Abstract
Background:
Hearing loss (HL) is the most frequent sensory birth defect in humans. Autosomal recessive non-syndromic HL (ARNSHL) is the most common type of hereditary HL. It is extremely heterogeneous and over 70 loci (known as DFNB) have been identified. This study was launched to determine the relative contribution of more frequent loci in a cohort of ARNSHL families.
Methods:
Thirty-seven Iranian families including 36 ARNSHL families and 1 family with Pendred syndrome each with ≥ 4 affected individuals, from seven provinces of Iran, were ascertained. DFNB1 contribution was initially studied by DNA sequencing of GJB2 and linkage analysis using the relative STR markers. The excluded families were then subjected to homozygosity mapping for fifteen ARNSHL loci.
Results:
Sixteen families were found to be linked to seven different known loci, including DFNB1 (6 families), DFNB4 (3 families +1 family with Pendred syndrome), DFNB63 (2 families), DFNB2 (1 family), DFNB7/11 (1 family), DFNB9 (1 family) and DFNB21 (1 family). DNA sequencing of the corresponding genes is in progress to identify the pathogenic mutations.
Conclusion:
The genetic causes were clarified in 43.2% of the studied families, giving an overview of the causes of ARNSHL in Iran. DFNB4 is ranked second after DFNB1 in the studied cohort. More genetic and epigenetic investigations will have to be done to reveal the causes in the remaining families.
Keywords: ARNSHL, Genetic linkage analysis, DFNB loci, Iran
Introduction
Hearing loss (HL) is the most common sensory deficit in humans with an incidence of about one in 650 newborns. The prevalence continues to increase during childhood and reaches a rate of 2.7 per 1000 children before the age of five years and 3.5 per 1000 during adolescence. HL is a major public health concern in developing countries. Two thirds of the people who have HL worldwide live in developing countries (1). A severe HL in early childhood prevents from proper speech acquisition and subsequent literacy and its later onset would negatively affect the quality of life (2). As sanitary indexes are improved, the figure would change in favor of the role of genetics (3, 4). HL can be classified by different criteria including severity (mild: 20 to 39 dB, moderate: 40 to 69 dB, severe: 70 to 89 dB, or profound: ≥90 dB), age of onset (pre-lingual or post-lingual), origin (conductive, sensorineural or mixed) and presence or absence of associated features (syndromic or non-syndromic) (5). It is estimated that at least 50% of pre-lingual HL has a genetic basis. The etiology for another 25% remains unclear, suggesting an additional role for genetics. Approximately 70% of genetic HL cases are non-syndromic (NSHL), where no other anomaly exists, whereas the remaining 30% are syndromic. Up to now, over 400 syndromic forms have been described; Usher syndrome and Pendred syndrome are the most common examples (6). Whilst in NSHL, autosomal-dominant (ADNSHL) comprises ∼20% and only a minority of the causes include X-linked (∼1%) and mitochondrial (<1%) forms, Autosomal-recessive forms (ARNSHL) encompasses ∼ 80% of cases (3, 4). Notably, the frequency of ARNSHL becomes even higher in countries with high rate of consanguineous marriage (3). ARNSHL forms are usually pre-lingual and more severe in degree and are almost exclusively sensorineural (4, 7). The loci for ARNSHL, ADNSHL and X-linked HL are represented by DFNB, DFNA and DFN, respectively (4). Up to 1% of human genes are estimated to be involved in hearing process. Over 130 loci have been identified for NSHL so far. Thus, HL is one of the most heterogeneous human genetic trait (6, 8), of which more than 70 DFNB loci have been identified for ARNSHL (9).
Iran, with the average consanguineous marriage rate of 38.6% (10) and with a heterogeneous population due to different ethnicities, can provide a good opportunity for genetic research on ARNSHL. However, further research has been highly recommended to obtain an insight into the contributing loci, some of which might be new (11). Most studies in Iran on ARNSHL have only addressed certain loci with a special focus on DFNB1 (GJB2), the most common cause of ARNSHL. Studying other loci in the Iranian ARNSHL patients would provide insight into the role of other loci in pathogenesis of ARNSHL in this population. The results of such studies could be applied to a more efficient genetic screening of the disease and the concomitant DNA diagnostics and genetic counseling. The present study was launched to determine the contribution of DFNB1 (GJB2, GJB6) and 14 other DFNB loci to ARNSHL in the study cohort.
Materials and Methods
Subjects and clinical evaluation
This study was approved by the Institutional Review Boards of Shahrekord University of Medical Sciences and the University of Antwerp in Belgium. In this descriptive study, a number of families with Iranian origin were collected from 7 provinces of Iran, including Charmahal va Bakhtiari, Fars, Guilan, Tehran, Khuzestan, Azerbaijan Sharghi and Kurdestan.
HL informational questionnaires were filled out for all families and the pedigrees were drawn based on the filled-out questionnaires and interview with the members of the families. Families with the possibility of exposure to known environmental risk factors (head trauma, the use of ototoxic drugs, etc.) were excluded from the study. In total, 37 families with at least 4 patients were included in this study. All, but one family with Pendred syndrome, were affected with ARNSHL. Participants signed an informed consent form before their inclusion into this study. Pure tone audiometric test for air and bone conduction at frequencies varying from 250 to 8000 Hz was performed.
DNA extraction
DNA was extracted from EDTA-containing blood samples of participating individuals using the phenol/ chloroform standard procedure (12). DNA qualities were checked on 1.2% agarose gel. A NanoDrop 1000 spectrophotometer (Thermo Scientific Inc., Wilmington, DE, USA) was used to determine DNA concentration and purity.
GJB2 mutation screening
At least, one patient from every pedigree was subjected to DNA sequencing. The following primers were used as described elsewhere (13) F: 5′-CTCCCTGTTCTGTCCTAGCT-3′ R: 5′-CTCATCCCTCTCATGCTGTC-3′. PCR condition was as follow: 2 μl MgCl2 (4 mM), 2.5 μl Taq PCR buffer (10X), 1μl of each of the primers (10 PM), 0.1 μl Taq DNA polymerase (5U/ul), 1 μl dNTP mix (10 mM) and 1μl DNA (about 70 ng). The reaction was adjusted to the volume of 25ul by ddH2O. Standard cycling conditions was performed in a thermocycler (ASTEC PC-818; ASTEC, Fukuka, Japan) as follows: 95° C for 2 min; 35 cycles of 94° C for 30″, 57° C for 45″, 72° C for 45″, and finally 72°C for 7 min. The PCR product of the GJB2 gene was quality controlled on the 1.5% agarose gel. A single PCR product of 809 bp was obtained.
Subsequently, DNA sequencing of the PCR-amplified product was carried out bi-directionally on an ABI 3130 automated sequencer (Applied Biosystems) (Macrogen, South Korea) using the same primers.
SLINK analysis and selection of DFNB loci
Power of the pedigrees for linkage analysis was simulated by calculating SLINK, using FastSLink (version 2.51) option of Easylinkage plus version 5.05 software to predict the potential LOD score in a given family (14). Families with SLINK scores above 2.5 were considered informative enough for linkage analysis by screening several known loci. The threshold SLINK value of 3.3 was considered significant for genome-wide scan (GWS). Based on the literature review of the most frequent loci, both globally and regionally, 15 loci were selected for screening. Screening STR markers were selected based on their physical distance found at NCBI UniSTS and NCBI Map Viewer. STR markers of each locus and their primer sequences are listed in Table 1.
