Abstract
Background:
Successful treatment to eliminate HCV RNA depends on the identified genotype. In the present study, we compared the frequency of different HCV genotypes, during four years study (2004 till 2008).
Methods:
Sera specimens were received from 16 provinces of Iran. We used High Pure Viral Nucleic Acid Purification kit for extraction and samples were tested with improved form of RT-PCR technique. HCV genotypes were determined using Amplisense PCR kit and Amplicor HCV Monitoring Version 2 test utilized a reverse transcription (RT)-PCR approach to quantitative HCV RNA. Two hundreds six HCV positive specimens were entered to the study out of 389 tested samples.
Results:
Type 3a was the most frequent type (46.6%), followed by type 1 (including 1a and 1b with 25.73% and 17.47% for each respectively) with 43.2%. Looking through collected results of the four years study confirmed the rate of HCV infection in those single genotypes 1b, 3a were slightly increased from 12.22% and 38.88% in the first year to 18.66 and 46.51% in the fourth year of the study period.
Conclusion:
The analyzed data proved that some patients were infected with two different types. High viral load was also more correlated to genotype 1 than other types.
Keywords: PCR, HCV genotype, Viral load, Iran
Introduction
HCV is classified into six major genotypes with more than 90 subtypes based on nucleotide sequence homology (1). Prevalence and incidence of HCV genotypes vary among geographic areas (2). It is believed that genotypes 1, 2 and 3 are found in Western Europe, North America and Japan, while genotype 4 has been reported in Middle East, Central and North Africa (3). Genotypes 5 and 6 are mostly reported in South Africa and Sought-East Asia (4). Recently it was reported that clinical importance of HCV genotype 4 is increasing in Europe (5, 6). This type has been identified mostly in patients with cirrhosis, hepatocellular carcinoma or in drug addicts. In USA, genotype 1 has been reported to be 79% (58% for 1a and 21% 1b) (2, 7). In France, the most observed genotype was genotype 1 with the frequency of 45% (18% subtype 1a, 27% subtype 1b). The next most frequent types were genotypes 3 with the frequency of 21%, 2 and 4 with the frequency of 9% for each (8). In Spain, the most observed HCV genotypes were: 1b (41.3%), 1a (24.1%) (9). In UK the majority of HCV genotype were types 1a (32%), 1b (15%), and 3a (37%) (10). Researchers believe that genotyping pattern of Iranian patients is similar to Europe and North America since all reports confirm that most distributed genotypes are 1 and genotype 3 in Iran (1, 11, 12). But when we look carefully to the frequency of subtypes of genotype 1 and 3, we find out that distribution of these subtypes are different in each region although the Iranian genotyping pattern looks similar to that of Europe and North America may be concluded.
In the present study, we evaluated the frequency of different HCV genotypes and subtypes to underline its status in Iran.
Materials and Methods
Patients and samples
Two hundreds and six HCV positive specimens were analyzed out of 389 tested. These specimens were received from referred patients or sent specimens of general population from over 300 laboratories among the country from 16 provinces (Table 1). All received samples were tested for HCV PCR from 2004 until December 2007. These samples screened for HCV-RNA by RT-PCR. Those negative HCV PCR and genotyping were excluded. Therefore, 206 HCV-RNA positive samples with HCV genotyping test (out of 389 requests) were entered in the study.
Table 1:
Distribution of received specimens from provinces of Iran during the study period
| Provinces | n | |
|---|---|---|
| 1 | Mazandaran | 17 |
| 2 | South Khorasan | 7 |
| 3 | Yazd | 47 |
| 4 | Hormozghan | 4 |
| 5 | Golestan | 2 |
| 6 | East Azarbayejan | 17 |
| 7 | Balouchestan | 2 |
| 8 | Central | 3 |
| 9 | Eilam | 2 |
| 10 | Ghilan | 9 |
| 11 | Lorestan | 3 |
| 12 | Kordestan | 2 |
| 13 | Ghazvin | 8 |
| 14 | Khoosestan | 6 |
| 15 | Kermanshah | 2 |
| 16 | Tehran | 75 |
Extraction Method and RT-PCR
All samples were extracted by High Pure Viral Nucleic Acid Purification kit (Roche Co.). Clinical samples were tested with improved form of RTPCR technique containing labeled probe, called Fluorescent Amplification-Based Specific Hybridization (DNA technology) with the use of classic thermocycler (DNA Technology) (CE approved) (13,14).
Genotyping Protocols
Positive samples were examined for identification of genotypes and subtypes. Those negative HCV PCR samples were excluded from the study. HCV genotypes were determined using PCR kits (CE approved) (Amplisense Co.) (15). The Kit can specify subtypes 1a, 1b, 3a and genotype 2 with different product sizes 388, 395, 286 and 227 bp, after electrophoresis.
