Abstract
Deregulations in translational control are critical features of cancer initiation and progression. Activation of key oncogenic pathways promote rapid and dramatic translational reprogramming, not simply by increasing overall protein synthesis, but also by modulating specific mRNA networks that promote cellular transformation. Additionally, ribosomopathies caused by mutations in ribosome components alter translational regulation leading to specific pathological features, including cancer susceptibility. Exciting advances in our understanding of translational control in cancer have illuminated a striking specificity innate to the translational apparatus. Characterizing this specificity will provide novel insights into how cells normally utilize translational control to modulate gene expression, how it is deregulated in cancer, and how these processes can be targeted to develop new cancer therapies.
Introduction
Decades of research into the molecular programs that govern cellular transformation have mainly focused on the cancer transcriptome. For example, the microarray era has made it possible to catalogue genome-wide variations in the repertoire of transcriptional outputs downstream of specific oncogenic signaling pathways. Ultimately, however, these studies have fallen short in interrogating the end product of gene expression at the level of protein synthesis. A new and critical pipeline into understanding cancer evolution is the growing body of evidence showing that key oncogenic pathways such as Myc and PI3K have monopolized the translational machinery to direct specific posttranscriptional changes in gene expression directly at the level of protein production (Figure 1). In addition, an entire class of inherited syndromes, collectively referred to as ribosomopathies, is characterized by increased cancer susceptibility and harbor mutations in distinct components of the translational apparatus. The realization that there is a post-genomic control mechanism in cancer development has fundamental implications in the design of new cancer therapies that may eradicate the abnormal translational program of cancer cells.
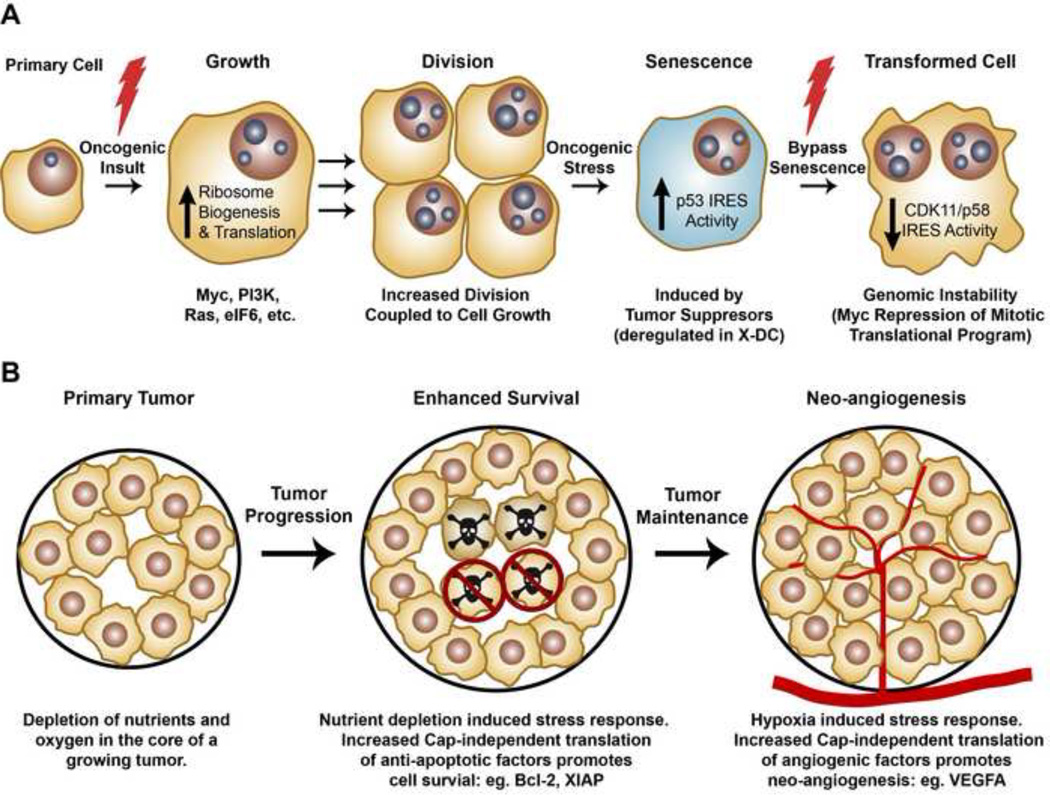
Figure 1. Deregulations in translational control can contribute to each step of cellular transformation and tumor progression.
A.) Upon receiving an oncogenic insult (represented by a lightning bolt), e.g. Myc overexpression or PI3K hyperactivation, cells induce ribosome biogenesis and global protein synthesis that leads to increased cell size, coupled to cell division. Upon oncogenic stress, cells initiate a tumor suppressive response, associated with increased IRES mediated translation, leading to cell cycle arrest and senescence. In order to overcome the barrier of oncogene-induced senescence, cells acquire additional mutations (secondary hits, represented by lightning bolt). One mechanism for this is decreased translation from the CDK11/p58 IRES during mitosis, resulting from Myc hyperactivation, that leads to genome instability. B.) Once established, a primary tumor will undergo unrestrained growth, leading to a stress response associated with depletion of oxygen and essential nutrients from the core of the tumor. Lack of key nutrients, such as growth factors, often leads to increased apoptosis (skulls). Tumor cells upregulate cap-independent translation of anti-apoptotic factors (such as Bcl-2 and XIAP) as a mechanism to promote survival (blocked skulls). To bypass stress caused by low levels of oxygen in the tumor, cells induce cap-independent translation of neo-angiogenesis promoting factors such as VEGF.
Do mutations in the ribosome cause cancer?
A number of mutations in ribosomal proteins or biogenesis factors have been identified that correlate with increased cancer incidence in humans (Table I). Animal models harboring mutations similar to those found in humans faithfully recapitulate these features and have shed light onto how specific mutations in the translational machinery lead to human pathologies.
Table.
