Abstract
Background:
Molecular epidemiology of measles virus (MV) is important, not only to measure the success of measles vaccination programs but also to monitor the circulation and elimination of the virus worldwide. In this study, we compared MV obtained from patients before the 2003 mass vaccination MR campaign and viruses detected after 2003 until 2008 in Iran.
Methods:
The nucleoprotein (N) gene of 29 MV strains circulating in Iran between 2002 and 2008 were amplified by RT-PCR and subjected to sequence and phylogenetic analysis.
Results:
Molecular characterization of MV studied here revealed that although the outbreaks in Iran were associated with MV genotype D4, the isolated viruses clearly belonged to several different lineages. Maximum and minimum homology within the 29 Iranian strains in our study was100% and 94.9% within the carboxyl terminus of the N gene, respectively. Using ClustalX program, the alignment of Iranian MV sequences showed nine lineages.
Conclusion:
This study provides the usefulness of MV sequence analysis for the demonstration of local interruption of indigenous strain transmission as well as providing a valuable means for monitoring the elimination processes of MV control.
Keywords: Measles, Genotype, Mass campaign, Iran
Introduction
Measles virus (MV), an enveloped RNA virus classified in the family Paramyxoviridae, genus Morbillivirus (1), is the most transmissible virus known in humans (2). Despite the vaccination programs, MV remains a heavy public health burden worldwide, particularly in developing countries (3, 4). Because of the high infectivity of measles, 95%–98% vaccine coverage is required to prevent virus circulation (2, 5). Serologically MV is known as a monotypic virus (6), however sequence analysis of the complete H gene and the 450 C-terminal nucleotides of the N gene (7, 8) classifies MV strains into eight clades (A–H) dividing into 23 genotypes (9,10). The MV clades have been recognized in different parts of the world however five of them (B1, D1, E, F, and G1) considered inactive since they have not been detected in the past 15 yr (11). In the Eastern Mediterranean region with established measles elimination goals by 2010 the objective has been to achieve and maintain interruption of indigenous measles transmission (3). Molecular epidemiology of MV is important for either measuring the success of measles vaccination programs or monitoring circulation and elimination of the virus throughout the world (12). Hence, genetic characterization of wild type MV can help to measure transmission pathways and to clarify epidemiological links during outbreaks, especially in the countries with an advanced elimination program, where most cases are imported (12, 9).
Measles vaccination started in Iran in 1967. In 1970 and 2003 the incidence of measles has been reported 346/100,000 (13) and 18/100,000 (11644 measles cases) respectively (14). Despite the reduction in the incidence of disease and the high vaccination coverage with routine measles vaccination program, including a two-dose vaccination regimen at nine months and 15 yr of age, measles cases have been reported in different areas in Iran (15). In this context, on December 2003, Iran conducted a measles/rubella (MR) catch-up campaign for individuals 5 to 25 yr olds that reached more than 33 million people with measles vaccine (16). The measles vaccination coverage has increased to 98% and the number of measles cases has clearly reduced after vaccination program. A post campaign serum-survey conducted in 2004 revealed >97.4% of the population aged between 5 and 40 yr had immunity to measles (15). Case-based surveillance for measles identified three children with laboratory confirmed disease in 2004, 35 in 2005, and 42 in 2006. The interruption of indigenous virus circulation by mass vaccination campaigns could be demonstrated by comparing the variability of pre and post campaign viruses (5). Importation of a new genotype into a country with high immunity to measles may lead to outbreaks in the susceptible population and genetic analysis will confirm the chain of transmission (17, 18).
In this study, we compared MV obtained from patients before the 2003 mass vaccination MR campaign and viruses detected after 2003 until 2008 in Iran.
Materials and Methods
This study was approved by the Disease Management Center in the Ministry of Health in Iran. Patients with rash signed informed consent. Blood (serum), throat swabs as well as nasopharyngeal specimens, and in some cases, urine that were obtained <7 d after onset of a rash from suspected cases. All collected samples were transported to the National Laboratory of Measles of Iran (NLM) in School of Public Health, Tehran University of Medical Sciences to process using standard procedures. The criteria used for the diagnosis of measles and sample collection were as described previously by WHO protocols for laboratory diagnosis of measles viral infection (19). MVs were detected from throat swabs, nasopharyngeal and urine specimens (that were serologically confirmed) using B95a and Vero-hSLAM cell lines or directly from clinical specimens. RNA was extracted by Nucleospin RNA Virus kit according to the manufacturer’s instructions (M & N, Germany).
