Summary
Human Serum Albumin (HSA) accounts for 60% of the total protein in blood serum and it is the most widely used intravenous protein in a number of human therapies. HSA, however, is currently extracted only from blood because of a lack of commercially feasible recombinant expression systems. HSA is highly susceptible to proteolytic degradation in recombinant systems and is expensive to purify. Expression of HSA in transgenic chloroplasts using Shine-Dalgarno sequence (SD), which usually facilitates hyper-expression of transgenes, resulted only in 0.02% HSA in total protein (tp). Modification of HSA regulatory sequences using chloroplast untranslated regions (UTRs) resulted in hyper-expression of HSA (up to 11.1% tp), compensating for excessive proteolytic degradation. This is the highest expression of a pharmaceutical protein in transgenic plants and 500-fold greater than previous reports on HSA expression in transgenic leaves. Electron micrographs of immunogold labelled transgenic chloroplasts revealed HSA inclusion bodies, which provided a simple method for purification from other cellular proteins. HSA inclusion bodies could be readily solubilized to obtain a monomeric form using appropriate reagents. The regulatory elements used in this study should serve as a model system for enhancing expression of foreign proteins that are highly susceptible to proteolytic degradation and provide advantages in purification, when inclusion bodies are formed.
Keywords: chloroplast genetic engineering, biopharmaceuticals, genetically modified crops, molecular farming, recombinant human blood proteins
Introduction
Availability of recombinant human proteins has revolutionized the use of therapeutically valuable proteins in clinical medicine. Plants offer a suitable alternative to microbial or animal expression of biopharmaceutical proteins because of their inexpensive production costs and absence of human pathogens. However, there are some limitations. In particular, expression of human proteins in nuclear transgenic plants has been disappointingly low, e.g. human serum albumin 0.02% of total soluble protein (tsp), human Interferon-β 0.000017% of fresh weight, human epidermal growth factor 0.001% of tsp and erythropoietin 0.0026% of tsp (Daniell et al., 2001d). Therefore, it is important to increase levels of expression in order to exploit plant production of pharmacologically important proteins.
As an alternative to nuclear expression, the chloroplast transgenic approach has been developed as an effective tool for the expression of biopharmaceutical proteins in plants (Daniell and Dhingra, 2002; Daniell et al., 2001a,b; DeGray et al., 2001; Guda et al., 2000; Staub et al., 2000). After the first demonstration of a protein based polymer expression with varied medical applications (Guda et al., 2000), transgenic chloroplasts have been shown to express very small antimicrobial peptides without fusion proteins (DeGray et al., 2001), assemble functional oligomers with disulphide bonds of the cholera toxin β-subunit (Daniell et al., 2001b), and express a monoclonal antibody with coordinated expression and assembly of heavy and light chain with proper folding and formation of disulphide bridges (Daniell et al., 2001a), suggesting that adequate redox environment or required chaperonins are present within chloroplasts. Expression of functional human somatotropin in transgenic tobacco chloroplasts established that chloroplasts are capable of proper folding of human proteins with disulphide bonds (Staub et al., 2000). The ability to express multiple genes in a single transformation event (Daniell and Dhingra, 2002; De Cosa et al., 2001), accumulation of exceptionally large quantities of foreign proteins (De Cosa et al., 2001), successful engineering of tomato chromoplasts for high level transgene expression in fruits (Ruf et al., 2001), coupled to hyper-expression of vaccine antigens (Daniell et al., 2001b), and the use of plant derived antibiotic free selectable markers (Daniell et al., 2001c), augur well for oral delivery of edible vaccines and biopharmaceuticals that are currently beyond the reach of those who need them most. In addition, chloroplast genetic engineering is an environmentally friendly approach, offering containment of transgenes and a solution to gene silencing and position effect encountered in nuclear transgenic plants (Bogorad, 2000; Daniell and Dhingra, 2002; Daniell et al., 2002; Daniell, 2002).
HSA is the most widely used intravenous protein and is prescribed in multigram quantities to replace blood volume in trauma and in various other clinical situations (Peters, 1995). HSA is a monomeric globular prepro-protein whose mature form consists of a single polypeptide chain of 585 amino acids (66.5 kDa with 17 disulphide bonds). The annual world need exceeds 500 tons, representing a market value of more than $1.5 billion. To date, albumin has been produced primarily by the fractionation of blood serum. Lack of glycosylation facilitates production of functional HSA in prokaryotic systems. Although the HSA gene and cDNA have been expressed in a wide variety of microbial systems, including E. coli (Latta et al., 1987), Bacillus subtilis (Saunders et al., 1987), Saccharomyces cerevisiae (Quirk et al., 1989), Kluyveromyces (Fleer et al., 1991) or Pichia pastoris (Ohtani et al., 1998), no system is yet commercially feasible. Sijmons et al. (1990) made the first reported attempt to express HSA in transgenic plants, but very low expression levels were attained (0.02% tsp). HSA could not be detected if expressed in the cytoplasm, suggesting that the protein is not stable in this compartment, due to high susceptibility to proteolytic degradation. A 10-fold increase in HSA accumulation has been reported recently by nuclear transformation of potato plants and targeting the HSA to the tuber apoplast (Farran et al., 2002). Estimates by industry, however, suggest that the cost-effective yield for pharmaceutical production is 0.1 mg of HSA per gram of fresh weight (Farran et al., 2002).
In addition, good recombinant systems are still not available for many human proteins that are expensive to purify or highly susceptible to proteolytic degradation. It is known that traditional purification of biopharmaceuticals using columns accounts for 30% of the production cost and 70% of the set up cost (Petrides et al., 1995). Proteolytic degradation is another serious concern for industrial bioprocessing. The increasing production of proteins in heterologous hosts through the use of recombinant DNA technology has brought this problem into focus; heterologous proteins appear to be more prone to proteolysis (Enfors, 1992). Recombinant proteins are often regarded by a cell as foreign and therefore degraded much faster than most endogenous proteins (Rozkov et al., 2000). Proteolytic stability of recombinant proteins is a significant factor influencing the final yield.
This study attempts to develop a more efficient method of recombinant HSA production, which may be used as a model system to enrich or purify biopharmaceutical proteins from transgenic plants, which are highly susceptible to proteolytic degradation.