Table 1:
The list of 15 DFNB loci screened in this study. The corresponding genes and details of screening markers are shown. Categories and functions of their encoded proteins are mentioned (52)
| Locus (gene), Physical location(bp), Category& function | Marker | Physical position (bp) | PCR product range | Forward primer (5′→3′) | Reverse primer (5′→3′) |
|---|---|---|---|---|---|
|
DFNB1(GJB2, GJB6) GJB2: 20761602..20767114 GJB6: 20796101..20806534) Ion homeostasis proteins: Connexins |
D13S1236 | 22696180–22696305 | 108–132 | GCACTTGGCCTGGGTAA | AAGGGGCTGGCTCTTCA |
| D13S1275 | - | 180–214 | ATCACTTGAATAAGAAGCCATTTG | CCAGCATGACCTTTACCAG | |
| D13S175 | 20848506–20848618 | 101–113 | TATTGGATACTTGAATCTGCTG | TGCATCACCTCACATAGGTTA | |
|
DFNB2(MYO7A) 76839310..76926286) Hair bundle morphogenesis proteins: Motor proteins |
D11S4179 | 76396260–76396495 | 200–256 | GGATGTAAGAGTAACTGGCTCCG | GAAAATGTTCTGCCTGAGGG |
| D11S4186 | 76968518–76968685 | 154–175 | ATTCTCCCAATCTATCGCTC | GGGCAGTAATGATGATGTG | |
| D11S4079 | 77119447–77119701 | 217–265 | CAGCAAGATCCTGTCTCAA | CTCCTTAAAGTGGGGGAGTT | |
| D11S911 | 77448583–77448769 | 159–203 | CTTCTCATGCTTGACCATTT | CTTCTGAACAATTGCCACAT | |
|
DFNB3(MYO15A) 18012020..18083116) Hair bundle morphogenesis proteins: Motor proteins |
D17S921 | 14260705–14260882 | 169–185 | CTTGGACTCCTACAAATCCTGGCA | GGCCACCATAATCATGTCAGACAAT |
| D17S953 | 16102497–16102619 | 119–131 | ACTATCCGCCCAATACA | AAGGGCTTGCTTTGAC | |
| D17S2196 | 17264482–17264618 | 139–163 | CCAACATCTAGAATTAATCAGAATC | ATATTTCAATATTGTAACCAGTCCC | |
|
DFNB4 (SLC26A4) 107301080..107358254) Ion homeostasis proteins: Ion channels |
D7S2420 | 106889928–106890211 | 240–290 | CCTGTATGGAGGGCAAACTA | AAATAATGACTGAGGCTCAAAACA |
| D7S496 | 107154713–107154849 | 129–141 | AACAACAGTCAACCCACAAT | GCTATAACCTCATAANAAACCAAAA | |
| D7S2459 | 107331501–107331642 | 140–152 | AAGAAGTGCATTGAGACTCC | CCGCCTTAGTAAAACCC | |
| D7S2456 | 107683218–107683460 | 238–252 | CTGGAAATTGACCTGAAACCTT | ACAGGGGTCTCTCACACATATTA | |
| D7S2425 | 108347079–108347322 | 234–246 | CTAGTCCTGAGAAGACATTACCC | CCTGTTTCAGATGTTTTATCCA | |
|
DFNB6 (TMIE) 46742823..46752413) Poorly understood function: integral membrane protein |
D3S3658 | 40903258–40903371 | 104–126 | AAAAGTTAGCAAACACAATCCTATC | CTGGACTAAATCTAAGTTGGTTATG |
| D3S2420 | 48067370–48067462 | 93–108 | ACAAGTGCGAAACTCTGCCT | CAGGAGCCTCTAAGTCAGCA | |
|
DFNB7/11(TMC1 75136717..75451267) poorly understood function:integral membrane protein |
D9S1837 | 75185129–75185367 | 205–251 | CATGATGGTGGTCTCTGG | GGTGGGGCTCAAAGAGTAG |
| D9S1806 | 74201357–74201620 | 216–266 | TTTTAGGTGTTCTCAGTACATGC | GGGAGCAACATTTTGACATT | |
| D9S1124 | 75224065–75224327 | 252–276 | GGTGCCCACCATACACTACT | TCTAATCCTTCCTTCCCTCG | |
| D9S1876 | 75232791–75232938 | 132–152 | GATGTACCCAGAGAAGTCTCG | AGTGGTTACCATTTACCCAAG | |
| D9S301 | 73802720–73802954 | 209–237 | AGTTTTCATAACACAAAAGAGAACA | ACCTAAATGTTCATCAAAAGAGG | |
| D9S1822 | 74930323–74930483 | 157–163 | AAGTTTGGCTTCTGCTGTAAGGGTC | AATTCCCCCAGGCTGAGTG | |
| D9S1799 | 73366891–73367055 | 139–178 | TTGCCAACTATTTTAGCCC | TGCAGTTTCAATCCACATC | |
|
DFNB8/10 (TMPRSS3) 43791999..43816200) Poorly understood function: Serine protease |
D21S1890 | 44848178–44848330 | 143–173 | GGTCTGACCACAGATTTCC | AAAAACACTCTGAACGATTAAGG |
| D21S1260 | 42796042–42796251 | 200–214 | TCCAAGGGGTTCATCC | CCCAAGGCACTGTTCC | |
|
DFNB9 (OTOF) 26680071..26781566) Poorly understood function: Exocytosis at auditory ribon synapse |
D2S365 | 28606342–28606533 | 164–204 | ATGATTTGTGTACCTTATGTATGTT | TCAATGGAGGAATCCTACTT |
| D2S2247 | 27303911–27304064 | 130–160 | TCCATCTTTTGCGTGC | CCGTGCTCTATGCCAG | |
| D2S174 | 26839873–26840075 | 203–221 | AGGCTGAATCCCACCTCC | TTAGAGCACACATGGTCACTCC | |
| D2S2223 | 26559144–26559325 | 182–200 | CACTGCGCCTAGCCTC | GGCGATTTATGAATAATCCTGC | |
|
DFNB12(CDH23) 73156704..73575704) Hair bundle morphogenesis proteins: Adhesion proteins |
D10S1432 | 74659396–74659569 | 157–185 | CAGTGGACACTAAACACAATCC | TAGATTATCTAAATGGTGGATTTCC |
| D10S1146 | 74659314–74659555 | 164–246 | ATTGCACTCCAGCCTGGGT | CACAATCCAATCACATGGATG | |
|
DFNB21(TECTA) 120973375..121061515) Extracellular matrix proteins:Structural component tectorial membrane |
D11S4107 | 121049124–121049321 | 172–212 | TCATTCTACAAGACTAGCATTACC | GCTTGATCATGGTGTATTATCTT |
| D11S925 | 120828264–120828438 | 172–199 | AGAACCAAGGTCGTAAGTCCTG | TTAGACCATTATGGGGGCAA | |
| D11S912 | 128624097–128624205 | 101–123 | TCGTGAGANTACTGCTTTGG | TTTTGTCTAGCCATGATTGC | |
| D11S4151 | 126292160–126292309 | 145–155 | GTCTTCCCACCTTGGATATGGGTA | AATGGGCACCTCCACCCTATTAGT | |
| D11S4089 | 120989673–120989875 | 199–213 | ATTCCTAGTTCCCTCATAAACACTG | TAATCAAAGGCTGTAGTGAATTGG | |
| D11S4110 | 126971672–126971780 | 93–107 | TGAGCCTCCCAGTACCTACC | GTTTGTGGCAGAGCCCTAAG | |
|
DFNB22(OTOA) 21689835..21772050) Extracellular matrix proteins: Anchoring protein between cellular gels and non-sensory cells |
D16S3046 | 20886507–20886610 | 84–108 | CCCAGAATAAACTGCGTG | TTCATGGACCCCCTATTG |
| D16S403 | 23037651–23037790 | 134–152 | GTTTTCTCCCTGGGACATTT | TATTCATTTGTGTGGGCATG | |
|
DFNB28 (TRIOBP) 38092995..38172563) Hair bundle morphogenesis proteins: Proteins of the cytoskeleton |
D22S1156 | 38381771–38381926 | 130–162 | TGAGGTAGTCACACGAGGCA | AATTCACTGGGCTCCGAGG |
| D22S1045 | 37536298–37536453 | 140–158 | GCTAGATTTTCCCCGATGAT | ATGTAAAGTGCTCTCAAGAGTGC | |
|
DFNB35(ESRRB) 76837726..76967208) Transcription factors |