Genotyping of the carboxyl terminus of the nucleoprotein (N) gene was performed by Reverse transcription (RT) and Polymerase Chain Reaction (PCR) according to the standard procedures for RT-PCR(10), which is based on a two-step semi-nested PCR. Briefly, following reverse transcription, specific primers targeting N gene were designed to amplify a 599 bp fragment through the first round primers (MV60-MV 63.3) using Qiagen RT-PCR kit. Second round of PCR were performed to amplify a 424 fragment using primers MV 61 and MV 63 (10). First round PCR products were sequenced using the ABI prism Big Dye Terminator Cycle Sequencing Ready Reaction Kit (PE Biosystems, Langen, Germany) by an Iranian Company (Genefanavaran Co). Genotype identification of the sequences was performed according to protocols by comparison to the WHO reference sequences, representative of the 23 established MV genotypes (3, 7, 19). The sequencing data was analyzed by Clustal X (20) and BioEdit version 7.0.0 DNA analysis softwars (21). Phylogenetic trees were constructed using Ttreecon package version 1.3 b (22) using the nucleotide Kimura-2 parameter and the neighbor- joining method with bootstrap analysis (1000 replicates).
Results
Clinical specimens from patients with acute measles infections were collected. Samples belonged to 2002–2008, one year before and 5 yr after the MR campaign in 2003. A two-step semi-nested RT-PCR was successfully performed and sequences were derived from the most variable region of the MV genome encoding the C-terminal part of the N-protein of the circulating viruses in Iran. Comparison of the analyzed 29 sequences with the sequences of the current WHO reference strains (7) clearly showed that all Iranian viruses in this study were members of genotype D4.
The derived sequences were registered in Gen-Bank (accession numbers: EU234465-EU234487 and FJ985774- FJ985779). Features of the 29 sequences are shown in Table 1.
Table 1:
Features of the sequences representative of 29 sequences obtained during 2002–2008
| Patients no | Age (year) | Sex | Days after rash onset | Source of specimen | Sequence name | Province of origin | Comments | Lineage | GenBank accession no. |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 20 | M | 4 | serum | MVs.Fars.IRA/42.02/1 | South | Johannesburg-like | 1 | EU234482 |
| 2 | 19 | M | 3 | serum | MVs.Fars.IRA/42.02/2 | South | Johannesburg-like | 1 | EU234468 |
| 3 | 19 | M | 3 | serum | MVs.Tabriz.IRA/43.02/ 1 | North-West | Johannesburg-like | 1 | EU234479 |
| 4 | 19 | M | 5 | serum | MVs.Tabriz.IRA/43.02/ 2 | North-West | Johannesburg-like | 3 | EU234471 |
| 5 | 21 | M | 3 | serum | MVs.Kordestan.IRA/42.02/ 3 | West | Johannesburg-like | 1 | EU234467 |
| 6 | 19 | M | 1 | serum | MVs.Kordestan.IRA/46.02/ 2 | West | Johannesburg-like | 1 | EU234466 |
| 7 | 19 | M | 4 | serum | MVs.Golestan.IRA/48.02/ 2 | North-Eastern | Johannesburg-like | 1 | EU234476 |
| 8 | 19 | M | 7 | serum | MVs.Kordestan.IRA/47.02 | West | Johannesburg-like | 3 | EU234469 |
| 9 | 1 | F | 6 | serum | MVs.Kordestan.IRA/45.02/ 3 | West | Johannesburg-like | 1 | EU234475 |
| 10 | 21 | M | 3 | serum | MVs.Tabriz.IRA/46.02/3 | North-Western | Johannesburg-like | 2 | EU234473 |