Results and discussion
Two chloroplast transformation vectors were designed with different 5′ regulatory sequences to direct HSA expression and maximize protein accumulation in transgenic chloroplasts. Basic pLD vector, developed in this laboratory for chloroplast transformation, was used (Daniell et al., 1998; Daniell et al., 2001b; De Cosa et al., 2001; Guda et al., 2000; Kota et al., 1999). In the plasmid pLDAsdHSA (Figure 1a), the aadA gene, which confers spectinomycin resistance, and the HSA gene are expressed as a polycistron from the plastid Prrn promoter. The Shine-Dalgarno (SD) consensus sequence GGAGG was placed upstream of both genes. High levels of foreign protein expression in chloroplasts (3–21% of tsp) have been shown for different proteins using this 5′ sequence (Daniell et al., 2001b; DeGray et al., 2001; Kota et al., 1999). In the pLDApsbAHSA vector (Figure 1a), the 204 bp tobacco chloroplast DNA fragment containing the promoter and the psbA 5′ UTR was inserted immediately upstream of the HSA coding sequence and downstream of the aadA gene. It is well known that foreign genes under the control of the psbA promoter and untranslated region are expressed at very high levels (Daniell et al., 1990). This enhancement of translation may be due to elements in the 5′UTR (Eibl et al., 1999). Vectors were bombarded into tobacco leaves as described previously (Daniell, 1997) and, after 5 weeks, several primary shoots appeared from each bombarded leaf as a result of independent transformation events. Putative transformed shoots were identified by growth on 500 μg/mL of spectinomycin.
Figure 1.
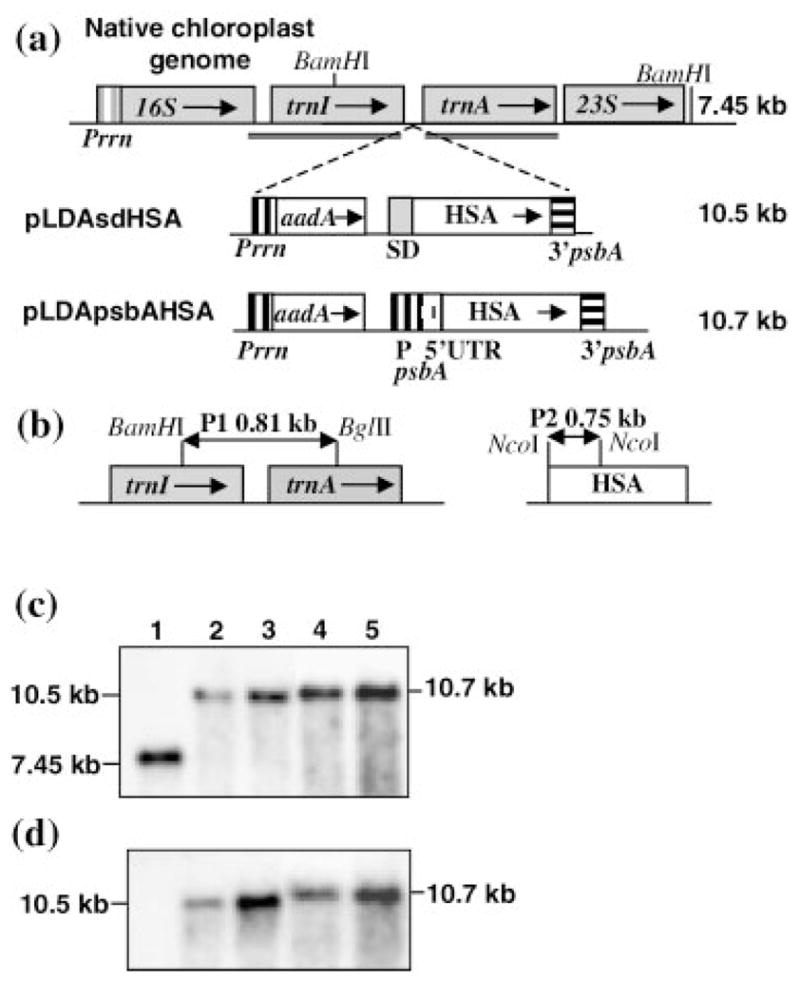
Integration of transgene cassettes into the chloroplast genome and study of homoplasmy. (a) Regions for homologous recombination are underlined in the native chloroplast genome. HSA is driven in all cassettes by the Prrn promoter upstream of the aadA gene for spectinomycin resistance with additional promoters and control elements as described in the text. Arrows within boxes show the direction of transcription. Numbers to the right indicate the predicted hybridizing fragments when total DNA digested with BamHI is probed with probe P1. (b) The 0.81 kb fragment (P1) flanking the cassette and 0.75 kb fragment containing HSA coding region (P2) were used as probes for the Southern blot analysis. (c, d) Southern blot analysis. 1: untransformed DNA; DNA from plants transformed with: 2,3: pLDAsdHSA; 4,5: pLDApsbAHSA. Plants for the first (T0) and second (T1) generation were analysed. 2,4: T0 generation. 3,5: T1 generation. Blots were probed with P1 (c) and P2 (d). AadA: aminoglycoside 3′-adenylyl transferase; kb: kilobases; P: promoter; Prrn: 16SrRNA promoter; SD: Shine-Dalgarno.
Integration of the foreign gene cassettes into the chloroplast genome was confirmed by PCR screening of primary shoots. The strategy employed lands one primer on the native chloroplast genome adjacent to the point of integration and the second primer on the aadA gene. This PCR product can not be obtained in nuclear transgenic plants or spontaneous mutants, thus both possibilities could be eliminated. It was found that 90% of total shoots obtained were true chloroplast transformants. Confirmed transformants were subjected to a second round of spectinomycin selection to achieve homoplasmy. They were rooted in the presence of spectinomycin and then transferred to pots for further characterization. Southern blot analysis was performed to select homoplasmic T0 lines and confirm stable maintenance of integrated transgenes in the T1 generation (Figure 1b–d). The flanking region probe (P1) identified a 7.45 kb fragment in the untransformed control plant, as expected (Figure 1c). In the chloroplast transgenic lines, only transformed genome copies are observed as evidenced by the 10.5 and 10.7 kb hybridizing fragments for pLDAsdHSA and pLDApsbAHSA transgenic lines, respectively. To confirm that the 10.5 and 10.7 kb fragments contained the HSA gene, the same blot was reprobed with the HSA P2 probe. As expected, hybridization was detected only in the chloroplast transgenic lines (Figure 1d). Absence of other hybridizing fragments eliminates nuclear and chloroplast integration events in the same transgenic line.