| Gene | Genetic Lesion |
Expression/ Activity Level |
Cancer Association | Ref | |
|---|---|---|---|---|---|
| Ribosomopathies | |||||
| X-DC | DKC1 | Mutation/ Deletion |
Decrease | Increased incidence of hematological and solid tumors | [1] |
| DBA | RPS 7,15,17,19, 24,27A RPL 5,11,35A,36 |
Mutation/ Deletion |
Decrease | Increased incidence of hematological and solid tumors | [60–65] |
| 5q-syndrome | RPS14 | Deletion | Decrease | Increased incidence of hematological tumors | [66] |
| SDS | SBDS | Mutation | Decrease | Increased incidence of hematological tumors | [67,68] |
| CHH | RMRP | Mutation | Decrease | Increased incidence of hematological and solid tumors | [69] |
|
Translation Initiation Factors |
eIF2α | ND | Increase | In hematological and solid tumors | [40,70] |
| eIF3a | ND | Increase | In solid tumors | [71,72] | |
| eIF3c | ND | Increase | In solid tumors | [73] | |
| eIF3e | ND | Increase | In solid tumors | [74] | |
| eIF3f | ND | Decrease | In solid tumors | [75] | |
| eIF3h | Amplification | Increase | In solid tumors | [76] | |
| eIF3i | ND | Increase | In solid tumors | [77,78] | |
| eIF4A | ND | Increase | In solid tumors | [79,80] | |
| eIF4E | ND | Increase | In hematological and solid tumors | [35] | |
| eIF4G | Amplification | Increase | In solid tumors | [81,82] | |
| eIF5A | ND | Increase | In hematological tumors | [83] | |
| eIF5A2 | Amplification | Increase | In solid tumors | [84] | |
| eIF6 | ND | Increase | In hematologic and solid tumors | [85,86] | |
|
Translation Elongation Factors |
eEF2 | ND | Increase | In solid tumors | [87] |
| eEF1A2 | Amplification | Increase | In solid tumors | [88,89] | |
|
IRES Trans-Acting Factors |
hnRNP A1 | ND | Increase | In solid tumors | [90] |
| hnRNP E1 | ND | Decrease | In solid tumors | [91,92] | |
| hnRNP E2 | ND | Decrease | In solid tumors | [93] | |
| La | ND | Increase | In solid tumors | [94] | |
| PTB | ND | Increase | In solid tumors | [95] | |
| YB1 | ND | Increase | Correlates with higher grade cancers | [96] | |
|
Ribosome Biogenesis Factors |
Ubf1 | ND | Increase | In solid tumors | [97] |
| Bop1 | Amplification | Increase | In solid tumors | [98] | |
| Npm | Mutation | Decrease | Associated with hematological tumors | [99] |
X-DC: x-linked dyskeratosis congenita; DBA: Diamond Blackfan anemia; SDS: Shwachman-Diamond syndrome; CHH: cartilage hair hypoplasia; RPS: ribosomal protein small subunit; RPL: ribosomal protein large subunit
One important example of how defects in the ribosome contribute to specific disease pathologies and cancer susceptibility is X-linked Dyskeratosis Congenita (X-DC). X-DC is the most common and severe form of Dyskeratosis Congenita (DC), and is invariably associated with mutations of the DKC1 gene, encoding for dyskerin (Table I)[1,2]. Although prominent features of X-DC pathogenesis include bone marrow failure and skin abnormalities, a wide variety of tumor types including carcinomas and hematopoietic malignancies are also manifest. Dyskerin is an evolutionarily conserved enzyme responsible for the modification of approximately 100 specific uridines into pseudouridines in ribosomal RNA[3,4]. The role of rRNA modifications in translational control has historically been poorly understood. Therefore, the unexpected human genetic mutations associated with a key enzyme responsible for site-specific rRNA modifications were enigmatic, especially with respect to the tissue-specific phenotypes present in X-DC.
The specific features of X-DC pathogenesis, including cancer susceptibility, can be explained, at least in part, by the finding that a specific subset of mRNAs are not efficiently translated in an X-DC mouse model and patient cells [5–8]. On the contrary, general protein synthesis appears to be unperturbed. These target mRNAs all share a common regulatory motif in their 5’untranslated region (5’UTR): an internal ribosome entry site (IRES) element that acts to initiate translation in a cap-independent manner. IRES elements are found in approximately 10% of mRNAs, including many tumor suppressor genes and anti-apoptotic factors (Figure 2) [9]. IRES-dependent translation acts to fine tune gene expression at the post-transcriptional level. Defects in the IRES mediated translation of a subset of mRNAs, including important tumor suppressors such as p53 and p27, in DKC1 mutant cells contributes to cellular transformation and tumor development [5,6,8]. These studies have also uncovered an unexpected requirement for rRNA modifications in a specific mode of translational control.
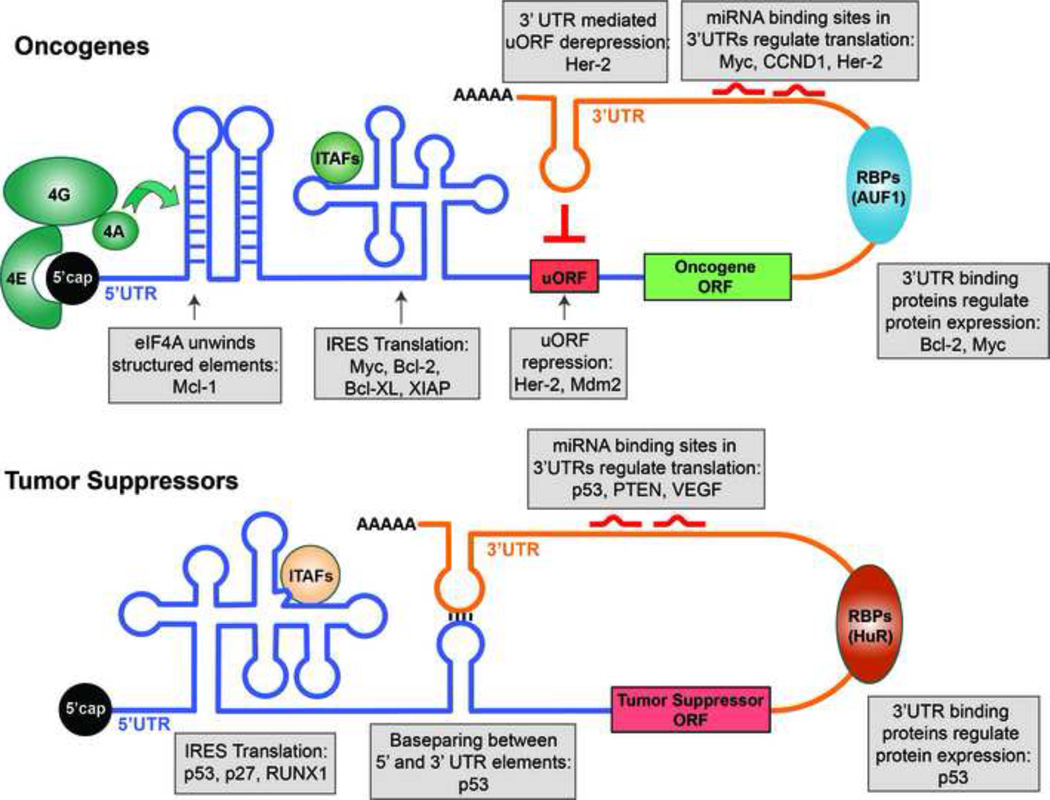
Figure 2. Oncogenes and Tumor Suppressors are translationally regulated through specific regulatory elements in their mRNAs.
Depictions of cis-regulatory elements present in oncogenes (top) and tumor suppressor (bottom) mRNAs. Examples of regulated mRNAs are given (grey boxes). The cap binding protein eIF4E binds the 5’ cap of mRNAs. Increased eIF4E activity recruits eIF4G and the eIF4A helicase to unwind (green arrow) structured elements in the 5’UTR of poorly translated mRNAs, increasing the expression of many growth promoting and pro-survival genes, such as Mcl-1. Another group of structural elements that direct translational regulation are IRES elements. IRES elements promote cap-independent protein synthesis of both tumor suppressors (p53) and pro-survival factors (XIAP) during different stages of tumor development (see Figure 1). IRES Trans-Acting Factors (ITAFs) modulate translational regulation directed by specific IRES elements. Upstream Open Reading Frames (uORF) are present in select mRNAs and inhibit protein synthesis by preventing the ribosome from scanning to the start codon. Translation of growth promoting factors, such as Mdm2 and Her-2, is limited by uORFs. Additionally, a region in the 3’UTR of the Her-2 mRNA can inhibit uORF mediated repression of translation. 3’UTRs contain many distinct elements that interact with RNA binding proteins (RBPs) and miRNAs to promote or inhibit translation. One interesting example is the regulation of p53 translation mediated by base pairing interactions between elements in the 5’ and 3’ UTRs of the p53.
