Abstract
Background:
BRCA1 and BRCA2 genes have been recognized to be responsible for 20–30% of hereditary breast cancers and approximately 50% of familial breast and ovarian cancers. Therefore, the demand for BRCA1 and BRCA2 mutation screening is rapidly increasing as their identification will affect medical management of people at increased risk. Because of high costs involved in analysis of BRCA1 and 2 genes, contribution of different mutation types in BRCA1 and 2 and not knowing who should be tested has hampered wide spread use of molecular testing of high –risk families. There is a need to identify the genes and types of mutations involved in breast or ovarian cancers at different age of onsets and polymorphism and polymorphic variations in our population.
Methods:
Twenty-seven patients with either early onset breast cancer (at age≤ 35 years) or a personal and/or family history of breast or ovarian cancer and 50 control subjects participated in this study. After collecting blood samples and extracting DNA, BRCA1 and BRCA2 genes were fully sequenced.
Results:
Thirteen missense substitutions in BRCA1 and BRCA2 (9 and 4, respectively) were revealed. Two nucleotide substitutions were novel (Gly1140Ser in BRCA1 and Glu1391Gly in BRCA2). The Glu1038Pro and Gly1140Ser were found in large series of breast and ovarian cancer and matched controls.
Conclusion:
Some nucleotide substitutions were seen only in single families and other in several. In other cases, mutations were seen in both BRCA1 and BRCA2 genes. Clinical significance of these mutations was evaluated comparing with normal controls.
Keywords: Breast cancer, BRCA1, BRCA2, Familial cancer
Introduction
Breast cancer is the most common forms of hereditary cancer worldwide. It is an important cause of morbidity and mortality. Based on the statistics released by the American Cancer Society in 2007, it is the second lethal cancer in women after lung cancer (1). Germ-line mutations in BRCA1 and BRCA2 genes cause hereditary predisposition to breast and ovarian cancer (2). These genes products play major role in DNA repair by homologous recombination, maintenance of chromosomal stability, activation of DNA damage checkpoints, transcription-coupled DNA repair, cell cycle regulation, and ubiquitylation (3–7). BRCA1 gene is located in 17q21 and consists of 24 exons coding for 1863 amino acids (8). BRCA2 in on 13q12 and consists 27 exons coding for 3418 amino acids (3). The majority of deleterious mutations found so far are frame-shift or nonsense mutations. They result in premature translation termination and are distributed throughout the genes, but splice-site mutations as well as single amino acid substitutions (missense mutations) in functionally important and well-defined domains have also been reported (7). 20–30% of individual with breast cancer have a family history of the cancer (3). By the age of 40, carrying a deleterious BRCA1 mutation confers a 20% risk of developing breast cancer and or 17% chance for ovarian cancer. The risk increases with age and lifetime risk reaches to 82% by age 80 (4) as well as for ovarian cancer increases to 39% by age 70 and 54% by age 80 (5–9). Breast cancer risk by age 70 for BRCA2 mutation carriers have been found to be 45%, and for ovarian cancer being 11% (9). Molecular studies have shown that the probability of finding mutations in BRCA1 and BRCA2 genes increases as age decreases. For example there is higher chance of finding mutation in either of these two genes at age 40 than 70 (10). Some mutations have been shown to have high penetration and they account for 16–25% of familial cases (11). These data strongly suggest that genomic rearrangements of BRCA1 and BRCA2 genes should be assessed in young probands with a strong family history of breast cancer. Genetic testing is currently used to determine if individuals with a personal and/or family history of breast cancer carry mutations or genomic rearrangements in these two genes. The results of these tests provide useful guidance in deciding how to follow these high-risk individuals in order to prevent the occurrence of breast cancer or even ovarian or permit early cancer detection. Worldwide prevalence of BRCA1 and BRCA2 mutations in breast cancer varies between 1.8–13.1% (12–13) but in Asian countries it varies from 0.8 to 8.6% (14).
In the present study, which is part of a major study in Iranian population, 25 breast cancers or patients or their relatives from four high risk families and 50 control (elderly women without breast or ovarian cancer) were analyzed for BRCA1 and BRCA2 genes mutations and describe the nature of genetic variations in these two genes.