HSA quantities in transgenic tobacco chloroplasts were determined by ELISA. More than a 360-fold difference in HSA accumulation was observed between plants transformed with the two different vectors (Figure 3a): 0.02% vs. 7.2% tp in pLDAsdHSA and pLDApsbAHSA transgenic lines, respectively. Chloroplast constructs with the SD sequence have been demonstrated to direct CTB expression very efficiently (up to 4% of tsp; Daniell et al., 2001b). Similar constructs, but inserted in other areas of the plastid genome, have also been successful (3–21% tsp; DeGray et al., 2001; Kota et al., 1999), demonstrating that high protein expression levels can be achieved by using this construct in an operon with a SD sequence. Thus, low levels of HSA expression in the pLDAsdHSA transgenic plants could not be due to the effect of regulatory signals in the construct. Differences in the amounts of HSA could be due to post-transcriptional, translational or post-translational effects. To study differences in HSA expression, transcript abundance was examined by Northern blots, which were performed using the 3′psbA region as the probe (Figure 2). The 5′psbA/HSA monocistron transcript is much more abundant than the aadA/SD/HSA dicistron, but such differences do not show a linear correlation with the 360-fold difference in HSA accumulation between both transgenic lines. Such a lack of correlation between transcript abundance and protein accumulation has been reported from several laboratories when the psbA 5′UTR is used (Mayfield et al., 1995; Staub and Maliga, 1993, 1994), suggesting an important role of the psbA 5′UTR in enhancement of translation. Eibl et al. (1999) also showed that deletion of the terminal sequences of the psbA 5′UTR decreased the ability of the UTR to enhance translation. Thus, efficient translation in the pLDApsbAHSA transgenic line might be an important factor in establishing high levels of HSA accumulation.
Figure 3.
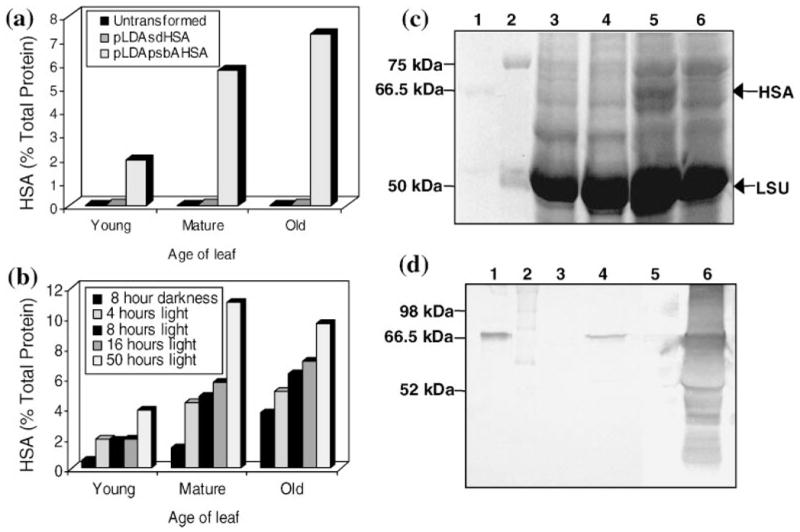
Analysis of HSA accumulation in transgenic chloroplasts. (a) ELISA of HSA accumulation in leaves of potted plants at different stages of development. Samples were collected from untransformed plants or transformed with pLDAsdHSA or pLDApsbAHSA. Expression levels are indicated as a percentage of total protein. (b) Study after different hours of illumination. Samples of leaves were collected from potted plants transformed with pLDApsbAHSA after the 8-h dark period or at indicated hours in the light. (c) Coomassie stained gel to study HSA accumulation in tobacco leaves of potted plants. Total protein extracts were loaded in the gel. 1: 500 ng pure HSA; 2: molecular weight marker; 3: untransformed plant; transformed with 4: pLDAsdHSA; 5: pLDApsbAHSA after 8 h of illumination; 6: pLDApsbAHSA after 8 h of darkness. Between 40 and 50 μg of plant protein were loaded per well. The positions of HSA and RuBisCO large subunit (LSU) are marked. (d) Colorimetric immunoblot detection of tobacco protein extracts from mature leaves in potted plants. Total protein extracts were loaded in the gel. 1: 40 ng pure HSA; 2: molecular weight marker; 3,5: untransformed plant extract; 4: pLDAsdHSA plant extract; 6: pLDApsbAHSA plant extract. Between 40 and 50 μg of plant protein were loaded per well. kDa: kiloDalton; LSU: RuBisCO large subunit.
Figure 2.