Diamond Blackfan anemia (DBA) is a similar case where mutations in several different ribosomal proteins lead to bone marrow failure and an increased risk of leukemia and solid tumors (Table I)[10]. This is also consistent with studies in zebrafish showing that mutations in ribosomal proteins cause cancer [11]. In DBA, alterations in rRNA processing have been described that are associated with defects in ribosome biogenesis. This may trigger a stress response, marked by p53 activation, leading to cell cycle arrest or programmed cell death [12–14]. While the induction of p53 may explain certain pathological features of DBA, the molecular basis for increased cancer susceptibility remains poorly understood. It is currently not clear whether ribosomal proteins may exert more specialized functions in translational control, either on or off the ribosome, that may underlie the cancer susceptibility associated with DBA [15, 16]. For example, very recent studies have shown that a single ribosomal protein, RPL38, regulates transcript-specific translational control of an important class of Hox mRNAs. Moreover, it appears that ribosomal protein expression may be dynamically regulated [17]. As such, it is tempting to speculate that mutations in ribosomal proteins may have more specific roles in translational control that underlie the increased cancer susceptibility observed in DBA.
The X-DC and DBA syndromes are specific examples of an entire class of inherited human cancer susceptibility disorders that are collectively referred to as “ribosomopathies”. Additional notable examples include Cartilage-Hair Hypoplasia syndrome, Shwachman Diamond Syndrome, and 5q deletion syndrome, which also show increased risk of developing particular cancers of the skin (basal cell carcinoma) as well as blood (leukemia and lymphoma) [18,19]. In all of these cases, mutations in regulatory factors important for ribosome biogenesis and translational control have been identified (Table I). In certain cases, extra-ribosomal functions exerted by these factors may also play a causative role in disease pathogenesis [14].
Collectively, these inherited ribosomopathies reveal that mutations in regulatory factors important for ribosome activity may produce mutant ribosomes lacking important constituents such as ribosomal proteins or rRNA modifications. Can these mutant forms of the translational machinery be referred to as “cancer ribosomes”? If so, an outstanding question is the mechanism by which the “cancer ribosome” could promote cancer development at the level of aberrant translational control. This is an important question to resolve, as at first glance it may appear counterintuitive that loss of ribosome function could cause cancer, especially given the important connection between increased protein synthesis and cell growth (see below). However, recent findings show surprising specificity in the classes of mRNAs that are specifically deregulated and underlie cancer susceptibility as a consequence of perturbations in ribosome function, for example, as illustrated by X-DC. This also reflects an emerging appreciation and increased knowledge of more specialized and dynamic regulation of translational control in vivo, at an organismal level. In the context of key tumor suppressors and proto-oncogenes, a common denominator may be the tight regulation in their expression levels, specifically at the level of translational control. A central principle emerging from detailed molecular studies is that the presence of important regulatory elements (e.g. IRES, complex 5’ and 3’UTRs, RNA binding protein or micro-RNA sites) within key tumor suppressors and proto-oncogenes may render these mRNAs exquisitely sensitive to perturbations in translational control (Figure 2).
How does manipulation of the translational machinery by oncogenic signaling promote cancer?
Ribosome biogenesis and global protein synthesis are tightly and dynamically regulated to accommodate the growth demands of a cell. Indeed, an increase in cell mass is a pre-requisite for accurate cell division. This is achieved by signaling pathways that simultaneously sense energy, stress, nutrient availability, as well as growth factors, and integrate these inputs to direct control of ribosome production and activity. One of the primary reasons for this cross talk is to integrate external stimuli such as energy and nutrient abundance, with the execution of cell growth and division that is directly coupled to protein synthesis. When these signaling pathways are altered, it may lead to unrestrained control of protein synthesis. Indeed, one of the earliest markers for cancer cells, discovered more than 100 years ago, is an increase in the size and number of nucleoli[20]. This may reflect the increasing demands for ribosome biogenesis in cancer cells in order to sustain their elevated rates of growth and division. Is this a cause or consequence of cellular transformation? This question has been answered, at least in part, by the identification of several oncogenic signaling pathways that directly modulate the activity of specific translational components. Notable examples include the PI3K-Akt-mTOR and RAS-MAPK signal transduction pathways, as well as transcriptional programs regulated by oncogenic Myc [21–25].
One of the best-studied examples of oncogenic signaling impinging on translational control is the PI3K-AKT pathway, which modulates translation initiation largely through activation of the kinase mammalian target of rapamycin complex 1 (mTORC1)[26]. mTORC1 phosphorylates ribosomal protein S6 kinase 1/2 (S6K1/2) and the 4EBPs, which negatively regulate the major cap-binding protein eIF4E[27,28]. The latter leads to a conformational change that releases 4EBPs from eIF4E and ultimately recruits the 40S ribosomal subunit to the 5’ end of mRNAs[29,30]. Overexpression of eIF4E promotes cancer and cooperates with c-Myc to drive lymphomagenesis in vivo in transgenic mice [31, 32]. 4EBPeIF4E exerts significant control over cap-dependent translation, cell growth, cancer initiation, and progression downstream of mTOR hyperactivation [33,34]. Molecularly, eIF4E hyperactivation is able to enhance the translation of select mRNAs[35]. The 5’UTR of these mRNAs are believed to harbor the regulatory elements that impart this selectivity, such as complex secondary structures. One example is Mcl-1, an anti-apoptotic factor containing a complex 5’UTR that is specifically translationally upregulated upon eIF4E hyperactivation leading to enhanced survival of cancer initiating cells [34,36]. While mounting evidence, including elegant genetic studies, have clearly shown a central role of eIF4E hyperactivation in cancer development, the repertoire of translational target mRNAs that are specifically sensitive to eIF4E hyperactivation remains poorly defined. There are now emerging technologies that may facilitate their identification. In particular, the ability to deep sequence ribosome protected mRNAs will enable codon-by-codon resolution of ribosome occupancy on specific mRNAs [37]. Furthermore, through deep sequencing, it is also now possible to determine the secondary structures of mRNAs by using a novel strategy termed parallel analysis of RNA structures (PARS) [38]. The combination of these two technologies may provide a very accurate portrait of how mRNA secondary structures control cap-dependent translation and impact on translation of the cancer genome.
Regulation of eIF4E is not the only node where information from signaling pathways is received by the translational machinery. It is now also clear that an entire repertoire of translational components may be co-opted to promote cancer initiation. For example, AKT hyperactivation also modulates translation elongation [39] (Table I). Additional regulated translational components include eIF2α, which is part of the ternary complex required to chaperone the initiator tRNA to the ribosome. eIF2α is commonly overexpressed in cancers and may thereby provide an uncontrolled stimulus leading to increased rates of protein synthesis [40]. Interestingly, even overexpression of the initiator tRNA itself is able to drive cellular transformation [41]. Another translation factor that promotes cellular transformation is eIF6. eIF6 regulates the joining of the 60S ribosomal subunit to the 48S pre-initiation complex to initiate translation [42]. Importantly, eIF6 has been shown to be rate limiting for translation, cell growth and transformation[42]. Interestingly, eIF6 interacts with RACK1, a ribosome associated scaffolding protein that coordinates signaling by PKC and src kinases [43]. Therefore, signaling through RACK1 to eIF6 may be another important node of oncogenic regulation.