D14S1015 | 92736133–92736396 | 224–264 | GAATGCCATTATTTTGTCCT | TTAGAAAACACCGAGCAGA |
| D14S77 | 73570540–73570772 | 203–251 | GCGTGAGTCACTGTGCC | CAGACAGAAATTAACCAGAGTTGAA | |
| D14S1045 | 0 | 240–246 | AGGGCTGGTGACAATG | GTAAGGNCTTGGGTGG | |
| D14S48 | 88428727–88429001 | 260–265 | CATAAAAGGCTTATTGGTTTG | CAAAACAGAGAACAGAGTAG | |
|
DFNB59 (PJVK) 179316163..179326113) Poorly understood function: Signaling of hair cells and neurons |
D2S2981 | 176173334–176173578 | 234–262 | AAAATATGCAGGTAATGACTTGG | CAAGCAAAACTGACAGGTAGG |
| D2S301 | 217887163–217887396 | 224–240 | CATAGGACTGAAGGGGTGTA | GGAAAATCTCGAATGTACCAAT | |
| D2S2173 | 178445536–178445768 | 201–243 | GGAGACAGAGAGTTTACATTTGAG | GCCACACTTTCCTGAATC | |
| D2S324 | 179656244–179656508 | 264–275 | TTACCCACCGGGACAGT | CAGCAAATGCTTCTAGGTCA | |
| D2S307 | 204654566–204654784 | 205–221 | CATGACCTGAAATAAACATAGACA | AGCTTTTCCTGTAGGCTGTC | |
| D2S2314 | 176862406–176862505 | 96–118 | GGTGTCAGTGAGACCCTGT | ATTTCTAGCGGCCCTAAAAC | |
|
DFNB63 (LRTOMT) 71791382..71821828) leucine-rich methyltransferase |
D11S1314 | 72323192–72323414 | 209–227 | TTGCTACGCACTCCTCTACT | GTGAAGGCAGGAAATGTGAC |
| D11S4132 | 119948397–119948596 | 176–214 | GTGCAAGTTTTGGCTTCGTC | ACTCCAGCCTGGGTGAAA | |
| D11S4162 | 70975752–70976016 | 263–269 | GTTCTCCAGAGAGACAGCAC | GAGAGCAACACTATTGCCC | |
| D11S4140 | 71945684–71945874 | 189–199 | TGCAACAAGGTTCCACACT | CTTATGGGTGAGGGCACAG | |
| D11S4196 | 0 | 200–240 | GAACGTTNTTCATGTAGGCGT | TAATGGTCGCTGTCCC | |
| D11S4184 | 72670843–72671103 | 263–277 | CCCAGCCTTACATATTCC | GCTGATGAGCAGAGGTAG |
Physical positions were determined from the National Center for Biotechnology Information (NCBI) Build 37.1 sequence-based physical map (International Human Genome Sequence Consortium 2009).
Pooling strategy
A subset of the samples was subjected to DNA pooling prior to genotyping by the following protocol: The concentration of DNA in the individual samples to be pooled was first estimated by measuring UV light absorption at 260 nm. Samples were then diluted to 30 ng/ul and re-adjusted. The purity of samples was estimated by the absorption ratios of 260/280 and 260/230 nm. Equal amounts of each sample were pooled. Separate pools for normal and affected samples were prepared for each nuclear family of the pedigree. Af (1, 2,…) and N (1, 2,…..), representing different affected and normal pools, were used to label the pooled samples. In addition, critical samples with poor DNA qualities were analyzed individually.
Genotyping STR markers and Linkage Analysis
Fluorescent PCR of STR markers was conducted according to the standard procedure. Fragment analysis was carried out by capillary electrophoresis with an ABI 3130 automated DNA sequencer (Applied Biosystems, California, USA). Alleles were assigned by Genescan software (Applied Biosystems, Foster City, CA, USA). At least, two screening markers were analyzed for every known locus. Upon encountering an uninformative marker or finding clues of linkage, further markers were genotyped. LOD score calculations were combined with haplotype analysis to confirm or exclude linkage. Two-point and multi-point parametric LOD scores were, respectively, calculated by Superlink version 1.6 (15) and Simwalk version 2.91, both options of Easylinkage plus version 5.05 software (14). While two-point LOD score is much faster than multi-point LOD score and provides an initial evaluation of the linkage status, multi-point LOD score is more comprehensive and is able to provide haplotype data. AR mode of inheritance, complete penetrance, disease-allele frequency of 0.001, existence of no phenocopy, equal allele frequencies for markers and identical meiotic recombination frequencies in both sexes were assumed for LOD score calculations. Haplotypes were reconstructed via Simwalk and were visualized by Haplopainter software version 029.5 (16).
Results
Families and clinical data
The majority of subjects of this study displayed bilateral, severe to profound sensorineural prelingual HL, whereas 3 families showed moderate to severe HL. Twenty-nine out of 37 families (78.4%) had at least one consanguinity loop within the pedigrees. Although the possibility of inbreeding was not completely ruled out for the other families, particularly those living in isolated villages.
Screening of GJB2
DNA sequencing of the coding region of GJB2 could reveal pathogenic variants in 6 out of 37 studied families. The homozygous GJB2 mutations included 35delG (3 families), R127H (2 families) and 167delT (two families). These families were excluded from further analysis. As the coding region sequencing of GJB2 cannot completely rule out the DFNB1 as the disease-causing locus, the remaining families were further analyzed for the linkage to DFNB1 using at least 3 informative markers. However, linkage analysis could not find any other family linked to DFNB1 locus.
SLINK calculation
Fifteen families were of SLINK values ≥3.3, Seven families had SLINK values of ≤2 .the rest of the families present value between 2–3.3. Later on, however, the lower families together with a subset of others were examined by DNA pooling strategy.
DNA pooling, genotyping, and linkage analysis
Screening loci for homozygosity mapping in this study were composed of: DFNB1 (GJB2), DFNB2 (MYO7A), DFNB3 (MYO15A), DFNB4 (SLC26A4), DFNB6 (TMIE), DFNB7/11 (TMC1), DFNB8/10 (TMPRSS3), DFNB9 (OTOF), DFNB12 (CDH23), DFNB21 (TECTA), DFNB22 (OTOA), DFNB28 (TRIOBP), DFNB35(ESRRB), DFNB59 (PJVK) and DFNB63 (LRTOMT).