Material and Methods
Families
After genetic counseling of index cases (mostly patients of first-degree relatives) with breast and/or ovarian cancer and obtaining a written informed consent, we screened them for BRCA1 and BRCA2 mutations. High-risk family defined if at least one of below exists:
1) at least 3 relative cases in a family, diagnosed at any age; 2) at least 2 patient first degree relatives (or second degree from the paternal side) with a personal history of breast and/or ovarian cancer; 3) an individual with either bilateral breast cancer, breast and ovarian cancer, or bilateral ovarian cancer, with at least one of the cancers diagnosed age of 50; 4) one affected men (15).
Mutation screening: DNA extraction and PCR amplification
Genomic DNA extraction from peripheral blood samples, were carried out using standard extraction protocols (Using a Promega DNA purification kit (catalogue no. LA1620). All BRCA1 and BRCA2 exons and exon-intron boundaries, except exons 1 and 4 in BRCA1 and exon 1 in BRCA2 were amplified by PCR using exon-specific primers. Primers were selected to expand 100-50 bases from the exon-intron boundaries and were checked using Gene runner software (16). Each 25 ul (microliters) PCR reaction contained ∼30 ng of genomic DNA, 2 ul 10X buffer, 2.5 ul dNTPs, 2.5 ul Mgcl2, 0.2 ul forward primer, 0.2 ul reverse primer, 0.2 ul Taq polymerase and 20 ul of deionized dH2O in 25 ul reaction mix. The PCR mixture was denatured at 93° C for 1 min (except the initial denaturing for 3 min), annealed for 1 min at a temperature varying from 56° C to 62° C depending on the melting temperature of the exon-specific primer pairs, and extended at 72° C for 1 min (except the extension for the final cycle for 2min) for 35 cycles. The presence of the PCR product for each PCR reaction was measured by agarose-gel electrophoresis. The amplified PCR products were purified using KBC pure kit (KBC co., Tehran, Iran) according to the manufacturer’s instructions, and eluted in 70–140 ul ddH2O prior to sequenicng.
Direct sequencing
We sequenced PCR products with an ABI PRISM DyeDeoxy Terminator Cycle Sequencing Kit and an ABI 3100 Genetic Analyzer (Applied Biosystems, Warrington, UK). The reactions were performed using 5–100 ng of purified PCR products, 10.4 pmoles of forward or reverse primers, Ready Reaction Premix and 1× reaction buffer in a total volume of 20 ul. Cycle sequencing reactions were performed in a Primus 96 cycler (ABI, UK) at 96° C for 60 s ,96° C for 10 s, 50° C for 5 s, and 60° C for 4 min for 28 cycles. Prior to capillary electrophoresis, unincorporated dye terminators were removed from the extension product using KBC pure kit and DNA was precipitated by using ethanol precipitation. The purified extension products were denatured at 95° C for 5 min and placed on ice for 3 min. Sequencing was performed on ABI Genetic Analyzer 3130 or 3130xl machines (Applied Biosystems Inc., USA) by KBC Co. (KBC Co., Tehran, Iran).
We processed the data collected from the ABI detection system using the Sequencing Analyzer version 5.2 software. Sequence alignments for each exon read were viewed in the Consed viewer Software (ABI, UK) and sequence variations were annotated and recorded. Negative controls were checked for the absence of analyzable sequence. Electrophorograms were checked for the quality of sequencing reactions and the results were checked against normal sequence in the Genbank (NCBI) using Gene Runner software. Any sequence variation was checked in the BIC and HGMD.
Results
The clinical information of patient and her family from 4 breast cancer families who had came to our center are summarized in Tables 1, 2 and 3. In family A (Pedigree 1), three sisters have had breast cancer, diagnosed at age 40, 38 and 28 yr, and breast cancer in their deceased mother had been diagnosed at age 38 yr. Several different mutations were found in this family (Table 1, 2). These were leu771leu (T2430G), leu871Pro (C27-31T), Glu1038Gly (A3232G), Ser1436Ser (T44-27C), Ser1613Gly (A4956G) in heterozygote form and Gly1140Ser (G3538A) in homozygote form in BRCA1 gene as well as leu1521leu (A4770G), Val2171Val (G6722C) in homozygote form in BRCA2 gene. All three affected sisters and their healthy brother had all the above mutations and SNPs.