A prominent example of an oncogenic signal that relies on the translational machinery for cellular transformation is the Myc oncogene, which is commonly deregulated in human cancers[44]. Myc directly increases protein synthesis rates by controlling the expression of multiple components of the protein synthetic machinery, including ribosomal proteins, initiation factors of translation, Pol III and rDNA[24,45,46]. Genetic strategies that restore increased protein synthesis in Myc transgenic mice to normal levels reveal that the oncogenic potential of Myc is suppressed in this context [47]. These findings also demonstrate that the ability of Myc to increase protein synthesis directly augments cell size and is sufficient to accelerate cell cycle progression[47]. Surprisingly, deregulations in mitotic translational control as a consequence of Myc hyperactivation also directly lead to genome instability by modulating the translation of specific mRNAs[47]. Thereby, Myc-dependent control of the translational machinery has a pleiotropic role in distinct steps of cancer initiation and progression.
The remarkable repertoire of translational components found deregulated in cancer, whose activity is directly controlled downstream of specific oncogenic signals, strongly supports a critical and causal role in cancer initiation and progression. What is also emerging from these studies is that perturbations in translational control provide a highly specific outcome for gene expression, genome instability, and distinct steps along the pathway towards cancer development.
How does aberrant translation promote cancer- it’s not all just growth?
In most representations of cancer development, deregulations in protein synthesis are simply depicted by a generic arrow that culminates in cell growth. While this may certainly be one of the mechanisms by which deregulations in protein synthesis lead to cancer development, it is not the full story. Recent studies are clearly showing that oncogenic signaling may monopolize the translational machinery at almost every stage of cancer initiation and development for very specific and distinct cellular outcomes (Figure 1).
For a primary cell to undergo cellular transformation, several tumor suppressive barriers must be overcome. As previously discussed, one of the earliest effects of oncogenic signals such as MYC or PI3K-AKT-mTOR hyperactivation is increased protein synthesis that leads to increased cell growth. This may provide a significant competitive advantage to a pre-neoplastic cell. For example, a larger cell may outcompete neighboring normal cells in uptake of nutrients and growth factors. Consequently, a somatic clone harboring such a cancerous lesion may possess an early competitive advantage through increased cell growth. The mechanism(s) by which the translational program links increases in cell growth to cell division remain poorly understood. Specifically, it is not clear whether proliferation is normally initiated due to an accumulation of total protein mass (cell size), or that this impacts directly on the translation of specific cell cycle mRNAs such as cyclins or replication enzymes. However, what is clear is that increases in protein synthesis set up a cell size threshold that is interpreted by the cell cycle machinery as a stimulus to commence cell division [47,48].
Increased cell division as a consequence of increased cell growth is not sufficient to induce cellular transformation. Indeed, a critical cellular response that counteracts cellular transformation is oncogene-induced senescence (OIS). OIS is characterized by cell cycle arrest and induction of p53, which restrains the proliferative potential of pre-neoplastic clones[49]. Our recent findings have shown that during OIS, a switch between cap- and IRES-dependent translation occurs [50]. During this switch, an IRES element positioned in the 5'UTR of p53 is engaged to promote p53 translation[5]. Therefore specialized translational control of mRNAs, such as p53, provides a molecular barrier for cellular transformation[5,8]. Interestingly, defects in rRNA modifications specifically perturb p53 IRES-dependent translation, resulting in defective OIS and expansion of pre-neoplastic clones[5]. This may be a critical mechanism that underlies cancer susceptibility in X-DC[5,8].
A subsequent mechanism that is critical to achieve full cellular transformation is the acquisition of additional genetic lesions, commonly referred to as secondary hits. Unexpectedly, deregulations in mitotic translational control have been shown to play an important role at this step, and may contribute to genomic instability[47]. During mitosis, only a small fraction of mRNAs are translated in a cap-independent manner via a switch to IRES-dependent translation[51,52]. CDK11/p58 is a well-characterized endogenous mRNA that is only translated during mitosis by an IRES element [53–55]. An aberrant increase in capdependent translation downstream of Myc hyperactivation specifically impairs this translational switch to IRES-dependent translation. This results in reduced mitotic-specific expression of CDK11/p58 that leads to cytokinesis defects and is associated with increased centrosome numbers and genome instability [47]. Thereby, deregulations in mitotic translation control may create additional genetic lesions that are required to achieve full cellular transformation.
Once a tumor has been established it will, again, progress through a number of stages and may ultimately become metastatic. An emerging concept is that tumor cells may survive stress-conditions such as nutrient and oxygen deprivation through their ability to promote cap-independent translation. This is achieved through translational control of specific anti-apoptotic factors such as Bcl-2 and XIAP, which contain IRES-elements, as well as neo-angiogenic factors, such as VEGF, that enhance blood flow to the tumor [56–59]. Thereby, tumor cells have co-opted specific modes of translational regulation that impact on the expression of key tumor suppressors/oncogenes for their continued survival and development (Figure 1).
In summary, it is clear that manipulating translational regulation is important during almost every phase of tumor development. It also appears that different modes of translational regulation are deployed during distinct stages of tumor progression. It is noteworthy that translational control impinges on each checkpoint established to prevent cellular transformation and tumor growth, highlighting its importance in cancer development.
Concluding Remarks
We are only beginning to understand the broad implications of translational regulation as it relates to cancer biology. Oncogenic signaling appears to monopolize translational control at almost every stage of cancer initiation and development for very specific and distinct cellular outcomes (Figure 1). Currently there is a shift towards a growing realization of the importance of specificity in translational regulation mediated by the core components of the translational apparatus. Key among these studies is the finding that loss of rRNA modification leads to a reduction in the translation of only specific mRNAs that harbor IRESelements. Mutant ribosomes lacking important constituents (such as ribosomal proteins or rRNA modifications) may be largely functionally active, but preferentially defective in specific aspects of translational control in a growing class of ribosomopathies. Moreover, studies of translational control in cancer have uncovered unexpected specificity in control of gene expression at the posttranscription level and highlight the dynamic nature of the translational machinery. There is a growing appreciation for the contributions of specific regulatory elements within mRNAs, such as the IRES elements, structured 5’UTRs, and sequences in 3’UTRs that are recognized by proteins or microRNAs, that control their expression during specific steps of cellular transformation and tumor development (Figures 1 and 2). Elucidating the relationship between these regulatory elements to oncogenic signaling pathways that feed into the translational machinery will be an exciting new area of research. The ultimate importance of this research is reflected by a number of novel and promising therapeutic approaches to target specific translational components in cancer that are currently in clinical trials.
Highlights.
Deregulation of translational control is critical for cancer development.
Mutations in ribosome components are associated with cancer susceptibility syndromes.
Alterations in translational control impinge on distinct steps of cellular transformation.
Translational deregulation of specific mRNAs contributes to tumor initiation and progression
Specific regulatory elements control translation of oncogenes and tumor suppressors.