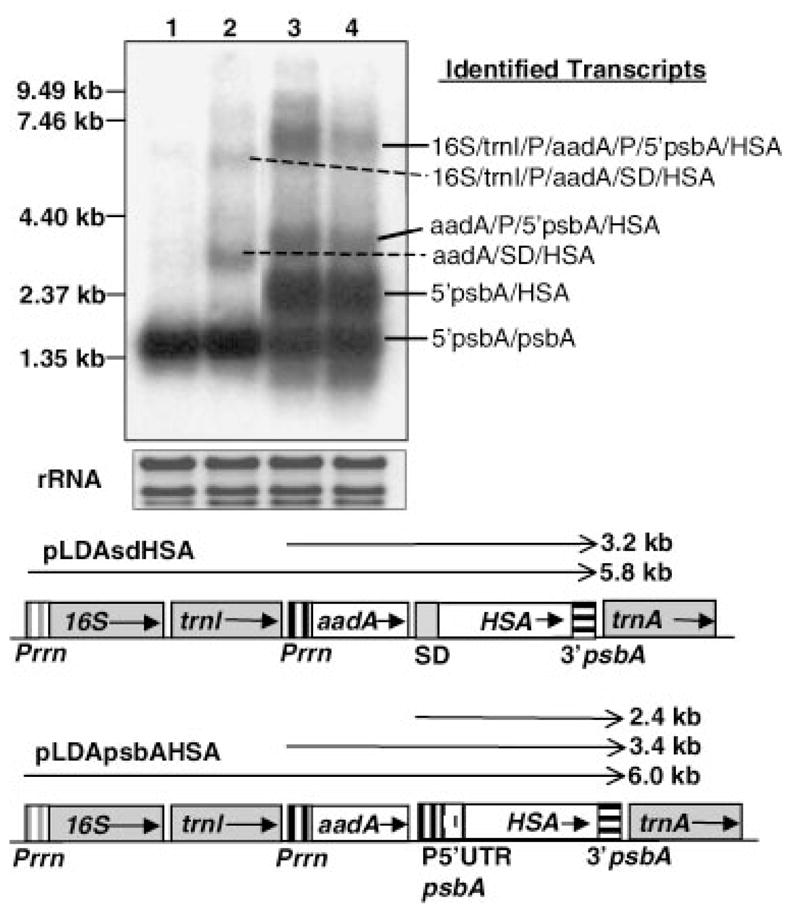
Transcription patterns of transgenic plants. A Northern blot analysis was performed with total RNA extracted from leaves of potted plants. The 3′ of the psbA gene was used as probe. 1: untransformed plant; 2: transformed with pLDAsdHSA; 3: transformed with pLDApsbAHSA after illumination or 4: in the dark. Ethidium bromide-stained rRNA was used to assess loading. Identified transcripts are indicated to the right. A scheme of transcription patterns expected for the different cassettes integrated into the chloroplast genome is shown at the bottom of the figure. Horizontal arrows above genes show anticipated transcripts. Arrows within boxes show the orientation of genes within the chloroplast genome. Read through transcripts are not shown in this figure. rRNA: ribosomal RNA.
There are several studies demonstrating that psbA 5′UTR confers light-dependent translation not only to the psbA gene (Zerges, 2000) but also to other heterologous proteins (Eibl et al., 1999; Staub and Maliga, 1993, 1994). Expression of HSA under the psbA 5′UTR control is therefore expected to be light dependent. Changes in HSA accumulation after different periods of illumination were monitored by ELISA (Figure 3b). HSA quantity was observed to be maximum up to 50 h of continuous illumination (11.1% of tp) in mature leaves and a 2–4-fold decrease was observed after the 8 h dark period. Such differences in HSA accumulation were so pronounced that it was detected by staining gels with Coomassie Brilliant Blue (Figure 3c). Staub and Maliga (1993, 1994) and Eibl et al. (1999) showed that although translation is arrested in the dark, the 5′psbA/uidA mRNA turnover was very low. This observation was confirmed for HSA by Northern blot analysis, which showed no major differences between light and dark amount of 5′psbA/HSA transcripts (Figure 2, lanes 3, 4). Therefore, differences in HSA accumulation between dark and light could not be due to differences in the rate of transcription or transcript stability, but due to the arrest of translation in the dark and the turnover of HSA in the chloroplast.
Proteins from transformed plants were separated to study the pattern of HSA accumulation within transgenic chloroplasts. Western blots confirmed differences in HSA quantities among transgenic lines (Figure 3d). In pLDApsbAHSA transgenic lines, HSA is partially solubilized with the standard buffer used for total protein extraction. This observation suggests formation of HSA aggregates inside transgenic chloroplasts in the pLDApsbAHSA transformants. Electron microscopy and immunogold labelling therefore were performed in transformed and untransformed plants to further investigate this. As expected, electron micrographs of leaf tissues showed formation of large aggregates or inclusion bodies within transgenic chloroplasts of pLDApsbAHSA mature transformed plants (Figure 4b–d). It is interesting to note that chloroplasts containing inclusion bodies increased in size to accommodate large accumulation of HSA (compare Figures 4a,d). However, the phenotype of these plants appeared normal (Figure 5). The amount of HSA in pLDAsdHSA transgenic chloroplasts was so low that it was not possible to detect immunogold labelling above the background. No significant changes in chloroplast size were observed in these plants.
Figure 4.
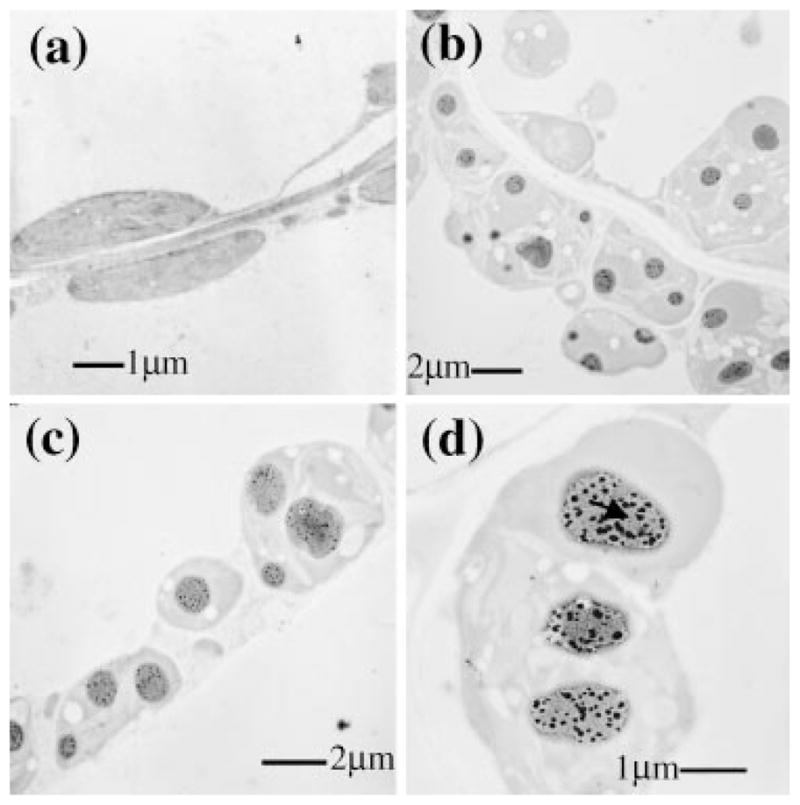
Study of HSA accumulation into inclusion bodies. (a–d) Electron micrographs of immunogold labelled tissues from untransformed (a) and transformed mature leaves with the chloroplast vector pLDApsbAHSA (b–d). Note presence of inclusion bodies (b–d) marked with an arrow in (d). Scale bars indicate μm. Magnifications are a × 10 000; b × 5000; c × 6300; d × 12 500.