Table 1:
The clinical information of patients from four breast cancer families in Iran
| Family | Sample ID | Age of onset | Disease | |
|---|---|---|---|---|
| A | A1 | I | 38 | Breast cancer |
| A2 | II | 28 | Breast cancer | |
| A3 | II | 38 | Breast cancer | |
| A4 | II | 40 | Breast cancer | |
| A5 | II | 37 | Carrier | |
| B | B1 | I | 53 | Breast cancer |
| B2 | II | 25 | Breast cancer | |
| B3 | II | 30 | Breast cancer | |
| B4 | III | 35 | Breast cancer | |
| B5 | III | 57 | Breast cancer | |
| B6 | II | 36 | Carrier | |
| B7 | II | 39 | Carrier | |
| C | C1 | I | 77 | Breast cancer |
| C2 | I | 60 | Breast cancer | |
| C3 | II | 35 | Breast cancer | |
| C4 | II | 42 | Carrier | |
| C5 | II | 39 | Carrier | |
| D | D1 | I | 75 | Rectorajy |
| D2 | I | 60 | Brain tumor | |
| D3 | II | 45 | Breast cancer | |
| D4 | II | 44 | Breast cancer | |
| D5 | II | 43 | Breast cancer | |
| D6 | II | 42 | Nose tumor | |
| D7 | II | 44 | Thyroidtumor | |
| D8 | II | 46 | femourtumor | |
Family: A–D; Generation: I–II.
Table 2:
Missense substitutions in the BRCA1 and BRCA2 genes of Iranian breast cancer families
| gene | exon | Nucleotide change | Amino acid change | Mutation effect |
|---|---|---|---|---|
| BRCA1 | 7 | IVS7+83(-TT) | - | Non-coding IVS |
| BRCA1 | 9 | IVS8 -70(-CATT) | - | Non-coding IVS |
| BRCA1 | 11 | TTG>CTG | Leu771 | Synonymous |
| BRCA1 | 11 | CTG>CCG | Leu871Pro | Missense |
| BRCA1 | 11 | GAA>GGA | Glu1038Gly | Missense |
| BRCA1 | 11 | AGC>AAC | Ser1040Asn | Missense |
| BRCA1 | 11 | GGT>AGT | Gly1140Ser | Missense |
| BRCA1 | 13 | TCT>TCC | Ser1436 | Synonymous |
| BRCA1 | 16 | AGT>GGT | Ser1613Gly | Missense |
| BRCA1 | 20 | GAA>GAG | Glu1735 | Synonymous |
| BRCA1 | 20 | GGA>GAA | Gly1738Glu | Missense |
| BRCA2 | 11 | GAA>GGA | Glu1391Gly | Missense |
| BRCA2 | 11 | CTA>CTG | Leu1521 | Synonymous |
| BRCA2 | 11 | GTG>GTC | VAL2171 | Synonymous |
| BRCA2 | 10 | CAG>CAA | Gln373 | Synonymous |
Table 3:
The BRCA1 and BRCA2 mutations were reported among population
| Gene | Exon | Sequence variant | Previously reported |
|---|---|---|---|
| BRCA1 | 11 | leu771leu | Yes |
| BRCA1 | 11 | leu871Pro | Yes |
| BRCA1 | 11 | Glu1038Gly | Yes |
| BRCA1 | 11 | Gly1140Ser | NO |
| BRCA1 | 13 | Ser1436Ser | Yes |
| BRCA1 | 16 | Ser1613Gly | Yes |
| BRCA2 | 11 | leu 1521leu | Yes |
| BRCA2 | 11 | Val2171Val | Yes |
| BRCA1 | 11 | Ser1040Asn | Yes |
| BRCA1 | 20 | Gly1738Glu | Yes |
| BRCA1 | 20 | Glu1735Glu | NO |
| BRCA2 | 11 | Glu1391Gly | NO |
| BRCA2 | 11 | Leu1521Leu | Yes |
| BRCA1 | 9 | IVS8-70(-CAAT) | NO |
| BRCA1 | 7 | IVS7+83(-TT) | NO |
Pedigree 1:

All three affected sisters and their healthy brother had leu871Pro, Glu1038Gly, Ser1613Gly, Gly1140Ser haplotype in BRCA1.