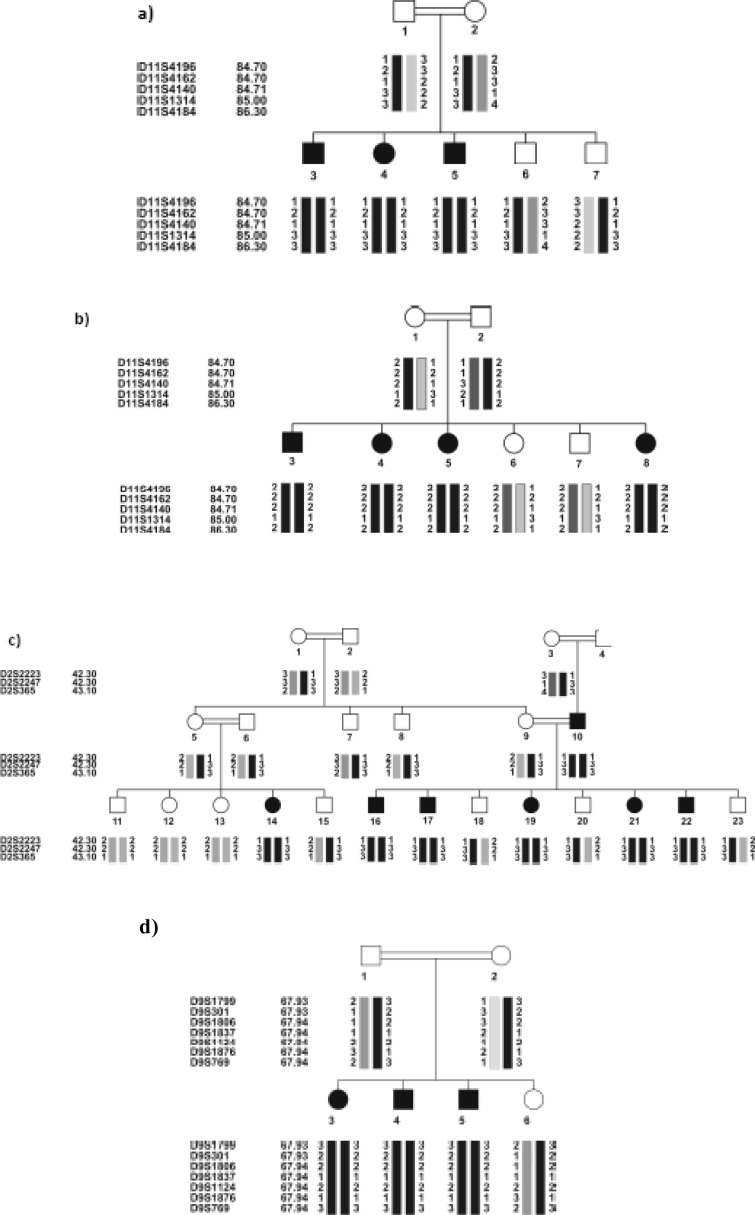
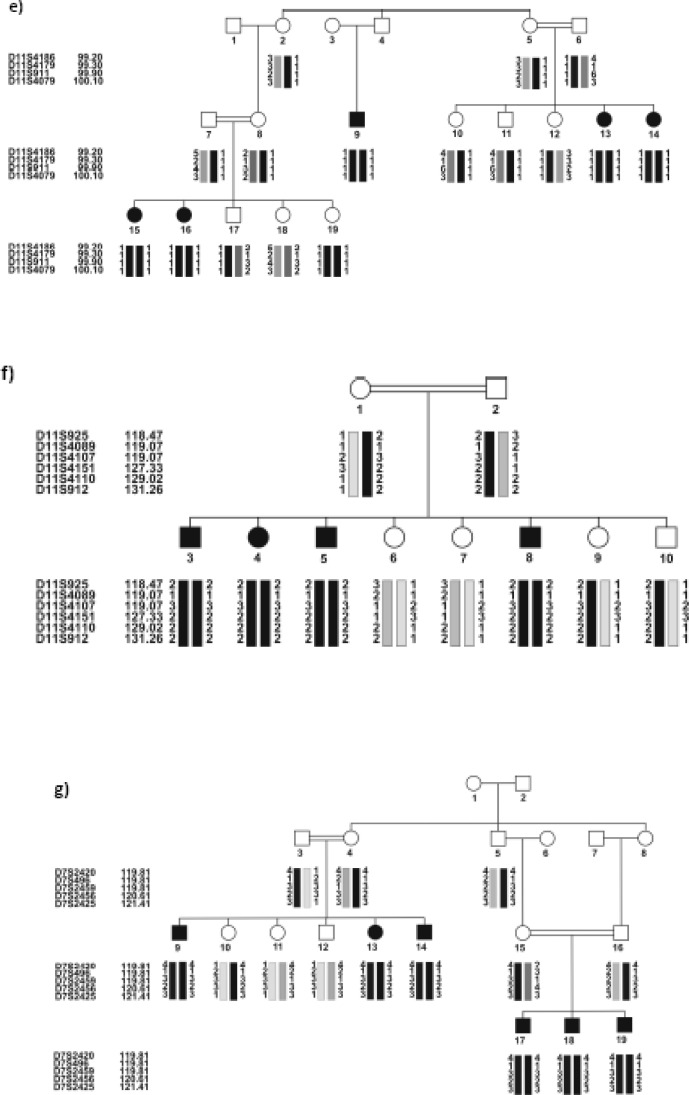
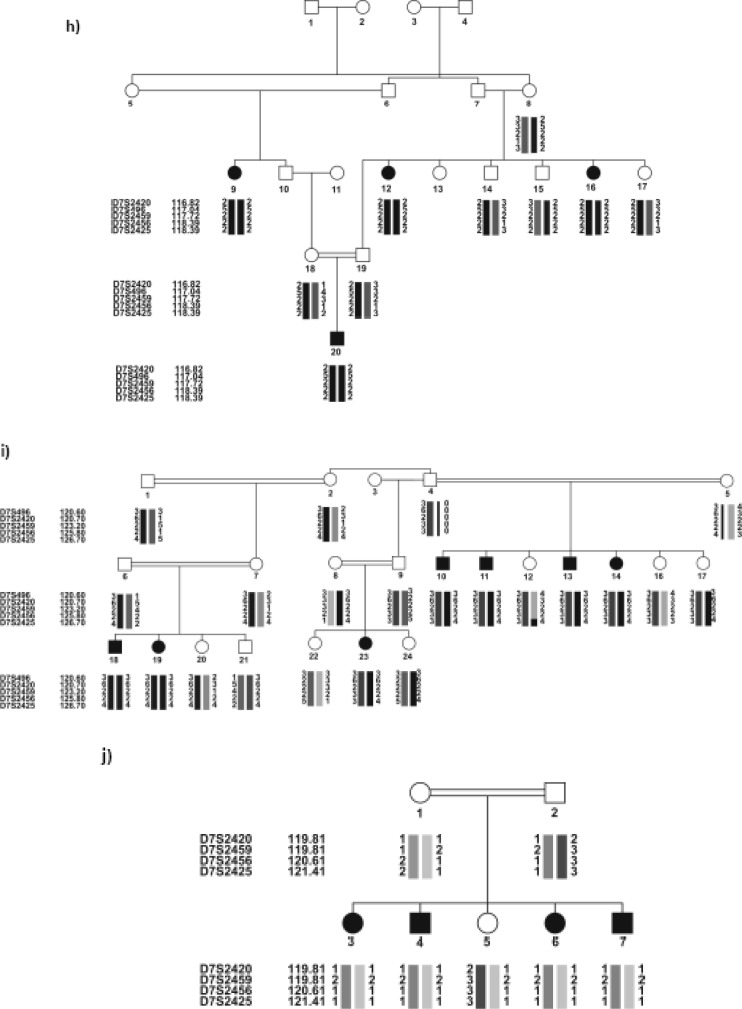
Ten out of the 31 families, all negative for DFNB1 locus, were linked to six different loci, 4 of which had been found by performing the locus screening on the pools of the related families. The results were confirmed by individually genotyping the family members for the same markers, as well as additional markers. Table 2 shows the linked families and the maximum values for SLINK, two-point and multi-point LOD scores. DFNB4 (4 families) was the most frequent locus, after DFNB1, in the studied ARNSHL series. The haplotypes of the linked families are shown in Figure 1 (a–J).