Acknowledgements
We would like to thank Maria Barna and members of the Ruggero lab for input and critical reading of this review. Thank you to Kimhouy Tong for editing the manuscript. We apologize to the many scientists whose work we were unable to cite. Dr. Davide Ruggero is a Leukemia & Lymphoma Society Scholar. This work is supported by NIH R01 HL085572 (D.R.) and NIH R01 CA140456 (D. R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Heiss NS, Knight SW, Vulliamy TJ, Klauck SM, Wiemann S, Mason PJ, Poustka A, Dokal I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet. 1998;19:32–38. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- 2.Ruggero D, Grisendi S, Piazza F, Rego E, Mari F, Rao PH, Cordon-Cardo C, Pandolfi PP. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science. 2003;299:259–262. doi: 10.1126/science.1079447. [DOI] [PubMed] [Google Scholar]
- 3.Lafontaine DL, Bousquet-Antonelli C, Henry Y, Caizergues-Ferrer M, Tollervey D. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 1998;12:527–537. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ni J, Tien AL, Fournier MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- **5. Bellodi C, Kopmar N, Ruggero D. Deregulation of oncogene-induced senescence and p53 translational control in X-linked dyskeratosis congenita. EMBO J. 2010:1–12. doi: 10.1038/emboj.2010.83. This paper demonstrated a translational switch during OIS leading to increased IRES translation. This switch is impaired when ribosomes are not fully modified, leading to decreased p53 function in vivo.
- *6. Bellodi C, Krasnykh O, Haynes N, Theodoropoulou M, Peng G, Montanaro L, Ruggero D. Loss of function of the tumor suppressor DKC1 perturbs p27 translation control and contributes to pituitary tumorigenesis. Cancer Res. 2010;70:6026–6035. doi: 10.1158/0008-5472.CAN-09-4730. This study identified a novel mutation in DKC1 from a human pituitary tumor and demonstrate cooperation between DKC and p27 in restraining pituitary tumor growth. A novel transgenic reporter mouse is used to illustrate the importance of DKC1 in p27 IRES mediated translation.
- 7.Yoon A, Peng G, Brandenburger Y, Zollo O, Xu W, Rego E, Ruggero D. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006;312:902–906. doi: 10.1126/science.1123835. [DOI] [PubMed] [Google Scholar]
- **8. Montanaro L, Calienni M, Bertoni S, Rocchi L, Sansone P, Storci G, Santini D, Ceccarelli C, Taffurelli M, Carnicelli D, et al. Novel dyskerin-mediated mechanism of p53 inactivation through defective mRNA translation. Cancer Res. 2010;70:4767–4777. doi: 10.1158/0008-5472.CAN-09-4024. The authors demonstrate that increased p53 activity in breast cancer is dependent on dyskerin-mediated increases in IRES-mediated translation and independent of effects on telomerase.
- 9.Graber TE, Holcik M. Cap-independent regulation of gene expression in apoptosis. Mol Biosyst. 2007;3:825–834. doi: 10.1039/b708867a. [DOI] [PubMed] [Google Scholar]
- 10.Narla A, Hurst SN, Ebert BL. Ribosome defects in disorders of erythropoiesis. Int J Hematol. 2011;93:144–149. doi: 10.1007/s12185-011-0776-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amsterdam A, Sadler KC, Lai K, Farrington S, Bronson RT, Lees JA, Hopkins N. Many ribosomal protein genes are cancer genes in zebrafish. Plos Biol. 2004;2:E139. doi: 10.1371/journal.pbio.0020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danilova N, Sakamoto KM, Lin S. Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood. 2008;112:5228–5237. doi: 10.1182/blood-2008-01-132290. [DOI] [PubMed] [Google Scholar]
- 13.McGowan KA, Li JZ, Park CY, Beaudry V, Tabor HK, Sabnis AJ, Zhang W, Fuchs H, de Angelis MH, Myers RM, et al. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat Genet. 2008;40:963–970. doi: 10.1038/ng.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *14. Dutt S, Narla A, Lin K, Mullally A, Abayasekara N, Megerdichian C, Wilson FH, Currie T, Khanna-Gupta A, Berliner N, et al. Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood. 2011;117:2567–2576. doi: 10.1182/blood-2010-07-295238. The authors demonstrate a mechanism leading from ribosomal protein deficiency to activation of p53 by inhibition of HDM2 in human erythroid progenitor cells.
- *15. Landry DM, Hertz MI, Thompson SR. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes & Development. 2009;23:2753–2764. doi: 10.1101/gad.1832209. This paper demonstrates that loss of RPS25 greatly impairs viral IRES mediated translation, while exhibiting minimal effect on global protein synthesis, in yeast.
- 16.Mazumder B, Sampath P, Seshadri V, Maitra RK, DiCorleto PE, Fox PL. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell. 2003;115:187–198. doi: 10.1016/s0092-8674(03)00773-6. [DOI] [PubMed] [Google Scholar]
- **17. Kondrashov N, Pusic A, Stumpf C, Shimizu K, Shieh A, Ishijima J, Shiroishi T, Barna M. Ribosome mediated specificity of Hox mRNA translation regulates vertebrate tissue patterning. Cell. doi: 10.1016/j.cell.2011.03.028. In Press. This study characterized a ribosomal protein mutation that caused a specific developmental phenotype due to decreased translation of specific mRNAs. Additionally, the expression of ribosomal proteins is tissue specific and dynamic during development.
- 18.Shimamura A, Alter BP. Pathophysiology and management of inherited bone marrow failure syndromes. Blood Rev. 2010;24:101–122. doi: 10.1016/j.blre.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barlow JL, Drynan LF, Trim NL, Erber WN, Warren AJ, McKenzie AN. New insights into 5q-syndrome as a ribosomopathy. Cell Cycle. 2010;9:4286–4293. doi: 10.4161/cc.9.21.13742. [DOI] [PubMed] [Google Scholar]
- 20.Pianese G. Beitragzur Histologie und Aetiologie der Carcinoma. Beitr. Pathol. Anat. Allgem. Pathol. 1896;142:1–193. [Google Scholar]
- **21. Furic L, Rong L, Larsson O, Koumakpayi IH, Yoshida K, Brueschke A, Petroulakis E, Robichaud N, Pollak M, Gaboury LA, et al. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc Natl Acad Sci USA. 2010;107:14134–14139. doi: 10.1073/pnas.1005320107. The authors show that eIF4E phosphorylation is important for tumorigenesis in a PTEN-deficient prostate cancer model. Additionally, eIF4E phosphorylation enhances the translation of a subset of mRNAs.
