrum β-lactamase detection methods. J Clin Microbiol. 2001;39(8):2864–72. doi: 10.1128/JCM.39.8.2864-2872.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nijssen S, Florijn A, Bonten MJM, Schmitz FJ, Verhoef J, Fluit AC. β-lactam susceptibilities and prevalence of ESBL-producing isolates among more than 5000 European Enterobacteriaceae isolates. Internat J of Antimicrob Agents. 2004;24:585–91. doi: 10.1016/j.ijantimicag.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 13.Reinert RR, Low DE, Rossi F, Zhang X, Wattal C, Dowzicky MJ. Antimicrobial susceptibility among organisms from the Asia/Pacific Rim, Europe and Latin and North America collected as part of test and the in vitro activity of tigecycline. J Antimicrob Agents Chemother. 2007;60(5):1018–29. doi: 10.1093/jac/dkm310. [DOI] [PubMed] [Google Scholar]
- 14.Aminzadeh Z, Sadat Kashi M, Sha’bani M. Bacteriuria by extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates in a governmental hospital in south of Tehran, Iran. Iran J Kidney Dis. 2008;2:197–200. [PubMed] [Google Scholar]
- 15.Feizabadi MM, Etemadi G, Yadegarinia D, Rahmati M, Shabanpoor S, Bokaei S. Antibiotic resistance patterns and frequency of extended-spectrum β-lactamase-producing isolates of Klebsiella pneumoniae in Tehran. Med Sci Monitor. 2006;12(11):362–5. [PubMed] [Google Scholar]
- 16.FeizabadIi MM, Farahani AS, Rahmati M, Asadi S. Phenotypic characterization and plasmid analysis of Klebsiella pne-umoniae strains from Iranian patients. R. Ci. méd. biol. Salvador. 2008;7(3):273–9. [Google Scholar]
- 17.Irajian G, Jazayeri-Moghadas A, Beheshti A. Prevalence of extended-spectrum β–lactamase positive and multidrug resistance pattern of Escherichia coli and Klebsiella pneumoniae isolates, Semnan, Iran. Iran J Microbiol. 2009;1:49–53. [Google Scholar]
- 18.Shahcheraghi F, Moezi H, Feizabadi MM. Distribution of TEM and SHV beta-lactamase genes among Klebsiella pneumoniae strains isolated from patients in Tehran. Med Sci Monitor. 2007;13(11):247–50. [PubMed] [Google Scholar]
- 19.National Committee for Clinical Laboratory Standards (NCCLS) 8th edition. Wayne, Pennsylvania, USA: 1999. Performance standards for antimicrobial disc susceptibility tests. Approved standard M2-A8. [Google Scholar]
- 20.National Committee for Clinical Laboratory Standards (NCCLS) Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Wayne, Pennsylvania, USA: 2002. Approved Standard M2/A7. [Google Scholar]
- 21.Health Protection Agency, Laboratory detection and reporting of bacteria with extended spectrum β-lactamase. National Standard Method QSOP 51. 2008;(2.2) [Google Scholar]
- 22.Lal P, Kapil A, Das BK, Sood S. Occurrence of TEM & SHV gene in extended spectrum β-lactamases (ESBLs) producing Klebsiella spp. isolated from a tertiary care hospital. Ind J Med Res. 2007;125:173–8. [PubMed] [Google Scholar]
- 23.Tribuddharat C, Srifuengfung S, Chiangjong W. A correlation between phenotypes and genotypes of extended-spectrum beta-lactamase (ESBL)-producing Klebsiella pneumoniae in a university hospital, Thailand. Dis Antimicrob Agents. 2007;24:117–23. [Google Scholar]
- 24.Jain A, Mondal R. TEM and SHV genes in extended spectrum β-lactamase producing Klebsiella species and their antimicrobial resistance pattern. Ind J Med Res. 2008;128:759–64. [PubMed] [Google Scholar]
- 25.Perilli M, Amico ED, Segatore B, Rosaria de Massis M, Bianchi C, Luzzaro F. Molecular characterization of extended-spectrum β-lactamases produced by nosocomial isolates of Enterobacteriaceae from an Italian Nationwide survey. J Clin Microbiol. 2002:611–4. doi: 10.1128/JCM.40.2.611-614.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown DFJ, Andrews J, King A, MacGowan AP. Detection of extended-spectrum β-lactamases with E-test and double disc potentiation method. J Antimicrob Chemother. 2000;46:323–42. doi: 10.1093/jac/46.2.327. [DOI] [PubMed] [Google Scholar]
- 27.Thomson KS. Controversies about extended-spectrum and AmpC β-lactamases. Emerg Infect Dis. 2001;7(2):333–6. doi: 10.3201/eid0702.010238. [DOI] [PMC free article] [PubMed] [Google Scholar]