| 11 | 12 | M | 2 | serum | MVs.Esfahan.IRA/44.03/2 | Center | Johannesburg-like | 2 | EU234483 |
| 12 | 21 | M | 2 | serum | MVs.Kordestan.IRA/46.02/ 1 | West | Johannesburg-like | 1 | EU234474 |
| 13 | 10 | F | 3 | serum | MVs.Kordestan.IRA/45.02/ 1 | West | Johannesburg-like | 1 | EU234472 |
| 14 | 25 | F | 6 | serum | MVs.Tabriz.IRA/45.02/2 | North-Western | Johannesburg-like | 1 | EU234477 |
| 15 | 20 | M | 5 | serum | MVs.Tabriz.IRA/42.02/4 | North-Western | Johannesburg-like | 1 | EU234487 |
| 16 | 19 | M | 1 | serum | MVs.Kordestan.IRA/44.02/ 1 | West | Johannesburg-like | 1 | EU234481 |
| 17 | 17 | M | 4 | NP | MVi/Golestan.IRA/48.05 | North-Eastern | Johannesburg-like | 6 | EU234465 |
| 18 | 17 | M | 5 | NP | MVi/Tehran.IRA/25.06 | Center | Johannesburg-like | 6 | EU234485 |
| 19 | 3 | F | 4 | TS | MVs/Sistan.IRA/30.07 | South-Eastern | Johannesburg-like | 7 | EU234486 |
| 20 | 6month | M | 2 | TS | MVs/Sistan.IRA/31.07/1 | South-Eastern | India-like | 8 | EU234480 |
| 21 | 4 | F | 3 | TS | MVs/Sistan.IRA/31.07/2 | South-Eastern | Johannesburg-like | 7 | EU234484 |
| 22 | 4month | M | 3 | TS | MVs/Sistan.IRA/31.07/3 | South-Eastern | Johannesburg-like | 7 | EU234470 |
| 23 | 6month | M | 2 | TS | MVs/Sistan.IRA/31.07/4 | South-Eastern | India-like | 8 | EU234478 |
| 24 | 30 | F | 4 | TS | MVs/Siatan. IRA /45.07/1 | South-Eastern | Johannesburg-like | 7 | FJ985774 |
| 25 | 6 | M | 3 | TS | MVs/Siatan. IRA /47.07/2 | South-Eastern | Johannesburg-like | 7 | FJ985775 |
| 26 | 31 | F | 3 | TS | MVi/Mazandaran. IRA /10.08 | North | Johannesburg-like | 9 | FJ985776 |
| 27 | 31 | M | 3 | Urine | MVi/Mazandaran. IRA /17.08 | North | Johannesburg-like | 9 | FJ985778 |
| 28 | 4 | F | 4 | TS | MVs/Khorasan.Iran/20.08 | North-Eastern | Johannesburg-like | 9 | FJ985777 |
| 29 | 1.4 | F | 1 | TS | MVs/Tehran.Iran/31.08 | Center | Johannesburg-like | 9 | FJ985779 |
NP: nasopharyngeal
TS: throat swabs
Analyzing of the 29 Iranian strains showed the maximum and minimum homology within the carboxyl terminus of the N gene was 100% and 94.9%, respectively. In the other genotypes (e.g. within clade H in China) similar variability was observed (23).
It is noteworthy that for phylogenetic analysis we used other five Iranian MV strains (lineage 4 and 5) which were isolated in 2003 before MR campaign and sequenced in Measles Regional Laboratory in Tunisia before measles mass vaccination. The result of that study was published by Djebbi et al in 2005 (14).
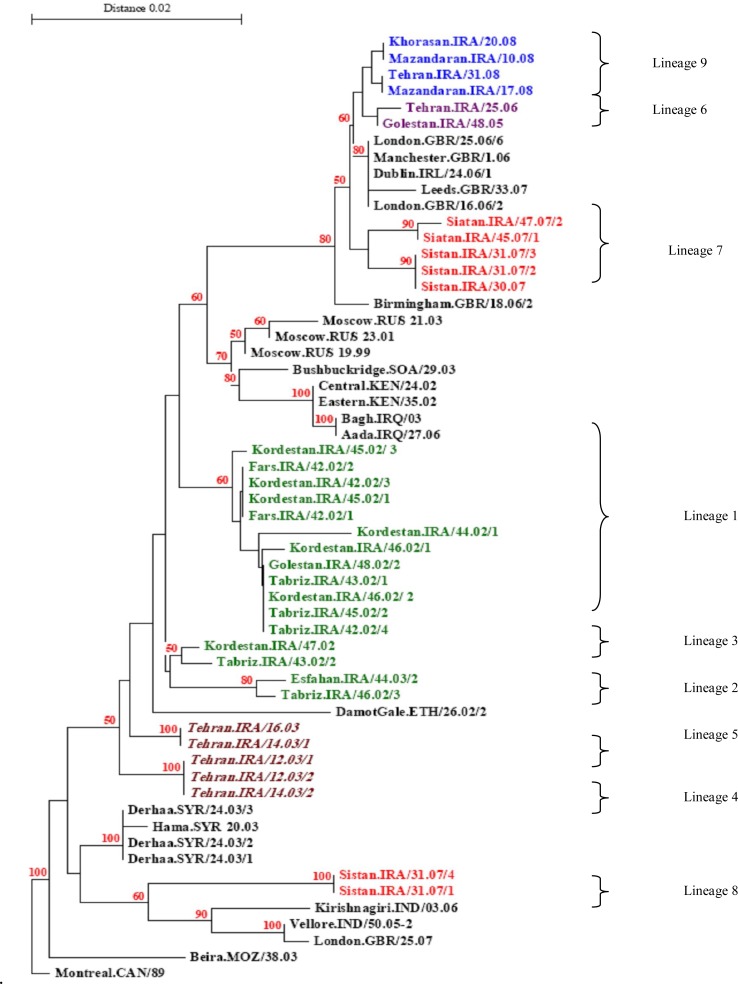
Using ClustalX program, the alignment of the 34 adequate sequences showed that the sequences from 2002 to 2008 represented in nine lineages. According to our findings, five lineages of the Iranian strains (lineages 1–5) were found before and 4 lineages (lineages 6–9) were detected after the MR campaign. The 9 lineages distinction (designated 1–9) defined by year of isolation, bootstrap values and long branch lengths are shown in Table 1 and Fig. 1.
Fig. 1:
