Abstract
Background
Hepatitis B virus (HBV) gene and protein variations are frequently been seen in chronic patients. The aims of study were to determine the genotypes as well as the patterns of variations distribution in chronically-infected patients from the central part of Iran.
Methods:
The surface gene was amplified, sequenced and subsequently aligned using international and national Iranian database.
Results:
All strains belonged to genotype D, subgenotype D1 and subtype ayw2. Of all 62 mutations occurred at 39 nucleotide positions, 31 (50%) were missense (amino acid altering) and 31 (50%) were silent (no amino acid changing). At the amino acid level, 30 substitutions occurred, however, 3 were in positions 122 and 127, corresponded to subtypic determination. 22 (73%) out of 30 amino acid mutations occurred in different immune epitopes within surface protein, of which 12 (54.54%) in B cell epitopes in 10 residues; 5 (45.45%) in T helper epitopes in positions; 5 (22.73%) in inside CTL epitopes in 4 residues.
Conclusion:
The distribution of amino acid mutations as well as the ratio between silent and missense nucleotide mutations showed a narrowly focused immune pressure had already been on the surface protein in these patients, led to the emergence of escape mutants in these patients.
Keywords: HBV genotypes, HBV genotype D, HBV genotype in Iran, HBV immune epitopes
Introduction
Hepatitis B virus (HBV) is a well-known agent of acute and chronic hepatitis, chronically infecting around 400 million individuals worldwide. The morbidity and mortality of persistent HBV infection are a major public health concern. More than one million deaths every year are due to end-stage HBV liver disease, such as decompensated liver cirrhosis and hepatocellular carcinoma (HCC).
Recent studies have shown that HBV surface protein (HBsAg) is more variable than initially thought, and amino acid exchanges are scattered over the whole molecule. These changes are classified as either “variants” (determined by host HLA amino acid arrangement over a long period) or “mutations” (arose after vaccine/drug therapy) (1). According to the former classification, HBV genome variability can usefully be classified into at least eight families (genotypes) based on surface protein variations with a characteristic geographic distribution (2–5). Further, Variation within a sub-component of the S gene within the major hydrophilic region (MHR) of HBsAg, the “a determinant”, is strongly associated with subtype variation (6).
On the other hand, HBV mutations define a different and a more complicated scenario. In fact, this phenomenon seemed to be the potential mechanism for the pathogenesis basis of chronicity and the clinical complications of this infection. Thanks to the invention of polymerase chain reaction (PCR) and other facilities for direct sequencing and molecular approaches, hundreds of reports have been published so far to reveal the relationship between correspondent mutations and the clinical/serological pictures of the chronic patients.
Although the Middle East region countries are recognized as high endemic areas of HBV infection (2%–20% prevalence of HBV surface antigen) (7–8) but there is lack of data on HBV genotypic prevalence in some parts of this region. Reports by Alavian revealed that the prevalence of HBV in Iran ranges between 1.7%–2.5% of the general population and according to the recent epidemiologic data, this prevalence has been decreased in recent years, due to the impact of global vaccination (9). However, molecular epidemiology findings, as HBV genotype, have also remained vague in many countries which are located in the Middle East. The aim of this study was to determine the genotypes of HBV in a central part region of Iran and to characterize the molecular variations of the chronic patients.
Materials and Methods
Sera
Nineteen HBsAg-positive patients who were referred to the Isfahan Hepatitis Centre (2004–2006), were enrolled in a cross-sectional study. To cover the whole province of Isfahan, we studied seven regions based on population and geographical zones. All patients were interviewed and examined by gastroenterologists to evaluate the clinical findings and the results of the investigative workup (liver histology, ultrasonography, and laboratory tests such as serologic, biochemical and virological tests) in order to determine the clinical status of the patient. Samples were tested by (ELISA) with commercial kits for HBsAg detection (Diapro, Milan, Italy). 2 ml of sera were taken from each patient and stored at −80° C for further investigations.
DNA extraction
HBV DNA was extracted from a 200 μl of aliquot of serum using Qiagen Mini Blood Kit (Qiagen, Hilden, Germany) according to manufacturer’s instruction. In brief, 20 μl of protease added to the serum in a 1.5 ml tube. Then, 200 μl of Al buffer added to each tube, vortexed and incubated for 10 min in 56°C. For DNA precipitation, 200 μl of ethanol was added to the mixture, centrifuged for 1 min. Components transferred to a collection tube contained filter tube. Trapped DNA was washed in two steps by AW1 and AW2 buffers to eliminate puririties together with centrifugation after each step. Finally, DNA was eluted using 100 μl of elution buffer, stored in −20 °C.