- 22.Waskiewicz AJ, Johnson JC, Penn B, Mahalingam M, Kimball SR, Cooper JA. Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol Cell Biol. 1999;19:1871–1880. doi: 10.1128/mcb.19.3.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuhmacher M, Kohlhuber F, Hölzel M, Kaiser C, Burtscher H, Jarsch M, Bornkamm GW, Laux G, Polack A, Weidle UH, et al. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Research. 2001;29:397–406. doi: 10.1093/nar/29.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeller KI, Zhao X, Lee CW, Chiu KP, Yao F, Yustein JT, Ooi HS, Orlov YL, Shahab A, Yong HC, et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci U S A. 2006;103:17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frederickson RM, Mushynski WE, Sonenberg N. Phosphorylation of translation initiation factor eIF-4E is induced in a ras-dependent manner during nerve growth factor-mediated PC12 cell differentiation. Mol Cell Biol. 1992;12:1239–1247. doi: 10.1128/mcb.12.3.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dennis PB, Pullen N, Kozma SC, Thomas G. The principal rapamycin-sensitive p70(s6k) phosphorylation sites, T-229 and T-389, are differentially regulated by rapamycin-insensitive kinase kinases. Mol Cell Biol. 1996;16:6242–6251. doi: 10.1128/mcb.16.11.6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hara K, Yonezawa K, Kozlowski MT, Sugimoto T, Andrabi K, Weng QP, Kasuga M, Nishimoto I, Avruch J. Regulation of eIF-4E BP1 phosphorylation by mTOR. J Biol Chem. 1997;272:26457–26463. doi: 10.1074/jbc.272.42.26457. [DOI] [PubMed] [Google Scholar]
- 29.Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N. Regulation of 4E-BP1 phosphorylation. a novel two-step mechanism. Genes Dev. 1999;13:1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruggero D, Sonenberg N. The Akt of translational control. Oncogene. 2005;24:7426–7434. doi: 10.1038/sj.onc.1209098. [DOI] [PubMed] [Google Scholar]
- 31.Ruggero D, Montanaro L, Ma L, Xu W, Londei P, Cordon-Cardo C, Pandolfi PP. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med. 2004;10:484–486. doi: 10.1038/nm1042. [DOI] [PubMed] [Google Scholar]
- 32.Wendel HG, De Stanchina E, Fridman JS, Malina A, Ray S, Kogan S, Cordon-Cardo C, Pelletier J, Lowe SW. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–337. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- **33. Dowling RJ, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, Petroulakis E, Wang X, Larsson O, Selvaraj A, Liu Y, et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science. 2010;328:1172–1176. doi: 10.1126/science.1187532. The authors show that mTORC1 promotes proliferation by inhibiting 4EBP mediated repression of specific mRNAs.
- **34. Hsieh AC, Costa M, Zollo O, Davis C, Feldman ME, Testa JR, Meyuhas O, Shokat KM, Ruggero D. Genetic Dissection of the Oncogenic mTOR Pathway Reveals Druggable Addiction to Translational Control via 4EBP-eIF4E. Cancer Cell. 2010;17:249–261. doi: 10.1016/j.ccr.2010.01.021. This paper demonstrated the requirement of 4EBP phosphorylation in mTOR-mediated tumor initiation and progression. Efficacy of a novel ATP site mTOR inhibitor is shown to be mediated through inhibition of oncogenic eIF4E hyperaction.
- 35.De Benedetti A, Graff JR. eIF-4E expression and its role in malignancies and metastases. Oncogene. 2004;23:3189–3199. doi: 10.1038/sj.onc.1207545. [DOI] [PubMed] [Google Scholar]
- 36.Mills JR, Hippo Y, Robert F, Chen SM, Malina A, Lin CJ, Trojahn U, Wendel HG, Charest A, Bronson RT, et al. mTORC1 promotes survival through translational control of Mcl-1. Proc Natl Acad Sci U S A. 2008;105:10853–10858. doi: 10.1073/pnas.0804821105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **37. Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. The authors describe a novel technique that allows for the analysis of translational regulation at the genome-wide level.
- 38.Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, Chang HY, Segal E. Genome-wide measurement of RNA secondary structure in yeast. Nature. 467:103–107. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Li W, Williams M, Terada N, Alessi DR, Proud CG. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J. 2001;20:4370–4379. doi: 10.1093/emboj/20.16.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang S, Rosenwald IB, Hutzler MJ, Pihan GA, Savas L, Chen JJ, Woda BA. Expression of the eukaryotic translation initiation factors 4E and 2alpha in non-Hodgkin's lymphomas. Am J Pathol. 1999;155:247–255. doi: 10.1016/s0002-9440(10)65118-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marshall L, Kenneth NS, White RJ. Elevated tRNA(iMet) synthesis can drive cell proliferation and oncogenic transformation. Cell. 2008;133:78–89. doi: 10.1016/j.cell.2008.02.035. [DOI] [PubMed] [Google Scholar]
- 42.Gandin V, Miluzio A, Barbieri AM, Beugnet A, Kiyokawa H, Marchisio PC, Biffo S. Eukaryotic initiation factor 6 is rate-limiting in translation, growth and transformation. Nature. 2008;455:684–688. doi: 10.1038/nature07267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ceci M, Gaviraghi C, Gorrini C, Sala LA, Offenhauser N, Marchisio PC, Biffo S. Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature. 2003;426:579–584. doi: 10.1038/nature02160. [DOI] [PubMed] [Google Scholar]
- 44.Ruggero D. The role of Myc-induced protein synthesis in cancer. Cancer Res. 2009;69:8839–8843. doi: 10.1158/0008-5472.CAN-09-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, Cocito A, Amati B. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gomez-Roman N, Grandori C, Eisenman RN, White RJ. Direct activation of RNA polymerase III transcription by c-Myc. Nature. 2003;421:290–294. doi: 10.1038/nature01327. [DOI] [PubMed] [Google Scholar]
- 47.Barna M, Pusic A, Zollo O, Costa M, Kondrashov N, Rego E, Rao PH, Ruggero D. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature. 2008;456:971–975. doi: 10.1038/nature07449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schuhmacher M, Staege MS, Pajic A, Polack A, Weidle UH, Bornkamm GW, Eick D, Kohlhuber F. Control of cell growth by c-Myc in the absence of cell division. Curr Biol. 1999;9:1255–1258. doi: 10.1016/s0960-9822(99)80507-7. [DOI] [PubMed] [Google Scholar]
- 49.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 50.Bellodi C, Kopmar N, Ruggero D. Deregulation of oncogene-induced senescence and p53 translational control in X-linked dyskeratosis congenita. EMBO J. 2010;29:1865–1876. doi: 10.1038/emboj.2010.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pyronnet S, Pradayrol L, Sonenberg N. A cell cycle-dependent internal ribosome entry site. Molecular Cell. 2000;5:607–616. doi: 10.1016/s1097-2765(00)80240-3. [DOI] [PubMed] [Google Scholar]
- 52.Qin X, Sarnow P. Preferential translation of internal ribosome entry site-containing mRNAs during the mitotic cycle in mammalian cells. J Biol Chem. 2004;279:13721–13728. doi: 10.1074/jbc.M312854200. [DOI] [PubMed] [Google Scholar]
- 53.Wilker EW, van Vugt MATM, Artim SA, Huang PH, Petersen CP, Reinhardt HC, Feng Y, Sharp PA, Sonenberg N, White FM, et al. 14-3-3sigma controls mitotic translation to facilitate cytokinesis. Nature. 2007;446:329–332. doi: 10.1038/nature05584. [DOI] [PubMed] [Google Scholar]
- 54.Cornelis S, Bruynooghe Y, Denecker G, Van Huffel S, Tinton S, Beyaert R. Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Molecular Cell. 2000;5:597–605. doi: 10.1016/s1097-2765(00)80239-7. [DOI] [PubMed] [Google Scholar]
- 55.Tinton SA, Schepens B, Bruynooghe Y, Beyaert R, Cornelis S. Regulation of the cell-cycle-dependent internal ribosome entry site of the PITSLRE protein kinase. roles of Unr (upstream of N-ras) protein and phosphorylated translation initiation factor eIF-2alpha. Biochem J. 2005;385:155–163. doi: 10.1042/BJ20040963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Holcik M, Korneluk RG. Functional characterization of the X-linked inhibitor of apoptosis (XIAP) internal ribosome entry site element. role of La autoantigen in XIAP translation. Mol Cell Biol. 2000;20:4648–4657. doi: 10.1128/mcb.20.13.4648-4657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Silvera D, Schneider RJ. Inflammatory breast cancer cells are constitutively adapted to hypoxia. Cell Cycle. 2009;8:3091–3096. doi: 10.4161/cc.8.19.9637. [DOI] [PubMed] [Google Scholar]
- **58. Silvera D, Arju R, Darvishian F, Levine PH, Zolfaghari L, Goldberg J, Hochman T, Formenti SC, Schneider RJ. Essential role for eIF4GI overexpression in the pathogenesis of inflammatory breast cancer. Nat Cell Biol. 2009;11:903–908. doi: 10.1038/ncb1900. This paper demonstrates that eIF4G is required for VEGF IRES-mediated translation to promote neo-angiogenesis during tumor progression.