Table 2:
Maximum SLINK and LOD score (two-point and multi-point) values for the linked families. Asterisk shows the family with Pendred syndrome
| Family ID | Linked locus | SLINK value | Two-point LOD score | Multi-point LOD score |
|---|---|---|---|---|
| IR-JOL | DFNB4 | 2.4 | 2.1 | 2.4 |
| IR-SH17* | DFNB4 | 5.2 | 3.3 | 4.1 |
| IR-SH9 | DFNB4 | 6.2 | 3.5 | 5.1 |
| IR-ABY | DFNB4 | 7.4 | 3.4 | 4.6 |
| IR-GHA | DFNB7/11 | 1.8 | 1.6 | 2.0 |
| IR-JAF | DFNB21 | 2.9 | 2.6 | 3.1 |
| IR-SH5 | DFNB9 | 3.9 | 3.2 | 3.3 |
| IR-SH6 | DFNB2 | 4.4 | 3.6 | 4.2 |
| IR-HEM | DFNB63 | 2.8 | 1.9 | 2.4 |
| IR-SH11 | DFNB63 | 2.1 | 2.1 | 2.1 |
Fig. 1:
Pedigree of the 10 Iranian families with ARNSHL, negative for GJB2 mutations, linked to 6 known loci. Black symbols indicate affected individuals. For conciseness, some of the pedigrees are partly shown. haplotypes are shown below each symbol, For individual families, the corresponding genetic map of the markers is shown in parenthesis. (a,b) linked to DFNB63: a) IR-HEM and b) IR-SH11 (LDB map), c) IR-SH5 linked to DFNB9 (LDB map), d) IR-GHA linked to DFNB7/ 11(Marshfiled map), e) IR-SH6 linked to DFNB2 (LDB map), f) IR-JAF linked to DFNB21 (Marshfiled map), (g-J) linked to DFNB4: g) IR-SH9 (Marshfiled map), h) IR-SH17 (decode mapand i) IR-ABY(LDB map) j) IR-JOL (Marshfiled map). In IR-ABY (i) a cross over must have happened in one of the upper generations of individual 1 between markers D7S2459 (which is an intragenic marker) and D7S2456, thus creating two haplotypes segregating with HL in two parts of the pedigree.
Discussion
The locus DFNB1, harboring GJB2 and GJB6, was the first to be excluded in our study cohort composed of 37 families. It plays a major role in the pathology of ARNSHL worldwide. GJB2 mutations have been estimated to account for up to 50% of ARNSHL cases in the North American, Mediterranean, and most of the European populations (17–19). Thus, GJB2 was tested by both gene mutation screening and linkage analysis to strictly rule out the cause in the studied families. Furthermore, GJB6 deletions were investigated (13) with no finding of the mutations. DFNB1 linkage analysis confirmed the results and showed no more positive family with DFNB1 involvement. Totally, 16.2% of the families were homozygous for GJB2 mutations which were set aside from further analysis. This fits well with the 16.6% (20) or 18.3% (11) rate of GJB2 involvement in ARNSHL in previous studies on the Iranian population and would emphasize the diversity of the Iranian population and the fact that the contribution of other loci should be quested. Interestingly, two mutations 35delG (3 families) and R127H (2 families) that had been previously reported as the first and second prevalent mutations in Iran (11), were found to cause HL in 5 of the 6 families with a mutation in GJB2 in this study. Two families (IR-JAF and IR-Sh11), both heterozygous for the GJB2 mutation V37I, were included in homozygosity mapping for two reasons. At first, the pathogenicity of this variant has been doubted by some investigators (21). Secondly, there is a possibility to be carrier for this mutation but the real disease-causing gene would be different. However, both families showed linkage to DFNB21 and DFNB63, respectively.
Three out of 36 (8.3%) ARNSHL families of the study cohort were linked to DFNB4. One additional family, diagnosed as Pendred syndrome due to co-segregation of hypothyroidism (goiter) in later ages was also linked to the region containing SLC26A4, also known as PDS, which encodes pendrin, a chloride and iodide transporter (22). Its molecular pathology has been linked to both ARNSHL and Pendred syndrome. Although the prevalence of Pendred syndrome is not known exactly, it seems to be the most common form of syndromic HL (23). Actually, distinction between Pendred syndrome and NSHL can be challenging since not all affected individuals may co-segregate thyroid disease (24).
About 5% of ARNSHL cases in South Asia have been tied to SLC26A4 mutations (25, 26). In a study, 12 families out of 80 (15%) Iranian families with 2 or more HL patients were linked to DFNB4 locus with clues for 5 families to be syndromic (27). In a recent study, out of 34 families negative for GJB2, 3 families (8.8%) were linked to DFNB4. Thus, DFNB4 contributes significantly to HL in Iran and is ranked second after DFNB1.
In our study, two families showed linkage to DFNB63. The locus which contains LRTOMT, has been reported in families of Turkish, Tunisian, and Pakistani origin (28, 29). In a study, a mutation in LRTOMT was identified in one out of 192 screened Iranian families (30). However, the true frequency of the locus might be more in the series since only catechol-O-methyltransferase (LRTOMT2) had been addressed and the possibility that mutations in isoforms of LRTOMT1 could lead to HL, can not be discounted. The finding of two DFNB63-linked families in our cohort may substantiate the above hypothesis.
One out of 36 families (2.7%) was linked to DFNB2. The related gene MYO7A, encoding myosin VIIA, is a cytoskeletal motor proteins facilitating the movement of cell components along actin filaments (31). DFNB2- linked families have been reported from Iran (32, 33) and one family out of 40 (2.5%) was found to be linked to DFNB2 (32).
In our study, one family (2.7%) was linked to DFNB7/11. The novel gene, called TMC1, is required for postnatal hair cell development. TMC1 mutations seem to be a rather common cause of ARNSHL in India and Pakistan (34). In a research in the North East and East of Turkey, four out of 65 (6.2%) families, negative for GJB2, were shown to be linked to DFNB7/11 (35). The locus could be one a common causes of ARNSHL in the Iranian population.
One family (2.7%) was linked to DFNB9 (OTOF). In a study from Lebanon, 3 out of 30 families (10%) were linked to the locus (36). Other studies have reported DFNB9 from India (37) and UAE (38). OTOF mutations cause ARNSHL, which may be accompanied by auditory neuropathy in about half of cases, with important implications in DNA diagnostics (39).
We found one family linked to DFNB21 in this study. The corresponding gene, TECTA, can cause both dominant and recessive HL TECTA encodes α-tectorin, a major non-collagenous constituent of the tectorial membrane that bridges the stereocilia bundles of the sensory hair cells (40). Interestingly, the DFNB21-linked family in our study, showed the distinctive audio profile (moderate to severe HL, more pronounced in the mid-frequencies) that has been suggested for DFNB21- linked families and is important in DNA diagnostics (41). In a study, linkage to DFNB21 was found in 3 (6.6%) out of 45 Iranian consanguineous families which were negative for GJB2 mutations (42). In another study on 75 Iranian families segregating ARNSHL, 1 family (1.33%) was linked to DFNB21 (41) and finally, in a genetic linkage study of forty ARNSHL families living in Markazi and Qom provinces of Iran with at least 3 affected individuals per family, no instance of linkage to DFNB21 was found (32). No linkage was found to DFNB3 in any of the study families. The corresponding gene, MYO15, codes for myosin XV that is necessary for actin organization in hair cells (43). It has been suggested that at least 5% of the studied Pakistani population are caused by the gene (44). Unlike our study, with no DFNB3-linked family, in a recent study on 40 Iranian ARNSHL families from Qom and Markazi provinces of Iran with 3 or more patients, 2 families were linked to DFNB3 (5.8%) (32). Thus, the prevalence of the DFNB3 HL may vary among Iranian populations. More ARNSHL families have to be studied before reaching any definite conclusion in this regard.