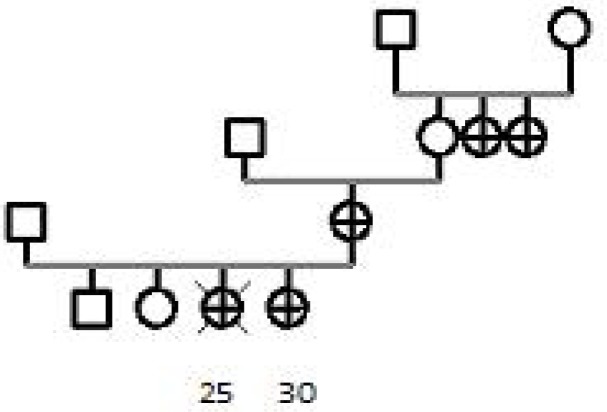
In Family B (Pedigree 2), the index case had a daughter who had developed breast cancer at age of 25 and died 6 mo later. The index case, her daughter, two aunts had breast cancer diagnosed at age 53, 30, 35 and 57 yr, respectively. She was homozygote for the leu871pro (C2731T) missense substitution. She was also heterozygote for Ser1040 Asn (G3238A), Gly1738Glu (Fig. 1) (G5331A) and Gly1140Ser (G3538A) mutations of the BRCA1 gene. Affected members had Gly1738Glu (G53 31A), leu871pro (C2731T) and Gly1140Ser (G35 38A) mutations. Her apparently healthy daughter, aged 38, had only leu871pro (C2731T) and Ser 1040Asn (G3238A) mutation. Three mutations namely Ser1040Asn (G3238A), Glu1035Gly (A32 21G) and Gly1140Ser (G3538A) were seen in her apparently healthy son.
Pedigree 2:
All affected members had the missense substitutions Gly1738Glu, leu871pro and Ser1040Asn in Brca1
Fig. 1:
Mut (G1738E) GGA/GAA.. GlY/Glu in BRCA1
The deceased mother and her sister, in family C, (Pedigree 3), have had breast cancer diagnosed at age77 and 60 respectively. Her niece (deceased sister’s daughter) had breast cancer at 35 yr, but her pathological data were not available. BRCA1 gene scanning showed that her two apparently healthy daughter’s at 39 and 42 had homozygous Glu1038Gly (A32-32G), Gly1140Ser (G3538A) missense substitutions and were heterozygote for Ser1436Ser (T44-27C), Ser1613Gly (A4956G) mutations. Both were heterozygote for IVS 9-70 CATT deletion.
Pedigree 3:

Two apparently healthy daughters at 39 and 42 are carrier of the missense substitutions Glu1038Gly, Gly1140Ser, Ser1613Gly and IVS 9-70 Del CATT in BRCA1.
In family D (Pedigree 4), three sisters had breast cancer diagnosed at 46, 42 and 44. Their mother, aged 70, is not affected with breast cancer. However their uncle had brain tumor and their cousins had gland in their nose, thyroid and femur and their father had rectorajy. The three sisters and their father have missense substitutions as Glu1038Gly (A3232G), Gly1140Ser (G3538A) and Ser1436Ser (T4427C) in heterozygote form in their BRCA1 gene and were homozygote for leu1521leu (A4770G), Val2171Val (G6722C) and Glu1391Gly A4350G to mutations in their BRCA2 gene.
Pedigree 4:

The three sisters and their father had the missense substitutions Glu1038Gly, Gly1140Ser in BRCA1 gene and Glu1391Gly in BRCA2 gene.
Discussion
This finding suggests, as has been documented by others, that early-onset alone is not show a good indicator of the presence of BRCA1/2 mutations, but the combination of early onset breast cancer and other risk factors, such as family history or bilateral breast cancer, are good indicators for being a mutation carrier.
Nine polymorphisms (Leu771, Leu871, Glu1038, Ser1436, Ser1613, Ser1040, Glu1735 in BRCA1 gene and Leu1521, Val2171 in brca2 genes) found to be present in more than one family and in >10% of control samples. Polymorphisms (variants with no obvious functional effect, or that appear reasonably often with a similar frequency incases as in controls) are not considered to have a major influence on cancer risk (such as relative risks less than 2) (17).