Phylogenetic comparison of 34 Iranian MV strains (C-terminus of N gene) collected from 2002–2008 with other wild-type measles genotype D4 detected in other countries. Bootstrap values (1000 replicates) >70% are indicated. The Iranian sequences before mass vaccination are shown in bold (green and brown) (Lineages 1–5). The Iranian sequences after mass vaccination are shown in bold (blue, purple and red)(Lineages 6–9). NOTE: The Iranian sequences that indicated in bold (brown) and italic were sequenced in Measles Regional Center Tunisia before mass vaccination (Lineage 4, 5) (14)
Discussion
The utility of sequence analysis of MV strains circulating in a population is a critical means in measles, which helps distinguish between endemic transmission and importation of the virus (24, 25). This study investigates the pattern of MV genotypes before and after mass vaccination campaign in Iran.
The endemic MV strains circulating in Iran were not known prior to 1999 and data regarding genotype analysis of MV in Iran is limited. Briefly, in 1999 during an outbreak in Tehran about 30 throat samples from suspected measles cases were tested in Centers for Disease Control and Prevention (Atlanta-Georgia), which two of them were positive by RT-PCR and MV isolated and identified as genotype D4 from only one sample (7). Before measles mass campaign in 2003 in Iran, the NLM in Tehran was (and still is) in charge of working on measles suspicious cases and virus isolation. From sporadic cases in Tehran, five isolated viruses were sent to Tunisia for genotyping and all of them were again identified as genotype D4.
Following improvement in the ability of surveillance and epidemiological investigation, we studied all IgM positive samples by RT-PCR. Additionally, nasopharyngeal, throat swabs and urine specimens were inoculated on B95a and Vero-hSLAM cell lines. Attempts also successfully were made to isolate MV, as we could isolate MV from 2 nasopharyngeal samples, 1 throat swabs and 1 urine specimen.
The RNA was directly extracted from 25 clinical specimens (16 serums and 9 throat specimens) and 4 viruses were isolated from cell cultures.
After MR campaign in Iran, measles surveillance is well established and no measles epidemic has been reported since 2003. Based on our investigations, the phylogenetic analysis clearly showed that all of Iranian measles virus strains were in genotype D4 and no other obvious patterns were observed. We found that within the same province (e.g. Kordestan), strains belonging to different lineages could circulate simultaneously in different regions at different periods. In addition, one given lineage could circulate in different provinces at the same time. However, before mass vaccination no evidence was available for geographic or temporal restriction, since identical sequences were detected in multiple provinces at the same time. Nevertheless, there are reports that multiple lineages could co-circulated at the same time (26).
In our study, high diversity of the detected D4 strains suggests the role of indigenous viruses behind several outbreaks in Iran before mass vaccination campaign. Additionally, it seems that genotype D4 (with five transmission chains) has been responsible for most measles cases in these regions before introducing mass vaccination campaign.