No family was linked to DFNB59. The locus has been mapped to 2q31.2, and the corresponding gene, PJVK encoding pejvakin, has been identified in 4 Iranian families (45). Pejvakin plays a role in action potential propagation or intracellular trafficking. Like OTOF, its defects can sometimes cause auditory neuropathy (45). Thus, in case of auditory neuropathy, both DFNB9 and DFNB59 are strong candidates from the DNA diagnostic standpoint. Screening of 67 Turkish ARNSHL families led to finding of a linked family. It was concluded that it was not playing a significant role in the pathogenesis of HL in the Turkish patients (46). In a study on 30 Iranian ARNSHL families, 2 families (6.7%) were found to be linked. The investigators proposed checking the locus in the Iranian ARNSHL families (47). Based on our study, It is possible that DFNB59 plays no major role in the pathogenesis of ARNSHL.
As a sub-goal of the study, we successfully performed the DNA pooling strategy for a subset of the families. In the DNA pooling, the DNA samples are segregated into pools based on the contrast phenotype. Thus, in an inbred family linked to a given locus, it is expected that affected individuals would be homozygous for a single marker allele closely linked to the given locus and is thus “hunted” (48).While the attraction of pooling lies in reducing time and expense for genotyping individuals, some information, that could have been obtained by individual genotyping, might be lost (49). This can specially occur if more than one cause of hearing loss is segregating in a family. Other pitfalls include differential amplification of alleles and the STR stutter bands, the PCR artifacts associated with STR markers that can sometimes complicate allele calling of individual pools (50). As to tackle with some of its limitations, for the pedigrees showing possible clues of digenic inheritance, based on the structure of the pedigrees or audiometric profiles, a number of screening markers were independently checked on the individuals. In this experiment, we applied a set of adjustments to successfully conduct the DNA pooling and could find 4 linked families which were confirmed by further genotyping in the corresponding family individuals. Our result shows that the strategy could easily be applied to the studies of genetic linkage covering the extraordinary big pedigrees or those with SLINK values lower than 2.5, which are often discarded from additional analyses.
In the present study, we could only clarify the genetic etiology of 16 out of 37 (43.2%) HL families, including 36 ARNSHL families and one with Pendred syndrome, and 15 out of 36 (41.6%) ARNSHL families. This could be explained by the extreme heterogeneity of the disease arising from the complexity of the auditory system, and the lower contribution of DFNB1 (GJB2) in Iran as compared to some other populations. The results of the present study confirm those of previous studies in Iran. The results would also give an overview of the most frequent loci: DFNB1, DFNB4, DFNB2, DFNB21 and DFNB7/11. It might be worthwhile including also DFNB63 in the locus-screening list of future studies on Iranian ARNSHL patients. The design and practice of similar studies on the different populations of Iran will provide a wealth of population-specific knowledge for genetic diagnosis and genetic counseling of the families.
The next phase of the study involves the DNA sequencing of the known DFNB genes we have already mapped in the linked families as to identify their underlying pathogenic mutations in the families. Several families with SLINK≥ 3.3 are currently being subjected to GWS by the llumina SNP array utilizing 6K SNP Linkage Panel IVb (Illumina Inc., San Diego, CA). Thus far, one novel locus (DFNB93) for hearing loss has been identified (51).
Ethical Considerations
Ethical issues including plagiarism, informed consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc. have been completely observed by the authors.
Acknowledgments
This research has been supported by the Flemish FWO (grant G.0138.07) and the Cellular and Molecular Research Center, School of Medicine, Shahrekord University of Medical Sciences, Shahrekord, Iran (grant numbers 557 and 683). We would like to sincerely thank the families for their cooperation in the process of this research. The authors declare that there is no conflict of interests.
References
- 1.Tucci D, Merson MH, Wilson BS. A summary of the literature on global hearing impairment: current status and priorities for action. Otol Neurotol. 31(1):31–41. doi: 10.1097/mao.0b013e3181c0eaec. [DOI] [PubMed] [Google Scholar]
- 2.Brink P, Stones M. Examination of the relationship among hearing impairment, linguistic communication, mood, and social engagement of residents in complex continuing-care facilities. Gerontologist. 2007;47(5):633–41. doi: 10.1093/geront/47.5.633. [DOI] [PubMed] [Google Scholar]
- 3.Smith RJ, Bale JF, Jr, White KR. Sensorineural hearing loss in children. Lancet. 2005;365(9462):879–90. doi: 10.1016/S0140-6736(05)71047-3. [DOI] [PubMed] [Google Scholar]
- 4.ACMG Genetics Evaluation Guidelines for the Etiologic Diagnosis of Congenital Hearing Loss. Genetic Evaluation of Congenital Hearing Loss Expert Panel. ACMG statement. Genet Med. 2002;4(3):162–71. doi: 10.1097/00125817-200205000-00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schrijver I. Hereditary non-syndromic sensorineural hearing loss: transforming silence to sound. J Mol Diagn. 2004;6(4):275–84. doi: 10.1016/S1525-1578(10)60522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hilgert N, Smith RJ, Van Camp G. Forty-six genes causing nonsyndromic hearing impairment: which ones should be analyzed in DNA diagnostics? Mutat Res. 2009;681(2–3):189–96. doi: 10.1016/j.mrrev.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morton NE. Genetic epidemiology of hearing impairment. Ann N Y Acad Sci. 1991;630:16–31. doi: 10.1111/j.1749-6632.1991.tb19572.x. [DOI] [PubMed] [Google Scholar]
- 8.Van Laer L, Cryns K, Smith RJ, Van Camp G. Nonsyndromic hearing loss. Ear Hear. 2003;24(4):275–88. doi: 10.1097/01.AUD.0000079805.04016.03. [DOI] [PubMed] [Google Scholar]
- 9.Van Camp G, Smith R. 2010. Hereditary Hearing Loss Homepage: http://hereditaryhearingloss.org/, updated July 2010.