In addition, the novel missense substitution Gly1140-Ser (G3538A) (Fig. 2) in BRCA1 gene was found in four of the studied breast cancer families. In addition the novel missense substitutions Glu1391Gly (Fig. 3) in BRCA2 genes was found in one family. In family B, the missense mutation, 5331 G > A (G1738E) (Fig. 1) was identified. The exact effect of G-1738E, on the protein function is unclear. The altered glycine is located on the surface of the coil structure of the BRCT linker region and therefore the mutation may disrupt the interface or affect protein interaction (18, 19). This mutation has been previously described in 4 unrelated Greek patients (20) and in few cases by Abkevick in 2000 and 2004, respectively (21, 22). In vitro studies have shown that G1738E, substitution causes loss of function of the protein (20, 23). According to this information, the family history of the individuals, and absence of the mutation in the proband’s unaffected sister and 50 control populations, therefore we hypothesize that this mutation probability is significant and more data are needed for its pathogencity.
Fig. 2:
Mut (G1140S) GGT/AGT.. GlY>Ser in BRCA1
Fig. 3:
Mut (E1391G) GAA>GGA.. Glu>Gly in BRCA2
In family A, with three women with early onset breast cancer missense substitutions Leu871Pro, G Lu1038Gly, Ser1613Gly, Gly1140Ser (Fig. 2) in BR-CA1 gene have been found, concomitantly. It is interesting to see that in family C, two members are also carriers of the above missense substitutions except Leu871Pro. In family D missense substitutions Glu1038Gly, Gly1140Ser in BRCA1 and Glu1391Gly in BRCA2 in three sisters have been found that two of which are similar to those in family A and family C (Glu1038Gly and Gly-1140Ser). Therefore, these results and those from the controls that may display haplotype at the BRCA1 locus defined by these alleles (i.e. Leu-871Pro, G Lu1038Gly, Ser1613Gly, Gly1140-Ser and Glu1038Gly, Ser1613Gly, Gly1140ser) are alert in carrier; therefore, carriers have to do necessary actions about it. In family C, the IVS 9–70 –Del CATT may not have pathogenic effect since it is located in the intron far from the splice junction site. We also checked any cryptic splice effect for this region by checking this sequence in software and no abnormality was observed. Novel missense substitutions Gly1140Ser in BRCA1 gene was found in members of four families with different haplotypes which indicate no founder effect. It was also seen in >10% of the control population. Therefore, it cannot be claimed with certainly that this variant has any effect on predisposing a person to breast cancer. In family D, missense substitutions Glu1038Gly, Gly1140-Ser in BRCA1 and Glu1391Gly (Fig. 3) in BRCA2 were seen in three affected sisters’ probability is pathogenic although additional data are needed. Heterozygosis and homozygosis of any of the examined nine BRCA1 and BRCA2 missense polymorphisms cannot explain the increased risk of breast and/or ovarian cancer observed in families with hereditary breast and/or ovarian cancer. Empirical predictive models such as Myriad II, Couch, and Manchester Scoring System are intended to estimate the possibility of a BRCA1 or BRCA2 mutation in a woman based on her family history and are used widely in clinical practice (24–26). Genetic counseling of such families safely can disregard findings of these missense polymorphisms (27, 28).
Ethical Considerations
Ethical issues including plagiarism, informed consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc. have been completely observed by the authors.
Acknowledgments
We thank the patients and their family members for cooperation. This work was supported by Genetics Laboratory of Dr.Zeinali, Kawsar Human Genetics Research Center, Tehran, Iran. The authors declare that there is no conflict of interests.