- 59.Sherrill KW, Byrd MP, Van Eden ME, Lloyd RE. BCL-2 translation is mediated via internal ribosome entry during cell stress. J Biol Chem. 2004;279:29066–29074. doi: 10.1074/jbc.M402727200. [DOI] [PubMed] [Google Scholar]
- 60.Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig TN, Dianzani I, Ball S, Tchernia G, Klar J, Matsson H, et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet. 1999;21:169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- 61.Gazda HT, Grabowska A, Merida-Long LB, Latawiec E, Schneider HE, Lipton JM, Vlachos A, Atsidaftos E, Ball SE, Orfali KA, et al. Ribosomal protein S24 gene is mutated in Diamond-Blackfan anemia. Am J Hum Genet. 2006;79:1110–1118. doi: 10.1086/510020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cmejla R, Cmejlova J, Handrkova H, Petrak J, Pospisilova D. Ribosomal protein S17 gene (RPS17) is mutated in Diamond-Blackfan anemia. Hum Mutat. 2007;28:1178–1182. doi: 10.1002/humu.20608. [DOI] [PubMed] [Google Scholar]
- 63.Farrar JE, Nater M, Caywood E, McDevitt MA, Kowalski J, Takemoto CM, Talbot CC, Jr, Meltzer P, Esposito D, Beggs AH, et al. Abnormalities of the large ribosomal subunit protein, Rpl35a, in Diamond-Blackfan anemia. Blood. 2008;112:1582–1592. doi: 10.1182/blood-2008-02-140012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gazda HT, Sheen MR, Vlachos A, Choesmel V, O'Donohue MF, Schneider H, Darras N, Hasman C, Sieff CA, Newburger PE, et al. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am J Hum Genet. 2008;83:769–780. doi: 10.1016/j.ajhg.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cmejla R, Cmejlova J, Handrkova H, Petrak J, Petrtylova K, Mihal V, Stary J, Cerna Z, Jabali Y, Pospisilova D. Identification of mutations in the ribosomal protein L5 (RPL5) and ribosomal protein L11 (RPL11) genes in Czech patients with Diamond-Blackfan anemia. Hum Mutat. 2009;30:321–327. doi: 10.1002/humu.20874. [DOI] [PubMed] [Google Scholar]
- 66.Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, Raza A, Root DE, Attar E, Ellis SR, et al. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 2008;451:335–339. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boocock GR, Morrison JA, Popovic M, Richards N, Ellis L, Durie PR, Rommens JM. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat Genet. 2003;33:97–101. doi: 10.1038/ng1062. [DOI] [PubMed] [Google Scholar]
- 68.Austin KM, Leary RJ, Shimamura A. The Shwachman-Diamond SBDS protein localizes to the nucleolus. Blood. 2005;106:1253–1258. doi: 10.1182/blood-2005-02-0807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ridanpaa M, van Eenennaam H, Pelin K, Chadwick R, Johnson C, Yuan B, vanVenrooij W, Pruijn G, Salmela R, Rockas S, et al. Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell. 2001;104:195–203. doi: 10.1016/s0092-8674(01)00205-7. [DOI] [PubMed] [Google Scholar]
- 70.Rosenwald IB, Hutzler MJ, Wang S, Savas L, Fraire AE. Expression of eukaryotic translation initiation factors 4E and 2alpha is increased frequently in bronchioloalveolar but not in squamous cell carcinomas of the lung. Cancer. 2001;92:2164–2171. doi: 10.1002/1097-0142(20011015)92:8<2164::aid-cncr1559>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 71.Bachmann F, Banziger R, Burger MM. Cloning of a novel protein overexpressed in human mammary carcinoma. Cancer Res. 1997;57:988–994. [PubMed] [Google Scholar]
- 72.Dellas A, Torhorst J, Bachmann F, Banziger R, Schultheiss E, Burger MM. Expression of p150 in cervical neoplasia and its potential value in predicting survival. Cancer. 1998;83:1376–1383. doi: 10.1002/(sici)1097-0142(19981001)83:7<1376::aid-cncr15>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 73.Rothe M, Ko Y, Albers P, Wernert N. Eukaryotic initiation factor 3 p110 mRNA is overexpressed in testicular seminomas. Am J Pathol. 2000;157:1597–1604. doi: 10.1016/S0002-9440(10)64797-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marchetti A, Buttitta F, Pellegrini S, Bertacca G, Callahan R. Reduced expression of INT-6/eIF3-p48 in human tumors. Int J Oncol. 2001;18:175–179. doi: 10.3892/ijo.18.1.175. [DOI] [PubMed] [Google Scholar]
- 75.Shi J, Kahle A, Hershey JW, Honchak BM, Warneke JA, Leong SP, Nelson MA. Decreased expression of eukaryotic initiation factor 3f deregulates translation and apoptosis in tumor cells. Oncogene. 2006;25:4923–4936. doi: 10.1038/sj.onc.1209495. [DOI] [PubMed] [Google Scholar]
- 76.Saramaki O, Willi N, Bratt O, Gasser TC, Koivisto P, Nupponen NN, Bubendorf L, Visakorpi T. Amplification of EIF3S3 gene is associated with advanced stage in prostate cancer. Am J Pathol. 2001;159:2089–2094. doi: 10.1016/S0002-9440(10)63060-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rauch J, Ahlemann M, Schaffrik M, Mack B, Ertongur S, Andratschke M, Zeidler R, Lang S, Gires O. Allogenic antibody-mediated identification of head and neck cancer antigens. Biochem Biophys Res Commun. 2004;323:156–162. doi: 10.1016/j.bbrc.2004.08.071. [DOI] [PubMed] [Google Scholar]
- 78.Ahlemann M, Zeidler R, Lang S, Mack B, Munz M, Gires O. Carcinoma-associated eIF3i overexpression facilitates mTOR-dependent growth transformation. Mol Carcinog. 2006;45:957–967. doi: 10.1002/mc.20269. [DOI] [PubMed] [Google Scholar]
- 79.Eberle J, Krasagakis K, Orfanos CE. Translation initiation factor eIF-4A1 mRNA is consistently overexpressed in human melanoma cells in vitro. Int J Cancer. 1997;71:396–401. doi: 10.1002/(sici)1097-0215(19970502)71:3<396::aid-ijc16>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 80.Shuda M, Kondoh N, Tanaka K, Ryo A, Wakatsuki T, Hada A, Goseki N, Igari T, Hatsuse K, Aihara T, et al. Enhanced expression of translation factor mRNAs in hepatocellular carcinoma. Anticancer Res. 2000;20:2489–2494. [PubMed] [Google Scholar]