- 10.Saadat M, Ansari-Lari M, Farhud DD. Consanguineous marriage in Iran. Ann Hum Biol. 2004;31(2):263–69. doi: 10.1080/03014460310001652211. [DOI] [PubMed] [Google Scholar]
- 11.Hashemzadeh Chaleshtori M, Farhud DD, Patton MA. Familial and Sporadic GJB2-Related Deafness in Iran: Review of Gene Mutations. Iranian J Publ Health. 2007;36:1–14. [Google Scholar]
- 12.Grimberg J, Nawoschik S, Belluscio L, McKee R, Turck A, Eisenberg A. A simple and efficient non-organic procedure for the isolation of genomic DNA from blood. Nucleic Acids Res. 1989;17(20):8390. doi: 10.1093/nar/17.20.8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tabatabaiefar MA, Montazer Zohour M, Shariati L, Saffari Chaleshtori J, Ashrafi K, Gholami A, et al. Mutation Analysis of GJB2 and GJB6 Genes and the Genetic Linkage Analysis of Five Common DFNB Loci in the Iranian Families with Autosomal Recessive Non- Syndromic Hearing Loss. J Sci I R Iran. 2010;21(2):105–12. [Google Scholar]
- 14.Lindner TH, Hoffmann K. Easy LINKAGE: a PERL script for easy and automated two-/multi-point linkage analyses. Bioinformatics. 2005;21(3):405–407. doi: 10.1093/bioinformatics/bti009. [DOI] [PubMed] [Google Scholar]
- 15.Fishelson M, Geiger D. Optimizing exact genetic linkage computations. J Comput Biol. 2004;11(2–3):263–75. doi: 10.1089/1066527041410409. [DOI] [PubMed] [Google Scholar]
- 16.Thiele H, Nurnberg P. HaploPainter: a tool for drawing pedigrees with complex haplotypes. Bioinformatics. 2005;21(8):1730–32. doi: 10.1093/bioinformatics/bth488. [DOI] [PubMed] [Google Scholar]
- 17.Marlin S, Garabedian EN, Roger G, Moatti L, Matha N, Lewin P, Petit C, Denoyelle F. Connexin 26 gene mutations in congenitally deaf children: pitfalls for genetic counseling. Arch Otolaryngol Head Neck Surg. 2001;127(8):927–33. doi: 10.1001/archotol.127.8.927. [DOI] [PubMed] [Google Scholar]
- 18.Denoyelle F, Weil D, Maw MA, Wilcox SA, Lench NJ, Allen-Powell DR, Osborn AH, Dahl HH, Middleton A, Houseman MJ, Dode C, Marlin S, Boulila-ElGaied A, Grati M, Ayadi H, BenArab S, Bitoun P, Lina-Granade G, Godet J, Mustapha M, Loiselet J, El-Zir E, Aubois A, Joannard A, Petit C, et al. Prelingual deafness: high prevalence of a 30delG mutation in the connexin 26 gene. Hum Mol Genet. 1997;6(12):2173–77. doi: 10.1093/hmg/6.12.2173. [DOI] [PubMed] [Google Scholar]
- 19.Frei K, Ramsebner R, Lucas T, Hamader G, Szuhai K, Weipoltshammer K, Baumgartner WD, Wachtler FJ, Kirschhofer K. GJB2 mutations in hearing impairment: identification of a broad clinical spectrum for improved genetic counseling. Laryngoscope. 2005;115(3):461–65. doi: 10.1097/01.mlg.0000157855.47143.71. [DOI] [PubMed] [Google Scholar]
- 20.Najmabadi H, Nishimura C, Kahrizi K, Riazalhosseini Y, Malekpour M, Daneshi A, et al. GJB2 mutations: passage through Iran. Am J Med Genet A. 2005;133A(2):132–37. doi: 10.1002/ajmg.a.30576. [DOI] [PubMed] [Google Scholar]
- 21.Wattanasirichaigoon D, Limwongse C, Jariengprasert C, Yenchitsomanus PT, Tocharoenthanaphol C, Thongnoppakhun W, Thawil C, Charoenpipop D, Pho-iam T, Thongpradit S, Duggal P. High prevalence of V37I genetic variant in the connexin-26 (GJB2) gene among non-syndromic hearing-impaired and control Thai individuals. Clin Genet. 2004;66(5):452–60. doi: 10.1111/j.1399-0004.2004.00325.x. [DOI] [PubMed] [Google Scholar]
- 22.Scott DA, Wang R, Kreman TM, Sheffield VC, Karniski LP. The Pendred syndrome gene encodes a chloride-iodide transport protein. Nat Genet. 1999;21(4):440–43. doi: 10.1038/7783. [DOI] [PubMed] [Google Scholar]
- 23.Iwasaki S, Tsukamoto K, Usami S, Misawa K, Mizuta K, Mineta H. Association of SLC26A4 mutations with clinical features and thyroid function in deaf infants with enlarged vestibular aqueduct. J Hum Genet. 2006;51(9):805–10. doi: 10.1007/s10038-006-0027-z. [DOI] [PubMed] [Google Scholar]
- 24.Reardon W, CF OM, Trembath R, Jan H, Phelps PD. Enlarged vestibular aqueduct: a radiological marker of pendred syndrome, and mutation of the PDS gene. QJM. 2000;93(2):99–104. doi: 10.1093/qjmed/93.2.99. [DOI] [PubMed] [Google Scholar]
- 25.Dahl HH, Wake M, Sarant J, Poulakis Z, Siemering K, Blamey P. Language and speech perception outcomes in hearing-impaired children with and without connexin 26 mutations. Audiol Neurootol. 2003;8(5):263–68. doi: 10.1159/000071998. [DOI] [PubMed] [Google Scholar]
- 26.Boublik J, Park H, Radisic M, Tognana E, Chen F, Pei M, Vunjak-Novakovic G, Freed LE. Mechanical properties and remodeling of hybrid cardiac constructs made from heart cells, fibrin, and biodegradable, elastomeric knitted fabric. Tissue Eng. 2005;11(7–8):1122–32. doi: 10.1089/ten.2005.11.1122. [DOI] [PubMed] [Google Scholar]
- 27.Kahrizi K, Mohseni M, Nishimura C, Bazazzadegan N, Fischer SM, Dehghani A, Sayfati M, Taghdiri M, Jamali P, Smith RJ, Azizi F, Najmabadi H. Identification of SLC26A4 gene mutations in Iranian families with hereditary hearing impairment. Eur J Pediatr. 2009;168(6):651–53. doi: 10.1007/s00431-008-0809-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tlili A, Masmoudi S, Dhouib H, Bouaziz S, Rebeh IB, Chouchen J, Turki K, Benzina Z, Charfedine I, Drira M, Ayadi H. Localization of a novel autosomal recessive non-syndromic hearing impairment locus DFNB63 to chromosome 11q 13.3–q13.4. Ann Hum Genet. 2007;71(Pt 2):271–75. doi: 10.1111/j.1469-1809.2006.00337.x. [DOI] [PubMed] [Google Scholar]
- 29.Khan SY, Riazuddin S, Tariq M, Anwar S, Shabbir MI, Riazuddin SA, Khan SN, Husnain T, Ahmed ZM, Friedman TB, Riazuddin S. Autosomal recessive nonsyndromic deafness locus DFNB63 at chromosome 11q13.2-q13.3. Hum Genet. 2007;120(6):789–93. doi: 10.1007/s00439-006-0275-1. [DOI] [PubMed] [Google Scholar]
- 30.Du X, Schwander M, Moresco EM, Viviani P, Haller C, Hildebrand MS, Pak K, Tarantino L, Roberts A, Richardson H, Koob G, Najmabadi H, Ryan AF, Smith RJ, Muller U, Beutler B. A catechol-O-methyltransferase that is essential for auditory function in mice and humans. Proc Natl Acad Sci USA. 2008;105(38):14609–614. doi: 10.1073/pnas.0807219105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tuxworth RI, Titus MA. Unconventional myosins: anchors in the membrane traffic relay. Traffic. 2000;1(1):11–18. doi: 10.1034/j.1600-0854.2000.010103.x. [DOI] [PubMed] [Google Scholar]
- 32.Sadeghi A, Sanati M, Alasti F, Hashemzadeh Chaleshtori M, Mahmoudian S, Ataei M. Contribution of GJB2 mutations and four common DFNB loci in autosomal recessive non-syndromic hearing impairment in Markazi and Qom provinces of Iran. Iran J Biotechnol. 2009;7(2):108–211. [Google Scholar]