Our results showed that the five Iranian viruses (Sistan.IRA/31.07/3, Sistan.IRA/31.07/2, Sistan. IRA/30.07, Siatan. Iran/45.07/1 and Siatan. Iran/47.07/2) as lineage 7 in 2007 and all four Iranian viruses (Mazandaran. Iran/10.08, Mazandaran. Iran/17.08, Khorasan. Iran/20.08 and Tehran. Iran/31.08) as lineage 9 in 2008 (after mass vaccination campaign) were closely related to other genotype D4 viruses which were detected in Iran as lineage 6 during 2005 and 2006 (Tehran. IRA/25.06 and Golestan. IRA/48.05) (Fig.1). Meanwhile they were very closely related to wild MV which have been reported in the UK in 2006 (Fig.1) (27, 28). In addition, our results showed that Iranian MVs, Sistan. IRA/31.07/4 and Sistan. IRA/31.07/1 were more closely related to MV from India (Fig.1).
Genotype D4 is one of the most widely detected genotype worldwide with an endemic circulation in southern and eastern Africa and in Asian countries such as India, Nepal and Pakistan (14).
In our study, Lineages 1, 4 and 5, the most frequent before mass vaccination campaign was and were present in six provinces throughout our study (Fig. 1).
It has already been shown that the isolates from Iran were divided into two lineages (Lineages 4 and 5) and were quite different from Syrian sequences with 1.3–1.8% nucleotide differences (14).
The co-circulation of the different lineages within genotype D4 in Iran suggests that multiple chains of transmission were present in Iran, as such a level of divergence within one genotype has already been observed in endemic countries, e.g. within genotype D4 in the Indian Ocean Islands; Mayotte and Seychelles (29), within genotype H2 in Vietnam (24), within genotype D8 in Nepal (30), within genotype D5 in Cambodia (26) and within genotype B3 in Sudan (31). The available data from Iran supports the minimum genetic heterogeneity within a single chain of transmission, regardless of the epidemiologic circumstances.
Analysis of the predicted amino acid sequences of the N proteins showed that the Iranian viruses contained several amino acid substitutions that have not yet been observed in any of the wild-type MV which have previously been sequenced (Data not shown).
As sequence diversity in the C-terminus of the N-gene of Iranian MV strains in our study was more than 3% before and after the MR campaign, the D4 genotype detected after the campaign could have been imported from neighboring countries or spread from imported cases which were present before the campaign, but remained undetectable as most of the laboratories in provinces lack the necessary equipment and supplies for virus isolation.
It should be mentioned that although the coverage of measles vaccination in Iran is high, it is probably not adequate to prevent infection from occurring at individual level in some regions and creates conditions that favor the rapid transmission of a newly introduced genotypes (32).
It should be noted that the limited registered measles virus strains in different databases makes the comparison difficult to make sure whether a newly detected strain is indigenous in origin or imported.
In conclusion, our study showed that different genetic variants of measles virus were circulating before and after the vaccination campaign in Iran in 2003, although all strains belonged to the same genotype (D4). The analysis of genetic distances between MV strains before and after 2003 suggests that indigenous MV transmission has been interrupted and the more recent strains were imported from other countries. Thus this study provides the usefulness of MV sequence analysis for the demonstration of local interruption of indigenous strain transmission as well as providing a valuable means for monitoring the elimination processes of virus control.
Ethical Considerations
Ethical issues including plagiarism, informed Consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc. have been completely observed by the authors.
Acknowledgments
This Investigation received technical and financial support partly from the joint WHO Eastern Mediterranean Region (No. SGS04/70), Division of Communicable Diseases (DCD) and the WHO Special Programmed for Research and Training in Tropical Diseases (TDR): The EMRO /TDR Small Grants scheme for Operational Research in Tropical and other Communicable Diseases. Also this study was supported in part (no, 3046) by the Office of Vice-Chancellor for Research, Tehran University of Medical Sciences.
We thank the staff of the Virology Division, School of Public Health, Tehran University of Medical Sciences, and Center for Diseases Management in the Ministry of Health, Iran for assistance in collection of data and serum samples. We also thank Dr. G. Tipples for his kind help in sequence analysis. The authors declare that they have no conflicts of interest.