- 81.Brass N, Heckel D, Sahin U, Pfreundschuh M, Sybrecht GW, Meese E. Translation initiation factor eIF-4gamma is encoded by an amplified gene and induces an immune response in squamous cell lung carcinoma. Hum Mol Genet. 1997;6:33–39. doi: 10.1093/hmg/6.1.33. [DOI] [PubMed] [Google Scholar]
- 82.Bauer C, Brass N, Diesinger I, Kayser K, Grasser FA, Meese E. Overexpression of the eukaryotic translation initiation factor 4G (eIF4G-1) in squamous cell lung carcinoma. Int J Cancer. 2002;98:181–185. doi: 10.1002/ijc.10180. [DOI] [PubMed] [Google Scholar]
- 83.Balabanov S, Gontarewicz A, Ziegler P, Hartmann U, Kammer W, Copland M, Brassat U, Priemer M, Hauber I, Wilhelm T, et al. Hypusination of eukaryotic initiation factor 5A (eIF5A): a novel therapeutic target in BCR-ABL-positive leukemias identified by a proteomics approach. Blood. 2007;109:1701–1711. doi: 10.1182/blood-2005-03-037648. [DOI] [PubMed] [Google Scholar]
- 84.Guan XY, Sham JS, Tang TC, Fang Y, Huo KK, Yang JM. Isolation of a novel candidate oncogene within a frequently amplified region at 3q26 in ovarian cancer. Cancer Res. 2001;61:3806–3809. [PubMed] [Google Scholar]
- 85.Harris MN, Ozpolat B, Abdi F, Gu S, Legler A, Mawuenyega KG, Tirado-Gomez M, Lopez-Berestein G, Chen X. Comparative proteomic analysis of all-trans-retinoic acid treatment reveals systematic posttranscriptional control mechanisms in acute promyelocytic leukemia. Blood. 2004;104:1314–1323. doi: 10.1182/blood-2004-01-0046. [DOI] [PubMed] [Google Scholar]
- 86.Sanvito F, Vivoli F, Gambini S, Santambrogio G, Catena M, Viale E, Veglia F, Donadini A, Biffo S, Marchisio PC. Expression of a highly conserved protein, p27BBP, during the progression of human colorectal cancer. Cancer Res. 2000;60:510–516. [PubMed] [Google Scholar]
- 87.Nakamura J, Aoyagi S, Nanchi I, Nakatsuka S, Hirata E, Shibata S, Fukuda M, Yamamoto Y, Fukuda I, Tatsumi N, et al. Overexpression of eukaryotic elongation factor eEF2 in gastrointestinal cancers and its involvement in G2/M progression in the cell cycle. Int J Oncol. 2009;34:1181–1189. [PubMed] [Google Scholar]
- 88.Anand N, Murthy S, Amann G, Wernick M, Porter LA, Cukier IH, Collins C, Gray JW, Diebold J, Demetrick DJ, et al. Protein elongation factor EEF1A2 is a putative oncogene in ovarian cancer. Nat Genet. 2002;31:301–305. doi: 10.1038/ng904. [DOI] [PubMed] [Google Scholar]
- 89.Lee JM. The role of protein elongation factor eEF1A2 in ovarian cancer. Reprod Biol Endocrinol. 2003;1:69. doi: 10.1186/1477-7827-1-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ma YL, Peng JY, Zhang P, Huang L, Liu WJ, Shen TY, Chen HQ, Zhou YK, Zhang M, Chu ZX, et al. Heterogeneous nuclear ribonucleoprotein A1 is identified as a potential biomarker for colorectal cancer based on differential proteomics technology. J Proteome Res. 2009;8:4525–4535. doi: 10.1021/pr900365e. [DOI] [PubMed] [Google Scholar]
- 91.Zhang T, Huang XH, Dong L, Hu D, Ge C, Zhan YQ, Xu WX, Yu M, Li W, Wang X, et al. PCBP-1 regulates alternative splicing of the CD44 gene and inhibits invasion in human hepatoma cell line HepG2 cells. Mol Cancer. 2010;9:72. doi: 10.1186/1476-4598-9-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang H, Vardy LA, Tan CP, Loo JM, Guo K, Li J, Lim SG, Zhou J, Chng WJ, Ng SB, et al. PCBP1 suppresses the translation of metastasis-associated PRL-3 phosphatase. Cancer Cell. 2010;18:52–62. doi: 10.1016/j.ccr.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 93.Roychoudhury P, Paul RR, Chowdhury R, Chaudhuri K. HnRNP E2 is downregulated in human oral cancer cells and the overexpression of hnRNP E2 induces apoptosis. Mol Carcinog. 2007;46:198–207. doi: 10.1002/mc.20265. [DOI] [PubMed] [Google Scholar]
- 94.Trotta R, Vignudelli T, Candini O, Intine RV, Pecorari L, Guerzoni C, Santilli G, Byrom MW, Goldoni S, Ford LP, et al. BCR/ABL activates mdm2 mRNA translation via the La antigen. Cancer Cell. 2003;3:145–160. doi: 10.1016/s1535-6108(03)00020-5. [DOI] [PubMed] [Google Scholar]
- 95.Jin W, McCutcheon IE, Fuller GN, Huang ES, Cote GJ. Fibroblast growth factor receptor-1 alpha-exon exclusion and polypyrimidine tract-binding protein in glioblastoma multiforme tumors. Cancer Res. 2000;60:1221–1224. [PubMed] [Google Scholar]
- 96.Stratford AL, Habibi G, Astanehe A, Jiang H, Hu K, Park E, Shadeo A, Buys TP, Lam W, Pugh T, et al. Epidermal growth factor receptor (EGFR) is transcriptionally induced by the Y-box binding protein-1 (YB-1) and can be inhibited with Iressa in basal-like breast cancer, providing a potential target for therapy. Breast Cancer Res. 2007;9:R61. doi: 10.1186/bcr1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huang R, Wu T, Xu L, Liu A, Ji Y, Hu G. Upstream binding factor up-regulated in hepatocellular carcinoma is related to the survival and cisplatin-sensitivity of cancer cells. FASEB J. 2002;16:293–301. doi: 10.1096/fj.01-0687com. [DOI] [PubMed] [Google Scholar]
- 98.Killian A, Sarafan-Vasseur N, Sesboue R, Le Pessot F, Blanchard F, Lamy A, Laurent M, Flaman JM, Frebourg T. Contribution of the BOP1 gene, located on 8q24, to colorectal tumorigenesis. Genes Chromosomes Cancer. 2006;45:874–881. doi: 10.1002/gcc.20351. [DOI] [PubMed] [Google Scholar]
- 99.Yun JP, Miao J, Chen GG, Tian QH, Zhang CQ, Xiang J, Fu J, Lai PB. Increased expression of nucleophosmin/B23 in hepatocellular carcinoma and correlation with clinicopathological parameters. Br J Cancer. 2007;96:477–484. doi: 10.1038/sj.bjc.6603574. [DOI] [PMC free article] [PubMed] [Google Scholar]