- 33.Hildebrand MS, Thorne NP, Bromhead CJ, Kahrizi K, Webster JA, Fattahi Z, Bataejad M, Kimberling WJ, Stephan D, Najmabadi H, Bahlo M, Smith RJ. Variable hearing impairment in a DFNB2 family with a novel MYO7A missense mutation. Clin Genet. 2010;77(6):563–71. doi: 10.1111/j.1399-0004.2009.01344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurima K, Yang Y, Sorber K, Griffith AJ. Characterization of the transmembrane channel-like (TMC) gene family: functional clues from hearing loss and epidermodysplasia verruciformis. Genomics. 2003;82(3):300–308. doi: 10.1016/s0888-7543(03)00154-x. [DOI] [PubMed] [Google Scholar]
- 35.Kalay E, Karaguzel A, Caylan R, Heister A, Cremers FP, Cremers CW, Brunner HG, de Brouwer AP, Kremer H. Four novel TMC1 (DFNB7/DFNB11) mutations in Turkish patients with congenital autosomal recessive nonsyndromic hearing loss. Hum Mutat. 2005;26(6):591. doi: 10.1002/humu.9384. [DOI] [PubMed] [Google Scholar]
- 36.Yasunaga S, Grati M, Cohen-Salmon M, El-Amraoui A, Mustapha M, Salem N, El-Zir E, Loiselet J, Petit C. A mutation in OTOF, encoding otoferlin, a FER-1-like protein, causes DFNB9, a nonsyndromic form of deafness. Nat Genet. 1999;21(4):363–69. doi: 10.1038/7693. [DOI] [PubMed] [Google Scholar]
- 37.Yasunaga S, Grati M, Chardenoux S, Smith TN, Friedman TB, Lalwani AK, Wilcox ER, Petit C. OTOF encodes multiple long and short isoforms: genetic evidence that the long ones underlie recessive deafness DFNB9. Am J Hum Genet. 2000;67(3):591–600. doi: 10.1086/303049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Houseman MJ, Jackson AP, Al-Gazali LI, Badin RA, Roberts E, Mueller RF. A novel mutation in a family with non-syndromic sensorineural hearing loss that disrupts the newly characterised OTOF long isoforms. J Med Genet. 2001;38(8):E25. doi: 10.1136/jmg.38.8.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodriguez-Ballesteros M, Reynoso R, Olarte M, Villamar M, Morera C, Santarelli R, Arslan E, Meda C, Curet C, Volter C, Sainz-Quevedo M, Castorina P, Ambrosetti U, Berrettini S, Frei K, Tedin S, Smith J, Cruz Tapia M, Cavalle L, Gelvez N, Primignani P, Gomez-Rosas E, Martin M, Moreno-Pelayo MA, Tamayo M, Moreno-Barral J, Moreno F, del Castillo I. A multicenter study on the prevalence and spectrum of mutations in the otoferlin gene (OTOF) in subjects with nonsyndromic hearing impairment and auditory neuropathy. Hum Mutat. 2008;29(6):823–31. doi: 10.1002/humu.20708. [DOI] [PubMed] [Google Scholar]
- 40.Legan PK, Lukashkina VA, Goodyear RJ, Kossi M, Russell IJ, Richardson GP. A targeted deletion in alpha-tectorin reveals that the tectorial membrane is required for the gain and timing of cochlear feedback. Neuron. 2000;28(1):273–285. doi: 10.1016/s0896-6273(00)00102-1. [DOI] [PubMed] [Google Scholar]
- 41.Alasti F, Sanati MH, Behrouzifard AH, Sadeghi A, de Brouwer AP, Kremer H, Smith RJ, Van Camp G. A novel TECTA mutation confirms the recognizable phenotype among autosomal recessive hearing impairment families. Int J Pediatr Otorhinolaryngol. 2008;72(2):249–55. doi: 10.1016/j.ijporl.2007.09.023. [DOI] [PubMed] [Google Scholar]
- 42.Meyer NC, Alasti F, Nishimura CJ, Imanirad P, Kahrizi K, Riazalhosseini Y, Malekpour M, Kochakian N, Jamali P, Van Camp G, Smith RJ, Najmabadi H. Identification of three novel TECTA mutations in Iranian families with autosomal recessive nonsyndromic hearing impairment at the DFNB21 locus. Am J Med Genet A. 2007;143A(14):1623–29. doi: 10.1002/ajmg.a.31718. [DOI] [PubMed] [Google Scholar]
- 43.Anderson DW, Probst FJ, Belyantseva IA, Fridell RA, Beyer L, Martin DM, Wu D, Kachar B, Friedman TB, Raphael Y, Camper SA. The motor and tail regions of myosin XV are critical for normal structure and function of auditory and vestibular hair cells. Hum Mol Genet. 2000;9(12):1729–38. doi: 10.1093/hmg/9.12.1729. [DOI] [PubMed] [Google Scholar]
- 44.Park HJ, Shaukat S, Liu XZ, Hahn SH, Naz S, Ghosh M, Kim HN, Moon SK, Abe S, Tukamoto K, Riazuddin S, Kabra M, Erdenetungalag R, Radnaabazar J, Khan S, Pandya A, Usami SI, Nance WE, Wilcox ER, Riazuddin S, Griffith AJ. Origins and frequencies of SLC26A4 (PDS) mutations in east and south Asians: global implications for the epidemiology of deafness. J Med Genet. 2003;40(4):242–48. doi: 10.1136/jmg.40.4.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Delmaghani S, del Castillo FJ, Michel V, Leibovici M, Aghaie A, Ron U, Van Laer L, Ben-Tal N, Van Camp G, Weil D, Langa F, Lathrop M, Avan P, Petit C. Mutations in the gene encoding pejvakin, a newly identified protein of the afferent auditory pathway, cause DFNB59 auditory neuropathy. Nat Genet. 2006;38(7):770–78. doi: 10.1038/ng1829. [DOI] [PubMed] [Google Scholar]
- 46.Collin RW, Kalay E, Oostrik J, Caylan R, Wollnik B, Arslan S, den Hollander AI, Birinci Y, Lichtner P, Strom TM, Toraman B, Hoefsloot LH, Cremers CW, Brunner HG, Cremers FP, Karaguzel A, Kremer H. Involvement of DFNB59 mutations in autosomal recessive nonsyndromic hearing impairment. Hum Mutat. 2007;28(7):718–23. doi: 10.1002/humu.20510. [DOI] [PubMed] [Google Scholar]
- 47.Hashemzadeh Chaleshtori M, Simpson MA, Farrokhi E, Dolati M, Hoghooghi Rad L, Amani Geshnigani S, Crosby AH. Novel mutations in the pejvakin gene are associated with autosomal recessive non-syndromic hearing loss in Iranian families. Clin Genet. 2007;72(3):261–63. doi: 10.1111/j.1399-0004.2007.00852.x. [DOI] [PubMed] [Google Scholar]
- 48.Pareek CS, Pareek RS, Walawski K. Novel linkage mapping approach using DNA pooling in human and animal genetics. I. Detection of complex disease loci. J Appl Genet. 2002;43(2):175–92. [PubMed] [Google Scholar]
- 49.Johnson T. Bayesian method for gene detection and mapping, using a case and control design and DNA pooling. Biostatistics. 2007;8(3):546–65. doi: 10.1093/biostatistics/kxl028. [DOI] [PubMed] [Google Scholar]
- 50.Khatib H, Darvasi A, Plotski Y, Soller M. Determining relative microsatellite allele frequencies in pooled DNA samples. PCR Methods Appl. 1994;4(1):13–18. doi: 10.1101/gr.4.1.13. [DOI] [PubMed] [Google Scholar]
- 51.Tabatabaiefar MA, Alasti F, Shariati L, Farrokhi L, Fransen E, Nooridaloii MR, Hashemzadeh Chaleshtori M, Van Camp G. DFNB93, a novel locus for autosomal recessive moderate-to-severe hearing impairment. Clin Genet. 2010. [DOI] [PubMed]
- 52.Hilgert N, Smith RJ, Van Camp G. Function and expression pattern of nonsyndromic deafness genes. Curr Mol Med. 2009;9(5):546–64. doi: 10.2174/156652409788488775. [DOI] [PMC free article] [PubMed] [Google Scholar]