Polymerase chain reaction
Polymerase chain reaction (PCR) was carried out in 100 μl of a mixture containing 5 μl of the extracted DNA, using standard methodology. The complete surface gene was amplified using S1, S2, S6 and S7 primers (Table 1) which includes the region of surface gene specifying HBV genotypes/subtype (amino acid positions 122–160). First round PCR was performed using 1 U of Taq DNA polymerase (HotStart Taq PCR, Qiagen, Hilden, Germany), 0.25 mM of each dNTP, 10x reaction buffer, 12.5 pmol of S1 and S2 primer. For the second round PCR, 1 μl of first round PCR product was added to 99 μl of the reaction mixture with the same composition as the first round except that S1/S2, were replaced by S6/S7. A quantity of 5 μl of the second round PCR products were analysed by electrophoresis in 1% agarose gel, stained by ethidium bromide, and visualized under u.v. light.
Table 1:
Oligonucleotide primers used for PCR and sequencing. Base positions numbered from the EcoRI site
| Primer | Sequence 5′ 3′ of Oligonucleotides | Base Position | Type |
|---|---|---|---|
| S1 | CCT GCT GGT GGC TCC AGT TC | 56–75 | Sense |
| S2 | CCA CAA TTC (K)TT GAC ATA CTT TCC A (K=G/T) | 1003–979 | Anti-sense |
| S6 | GCA CAC GGA ATT CCG AGG ACT GGG GAC CCT G | 113–146 | Sense |
| S7 | GAC ACC AAG CTT GGT TAG GGT TTA AAT GTA TAC C | 857–823 | Anti-sense |
DNA sequencing
The HBsAg subtype of the sequences was defined by substitutions in the 'a' determinant between codons 122 and 160 inclusive. Direct sequencing of surface gene was carried out (Perkin Elmer ABI-PRISMTM 377 DNA Sequencer, Fostercity, CA, USA) using 2 pmol of appropriate primers: S6C and S7D for surface gene. The results were analyzed using Chromas software.
Sequence analysis
After allocating a sequence to an HBV genotype by analysis of the S gene, the surface gene amino acid/nucleotide variations that were found were compared with a reference sequence obtained from Okamoto (1988, accession mumber, AB033559) and HBsAg sequences from Iranian isolates obtained from GenBank and NCBI and from our own laboratory reports. Comparing to the former, any amino acid changes defined as “variant” (host HLA-determined). With regards to the latter (Iranian database sequences), amino acid differences defined as “mutation”. Sequences have been submitted to GenBank, numbered from GU938323 to GU938341.
Phylogenetic analysis
The evolutionary history was inferred using the Neighbor-Joining method. The bootstrap consensus tree inferred from 1000 replicates was taken to represent the evolutionary history of the taxa analyzed. The evolutionary distances were computed using the Kimura 2-parameter method and were in the units of the number of base substitutions per site. Codon positions included were 1st+2nd+3rd+Noncoding. All positions containing gaps and missing data were eliminated from the dataset (Complete deletion option). There were a total of 681 positions in the final dataset. Phylogenetic analyses were conducted in MEGA4 (10).
Results
In this study, nineteen HBsAg-positive patients infected with HBV were enrolled who all were native residents of Isfahan Province (center of Iran). The group studied consisted of inactive hepatitis B carriers. 6 (31.6%) were female and 13 (68.4%) were male with a mean age of 39 years (results not shown). All cases were HBsAg positive.