Figure 5.

Plant T1 phenotypes. 1,2: untransformed plants; 3: plant transformed with pLDAsdHSA; 4: plant transformed with pLDApsbAHSA.
Inclusion bodies have been often observed in the cytosol of prokaryotes and eukaryotes when heterologous proteins are overexpressed. The occurrence of this feature in the chloroplast was first reported by Ketchner et al. (1995). It is widely known that protein aggregation into inclusion bodies mostly involves intermolecular associations of partially folded intermediates (Mitraki and King, 1989). High protein concentrations usually lead to conditions that frequently exceed the normal solubility limit. Even the most abundant protein in photosynthetic cells, RuBisCO, forms inclusion bodies in some cases. Many autotrophic bacteria and all cyanobacteria package much of the RuBisCO into inclusion bodies actively involved in the fixation of CO2, known as carboxysomes (Shively and English, 1991). Our hypothesis, based on the process of formation of inclusion bodies, is that in contrast to pLDAs-dHSA transgenic lines, HSA synthesized under the psbA 5′UTR forms large aggregates mainly due to the high local concentration of the protein.
Formation of inclusion bodies is one of the strategies for reducing the proteolysis of unstable recombinant proteins (Enfors, 1992). The majority of recombinant proteins studied have been shown to be highly resistant to proteolysis inside inclusion bodies. Although there is protection from proteases within inclusion bodies, some proteolysis can also take place directly on the aggregated protein (Carrio et al., 1999). HSA also appears to be susceptible to some proteolytic degradation within transgenic chloroplasts. However, the net balance between synthesis and degradation is highly favourable, especially after several hours of continuous illumination.
Properly folded HSA can be recovered from inclusion bodies after denaturation for complete solubilization and in vitro refolding. Proper refolding of HSA from inclusion bodies is a routine procedure that has been previously demonstrated in several studies with E. coli (Latta et al. (1987) and Saccharomyces cerevisiae (Dodsworth et al., 1996; Quirk et al., 1989). In these cases, human and recombinant refolded HSA were compared and it was shown that the two proteins were structurally equivalent, demonstrating that HSA may be recovered from inclusion bodies and properly folded. Following the guidelines from these protocols, HSA was extracted from transgenic chloroplasts. Figure 6a shows a silver stained SDS-PAGE gel in which HSA inclusion bodies could be separated from the soluble fraction (lane 3), where most of the cellular proteins are found. After solubilization of inclusion bodies and subsequent refolding, HSA could be completely converted into monomeric forms (Figure 6a, lane 5; Figure 6b, lane 5). Our estimations of HSA yields at the end of the protocol are about 20% of the initial quantities in leaves, although the reported protocol has been performed at the laboratory scale and may be further optimized for industrial production. Expression of HSA in transgenic plants has been estimated to be cost effective with levels of expression as low as 0.1 mg HSA/g fresh weight (Farran et al., 2002). The recoveries after solubilizing the inclusion bodies and refolding the HSA are about 0.25 mg HSA/g fresh weight (excluding soluble HSA in transgenic chloroplasts), which exceeds cost effective estimations of pharmaceutical industries.
Figure 6.
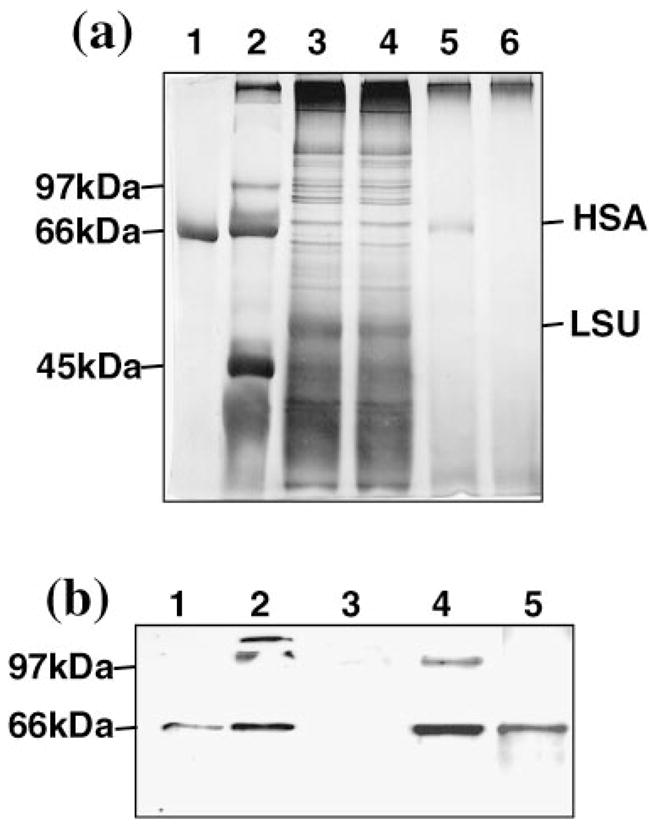
HSA extraction from inclusion bodies. (a) Silver stained SDS-PAGE gel showing 1: 500 ng pure HSA; 2: molecular weight marker; soluble fraction obtained after centrifugation of pLDApsbAHSA transformed plant extract (lane 3) or untransformed plant extract (lane 4); 5: HSA after solubilization from the pellet; 6: proteins from untransformed plant, which followed the same process as the proteins of lane 5. Amounts of protein loaded per well were 10 μg in lanes 3 and 4, 550 ng in lane 5 and 450 ng in lane 6. (b) Chemiluminiscent immunoblot detection of protein extracts. 1: 40 ng pure HSA; 2: HSA from a plant transformed with pLDApsbAHSA during the solubilization process, showing mono, di and trimeric forms; 3: proteins from an untransformed plant that followed the same process as the proteins for lane 2; 4: same HSA from lane 2 but in a more advanced stage of solubilization; 5: completely monomerized HSA after the end of the solubilization treatment (the sample of this lane corresponds with lane 5 in (a)).