Quantitative-PCR
The Amplicor HCV test (version 2.0) was used for reverse transcription RT-PCR approach to quantitation of HCV RNA in patient’s samples. All amplification and detection steps were performed by the automated Amplicor instrument (Roche Co.). Results from the Amplicor assays were reported in HCV RNA copies per milliliter or international units of HCV RNA per milliliter (1 copy/ml= IU/ml × 2.7). 8×105 IU/ml of HCV has been accepted as cut off for High HCV viral load.
Results
Totally 389 samples were screened for HCV RTPCR (293 men and 96 women). 206 cases were positive, which were included for genotyping (172 men and 34 women). The highest frequency of HCV positive patients was observed in 30–39 yr old (32.05%) with the mean of 41.29 (SD+22.01).
Results revealed that 195 patients had single genotype only while mixed genotype pattern was observed in 11 cases.
The most identified HCV type was genotype 3a (46.60%). The next frequent ones were 1a (25.73%), 1b(17.47%) cases and 2(1.95%) respectively. Patients with mixed genotype were as follows, 1b and 2 cases, 1a and 3a six cases. The rest three cases were 1a and 1b; 1a and 2; 2 and 3a one case for each). HCV genotype was not identified in 4 cases (Table 2).
Table 2:
Frequency of Various HCV genotypes in the study period
| Genotype | 2004 n (%) | 2005 n (%) | 2006 n (%) | 2007 n (%) | Total (%) |
|---|---|---|---|---|---|
| 1a | 5 (27.77) | 8 (22.85) | 17 (25.37) | 23 (26.7) | 53 (25.73) |
| 1b | 4 (12.22) | 5 (14.28) | 11 (16.41) | 16 (18.66) | 36 (17.47) |
| 2 | 1 (5.05) | - | 2 (2.98) | 1 (1.16) | 4 (1.95) |
| 3a | 7 (38.88) | 16 (45.71) | 33 (49.25) | 40 (46.51) | 96 (46.60) |
| Mixed genotype | - | 2 (5.715) | 4 (2.98) | 5 (5.81) | 11 (5.34) |
| Undetectable genotypes | 1 (5.55) | 4 (11.42) | 0 | 1 (1.16) | 6 (2.91) |
| Total | 18 | 35 | 67 | 86 | 206 |
Prevalence of HCV genotypes in different age-groups
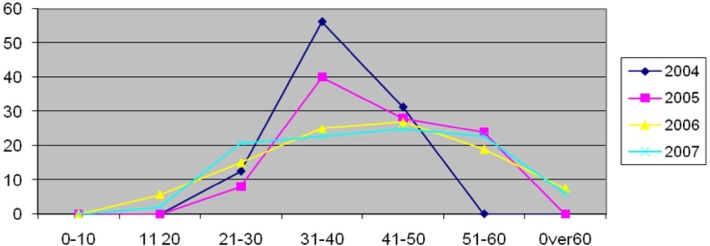
Prevalence of identified genotypes was determined for different age-groups of studied patients. The highest rate was observed in 31 to 50 yr old patients in this period (Table 3 & Fig. 1).
Table 3:
Frequency of HCV positive in different age groups from 2004 till end of 2007(%)
| Age | 2004 | 2005 | 2006 | 2007 |
|---|---|---|---|---|
| 0–10 | 0 | 0 | 0 | 0 |
| 11–20 | 0 | 0 | 5.7 | 2.07 |
| 21–30 | 12.5 | 8 | 15 | 20.84 |
| 31–40 | 56.25 | 40 | 25 | 22.92 |
| 41–50 | 31.25 | 28 | 26.9 | 25 |
| 51–60 | 0 | 24 | 19 | 22.92 |
| 0ver60 | 0 | 0 | 7.6 | 6.25 |
Fig. 1:
Frequency of genotypes 1a, 1b, and 3a in different age groups of HCV infected patients
Changes of HCV genotypes prevalence during studied years
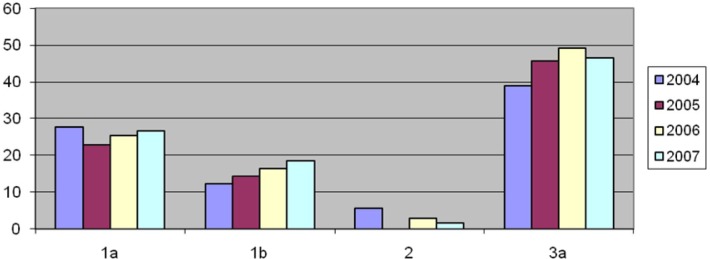
Analysis of the results revealed that the frequency of the HCV genotypes 1b and 3a were gradually increased in each year (1b from 12.22% to 18.66%, 3a from 38.88% to 46.51%) (Fig. 2).