References
- 1.Griffin DE. Measles virus. In: Knipe DM, Howley PM, editors. Fields Virology. 4th ed. Lippincott, Williams and Wilkins; Philadelphia, PA: 2001. pp. 1401–41. [Google Scholar]
- 2.Meissner HC, Strebel PM, Orenstein WA. Measles vaccines and the potential for worldwide eradication of measles. Pediatrics. 2004;114:1065–69. doi: 10.1542/peds.2004-0440. [DOI] [PubMed] [Google Scholar]
- 3.WHO State of the art of new vaccines Research & Development. 2003. Initiative for Vaccine Research. Geneva.
- 4.Rota PA, Bellini WJ. Update on the Global Distribution of Genotypes of Wild Type Measles Viruses. J Infect Dis. 2003;187:270–76. doi: 10.1086/368042. [DOI] [PubMed] [Google Scholar]
- 5.Mulders MN, Truong AT, Muller CP. Monitoring of measles elimination using molecular epidemiology. Vaccine. 2001;19:2245–49. doi: 10.1016/s0264-410x(00)00453-9. [DOI] [PubMed] [Google Scholar]
- 6.Bellini WJ, Rota PA. Genetic diversity of wild-type measles viruses: implications for global measles elimination programs. Emerg Infect Dis. 1998;4:29–35. doi: 10.3201/eid0401.980105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.WHO Nomenclature for describing the genetic characteristics of wild-type measles viruses (update), Part I. Wkly Epidemiol Rec. 2001a;76:242–47. [PubMed] [Google Scholar]
- 8.WHO Nomenclature for describing the genetic characteristics of wild-type measles viruses (update), Part II. Wkly Epidemiol Rec. 2001b;76:249–51. [PubMed] [Google Scholar]
- 9.WHO New genotype of measles virus and update on global distribution of measles genotype. Wkly Epidemiol Rec. 2005;80:341–52. [PubMed] [Google Scholar]
- 10.CDC Global measles and rubella laboratory network, MMR weekly of Center for Disease Prevention and Control. 2005;54:1100–04. January 2004–June 2005. [PubMed] [Google Scholar]
- 11.Rota PA, Rota JS, Redd SB, Papania MJ, Bellini WJ. Genetic analysis of measles viruses isolated in the United States between 1989 and 2001: absence of an endemic genotype since 1994. J Infect Dis. 2004;189(Suppl 1):160–64. doi: 10.1086/374607. [DOI] [PubMed] [Google Scholar]
- 12.Rota JS, Heath JL, Rota PA, King GE, Celma ML, Carabana J, et al. Molecular epidemiology of measles virus: Identification of pathways of transmission and the implications for measles elimination. J Infet Dis. 1996;173:32–37. doi: 10.1093/infdis/173.1.32. [DOI] [PubMed] [Google Scholar]
- 13.Khodabandeh Loo, Sabahi F, Soleimanjdhi H, Kazemnejad A, Roustai MH. Seroprevalence of neutralizing antibodies to measles virus in a vaccinated population in Iran, 1998. Eur J Epidemiol. 2003;18:1085–89. doi: 10.1023/a:1026104522780. [DOI] [PubMed] [Google Scholar]
- 14.Djebbi A, Bahri O, Mokhtariazad T, Alkhatib M, Ben Yahia A, Rezig D, et al. Identification of measles virus genotypes from recent outbreaks in countries from the Eastern Mediterranean Region. J Clin Virol. 2005;34:1–6. doi: 10.1016/j.jcv.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 15.Esteghamati A, Gouya MM, Zahraei SM, Dadras MN, Rashidi A, Mahoney F. Progress in Measles and Rubella Elimination in Iran. Pediatr Infect Dis J. 2007;26:1137–41. doi: 10.1097/INF.0b013e3181462090. [DOI] [PubMed] [Google Scholar]
- 16.Hamkar R, Jalilvand S, Mokhtari-Azad T, Jelyani KN, Nategh R. Evaluation of immunity against rubella in Iranian after mass campaign for measles-rubella vaccination on December 2003. Am J Infect Control. 2006;34:588–92. doi: 10.1016/j.ajic.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 17.Rima BK, Earle JA, Baczko K, ter Meulen V, Liebert UG, Carstens C, Carabana J, Caballero M, Celma ML, Fernandez-Munoz R. Sequence divergence of measles virus haemagglutinin duringnatural evolution and adaptation to cell culture. J Gen Virol. 1997;78:97–106. doi: 10.1099/0022-1317-78-1-97. [DOI] [PubMed] [Google Scholar]