Phylogenetic Analysis
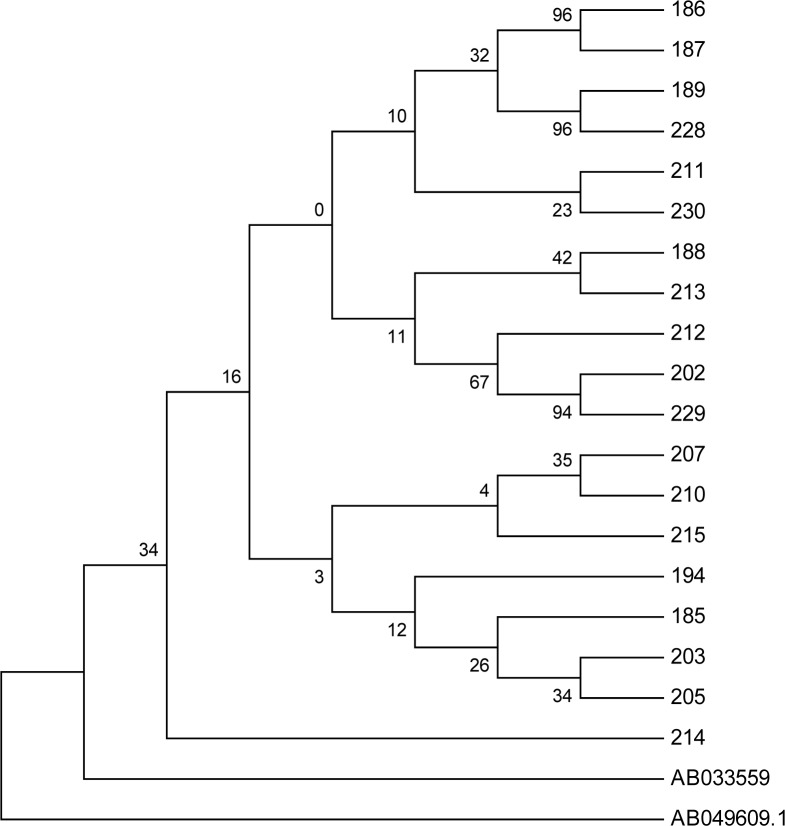
The results of the phylogenetic tree revealed that Iranian HBV isolates from Isfahan were of genotype D, supported by 95% and 97% bootstrap value (1,000 replicates), respectively (Fig 1). In the phylogenetic tree, a genotype C sequence (accession number AB049609) was chosen for out grouping. It is noteworthy that all Iranian isolates clustered in a distinct branch that separated them from the reference Okamoto genotype D. In total, with exception of samples 211 and 230, the rest of isolates phylogenetically showed a homogenous pattern.
Fig. 1:
Neighbour joining phylogenetic trees of surface genes sequences from 19 samples. Note: S gene tree rooted with sequence AB049609 (reference genotype C).All Iranian isolates were compared to sequence AB033559 (reference genotype D, see the text). The figure shows bootstrap values of ≥70% and scale denotes percent diversity. Coding numbers indicate samples that have been analysed in the figure. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsedAlignment of complete nucleotide sequences of HBsAg from 19 sera.
Note: Nucleotides are numbered from the beginning of the HBsAg using the single letter code
Substitutions in comparison with reference genotype D (Okamoto, AB033559)
Overall, comparing with reference sequence (Okamoto, 1988), at the nucleotide level, of a total of 168 changes, 118 (70.2%) and 50 (29.8%) were silent and missense, respectively (results not shown). At the amino acid levels, all contained A70P compared to Okamoto reference (Table-2). We believe that this substitution was assigned as “variant” (see material and methods). According to the above mentioned description, 19 out of 49 amino acid changes were variants and the other 30 changes were mutations (see below).
Table 2:
Alignment of complete amino acid sequences of HBsAg which shows genotype/subtype identification and other variations of 19 sera.
Note: Amino acid residues are numbered from the beginning of the HBsAg using the single letter code
Genotyping
Analysis of variation within the S gene of 19 patients with chronic HBV infection demonstrated that the only detected subtype was D (100%), subgenotype D1 (100%) and subtype ayw2 (100%) (Fig. 1 and Table 2).
Nucleotide and amino acid substitutions
In comparison with Iranian sequences obtained from the database as well as from our unpublished data, in addition to the genotypic characterization described above, the sequences of the strains showed a few variability over the regions sequenced. In all, 62 “mutations” occurred at 39 nucleotide positions, of them, 31 (5 %) were missense (amino acid altering) and 31 (50%) were silent (no amino acid changing) (Table 3). At amino acid level, 30 substitutions occurred, however, 3 were in positions 122 and 127 (Table 2–3). Amino acid variations in these positions were allocated for the subtype determination, thus, they were variants instead of true mutations. Table 3 shows the comparison between nucleotide and amino acid variations from the isolates. Further, it was possible to identify the level of S proteins evolution between isolates by measuring the mutation rate of individual sequences (Table 3). The average mutation rate of all sequences was 3.26 (0.48%) according to the number of mutations per site.