Fig. 2:
Frequency of HCV genotypes in studied years
Viral load of HCV genotypes
Viral load of all positive patients was >10000 IU/ml. Genotype 1 (including 1a and 1b) showed a high viral load of over than 800,000 IU/ml (69.22%). Patients with genotype 2 showed viral load lower than 800,000 IU/ml, while type 3a had a 30.76% frequency of high viral load (Table 4).
Table 4:
Frequency of HCV positive with viral load higher than 800,000 IU/ml in different identified genotypes
| Genotype | % |
|---|---|
| 1a | 38.46 |
| 1b | 30.76 |
| 3a | 30.76 |
| 2 | 0 |
Discussion
Today, for better treatment and clinical out-come, HCV genotyping became a necessary job in clinical management. We have analyzed 206 HCV positive Iranian patients for genotyping. Results indicate that genotype 3a is the dominant type with prevalence of 46.6%, while the dominant one was 1a with 47% in report of Samimi-Rad (11). It could be because of different source of studied patients in these two researches.
The frequency of other genotypes were 53 (25.73%), 36 (17.47) and 4 (1.95) cases for genotypes 1a, 1b, and 2 respectively. However, obtained results shows that 89.8% of HCV genotypes in Iran were1 and 3a that is comparable with Samimi-Rad of report (91%) (11). According to the report of Samimi-Rad, the rate of 1a, 1b and 3a were 47%, 8% and 36%, respectively.
Mixed genotypes were observed in 11 cases (5.32%) that were mentioned in the previous reports from Iran (12). The most frequent mixed genotypes were 1a and 3a. These two types were each the most dominant types, meaning possibility of the new infection.
HCV genotype was not indicated in the rest 6 cases (2.92%), since detection of other genotypes was not considered, in our study. The low frequency rate of undetected types showed minor role of other genotypes that had not been included in this study.
Analysis of the results revealed that the rate of HCV genotypes seems to be changed slightly in the study period but still needs more study in higher population. The rate of genotypes 1b and 3a was increased from 12.22% and 38.88% in first year to 18.66 and 46.51% in fourth year of the study period (Table 1). This finding is important in genotype 1 because of its role in severity of the disease. Patients with genotype 1 and 4 have about 50% chance of successful treatment. On the other hand the chance of a successful treatment is 80–90% for genotypes 2 or 3. Patients infected with these two types are prescribed with a lower dose of ribavirin than patients infected with genotype 1 or 4.
It is reported that patients with a lower viral load have better response to anti-viral therapy compared to those with higher viral load and has a better chance of HCV eradication (13). Moreover, a direct association has been observed between serum viral load of HCV and transmission rate of the virus. It has been reported that 800,000 IU/ml of HCV is a cut off for High and Low viral load that is equivalent to 2 million copies/ml (16). Analysis of our data revealed that type 1a was the type with highest rate among patients having above the cut off viral load that is consistent with other studies.
It is reported that women may respond better to current HCV medications than men (17). The reason is not clearly understood, but some experts believe that may be due to estrogen effects (17). In this research, 23 out of 155 patients requested for genotyping were women (14.83%) that are about eight times less than men.
In conclusion, the increased prevalence of 1b and 3a with observing mixed genotypes underline requirement for more attention in HCV medication, although it needs more study in higher populations.
Ethical Consideration
All Ethical issues (such as informed consent, conflict of interest, plagiarism, misconduct, co-authorship, double submission, etc) have been considered carefully.
Acknowledgments
This research was funded by Noor Pathobilogy Laboratory and we appreciate all colleagues especially those of this laboratory for providing the facilities to access the patients’ data and re-examinations in some necessary cases.