One of the primary goals of this study was to develop a more efficient expression system for human serum albumin, an important human therapeutic protein that is highly susceptible to degradation. Expression of HSA in mature plants under the translational control of SD sequence resulted in very low levels of HSA accumulation, probably due to excessive proteolytic degradation and poor rates of translation. However, when expressed under the control of psbA promoter and 5′UTR, up to a 500-fold increase in HSA accumulation was observed in mature plants compared to other regulatory sequences tested. HSA was observed to form large inclusion bodies, resulting even in a noticeable increase in the size of transgenic chloroplasts and presumably offering protection to HSA from proteolytic degradation. Inclusion bodies facilitated purification of HSA from other cellular proteins. The HSA molecule has a chemical and structural function rather than an enzymatic activity, therefore complex studies are necessary to fully demonstrate the functionality of the molecule (see Dodsworth et al., 1996; Ohtani et al., 1998; Petersen et al., 2000; Tarelli et al., 1998; Watanabe et al., 2001a,b). Such functional studies using in vitro refolded HSA are in progress.
Experimental procedures
Chloroplast expression vectors
pLDAsdHSA was constructed by inserting the HSA 1.8 kb EcoRI/NotI fragment into the multiple cloning site of the pLD vector (Daniell et al., 1998; Daniell et al., 2001b; De Cosa et al., 2001; Guda et al., 2000; Kota et al., 1999). This fragment contains the mature HSA coding sequence preceded by a Shine-Dalgarno (GGAGG) and it has an ATG as the initiation codon. These sequences were introduced by using the primer: 5′-GGAGGCAACCATGGATGCACACAAGAGTGAAGG-3′. For the pLDApsbAHSA vector, the 204 bp sequence including the promoter and the psbA 5′UTR, was amplified by PCR using tobacco DNA as template. The following primers were used: 5′-CCGTCGACGTAGAGAAGTCCGTATT-3′ and 5′-GCCCATGGTAAAATCTTGGTTTATTTA-3′. The fusion with the HSA gene was made at the NcoI site placed at the 3′ end of the psbA 5′UTR and then inserted into the pLD vector as a EcoRI/NotI fragment. Before proceeding with the bombardment, vectors were tested by Western blot analysis in E. coli.
Bombardment and regeneration
Sterile tobacco (cv. Petit Havana) leaves were bombarded using the Bio-Rad PDS-1000/He biolistic device as described previously (Daniell, 1997). Bombarded leaves were subjected to two rounds of selection on the RMOP medium containing 500 μg/mL of spectinomycin to regenerate transformants (Daniell, 1997). After regeneration, plants were rooted on 500 μg/mL of spectinomycin (Daniell et al., 2001b) and transferred to pots in growth chambers. Photoperiod was 16 h light and 8 h dark.
PCR and Southern blot analysis
PCR was used to analyse integration of different cassettes in the transformed plants as described (Daniell et al., 2001b,c; De Cosa et al., 2001; Kota et al., 1999). For Southern blot analysis, total DNA was extracted from leaves of transformed and untransformed plants (Qiagen Dneasy Kit). Total DNA (5 μg) was digested with BamHI, electrophoresed on 0.7% agarose gels and transferred to nylon membranes (Duralon-UV Stratagene). The template for probing flanking sequences was a 0.81 kb BglII/BamHI fragment and for HSA a 0.75 kb NcoI fragment. The probes were labelled with 32P-dCTP using the oligolabelling procedure (Ready To Go, Amersham). Probes were hybridized to the membranes following the QUICK-HYB protocol (Duralon-UV, Stratagene).
Northern blot analysis
Total RNA was extracted from leaves of transformed and untransformed plants (Rneasy Plant Kit, Qiagen). RNA 2.5 μg was electrophoresed on 1.2% agarose/formaldehyde gels and then transferred to nylon membranes (Stratagene). A 0.21 kb XbaI/PstI fragment of the 3′psbA gene was used as probe and labelled with 32P-dCTP using the oligolabelling procedure (Amersham).
HSA quantification
The ELISA Human Albumin Quantification Kit (Bethyl Laboratories) was used. Transformed and untransformed leaves (100 mg) from potted plants grown under a 16 h photoperiod were ground in liquid nitrogen, resuspended in 700 μL of 50 mM NaOH and analysed following the manufacturer’s protocol. Transgenic leaf extracts were diluted to fit in the linear range of the provided HSA standard. Absorbance was read at 450 nm. The DC protein assay (Bio-Rad) was used to determine total solubilized protein.
SDS-PAGE and immunoblot analysis
Transformed and untransformed leaves (100 mg) were ground in liquid nitrogen and resuspended in 200 μL of protein extraction buffer (200 mM Tris-HCl pH 8.0, 100 mM NaCl, 400 mM Sucrose, 14 mM βME, 0.05% Tween20, 0.1% SDS, 10 mM EDTA, 2 mM PMSF). Leaf extracts were boiled in sample buffer (Bio-Rad) and electrophoresed in a 10% polyacrylamide gel. Separated proteins were stained with Coomassie Brilliant Blue G-250 or transferred to a nitrocellulose membrane for immunoblotting. The primary antibody (rabbit anti-HSA, Nordic Immunology) was used at 1 : 10 000 dilution, and the secondary antibody (alkaline phosphatase conjugated mouse antirabbit, Sigma or goat antirabbit HRP conjugated, Southern Biotechnology) at 1 : 15 000. Alkaline phosphatase colour development reagents, BCIP/NBT, in AP Color Development Buffer (Bio-Rad) or the ECL kit (Amersham) were used for detection.
Solubilization of inclusion bodies
Soluble proteins were removed with a first extraction in 0.2 M NaCl, 25 mM Tris-HCl pH 7.4, 2 mM PMSF and 0.1% Triton X-100. After centrifugation for 60 min at 20 000 g, the pellet was solubilized for 16 h at 4 °C in 6 M Gu-HCl, 0.1 M βME and 0.25 mM Tris-HCl pH 7.4. After centrifugation for 60 min at 20 000 g, the supernatant was then slowly diluted 100-fold in 100 mM NaCl, 50 mM Tris-HCl pH 8.5 and 1 mM EDTA for 24 h at 4 °C. Fractions were electrophoresed in a SDS-PAGE 10% gel and silver stained with Bio-Rad reagents and protocol.