Table 3:
The levels of mutation rates between isolates deduced from the number and the percentage of individual sequences
| Sample Code | Nucleotide Mutation | Missense Mutation | Amino Acid Change | No. | Mutation Rate |
|---|---|---|---|---|---|
| 185 | T284G, T345C, G492A, T647A | T284G, T647A | L95W, L216Stop | 4 | 0.59% |
| 186 | T213C, C428T, G492T | C428T, G492T | S143L, E164D | 3 | 0.44% |
| 187 | T213C, C428T, G492T | C428T, G492T | S143L, E164D | 3 | 0.44% |
| 188 | T135C, T207A, A303C, A328G | T207A, A303C, A328G | C69Stop, Q101H, I110V | 4 | 0.59% |
| 189 | T213G, C246A, C369T, A386G, C432T, G587T | A386G, G587T | Q129R, W196L | 6 | 0.88% |
| 194 | T345C, A360C, G365A | G365A | R122K | 3 | 0.44% |
| 202 | T135C, A256T, G611T, C618T, A619C, T626A | A256T, G611T, A619C, T626A | I86F, S204I, S207R, L209Stop | 6 | 0.88% |
| 203 | T345C | 1 | 0.15% | ||
| 205 | C88A, T345C, T623C | C88A, T623C | Q30K, I208T | 3 | 0.44% |
| 207 | G36T, C192T, T339C, T345A, A364C, A366G, C379A, C383T | C379A, C383T | P127T, A128V | 8 | 1.17% |
| 210 | T345C, A352G, C379A | A352G, C379A | T118A, P127T | 3 | 0.44% |
| 211 | - | - | 0 | 0.00% | |
| 212 | A401T, C513T, C618T, A619C | A401T, A619C | Y134F, S207R | 4 | 0.59% |
| 213 | T207A, T345A | T207A | C69Stop | 2 | 0.29% |
| 214 | C420A, A534C | 2 | 0.29% | ||
| 215 | T345A, C358T | C358T | P120S | 2 | 0.29% |
| 228 | T213A, C246A, G587T | G587T | W196L | 3 | 0.44% |
| 229 | T22C, [C134A, T135C], C618T, A619C | T22C, [C134A, T135C], A619C | F8L, T45N, S207R | 5 | 0.73% |
| 230 | 0 | 0.00% | |||
| Average | - | - | - | 3.26 | 0.48% |
Amino acid mutations within the surface protein immune epitopes
22 (73%) out of 30 amino acid mutations occurred in different immune epitopes within surface protein, of which 12 (54.54%) in B cell epitopes in 10 residues; 5 (45.45%) in T helper epitopes in positions; 5 (22.73%) in inside CTL epitopes in 4 residues (Table 4).
Table 4:
Amino acid mutations within HBsAg of patient groups. B cell, T helper and CTL epitopes. Samples arranged accordance to the arrangement of immune epotopes. Amino acids are described by single letter code and numbered from the beginning of HBsAg
| Isfahan | Th Epitope | CTL Epitope | B Epitope | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||||||
| Sample Code | Amino Acid Position | 30 | 45 | 95 | 196 | 216 | 207 | 208 | 209 | 101 | 110 | 118 | 120 | 122 | 127 | 128 | 129 | 134 | 143 |
|
| |||||||||||||||||||
| Wild Type | Q | T | L | W | L | S | I | L | Q | I | T | P | R | P | A | Q | Y | S | |
| 185 | W | Stop | |||||||||||||||||
| 186 | L | ||||||||||||||||||
| 187 | L | ||||||||||||||||||
| 188 | H | V | |||||||||||||||||
| 189 | L | R | |||||||||||||||||
| 194 | K | ||||||||||||||||||
| 202 | R | Stop | |||||||||||||||||
| 203 | |||||||||||||||||||
| 205 | K | T | |||||||||||||||||
| 207 | T | V | |||||||||||||||||
| 210 | A | T | |||||||||||||||||
| 211 | |||||||||||||||||||
| 212 | R | F | |||||||||||||||||
| 213 | |||||||||||||||||||
| 214 | |||||||||||||||||||
| 215 | S | ||||||||||||||||||
| 228 | |||||||||||||||||||
| 229 | N | R | |||||||||||||||||
| 230 | |||||||||||||||||||
Discussion
It was possible to sequence the S gene for 19 HBV strains from Isfahan province. Genotype D, subgenotype D1 and subtype ayw2 accounted for 100% of isolates. Published and unpublished data from our laboratory indicated that in Iran, there has been an obvious predominance of these virus genetic pattern (11–12). We already hypothesized that this unique pattern of homology is related to the relative recent distribution and circulation of HBV in Iran compared to other countries in the region (12).