References
- 1.Zali MR, Mayumi M, Raoufi M, Nowroozi A. Hepatitis C virus genotypes in the Islamic Republic of Iran: a preliminary study. East Mediterr Health J. 2000;6(2–3):372–77. [PubMed] [Google Scholar]
- 2.Zein NN. Clinical significance if hepatitis C virus genotypes. Clinical Microbiology Review. 2000;12:223–35. doi: 10.1128/cmr.13.2.223-235.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agha S, Tanaka Y, Saudy N, Kurbanov F, Abo-Zeid M, EL-Malky M, Khalaf M, Ohta N, Yoshizawa H, Mizokamil M. Reliability of hepatitis C virus core antigen assay for detection of viremia in HCV genotypes 1, 2, 3, and 4 infected blood donors: a collaborative study between Japan, Egypt, and Uzbekistan. J Med Virol. 2004;73(2):216–22. doi: 10.1002/jmv.20078. [DOI] [PubMed] [Google Scholar]
- 4.Nousbaum JB. Genomic subtypes of hepatitis C virus: epidemiology, diagnosis and clinical consequences. Bull Soc Pathol Exot. 1998;91(1):29–33. [Article in French] [PubMed] [Google Scholar]
- 5.Matera G, Lamberti A, Quirino A, Focà D, Giancotti A, Barreca GS, Guadagnino V, Liberto MC. Changes in the prevalence of hepatitis C virus (HCV) genotype 4 in Calabria, Southern Italy. Diagn Microbiol Infect Dis. 2002;42(3):169–73. doi: 10.1016/s0732-8893(01)00350-9. [DOI] [PubMed] [Google Scholar]
- 6.Abdo AA, Lee SS. Management of hepatitis C virus genotype 4. J Gastroenterol Hepatol. 2004;19(11):1233–39. doi: 10.1111/j.1440-1746.2003.03325.x. [DOI] [PubMed] [Google Scholar]
- 7.Campiotto S, Pinho JR, Carrilho FJ, Da Silva C, Souto FJD, Spinelli V, et al. Geographic distribution of hepatitis C virus genotypes in Brazil. Braz J Med Biol Res. 2005;38(1):41–9. doi: 10.1590/s0100-879x2005000100007. [DOI] [PubMed] [Google Scholar]
- 8.Payan C, Roudot-Thoraval F, Marcellin P, et al. Changing of hepatitis C virus genotype patterns in France at the beginning of the third millenium: The GEMHEP Geno CII Study. J Viral Hepat. 2007;12(4):405–13. doi: 10.1111/j.1365-2893.2005.00605.x. [DOI] [PubMed] [Google Scholar]
- 9.Echevarria JM, Leon P, Pozo F, Avellón A. Follow-up of the prevalence of hepatitis C virus genotypes in Spain during a nine-year period (1996–2004) Enferm Infecc Microbiol Clin. 2006;24(1):20–5. doi: 10.1157/13083370. [DOI] [PubMed] [Google Scholar]
- 10.Harris KA, Gilham C, Mortimer PP, Teo CG. The most prevalent hepatitis C virus genotypes in England and Wales are 3a and 1a. J Med Virol. 1999;58(2):127–31. doi: 10.1002/(sici)1096-9071(199906)58:2<127::aid-jmv5>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 11.Samimi-Rad K, Nategh R, Malekzadeh R, Norder H, Magnius L. Molecular epidemiology of hepatitis C virus in Iran as reflected by phylogenetic analysis of the NS5B region. J Med Virol. 2004;74(2):246–52. doi: 10.1002/jmv.20170. [DOI] [PubMed] [Google Scholar]
- 12.Sammi-Rad K, Shahbaz B. Hepatitis C virus genotypes among patients with thalassemia and inherited bleeding disorders in Markazi province, Iran. Haemophilia. 2007;13(2):156–63. doi: 10.1111/j.1365-2516.2006.01415.x. [DOI] [PubMed] [Google Scholar]
- 13.Franciscus A. HCV Treatment: predictors of Treatment Response. Hepatitis C Support Project. 2006. www.hcvadvocate.org.
- 14.Hajia M, Shahrokhi N, Amirzargar AA, Farzanehkhah M, Biglari S, Ghorishy M, Sarafnejad A. Prevalence of Hepatitis C virus among Out-Patients of a Private Laboratory in Tehran. Iranian J Publ Health. 2007;36(1):79–84. [Google Scholar]
- 15.Gushchin AE, Hoskova OM, Shipulin GA. Development of “Amplisens-HCV-genotype” reagent set for identification of hepatitis C virus genotypes 1a, 1b, 2a and 3a. Vopr Virusol. 2003;48(3):45–8. [Article in Russian] [PubMed] [Google Scholar]
- 16.Martinot-Peignoux M, Boyer N, Le Breton V, Le Guludec G, Castelnau C, Akremi R, Marcellin P. A new step toward standardization of serum hepatitis C virus-RNA quantification in patients with chronic hepatitis C. Hepatology. 2000;31(3):788–89. doi: 10.1002/hep.510310324. [DOI] [PubMed] [Google Scholar]
- 17.Di Martino V, Lebray P, Myers RP, Pannier E, Paradis V, Charlotte F, Moussalli J, Thabut D, Buffet C, Poynard T. Progression of liver fibrosis in women infected with hepatitis C: long-term benefit of estrogen exposure. Hepatology. 2004;40(6):1426–33. doi: 10.1002/hep.20463. [DOI] [PubMed] [Google Scholar]