Transmission electron microscopy and immunogold labelling
Seedlings and mature leaves from untransformed and transgenic plants were analysed. Fixation and immunogold labelled electron microscopy were performed as described by Vrekleij and Leunissen (1989). Sections were first blocked, incubated for 1 h with a goat antihuman albumin polyclonal antibody (Nordic Immunology; dilution range from 1 : 1000 to 1 : 10 000) and then incubated for 2 h with a rabbit antig-oat IgG secondary antibody conjugate to 10 nM gold diluted 1 : 40 in blocking solution. Sections were examined in a Zeiss EM 10 transmission electron microscope at 60 kV.
Acknowledgments
This study was supported in part by grants from NIH RO1 GM 63879 and Chlorgen Inc. to H.D., and ‘Dirección General de Industria, Gobierno de Navarra’ (Spain) to A.M.C.
References
- Bogorad L. Engineering chloroplasts: an alternative site for foreign genes, proteins, reactions and products. Trends Biotechnol. 2000;18:257–263. doi: 10.1016/s0167-7799(00)01444-x. [DOI] [PubMed] [Google Scholar]
- Carrio MM, Corchero JL, Villaverde A. Proteolytic digestion of bacterial inclusion body proteins during dynamic transition between soluble and insoluble forms. Biochim Biophys Act. 1999;1434:170–176. doi: 10.1016/s0167-4838(99)00177-6. [DOI] [PubMed] [Google Scholar]
- Daniell H. Transformation and foreign gene expression in plants mediated by microprojectile bombardment. Meth Mol Biol. 1997;62:463–489. doi: 10.1385/0-89603-480-1:463. [DOI] [PubMed] [Google Scholar]
- Daniell H. Molecular strategies for gene containment in transgenic crops. Nat Biotechnol. 2002;20:581–586. doi: 10.1038/nbt0602-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Datta R, Varma S, Gray S, Lee SB. Containment of herbicide resistance through genetic engineering of the chloroplast genome. Nat Biotechnol. 1998;16:345–348. doi: 10.1038/nbt0498-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Dhingra A. Multigene engineering: Dawn of an exciting new era in biotechnology. Curr Opin Biotechnol. 2002;13:136–141. doi: 10.1016/s0958-1669(02)00297-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Dhingra A, Fernandez-San Millan A. Chloroplast transgenic approach for the production of antibodies, biopharmaceuticals and edible vaccines. Proceedings of the International Congress on Photosynthesis, S40-04; Brisbane, Australia. Colling wood, Australia: CSIRO Publishing; 2001a. [Google Scholar]
- Daniell H, Khan MS, Allison L. Milestones in chloroplast genetic engineering: an environmentally friendly era in biotechnology. Trends Plant Sci. 2002;7:84–91. doi: 10.1016/s1360-1385(01)02193-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Lee SB, Panchal T, Wiebe PO. Expression of the native cholera toxin B subunit gene and assembly as functional oligomers in transgenic tobacco chloroplasts. J Mol Biol. 2001b;311:1001–1009. doi: 10.1006/jmbi.2001.4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Muthukumar B, Lee SB. Marker free transgenic plants: Engineering the chloroplast genome wihout the use of antibiotic selection. Curr Genet. 2001c;39:109–116. doi: 10.1007/s002940100185. [DOI] [PubMed] [Google Scholar]
- Daniell H, Streatfield S, Wycoff K. Medical molecular farming: production of antibodies, biopharmaceuticals and edible vaccines in plants. Trends Plant Sci. 2001d;6:219–226. doi: 10.1016/S1360-1385(01)01922-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Vivekananda J, Neilsen B, Ye GN, Tewari KK, Sanford JC. Transient foreign gene expression in chloroplasts of cultured tobacco cells after biolistic delivery of chloroplast vectors. Proc Natl Acad Sci USA. 1990;87:88–92. doi: 10.1073/pnas.87.1.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cosa B, Moar W, Lee SB, Miller M, Daniell H. Overexpression of the Bt cry2Aa2 operon in chloroplasts leads to formation of insecticidal crystals. Nat Biotechnol. 2001;19:71–74. doi: 10.1038/83559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGray G, Rajasekaran K, Smith F, Sanford J, Daniell H. Expression of an antimicrobial peptide via the chloroplast genome to control phytopathogenic bacteria and fungi. Plant Physiol. 2001;127:852–862. [PMC free article] [PubMed] [Google Scholar]
- Dodsworth N, Harris R, Denton K, Woodrow P, Wood C, Quirk A. Comparative studies of recombinant human albumin and human serum albumin derived by blood fractionation. Biotechnol Appl Biochem. 1996;24:171–176. [PubMed] [Google Scholar]
- Eibl C, Zou Z, Beck A, Kim M, Mullet J, Koop HU. In vivo analysis of plastid psbA, rbcL and rpl32 UTR elements by chloroplast transformation: tobacco plastid gene expression is controlled by modulation of transcript levels and translation efficiency. Plant J. 1999;19:333–345. doi: 10.1046/j.1365-313x.1999.00543.x. [DOI] [PubMed] [Google Scholar]
- Enfors SO. Control of in vivo proteolysis in the production of recombinant proteins. Trends Biotechnol. 1992;10:310–315. doi: 10.1016/0167-7799(92)90256-u. [DOI] [PubMed] [Google Scholar]
- Farran I, Sanchez-Serrano JJ, Medina JF, Prieto J, Mingo-Castel AM. Targeted expression of human serum albumin to potato tubers. Transgenic Res. 2002;11:337–346. doi: 10.1023/a:1016356510770. [DOI] [PubMed] [Google Scholar]
- Fleer R, Yeh P, Amellal N, Maury I, Fournier A, Bacchetta F, Baduel P, Jung G, Lhote H, Becquart J, Fukuhara H, Mayaux JF. Stable multicopy vectors for high-level secretion of recombinant human serum albumin by Kluyveromyces yeasts. Bio/Technol. 1991;9:968–975. doi: 10.1038/nbt1091-968. [DOI] [PubMed] [Google Scholar]