References
- 1.Falagas ME, Zarkadoulia EA, Ioannidou EN, Peppas G, Christodoulou C, Rafailidis PI. The effect of psychosocial factors on breast cancer outcome: a systematic review. Breast Cancer Research. 2007;9:R44. doi: 10.1186/bcr1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nathanson KN, Wooster R, Webber BL. Breast cancer genetics: What we know and what we need. Nature Medicine. 2001;7(3):552–56. doi: 10.1038/87876. [DOI] [PubMed] [Google Scholar]
- 3.Fackenthal JD, Olopade OI. Breast cancer risk associated with BRCA1 and BRCA2 in diverse populations. Nat Rev Cancer. 2007;7(4):937–48. doi: 10.1038/nrc2054. [DOI] [PubMed] [Google Scholar]
- 4.King MC, Marks JH, Mandell JB. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302:643–46. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- 5.Le Page F, Randrianarison V, Marot D, Cabannes J, Perricaudet M, Feunteun J, Sarasin A. BRCA1 and BRCA2 Are Necessary for the Transcription-Coupled Repair of the Oxidative 8-Oxoguanine Lesion in Human Cells. Cancer Res. 2000;60:5548–52. [PubMed] [Google Scholar]
- 6.Starita LM, Parvin JD. The multiple nuclear functions of BRCA1: transcription, ubiquitination and DNA repair. Curr Opin Cell Biol. 2003;15(3):345–50. doi: 10.1016/s0955-0674(03)00042-5. [DOI] [PubMed] [Google Scholar]
- 7.Narod SA, Foulkes WD. BRCA1 and BRCA2: 1994 and beyond. Nat Rev Cancer. 2004;4:665–76. doi: 10.1038/nrc1431. [DOI] [PubMed] [Google Scholar]
- 8.Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72(4):456–876. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Unger MA, Nathanson KL, Calzone K, Antin-Ozerkis D, Shih HA, Martin AM, Lenoir GM, Mazoyer S, Weber BL. Screening for genomic rearrangements in families with breast and ovarian cancer identifies BRCA1 mutations previously missed by conformation-sensitive gel electrophoresis or sequencing. Am J Hum Genet. 2000;67(3):841–50. doi: 10.1086/303076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walsh T, Casadei S, Coats KH, Swisher E, Stray SM, Higgins J, Roach KC, Mandell J, Lee MK, Ciernikova S, Foretova L, Soucek P, King MC. Spectrum of mutations in BRCA1, BRCA2, CHEK2 and TP53 in families at high risk of breast cancer. JAMA. 2006;296(17):1379–88. doi: 10.1001/jama.295.12.1379. [DOI] [PubMed] [Google Scholar]
- 11.Beggs AD, Hodgson SV. Genomics and breast cancer: the different levels of inherited susceptibility. European Journal of Human Genetics. 2009;17:855–56. doi: 10.1038/ejhg.2008.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonadona V, Sinilnikova OM, Chopin S, Antoniou AC, Mignotte H, Mathevet P, Bremond A, Martin A, Bobin JY, Romestaing P, Raudrant D, Rudigoz RC, Leone M, Chauvin F, Easton DF, Lenoir GM, Lasset C. Contribution of BRCA1 and BRCA2 germ-line mutations to the incidence of breast cancer in young women: Results from a prospective population-based study in France. Genes Chromosomes Cancer. 2005;43(2):404–13. doi: 10.1002/gcc.20199. [DOI] [PubMed] [Google Scholar]
- 13.Tommasi S, Crapolicchio A, Lacalamita R, Bruno M, Monaco A, Petroni S, Schittulli F, Longo S, Digennaro M, Calistri D, Mangia A, Paradiso A. BRCA1 mutations and polymorphisms in a hospital based consecutive series of breast cancer patients from Apulia, Italy. Mutat Res. 2005;578(1–2):395–405. doi: 10.1016/j.mrfmmm.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 14.De Silva W, Karunanayake EH, Tennekoon KH, Allen M, Amarasinghe I, Angunawala P. Novel sequence variants and a high frequency of recurrent polymorphisms in BRCA1 gene in Sri Lankan breast cancer patients and at risk individuals. BMC Cance. 2008;214(8):2407–13. doi: 10.1186/1471-2407-8-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Capalbo C, Ricevuto E, Vestri A, Ristori E, Sidoni T, Buffone O, et al. BRCA1 and BRCA2 genetic testing in Italian breast and/or ovarian cancer families: mutation spectrum and prevalence and analysis of mutation prediction models. Annals of Oncology. 2006;17:34–40. doi: 10.1093/annonc/mdl947. [DOI] [PubMed] [Google Scholar]