The ratio between silent and missense nucleotide mutations in our patients was about 0.48%. This indicated that the proportion of deduced amino acid changes in these chronically infected patients was high. It is not unexpected, as with prolonged period of chronicity, the emergence of frequent mutations in these patients is inevitable (see below).
The distribution of the mutations within known surface protein immune epitopes reflects the virus-host interaction with a prolonged infection period. Being a structural protein, HBsAg in an immune target. The consequence of selection pressure posed by anti-S antibodies would be the emergence of immune escape mutations in this protein which no longer could be recognized by the host immune system. The results would be the presence of virus (and sometimes with a high level of viral load) in a chronically-infected patient. The occurrence of Th and CTL epiotpe mutations indicates an ineffective T cell response, as already shown that these responses are weak and sometimes undetectable during the chronic state of the infection (13).
In conclusion, it appears from this work that there are variations in the structural protein of HBV in chronic patients. For a better interpretation, the allocation of such molecular variations to the clinical, serological and biochemical pictures needs to be explored. In this scenario, even an individual variation must be taken into account.
Ethical considerations
Ethical issues (Including plagiarism, Informed Consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc) have been completely observed by the authors.
Acknowledgments
This study was supported by Tehran University of Medical Sciences (Grant No: 860227-5816). The authors declare that there is no conflict of interests.
References
- 1.Carman WF, Tomas H, Zuckerman AJ, Harrison TJ. Molecular variants. 2nd ed. Churchill Livingston; Hong Kong: 1998. pp. 141–172. [Google Scholar]
- 2.Naumann H, Schaefer S, Yoshida CF, Gaspar AM, Repp R, Gerlich WH. Identification of a new hepatitis B virus (HBV) genotype from Brazil that expresses HBV surface antigen subtype adw4. J General Virology. 1993;74(8):1627–32. doi: 10.1099/0022-1317-74-8-1627. [DOI] [PubMed] [Google Scholar]
- 3.Norder H, Courouce AM, Magnius LO. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology. 1994;198:489–503. doi: 10.1006/viro.1994.1060. [DOI] [PubMed] [Google Scholar]
- 4.Stuyver L, De Gendt S, Van Geyt C, Zoulim F, Fried M, Schinazi RF, Rossau R. A new genotype of hepatitis B virus: complete genome and phylogenetic relatedness. J General Virology. 2000;81:67–74. doi: 10.1099/0022-1317-81-1-67. [DOI] [PubMed] [Google Scholar]
- 5.Arauz-Ruiz P, Norder H, Robertson BH, Magnius LO. Genotype H: a new Amerindian genotype of hepatitis B virus revealed in Central America. J General Virology. 2002;83:2059–73. doi: 10.1099/0022-1317-83-8-2059. [DOI] [PubMed] [Google Scholar]
- 6.Okamoto H, Imai M, Kametani M, Nakamura T, Mayumi M. Genomic heterogeneity of hepatitis B virus in a 54-year-old woman who contracted the infection through maternofetal transmission. Jpn J Exp Med. 1987;57:231–6. [PubMed] [Google Scholar]
- 7.Malekzadeh R, Khatibian M, Rezvan H. Viral hepatitis in the world and Iran. J Iranian Med Council. 1997;15:183–200. [Google Scholar]
- 8.André F. Hepatitis B epidemiology in Asia, the Middle East and Africa. Vaccine. 2000;18(Suppl 1):S20–2. doi: 10.1016/s0264-410x(99)00456-9. [DOI] [PubMed] [Google Scholar]
- 9.Alavian SM, Fallahian F, Bagheri Lankarani K. The Changing Epidemiology of Viral Hepatitis B in Iran. J Gastrointestin Liver Dis. 2007;16(4):403–406. [PubMed] [Google Scholar]
- 10.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24:1596–99. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 11.Jazayeri SM, Basuni AA, Sran N, Gish R, Cooksley G, Locarnini S, Carman WF. HBV core sequence: definition of genotype-specific variability and correlation with g geographical origin. Journal of Viral Hepatitis. 2004;11:488–501. doi: 10.1111/j.1365-2893.2004.00534.x. [DOI] [PubMed] [Google Scholar]
- 12.Jazayeri SM, Carman WF. HBV Genotype D Evolution in Middle East & South Asia. Hepatitis Monthly. 2009;9(1):9–11. [Google Scholar]
- 13.Chisari FV, Ferrari F. Hepatitis B virus immunopathology. Springer Semin Immunopathol. 1995;17:261–81. doi: 10.1007/BF00196169. [DOI] [PubMed] [Google Scholar]