- 18.Oliveira MI, Rota PA, Curti SP, Figueiredo CA, Afonso AM, Theobaldo M, et al. Genetic homogeneity of measles viruses associated with a measles outbreak, Sao Paulo, Brazil, 1997. Emerg Infect Dis. 2002;8:808–13. doi: 10.3201/eid0808.020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.WHO . Geneva: 1999. World Health Organization Manual for the laboratory diagnosis of measles viral infection. [Google Scholar]
- 20.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25:4876–82. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall TA. Bio Edit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 22.Van de Peer Y, De Wachter R. Construction of evolutionary distance trees with TREECON for Windows: accounting for variation in nucleotide substitution rate among sites. Comput Applic Biosci. 1997;13:227–30. doi: 10.1093/bioinformatics/13.3.227. [DOI] [PubMed] [Google Scholar]
- 23.Liffick SL, Thi Thoung N, Xu W, Li Y, Phoung Lien H, Bellini WJ, et al. Genetic characterization of contemporary wild-type measles viruses from Vietnam and the People’s Republic of China: identification of two genotypes within clade H. Virus Res. 2001;77:81–87. doi: 10.1016/s0168-1702(01)00268-4. [DOI] [PubMed] [Google Scholar]
- 24.Rota Paul A, Liffick Stephanie L, Rota Jennifer S, Katz Russell S, Redd Susan, Papania Mark, Bellini William J. Molecular Epidemiology of Measles Viruses in the United States, 1997–2001. Emerg Infect Dis. 2002;8(9):902–08. doi: 10.3201/eid0809.020206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Atrasheuskayaa AV, Kulaka MV, Neverova AA, Rubinb S, Ignatyeva GM. Measles cases in highly vaccinated population of Novosibirsk, Russia, 2000–2005. Vaccine. 2008;26:2111–18. doi: 10.1016/j.vaccine.2008.02.028. [DOI] [PubMed] [Google Scholar]
- 26.Horm SV, Dumas C, Svay S, Feldon K, Reynes JM. Genetic characterization of wild-type measles viruses in Cambodia. Virus Res. 2003;97:31–37. doi: 10.1016/s0168-1702(03)00219-3. [DOI] [PubMed] [Google Scholar]
- 27.Kremer JR, Brown KE, Jin L, Santibanez S, Shulga SV, Aboudy Y, et al. High Genetic Diversity of Measles Virus, World Health Organization European Region, 2005–2006. Emerg Infect Dis. 2008;4(1):107–14. doi: 10.3201/eid1401.070778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chironna M, Prato R, Sallustio A, Martinelli D, Germinario C, Lopalco P, Quarto M. Genetic characterization of measles virus strains isolated during an epidemic cluster in Puglia, Italy 2006–2007. Virol J. 2007;4:90. doi: 10.1186/1743-422X-4-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waku-Kouomou D, Landreau D, Olivier S, Palmyre P, Benoit-Catin T, Freymuth F, et al. Molecular Characterization of Measles Virus Circulating in the Indian Ocean Islands during 2005–2006 and in France in 2006. J Med Virol. 2007;79:1381–1387. doi: 10.1002/jmv.20959. [DOI] [PubMed] [Google Scholar]
- 30.Truong AT, Mulders MN, Gautam DC, Ammerlaan W, de Swart RL, King CC, Osterhaus AD, Muller CP. Genetic analysis of Asian measles virus strains new endemic genotype in Nepal. Virus Res. 2001;76:71–78. doi: 10.1016/s0168-1702(01)00255-6. [DOI] [PubMed] [Google Scholar]
- 31.El Mubarak HS, van de Bildt MW, Mustafa OA, Vos HW, Mukhtar MM, Ibrahim SA, et al. Genetic characterization of wild-type measles viruses circulating in suburban Khartoum, 1997–2000. J Gen Virol. 2002;83:1437–43. doi: 10.1099/0022-1317-83-6-1437. [DOI] [PubMed] [Google Scholar]
- 32.Muscat M, Vinner L, Christiansen AH, Glismann S, Bottiger BE. The benefit of molecular characterization during a measles upsurge in Denmark. Vaccine. 2007;25:6232–36. doi: 10.1016/j.vaccine.2007.05.063. [DOI] [PubMed] [Google Scholar]