- 16.R Noor M, Goyal S, M Christensen S, M Iqbal S. Electrical detection of single-base DNA mutation using functionalized nanoparticles. Appl Phys Lett. 2009;4:231–38. [Google Scholar]
- 17.Bishop DT, Hopper JL. AT-tributable risks? Nature Genet. 1997;15:226. doi: 10.1038/ng0397-226. [DOI] [PubMed] [Google Scholar]
- 18.Huyton T, Bates PA, Zhang X, Sternberg MJ, Freemont PS. The BRCA1 C-terminal domain: structure and function. Mutat Res. 2000;460(3–4):319–32. doi: 10.1016/s0921-8777(00)00034-3. [DOI] [PubMed] [Google Scholar]
- 19.Konstantopoulou I, Kroupis C, Ladopoulou A, Pantazidis A, Boumba D, Lianidou ES, Petersen MB, Florentin L, Chiotellis E, Nounesis G, Efstathiou E, Skarlos D, Tsionou C, Fountzilas G, Yannoukakos D. BRCA1 mutation analysis in breast/ovarian cancer families from Greece. Hum Mutat. 2000;16(3):272–73. doi: 10.1002/1098-1004(200009)16:3<272::AID-HUMU17>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 20.Vallon-Christersson J, Cayanan C, Haraldsson K, Loman N, Bergthorsson JT, Brondum-Nielsen K, Gerdes AM, Moller P, Kristoffersson U, Olsson H, Borg A, Monteiro AN. Functional analysis of BRCA1 C-terminal missense mutations identified in breast and ovarian cancer families. Hum Mol Genet. 2001;10(4):353–60. doi: 10.1093/hmg/10.4.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abkevich V, Zharkikh A, Deffenbaugh AM, Frank D, Chen Y, Shattuck D, Skolnick MH, Gutin A, Tavtigian SV. Analysis of missense variation in human BRCA1 in the context of interspecific sequence variation. J Med Genet. 2004;41(7):492–507. doi: 10.1136/jmg.2003.015867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakayori M, Kawahara M, Shiraishi K, Nomizu T, Shimada A, Kudo T, Abe R, Ohuchi N, Takenoshita S, Kanamaru R, Ishioka C. Evaluation of the diagnostic accuracy of the stop codon (SC) assay for identifying protein-truncating mutations in the BRCA1 and BRCA2genes in familial breast cancer. J Hum Genet. 2003;48(3):130–37. doi: 10.1007/s100380300020. [DOI] [PubMed] [Google Scholar]
- 23.Couch FJ, DeShano ML, Blackwood MA, Calzone K, Stopfer J, Campeau L. BRCA1 mutations in women attending clinics that evaluate the risk of breast cancer. N Engl J Med. 1997;336:1409–15. doi: 10.1056/NEJM199705153362002. [DOI] [PubMed] [Google Scholar]
- 24.Frank TS, Deffenbaugh AM, Reid JE. Clinical characteristics of individuals with germ line mutations in BRCA1 and BRCA2: analysis of 10,000 individuals. J Clin Oncol. 2002;20:1480–90. doi: 10.1200/JCO.2002.20.6.1480. [DOI] [PubMed] [Google Scholar]
- 25.Evans DG, Lalloo F, Wallace A, Rahman N. Update on the Manchester Scoring System for BRCA1 and BRCA2 testing. J Med Genet. 2005;6:42–39. doi: 10.1136/jmg.2005.031989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Antoniou AC, Pharoah PP, Smith P, Easton DF. The BOADICEA model of genetic susceptibility to breast and ovarian cancer. Br J Cancer. 2004;91:1580–90. doi: 10.1038/sj.bjc.6602175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palma MD, Domchek SM, Stopfer J, Erlichman J, Siegfried JD, Tigges-Cardwell J, Mason BA, Rebbeck TR, Nathanson KL. The Relative Contribution of Point Mutations and Genomic Rearrangements in BRCA1 and BRCA2 in High-Risk Breast Cancer Families. Cancer Res 1. 2008;68(17):7006–14. doi: 10.1158/0008-5472.CAN-08-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin YP, Chen YL, Chang HT, Li S. Nature of genetic variants in the BRCA1 and BRCA2 genes from breast cancer families in Taiwan. Life Science Journal. 2009;6:99–103. [Google Scholar]