- Guda C, Lee SB, Daniell H. Stable expression of a biodegradable protein-based polymer in tobacco chloroplasts. Plant Cell Report. 2000;19:257–262. doi: 10.1007/s002990050008. [DOI] [PubMed] [Google Scholar]
- Ketchner SL, Drapier D, Olive J, Gaudriault S, Girard-Bascou J, Wollman FA. Chloroplasts can accomodate inclusion bodies. J Biol Chem. 1995;270:15299–15306. doi: 10.1074/jbc.270.25.15299. [DOI] [PubMed] [Google Scholar]
- Kota M, Daniell H, Varma S, Garczynski F, Gould F, Moar WJ. Overexpression of the Bacillus thuringiensis (Bt) Cry2Aa2 protein in chloroplasts confers resistance to plants against susceptible and Bt-resistant insects. Proc Natl Acad Sci USA. 1999;96:1840–1845. doi: 10.1073/pnas.96.5.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latta L, Knapp M, Sarmientos P, Brefort G, Becquart J, Guerrier L, Jung G, Mayaux J. Synthesis and purification of mature human serum albumin from E. coli. Bio/Technol. 1987;5:1309–1314. [Google Scholar]
- Mayfield SP, Yohn CB, Cohen A, Danon A. Regulation of chloroplast gene expression. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:147–166. [Google Scholar]
- Mitraki A, King J. Protein folding intermediates and inclusion body formation. Bio/Technol. 1989;7:690–697. [Google Scholar]
- Ohtani W, Ohda T, Sumi A, Kobayashi K, Ohmura T. Analysis of Pichia pastoris components in recombinant human serum albumin by immunological assays and by HPLC with pulsed amperometric detection. Anal Chem. 1998;70:425–429. doi: 10.1021/ac970596h. [DOI] [PubMed] [Google Scholar]
- Peters T. All About Albumin: Biochemistry, Genetics and Medical Applications. Chap 6. San Diego, CA: Academic Press; 1995. The Albumin molecule; pp. 251–284. [Google Scholar]
- Petersen CE, Ha C, Harohalli K, Feix JB, Bhagavan NV. A dynamic model for bilirrubin binding to human serum albumin. J Biol Chem. 2000;275:20985–20995. doi: 10.1074/jbc.M001038200. [DOI] [PubMed] [Google Scholar]
- Petrides D, Sapidou E, Calandranis J. Computer aided process analysis and economic evaluation for biosynthetic human insulin production – a study case. Biotechnol Bioeng. 1995;48:529–541. doi: 10.1002/bit.260480516. [DOI] [PubMed] [Google Scholar]
- Quirk AV, Geisow MJ, Woodrow JR, Burton SJ, Wood PC, Sutton AD, Johnson RA, Dodsworth N. Production of recombinant human serum albumin from Saccharomyces cerevisiae. Biotechnol Appl Biochem. 1989;11:273–287. [PubMed] [Google Scholar]
- Rozkov A, Schweder T, Veide A, Enfors SO. Dynamics of proteolysis and its influence on the accumulation of intracellular recombinant proteins. Enzyme Microbiol Technol. 2000;27:743–748. doi: 10.1016/s0141-0229(00)00294-5. [DOI] [PubMed] [Google Scholar]
- Ruf S, Hermann M, Berger IJ, Carrer H, Bock R. Stable genetic transformation of tomato plastids and expression of a foreign protein in fruit. Nat Biotechnol. 2001;19:870–875. doi: 10.1038/nbt0901-870. [DOI] [PubMed] [Google Scholar]
- Saunders CW, Schmidt BJ, Mallonee RL, Guyer MS. Secretion of human serum albumin from Bacillus subtilis. J Bacteriol. 1987;169:2917–2925. doi: 10.1128/jb.169.7.2917-2925.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shively JM, English RS. The carboxysome, a prokaryotic organelle: a mini review. Can J Bot. 1991;69:957–962. [Google Scholar]
- Sijmons PC, Dekker BM, Schrammeijer B, Verwoerd TC, van den Elzen PJ, Hoekema A. Production of correctly processed human serum albumin in transgenic plants. Bio/Technol. 1990;8:217–221. doi: 10.1038/nbt0390-217. [DOI] [PubMed] [Google Scholar]
- Staub JM, Garcia B, Graves J, Hajdukiewicz PT, Hunter P, Nehra N, Paradkar V, Schlittler M, Carroll JA, Spatola L, Ward DYeG, Russell DA. High-yield production of a human therapeutic protein in tobacco chloroplasts. Nat Biotechnol. 2000;18:333–338. doi: 10.1038/73796. [DOI] [PubMed] [Google Scholar]
- Staub JM, Maliga P. Accumulation of D1 polypeptide in tobacco plastid is regulated via the untranslated region of the psbA mRNA. EMBO J. 1993;12:601–606. doi: 10.1002/j.1460-2075.1993.tb05692.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub JM, Maliga P. Translation of psbA mRNA is regulated by light via the 5′-untranslated region in tobacco plastids. Plant J. 1994;6:547–553. doi: 10.1046/j.1365-313x.1994.6040547.x. [DOI] [PubMed] [Google Scholar]
- Tarelli E, Mire-Sluis A, Tivnann HA, Bolgiano B, Crane DT, Gee C, Lemercinier X, Athayde ML, Sutcliffe N, Corran PH, Rafferty B. Recombinant human albumin as a stabilizer for biological materials and for the preparation of international reference reagents. Biologicals. 1998;26:331–346. doi: 10.1006/biol.1998.0163. [DOI] [PubMed] [Google Scholar]
- Vrekleij AJ, Leunissen JM, editors. Immuno-Gold Labeling in Cell Biology. Boca Raton, FL: CRC Press; 1989. [Google Scholar]
- Watanabe H, Kragh-Hansen U, Tanase S, Nakajou K, Iwao Y, Maruyama T, Otagiri M. Conformational stability and warfarin-binding properties of human serum albumin studied by recombinant mutants. Biochem J. 2001a;357:269–274. doi: 10.1042/0264-6021:3570269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H, Yamasaki K, Kragh-Hansen U, Tanase S, Harada K, Suenaga A, Otagiri M. In vitro and in vivo properties of recombinant human serum albumin from Pichia pastoris purified by a method of short processing time. Pharm Res. 2001b;18:1775–1781. doi: 10.1023/a:1013391001141. [DOI] [PubMed] [Google Scholar]
- Zerges W. Translation in chloroplasts. Biochimie. 2000;82:583–601. doi: 10.1016/s0300-9084(00)00603-9. [DOI] [PubMed] [Google Scholar]


